Abstract
Neural stem cells (NSCs) persist in the dentate gyrus of the hippocampus into adulthood and are essential for both neurogenesis and neural circuit integration. Exosomes have also been shown to play vital roles in regulating biological processes of receptor cells as a medium for cell-to-cell communication signaling molecules. The precise molecular mechanisms of exosome-mediated signaling, however, remain largely unknown. Here, we found that exosomes produced by denervated hippocampi following fimbria–fornix transection could promote the differentiation of hippocampal neural precursor cells into cholinergic neurons in coculture with NSCs. Furthermore, we found that 14 circular RNAs (circRNAs) were upregulated in hippocampal exosomes after fimbria–fornix transection using high-throughput RNA-Seq technology. We further characterized the function and mechanism by which the upregulated circRNA Acbd6 (acyl-CoA-binding domain–containing 6) promoted the differentiation of NSCs into cholinergic neurons using RT–quantitative PCR, Western blot, ELISA, flow cytometry, immunohistochemistry, and immunofluorescence assay. By luciferase reporter assay, we demonstrated that circAcbd6 functioned as an endogenous miR-320-5p sponge to inhibit miR-320-5p activity, resulting in increased oxysterol-binding protein–related protein 2 expression with subsequent facilitation of NSC differentiation. Taken together, our results suggest that circAcbd6 promotes differentiation of NSCs into cholinergic neurons via miR-320-5p/oxysterol-binding protein–related protein 2 axis, which contribute important insights to our understanding of how circRNAs regulate neurogenesis.
Keywords: exosomes, circAcbd6, neural stem cells, cholinergic neurons, hippocampus
Abbreviations: Acbd6, acyl-CoA-binding domain–containing 6; AChE, acetylcholinesterase; AD, Alzheimer’s disease; AP, anteroposterior; cDNA, complementary DNA; ceRNA, competitive endogenous RNA; circRNA, circular RNA; DG, dentate gyrus; DLS, dynamic light scattering; DMEM, Dulbecco's modified Eagle's medium; FF, fimbria–fornix; LV, lentiviral vector; MAP2, microtubule-associated protein 2; ML, mediolateral; NC, negative control; ncRNA, noncoding RNA; NSC, neural stem cell; Osbpl2, oxysterol-binding protein–like 2; PFA, paraformaldehyde; qPCR, quantitative PCR; SD, Sprague–Dawley; SGZ, subgranular zone; SSC, saline sodium citrate; TEM, transmission electron microscopy; VAChT, vesicular acetylcholine transporter
Neural stem cells (NSCs) have been now established to be mostly quiescent in the adult mammalian brain and play important roles in plasticity, aging, disease, and regeneration of the nervous system (1). There are two major neurogenic niches in the adult mammalian brain where endogenous NSCs reside, the subventricular zone of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (DG) of the hippocampus (2). Experimental evidence has demonstrated that the hippocampus and its function are significantly affected by cholinergic dysfunction. It has been shown that degeneration of the cholinergic neurons often manifests in patients with Alzheimer’s disease (AD) and contributes to the memory loss exhibited by patients with AD (3). Hence, NSCs are generally considered to be a focal point for cell replacement therapy. Nevertheless, because of the limited number of NSCs residing in adult brain, only a small number of neurons can be produced, which is far from enough to totally supplement the lost neurons (4). Therefore, either the promotion of endogenous NSC differentiation into cholinergic neurons or supplementation of the lost cholinergic neurons through exogenous means had been the concern for researchers.
Exosomes are a class of extracellular microsized vesicles with a diameter range of 40 to 150 nm and have received a lot of scientific attention over the past few decades (5). Recently, exosomes released from mammalian cells have been reported as a new tool for cell-to-cell communication as it transports lipids, proteins, mRNAs, noncoding RNAs, and even DNA to other cells (6). The transfer of biological information by exosomes to the neighboring cells has been identified to be involved in the pathogenesis of many diseases, including cancer and neurodegenerative disorders (7). It has been proposed that cellular levels of circular RNAs (circRNAs) may be regulated by either endonucleic activity or exosomal transport. Previously, Li et al. (8) had demonstrated that circRNAs were abundant in the exosomes compared with their corresponding producer cells. Furthermore, exo-circRNAs may be regulated by associated gene expression and may transfer biological activity to recipient cells (8).
Current studies reveal that some of circRNAs harbor miRNA response elements, suggesting a potential role as competitive endogenous RNAs (ceRNAs) to compete for miRNA-binding sites (9). For example, CiRS-7, as a human circRNA cerebellar degeneration–related protein 1 transcript, acts as a natural miRNA sponge for miR-7 and regulates the expression of ubiquitin-conjugating enzyme E2A in AD hippocampal CA1 (10). Zhang et al. (11) in their study characterizes circRNA-associated ceRNA networks in the cerebral cortex of senescence-accelerated mouse prone 8 and discovers that the networks in this AD mouse model were mainly involved in the regulation of Aβ clearance and myelin function.
According to reports, transection of the fimbria–fornix (FF) blocks the major cholinergic input to the hippocampus, resulting in the removal of most cholinergic activity in the hippocampus (12, 13). We have previously found that NSCs proliferated significantly and migrated along the subgranular layer, and more newborn cells differentiated into neurons or astrocytes in the denervated hippocampus (14). Moreover, NSCs incubated with the denervated hippocampal extract differentiated significantly into the microtubule-associated protein 2 (MAP2) or acetylcholinesterase (AChE)-positive neurons than those incubated with the normal hippocampal extract (15, 16). It suggested that FF transection provided proper microenvironment for the survival and neuronal differentiation of hippocampal NSCs.
In this study, we focused on exosomes derived from denervated and normal hippocampus and observed their effects on NSCs in vitro. Our results revealed that exosomes derived from denervated hippocampus cocultured with NSCs promoted hippocampal neural progenitor cells to differentiate into neurons and cholinergic neurons. Hence, we generated and screened RNA-Seq data of exosomes from hippocampus. We identified a candidate circAcbd6 (acyl-CoA-binding domain–containing 6) and characterized its function in neuron development. We found that circAcbd6 acted as a sponge and interacted with miR-320-5p-oxysterol-binding protein–like 2 (Osbpl2) to promote NSC differentiation into neurons and cholinergic neurons. These results contributed to our current understanding of circRNA function in neurogenesis.
Results
Identification of exosomes
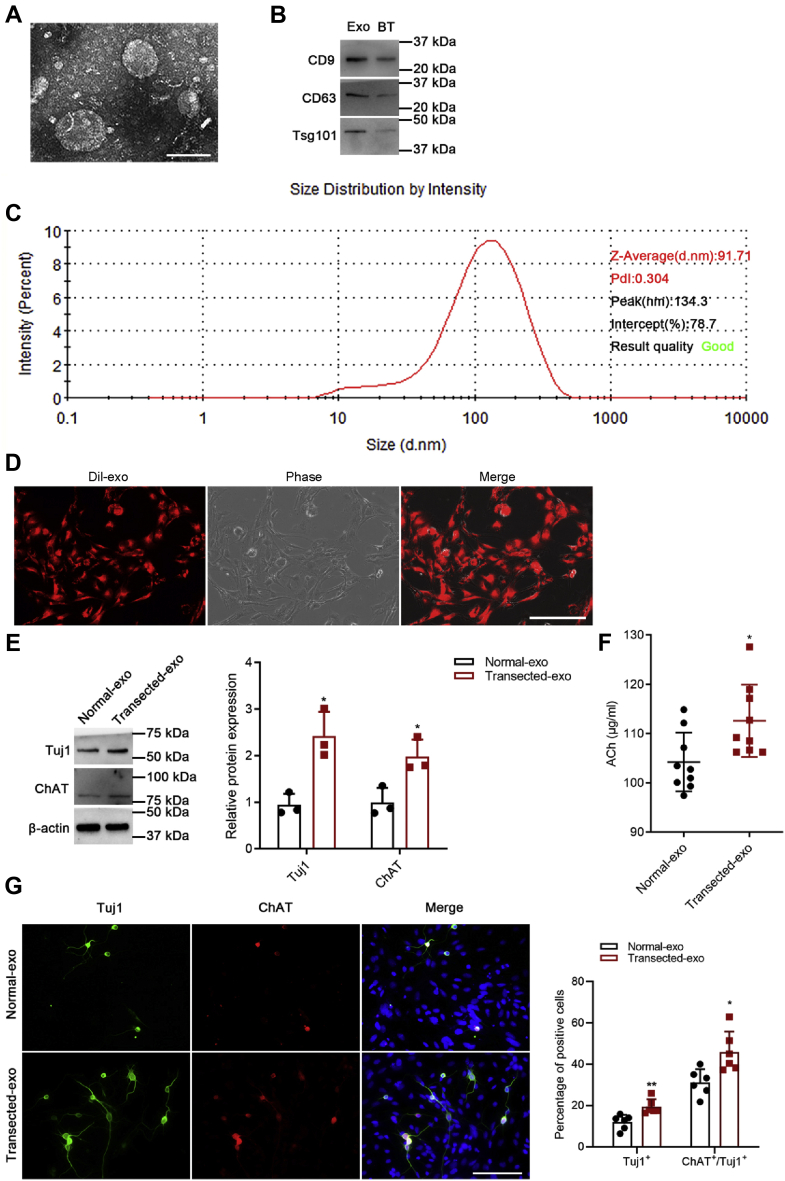
To identify and characterize the isolated exosomes, we applied three different techniques: transmission electron microscopy (TEM), Western blot, and dynamic light scattering (DLS). As shown in Figure 1A, hippocampi-derived exosomes were lightly stained with diameters within 40 to 150 nm under TEM. Western blot analysis revealed that several exosome-specific markers, such as CD9, CD63, and Tsg101, were detectable in all samples (Fig. 1B). For DLS, the result showed a single peak at 134.3 nm with Z-average (d. nm) of 91.71 (Fig. 1C).
Figure 1.
Effects of exosomes derived from hippocampus on NSC differentiation.A, TEM images of exosomes. The bar represents 100 nm. B, identification of CD9, CD63, and Tsg101 by Western blot. C, size distribution of exosomes decided by DLS. D, representative data showing the presence of CM-Dil in NSCs after adding exosomes purified from hippocampus. The bar represents 200 μm. E, Western blot analysis of Tuj1 and ChAT protein levels with β-actin normalization. (Normal-exo) NSCs treated with normal hippocampal exosomes; (Transected-exo) NSCs treated with deafferented hippocampal exosomes. F, ELISA of ACh in cell supernatants. G, immunofluorescence analysis of Tuj1-positive (green) and ChAT-positive (red) cells of normal-exo and transected-exo group. Nuclei were stained with Hoechst. The bar represents 100 μm. The data are presented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01. ACh, acetylcholinesterase; BT, brain tissue; DLS, dynamic light scattering; Exo, exosome; NSC, neural stem cell; TEM, transmission electron microscopy.
Effects of exosomes derived from hippocampus on NSC differentiation
To confirm that exosomes could be transferred to NSCs, NSCs were cocultured with exosomes that were labeled with CM-Dil. As shown in Figure 1D, after incubation with exosomes, the CM-Dil fluorescence signal was observed in NSCs. As shown in Figure 1E, Western blot of Tuj1 and ChAT expression was upregulated significantly. Similarly, ELISA showed that ACh expression was upregulated significantly in transected-exo group (Fig. 1F). Immunofluorescence staining of Tuj1 and ChAT differentiated cells indicated that there were more Tuj1-positive neurons, and more Tuj1-positive neurons differentiated into ChAT-positive neurons in transected-exo group (Fig. 1G). Our results revealed that exosomes derived from denervated hippocampus facilitated differentiation of NSCs into neurons and cholinergic neurons.
Overview of RNA-Seq data
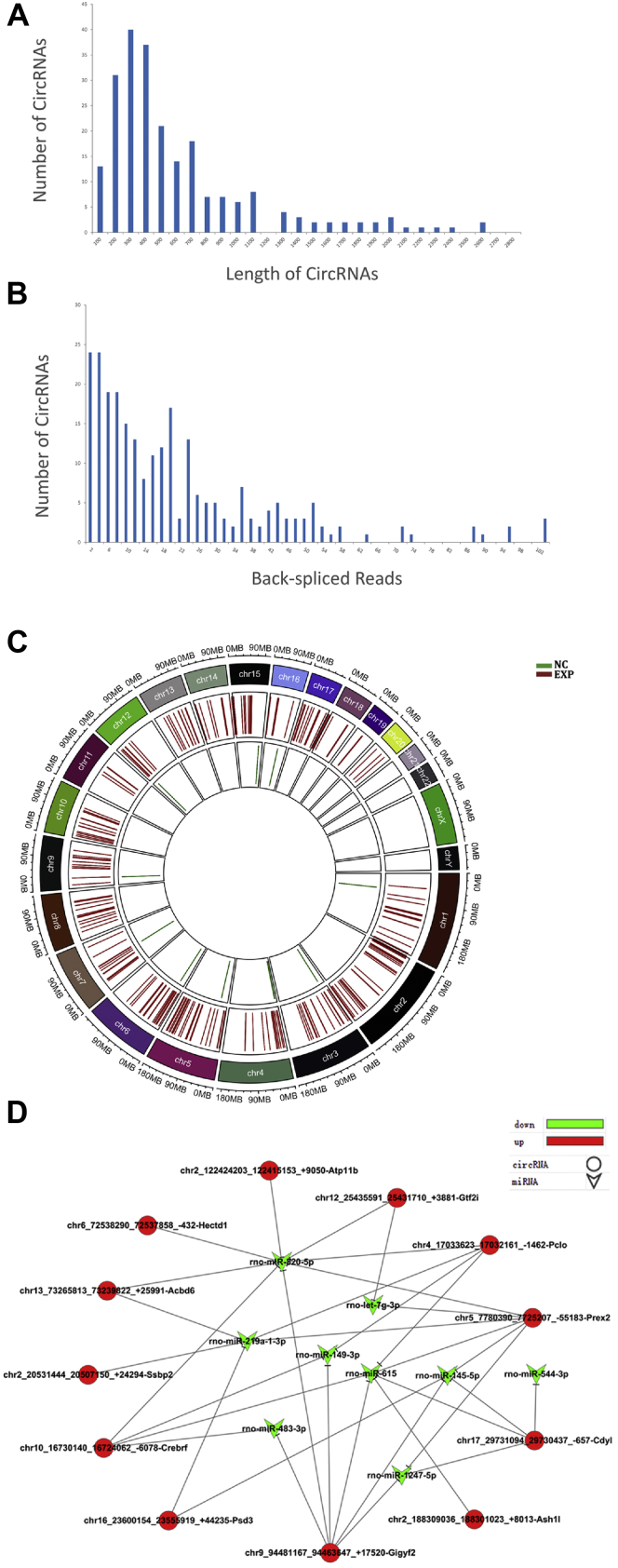
To characterize the circRNA profile involved in from hippocampal exosomes, bioinformatics analysis was performed. As shown in Figure 2A, the length of identified circRNAs was mostly around 100 to 1100 nucleotides. Multiple circRNAs could be produced from one host gene, and a total of 267 circRNAs were identified in samples with more than two unique back-spliced reads (Fig. 2B). In addition, based on the gene location, these circRNAs were dispersedly distributed on most chromosomes, showing significant differences between normal-exo group and transected-exo group (Fig. 2C). Some circRNAs have been shown to have multiple target sites to bind with miRNAs to play a regulatory role in organisms (17). To investigate the circRNA–miRNA interactions, the binding sites of circRNAs were predicted using online bioinformatics databases (RNhybrid); the network of circRNAs and their targeted relationship with miRNAs was presented in Figure 2D.
Figure 2.
Overview of RNA-Seq data.A, the length distribution of circRNAs. B, the number of circRNAs and junction reads identified in exosomes. C, the distribution of identified circRNAs in chromosomes. The red bars represent transected-exo group, and the green bars represent normal-exo group. D, the circRNA–miRNA network diagram. The v-shaped nodes represent miRNAs, the circular nodes represent circRNAs, the red represent upregulated genes, the green represent downregulated genes, and straight lines indicate interactions between miRNAs and circRNAs. circRNA, circular RNA.
Identification of circAcbd6
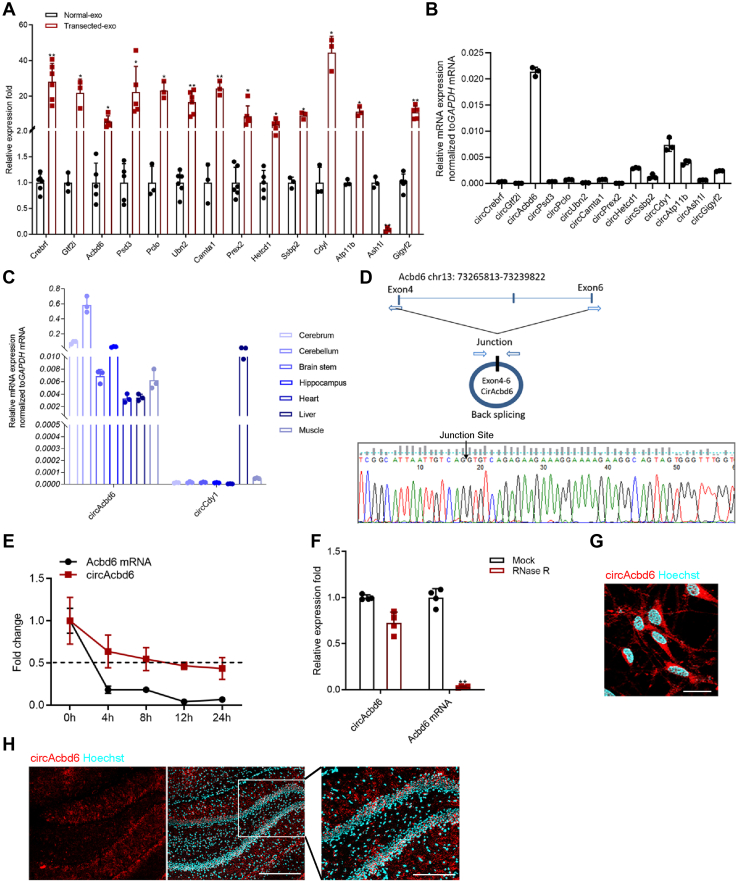
As shown in Figure 3A, except for circRNA Ash1l, the rest of differentially expressed circRNAs was consistent with the trends observed using RNA-Seq. Of the 14 circRNAs, we initially focused on two abundant circRNAs in NSCs, circAcbd6 and circCdy1 (Fig. 3B). To explore the expression pattern of validated differentially expressed circRNAs, we extracted RNA from tissues derived from ectoderm (cerebrum, cerebellum, brain stem, and hippocampus), mesoderm (heart and muscle), and endoderm (liver) and then carried on RT–quantitative PCR (qPCR) analysis. As shown in Figure 3C, circAcbd6 was highly significantly expressed in nervous tissues compared with other tissues. Finally, circAcbd6 was selected for deeper analysis. To validate the existence of circAcbd6, junction primers were designed to amplify the circAcbd6 junction expression in complementary DNA (cDNA), followed by Sanger sequencing (Fig. 3D). After treated with actinomycin D, an inhibitor of transcription, we found that a half-life of circAcbd6 was approximately 12 h, whereas the linear Acbd6 mRNA exhibited a half-life that was less than 4 h (Fig. 3E). To further confirm the circular characteristics of circAcbd6 in NSCs, the enzyme RNase R, a highly processive 3′ to 5′ exoribonuclease, was used to digest total RNA (18) and indicated that circAcbd6 was more stable than linear Acbd6 (Fig. 3F). Finally, FISH assay showed that circAcbd6 was predominantly localized in the cytoplasm of NSCs (Fig. 3G). FISH was performed to explore the location of circAcbd6 to more directly observe the expression trend in the DG. It was demonstrated that circAcbd6 was specially expressed in the SGZ, which contained a population of adult NSCs (Fig. 3H).
Figure 3.
Identification of circAcbd6.A, candidate circRNA verification by RT–qPCR panel. (Normal-exo) exosomes of normal hippocampi; (Transected-exo) exosomes of hippocampi after FF transection. B, basal expression levels of the candidate circRNAs in NSCs. C, circAcbd6 and circCdy1 expression in different tissues of SD rats. D, the presence of circAcbd6 was validated by RT–qPCR followed by Sanger sequencing. E, the expression levels of circAcbd6 and linear Acbd6 expression in NSCs treated with actinomycin D. F, the mRNA levels of circAcbd6 and linear Acbd6 in NSCs treated with RNase R. G, FISH assay showing the location of circAcbd6 in NSCs. The bar represents 25 μm. H, FISH assay showing the location of circAcbd6 in hippocampus. Nuclei were stained with Hoechst. The bar represents 400 μm. The rectangular part showed a higher magnification view of circAcbd6. The bar represents 200 μm. The data are presented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01. Acbd6, acyl-CoA-binding domain–containing 6; circRNA, circular RNA; FF, fimbria–fornix; NSC, neural stem cell; qPCR, quantitative PCR; SD, Sprague–Dawley.
Effects of circAcbd6 overexpression on differentiation of NSCs
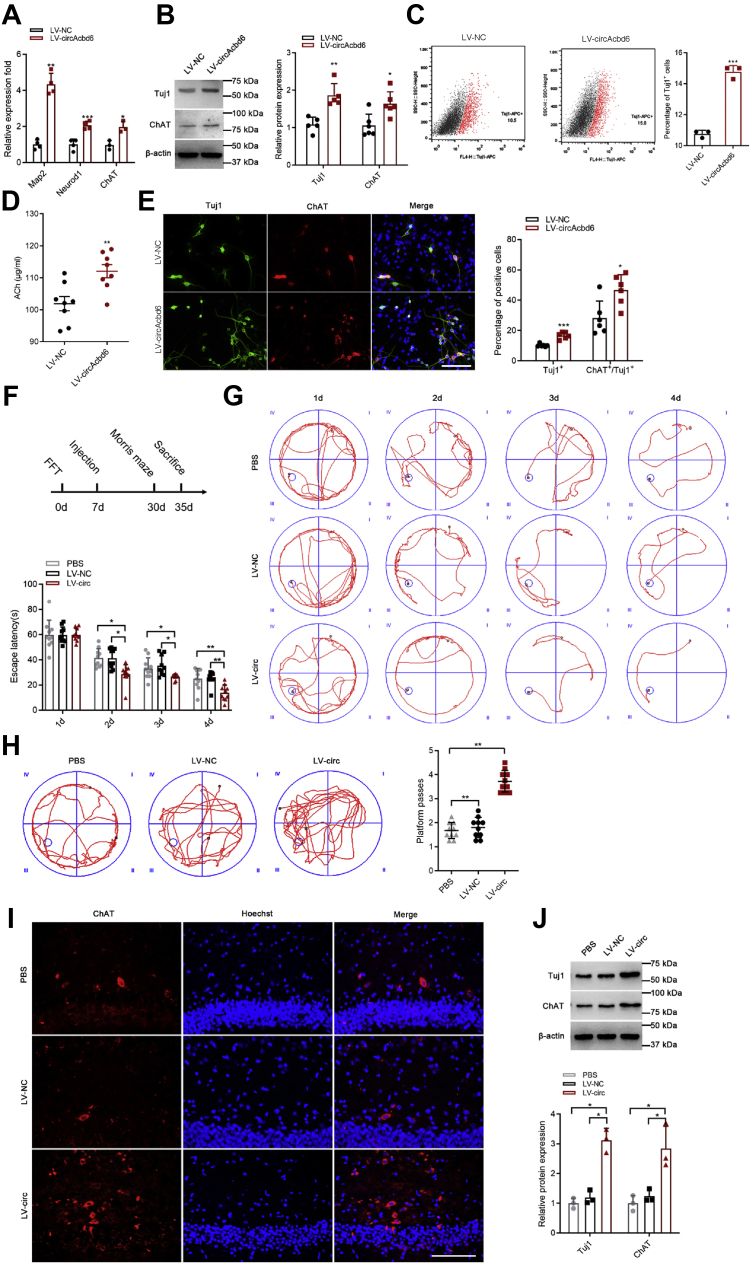
To investigate whether circAcbd6 regulated the differentiation of NSCs, we transfected NSCs with lentiviral vectors (LVs) overexpressing circAcbd6 (Fig. S1A). To characterize the roles of circAcbd6 on NSC differentiation, we performed RT–qPCR and Western blot analysis of three neuronal markers, Map2, Neurod1, and ChAT (19, 20, 21). The results showed that Map2, Neurod1, and ChAT in LV-circAcbd6 group were upregulated significantly compared with LV–negative control (NC) group (Fig. 4A). Western blot showed that Tuj1 and ChAT proteins were upregulated significantly (Fig. 4B). Flow cytometry assay showed that the percentage of Tuj1-positive cells notably increased (Fig. 4C). ELISA showed that the secretion level of ACh was upregulated significantly (Fig. 4D). Immunofluorescence staining indicated that there were more Tuj1-positive neurons, and more Tuj1-positive neurons differentiated into ChAT-positive neurons in LV-circAcbd6 group (Figs. 4E and S1B). Immunohistochemistry staining revealed that there were more vesicular acetylcholine transporter (VAChT)–immunopositive cells in LV-circAcbd6 group (Fig. S3A). Together, these results implied that circAcbd6 overexpression promoted NSC differentiation into neurons and cholinergic neurons.
Figure 4.
Effects of circAcbd6 overexpression on differentiation of NSCs.A, expression of Map2, Neurod1, and ChAT detected by RT–qPCR. LV–NC NSCs treated with NC of overexpression lentivirus; (LV-circAcbd6) NSCs treated with overexpression lentivirus of circAcbd6. B, expression of Tuj1 and ChAT detected by Western blot. C, the percentage of Tuj1-positive cells detected by flow cytometry assay. D, the secretion level of ACh measured with ELISA. E, immunofluorescence analysis of Tuj1 (green) and ChAT (red) double-positive cells. Nuclei were stained with Hoechst. The bar represents 100 μm. F, the schedule of in vivo experiments. G, representative trajectory diagrams and the time to reach the platform between the PBS or LV–NC group and LV-circAcbd6 group. (PBS) PBS injected into the hippocampus; (LV–NC) NC lentivirus injected into the hippocampus; (LV-circAcbd6) overexpression lentivirus of circAcbd6 injected into the hippocampus. H, representative trajectory diagrams and the number of platform location crosses during a single 120 s probe trial. I, immunofluorescence analysis of ChAT-positive (red) cells of hippocampus. Nuclei were stained with Hoechst. The bar represents 100 μm. J, the expression of Tuj1 and ChAT in hippocampus measured by Western blot. The data are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Acbd6, acyl-CoA-binding domain–containing 6; ACh, acetylcholinesterase; LV, lentiviral vector; NC, negative control; NSC, neural stem cell; qPCR, quantitative PCR.
In vivo circAcbd6 enhanced spatial learning in rats after FF transection
We used a rat model of cholinergic injury to investigate the effect of circAcbd6 on cognitive function recovery, and the schedule of the experiments in vivo is shown in Figure 4F. GFP detection in the hippocampus proved that the lentivirus successfully infected the target tissues (Fig. S1C). The escape latency to reach the platform in LV-circAcbd6 group rats was significantly shorter compared with PBS and LV–NC rats (Fig. 4G). LV-circAcbd6 group rats crossed the platform more frequently when compared with the PBS and LV–NC rats (Fig. 4H). Immunofluorescence assay revealed that there were more ChAT-positive cells in LV-circAcbd6 group (Fig. 4I). As shown in Figure 4J, the expression levels of Tuj1 and ChAT in hippocampus were upregulated significantly. Therefore, circAcbd6 could significantly improve learning and memory ability.
CircAcbd6 serves as a competing endogenous miR-320-5p sponge to mediate differentiation of NSCs
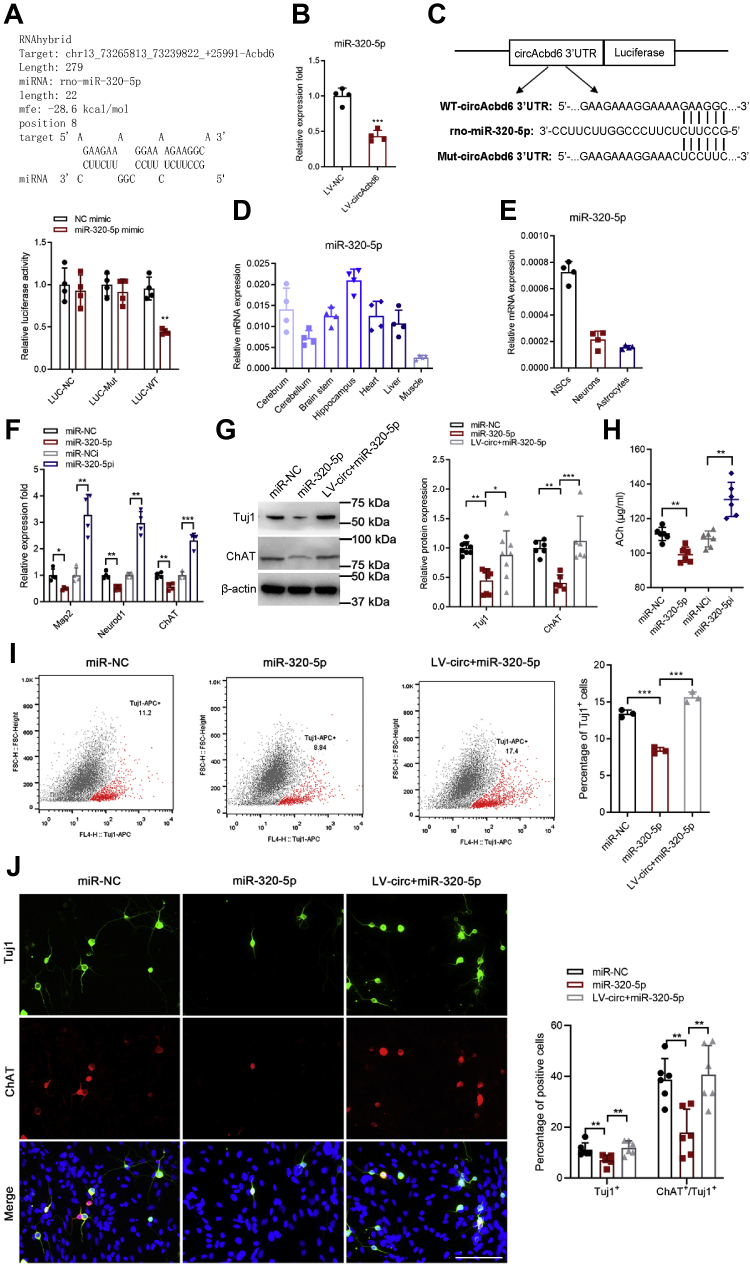
CircRNAs play important roles in regulating gene expression. However, the biological functions of most circRNAs are unknown. Database of RNAhybrid prompted that circAcbd6 possessed a complementary sequence to the miR-320-5p seed region (Fig. 5A). RT–qPCR analysis showed that circAcbd6 overexpression significantly decreased the relative abundance of miR-320-5p (Fig. 5B). Luciferase assay revealed direct binding between circAcbd6 and miR-320-5p (Fig. 5C). As shown in Figure 5D, miR-320-5p was significantly expressed in nervous tissues especially in the hippocampus. Intriguingly, we discovered that miR-320-5p was most expressed in NSCs, followed by neurons, and minimally in astrocytes (Fig. 5E). These discoveries suggested that miR-320-5p may play an important role in neurogenesis.
Figure 5.
Effects of circAcbd6-miR-320-5p on differentiation of NSCs.A, RNAhybrid predicted miR-320-5p-binding sites in circAcbd6. B, effect of circAcbd6 on the abundance of miR-320-5p. (LV–NC) NSCs treated with NC of overexpression lentivirus; (LV-circAcbd6) NSCs treated with overexpression lentivirus of circAcbd6. C, luciferase reporter assay in HEK-293A cotransfected with Luciferase-miR-320-5p fusion and circAcbd6-Wild or circAcbd6-Mut, 72 h post-transfection. D, miR-320-5p expression in different tissues. E, miR-320-5p expression in NSCs, neurons, and astrocytes. F, expression of Map2, Neurod1, and ChAT detected by RT–qPCR. (miR-NC) NSCs treated with NC of miRNA mimic; (miR-320-5p) NSCs treated with miR-320-5p mimic; (miR-NCi) NSCs treated with NC of miRNA inhibitor; (miR-320-5pi) NSCs treated with miR-320-5p inhibitor. G, expression of Tuj1 and ChAT detected by Western blot. (miR-NC) NSCs treated with NC of miRNA mimic; (miR-320-5p) NSCs treated with miR-320-5p mimic; (LV-circ + miR-320-5p) NSCs treated with overexpression lentivirus of circAcbd6 and miR-320-5p mimic. H, the secretion level of ACh measured with ELISA. I, the percentage of Tuj1-positive cells was detected by flow cytometry assay. J, immunofluorescence analysis of Tuj1 (green) and ChAT (red) double-positive cells. Nuclei were stained with Hoechst. The bar represents 100 μm. The data are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Acbd6, acyl-CoA-binding domain–containing 6; ACh, acetylcholinesterase; HEK-293A, human embryonic kidney 293A cell line; LV, lentiviral vector; NC, negative control; NSC, neural stem cell; qPCR, quantitative PCR.
To investigate whether circAcbd6-miR-320-5p regulated the differentiation of NSCs, we had transfected NSCs with miR-320-5p mimic/inhibitor (Fig. S1D). The RT–qPCR results showed that Map2, Neurod1, and ChAT in miR-320-5p group were downregulated significantly compared with miR-NC group, whereas the expression of Map2, Neurod1, and ChAT in miR-320-5pi group was upregulated significantly compared with miR-NCi group (Fig. 5F). Western blot showed that miR-320-5p overexpression significantly downregulated Tuj1 and ChAT proteins, whereas their expression was partially increased after overexpression of circAcbd6 (Fig. 5G). ELISA showed that the secretion level of ACh was downregulated significantly in miR-320-5p group, whereas inhibition of miR-320-5p significantly increased their expression (Fig. 5H). Flow cytometry indicated that Tuj1-positive cells were decreased in miR-320-5p group (Fig. 5I). Immunofluorescence staining indicated that there were less Tuj1 and ChAT double-positive cells in miR-320-5p group (Fig. 5J). Immunohistochemistry staining revealed that there were less VAChT-immunopositive cells in miR-320-5p group, whereas inhibition of miR-320-5p overexpression significantly increased their expression (Fig. S3B). Together, these results implied that circAcbd6-miR-320-5p regulated NSC differentiation into neurons and cholinergic neurons.
CircAcbd6 promoted the differentiation of NSCs by sponging miR-320-5p to regulate Osbpl2
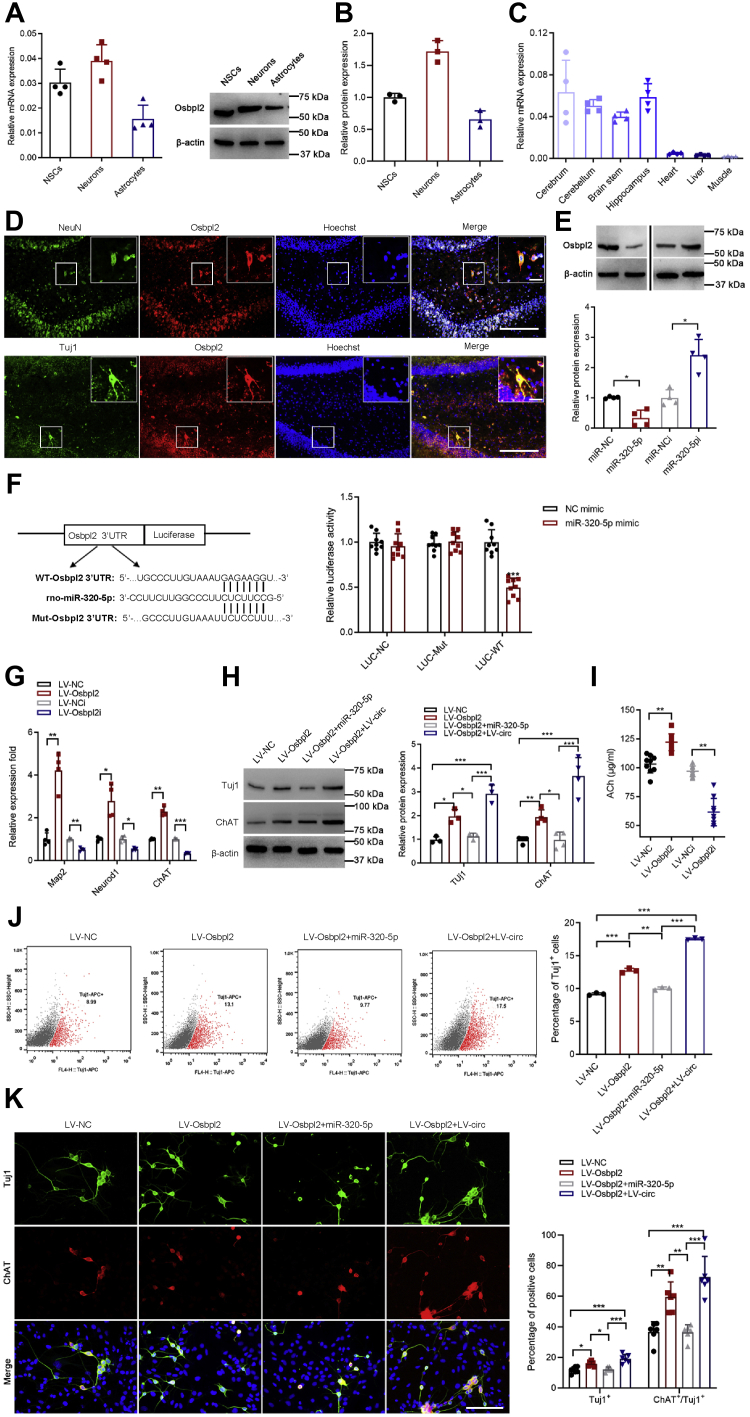
The target genes of miR-320-5p were determined by the union of target prediction sites from TargetScan 7.2 (https://www.targetscan.org/), miRWalk 2.0 (http://129.206.7.150/), miRDB (http://www.mirdb.org/), and RNA-Seq of significantly upregulated differentially expressed mRNA (Fig. S4A). As shown in Fig. S4B, circAcbd6 was a ceRNA network of miR-320-5p that targeted 10 mRNAs. According to the RNA-Seq data, the top five mRNAs with the largest differences were selected for further verification. RT–qPCR analysis demonstrated a negative correlation between miR-320-5p and Padi2, Traf3, Osbpl2 expression (Fig. S4C). Moreover, we discovered that Osbpl2 was most expressed in neurons, followed by NSCs, and minimally in astrocytes (Fig. 6, A and B). Intriguingly, Osbpl2 was significantly expressed in nervous tissues (Fig. 6C). Immunofluorescence staining showed that Osbpl2 was colocalized with NeuN and Tuj1 and expressed in the SGZ of DG (Fig. 6D). These discoveries suggested that Osbpl2 may play an important role in neurogenesis. Western blot analysis further demonstrated that miR-320-5p overexpression significantly decreased the relative abundance of Osbpl2, whereas miR-320-5p knockdown upregulated the expression of Osbpl2 significantly compared with miR-NCi group (Fig. 6E). Luciferase assay revealed direct binding between Osbpl2 and miR-320-5p (Fig. 6F).
Figure 6.
Effects of circAcbd6-miR-320-5p-Osbpl2 on differentiation of NSCs.A and B, Osbpl2 expression in NSCs, neurons, and astrocytes. C, Osbpl2 expression in different tissues. D, localization of Osbpl2 in the hippocampus. The bar represents 200 μm. The image in the upper right corner of the figure is an enlargement of the typical double-labeled neuron area in the small box of the figure. The bar represents 50 μm. E, effect of miR-320-5p on the abundance of Osbpl2 measured with Western blot. (miR-NC) NSCs treated with NC of miRNA mimic; (miR-320-5p) NSCs treated with miR-320-5p mimic; (miR-NCi) NSCs treated with NC of miRNA inhibitor; (miR-320-5pi) NSCs treated with miR-320-5p inhibitor. F, luciferase reporter assay in HEK-293A cotransfected with Luciferase-miR-320-5p fusion and Osbpl2-Wild or Osbpl2-Mut, 72 h post-transfection. G, expression of Map2, Neurod1, and ChAT detected by RT–qPCR. (LV–NC) NSCs treated with NC of overexpression lentivirus; (LV-Osbpl2) NSCs treated with overexpression lentivirus of Osbpl2; (LV–NCi) NSCs treated with lentivirus-mediated silencing of control; (LV-Osbpl2i) NSCs treated with overexpression lentivirus-mediated silencing of Osbpl2. H, expression of Tuj1 and ChAT detected by Western blot. (LV–NC) NSCs treated with NC of overexpression lentivirus; (LV-Osbpl2) NSCs treated with overexpression lentivirus of Osbpl2; (LV-Osbpl2 + miR-320-5p) NSCs treated with overexpression lentivirus of Osbpl2 and miR-320-5p mimic; (LV-Osbpl2 + LV-circ) NSCs treated with overexpression lentivirus of Osbpl2 and circAcbd6. I, the secretion level of ACh measured with ELISA. J, the percentage of Tuj1-positive cells detected by flow cytometry assay. K, immunofluorescence analysis of Tuj1 (green) and ChAT (red) double-positive cells. Nuclei were stained with Hoechst. The bar represents 100 μm. The data are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Acbd6, acyl-CoA-binding domain–containing 6; ACh, acetylcholinesterase; HEK-293A, human embryonic kidney 293A cell line; LV, lentiviral vector; NC, negative control; NSC, neural stem cell; Osbpl2, oxysterol-binding protein–like 2; qPCR, quantitative PCR.
To investigate whether circAcbd6-miR-320-5p-Osbpl2 regulated the differentiation of NSCs, we transfected NSCs with LV Osbpl2 (Fig. S1, E and F). As shown in Figures 6, G–K, S3C, S4, D and E, the results demonstrated that Osbpl2 overexpression significantly promoted NSC differentiation into neurons and cholinergic neurons, whereas silencing Osbpl2 inhibited their differentiation. Transfection of miR-320-5p mimic after overexpression of Osbpl2 significantly reduced the differentiation ratio of NSCs into neurons and cholinergic neurons. In addition, overexpression of Osbpl2 and circAcbd6 showed further enhanced NSC differentiation into neurons and cholinergic neurons. Overall, these results demonstrated that the circAcbd6-miR-320-5p-Osbpl2 axis was an important regulator in the progression of NSC differentiation into neurons and cholinergic neurons.
Discussion
In adult mammals, neurogenesis still persists in specific brain regions, is highly dynamic, and modulated by a variety of physiological stimuli and pathological states. Cholinergic input plays a critical role in memory function, which was strongly manifested in the impairment of hippocampus-dependent learning (22, 23). In our previous experiments, NSCs were transplanted into the denervated hippocampus and the contralateral side, and it was found that more MAP2- or AChE-positive neurons appeared in the denervated hippocampus (15). These detected AChE-positive neurons may be from local NSCs or the implanted NSCs. Besides, in the study of Parkinson's Disease model, more grafted cells in the denervated striatum were found to survive and differentiate into tyrosine hydroxylase–positive neurons (24). It implicated that microenvironment in brain induced the local or grafted NSCs to differentiate into specific phenotypes required for complementing the shortage of neurotransmitters or neural circuits.
According to research reports, in AD, exosomes have been shown to transmit toxic amyloid-beta and hyperphosphorylated tau between cells, and they have been suspected to contribute to neuronal loss (25). It indicated that exosomes might play an important role in cell-to-cell communication and influence the pathological processes of many neurodegenerative diseases. In our study, we found that incubated with the denervated hippocampal exosomes, more NSCs differentiated into neurons and cholinergic neurons. It suggested that the denervated hippocampal exosomes provided supportive microenvironment for the survival and neuronal differentiation of NSCs, where some substances expressed upregulation or downregulation.
Exosomes are released into the extracellular environment by the majority of cell types in the body and contain the molecular constituents of a cell, including proteins, RNAs, and DNAs (26). Previous studies have shown that the RNA profile of exosome-derived cells and its donor cells differs substantially (27). Total exosomal RNA typically lacks the 18S and 28S ribosomal RNA peaks compared with the total RNA of its donor cell. Detection of the specific components secreted by cell-derived exosomes, such as noncoding RNA (ncRNA), may possibly become new biomarkers for human diseases.
Based on these reports, we performed wide transcriptomic sequencing and obtained the differentially expressed ncRNAs between transected-exo and normal-exo group. It was shown that the upregulated ncRNAs were more than downregulated ncRNAs upon transection compared with normal hippocampus. It may be because neurogenesis occurs in the hippocampal DG after transection, which suggests that certain genes are activated and expressed. At this time, ncRNAs, protein, and lipid carried by exosomes that regulate neurogenesis increased significantly. After validation, we confirmed that most circRNAs could be upregulated in exosomes after transection, which may be correlated with gene activation. To speculate the function of these circRNAs, we conducted preliminary bioinformatics analysis and predicted their targets through the website. The data indicated that Gene Ontology analysis was enriched in spongiotrophoblast differentiation, trophoblast giant cell differentiation, and embryonic placenta development, which was in accord with the promotion of embryonic development. The enriched pathway analysis showed that they were involved in SNARE interactions in vesicular transport, and synaptic vesicle cycle, which was consistent with the research reported by Sudhof (28).
We analyzed the expression pattern of the significantly differentially expressed circRNAs in various tissues. As a result, circAcbd6 was highly expressed in nervous tissues. Although circRNAs with lower expression level in nervous system may also have functions, we believed that highly expressed circRNAs were more likely to play key roles in neural biological processes, thus we mainly concentrated on circAcbd6. Intriguingly, we also discovered that circAcbd6 was most expressed in NSCs. Thus, we hypothesized that circAcbd6 might be involved in neurogenesis.
There were almost no reports on the relationship between circAcbd6 and NSC differentiation, and there have been only few studies on the host gene of circAcbd6 so far. The members of ACBD family regulate various biological processes, including NSC self-renewal, neurodegeneration, stress resistance, lipid homeostasis, intracellular vesicle trafficking, organelle formation, viral replication, and apoptotic response (29, 30, 31, 32). For instance, expression of the hACBD6 protein was found to be elevated in bone marrow, spleen, placenta, cord blood, circulating CD34+ progenitors, and embryonic-like stem cells derived from placenta (33). Soupene and Kuypers (34) found that ACBD6 proteins promote N-myristoylation in mammalian cells and in one of their intracellular parasites under unfavorable substrate-limiting conditions. Now, following circAcbd6 overexpression, more NSCs differentiated into the Tuj1-positive and ChAT-positive cells. It provided new evidence for the role of circAcbd6 in the regulation of NSC differentiation.
Recent studies have revealed that circRNAs are rich in miRNA-binding sites and act as an miRNA sponge, thereby regulate the effect of the miRNAs on their target genes (35). Thus, we constructed ceRNA networks of circAcbd6 and further confirmed that miR-320-5p was a direct target of circAcbd6. It was reported that miR-320 enhanced cell apoptosis of the injury side cortical infarcted peripheral zone and increased brain infarction volume and edema volume in middle cerebral artery occlusion/reperfusion mice (36). Shen et al. (37) indicated that overexpressed miR-320 affected the proliferation, apoptosis, and oxidative stress injury of ischemic cerebral neuron by inhibiting Nox2/reactive oxygen species pathway. Based on these reports, we found that circAcbd6 acted as a modulator of cell differentiation by sponging miR-320-5p in NSCs.
To further elucidate the mechanisms underlying circAcbd6/miR-320-5p-mediated NSC differentiation, we explored the downstream effectors. According to the union of target prediction sites and RNA-Seq, we found that 10 genes are clustered, of which Osbpl2 was highly expressed in nervous tissues. Osbpl2 is a member of the oxysterol-binding protein–related protein family, known as a sterol sensor and transporter, which can regulate lipid/cholesterol metabolism, steroid hormone synthesis, cell signal transduction, vesicular transport, and cytoskeleton formation (38, 39, 40, 41, 42). In addition, Yao et al. (43) found that Osbpl2 deficiency might lead to the loss of tightly regulated cholesterol-homeostasis response in inner ear and cause auditory dysfunction. At present, there were almost no reports on Osbpl2 in the nervous system, and the potential molecular function of Osbpl2 still needed to be further investigated. In this study, we found that circAcbd6-miR-320-5p-Osbpl2 axis could regulate NSC differentiation into neurons and cholinergic neurons.
There are still several issues to be further explored. First, whether circAcbd6 could regulate the proliferation and apoptosis of NSCs. Second, whether circAcbd6 affects the differentiation of NSCs into glial cells. Third, whether circAcbd6 induces the differentiation of NSCs into other types of neurons. Fourth, whether circAcbd6 could regulate the differentiation of NSCs through other mechanisms. Finally, the relevant signaling pathway still needs to be elucidated. Clarifying these issues will provide clues and new ideas for uncovering the NSC differentiation mechanism and applying circRNAs in the treatment of neurodegenerative diseases.
Experimental procedures
Animals
We used pregnant Sprague–Dawley (SD) rats, 1-day-old neonatal SD rats, and adult SD rats (220–250 g) purchased from the experimental animal center of Nantong University (certificate no.: SYXK [SU] 2017-0046). All experimental protocols were approved by the Animal Ethics Committee of Nantong University. All efforts were made to minimize the number and suffering of animals used in this study.
Rat model of cholinergic nerve injury
Lesion of FF transections was performed as described by Hefti (44). Briefly, after chlorpent anesthesia (2 ml/kg body weight, i.p.), adult SD rats were transferred to the stereotaxic apparatus, and then transection of FF was performed with a wire knife on the dorsal side of hippocampal CA1 layer, at coordinates of bregma: anteroposterior (AP) = 1.4, mediolateral (ML) = 1.0 and AP = 1.4, ML = 4.0 (right); AP = 1.4, ML = −1.0 and AP = 1.4, ML = −4.0 (left), and depth 5.4 to 5.6 mm. There were no restrictions on the gender of the animals. Nissl staining was used to check whether bilateral side FF was completely disconnected (45).
Surgery and treatment
Sixty SD rats in total were used for surgery and treatment. Briefly, after chlorpent anesthesia, adult SD rats were transferred to the stereotaxic apparatus. At day 7 after FF transection, injections of virus into the left and right hippocampal dentate gyri at two points were performed at the following coordinates: 3.6 mm to bregma, 1.39 mm to the right or left of the midline, and 3.9 mm in depth. Five microliters of virus were loaded into an internal cannula needle with cannula tubing connected to a Hamilton syringe mounted onto a microinjection pump (Harvard Apparatus). The speed of the injection was 0.5 μl/min. The needle was kept in the position for an additional 10 min after completion of the injection and retrieved slowly out of the brain.
Exosome isolation
After 7 days of FF transection, denervated and normal hippocampi were quickly dissected and homogenized into ice-cold PBS. Exosomes were precipitated using Total Exosome Isolation Reagent (Invitrogen) as per manufacturer’s instructions. Homogenates were centrifuged at 2000g at 4 °C for 30 min to remove cells and debris, and the supernatants were passed through 0.22 μm filter to remove extracellular vesicles larger than the exosomes. The supernatants were transferred to a new tube without disturbing the pellet and were mixed with 0.5 volumes of the Total Exosome Isolation Reagent and incubated overnight at 4 °C. The mixture was centrifuged at 10,000g for 30 min, and the exosome pellet was resuspended in PBS.
Cell culture
Isolation, culture, and differentiation of NSCs were performed as previously described with some modifications (46). Briefly, pregnant SD rats were anesthetized, and the embryos were removed by cesarean section. Hippocampi were dissected out from E14.5 embryos, mechanically dissociated into single-cell suspension. After centrifugation and resuspension, the cell suspensions were plated in flasks with Dulbecco's modified Eagle's medium and Ham F-12 nutrient mixture (1:1, DMEM/F-12; Gibco) containing 2% B27 (Gibco), 20 ng/ml epidermal growth factor (Sigma–Aldrich), and 20 ng/ml basic fibroblast growth factor (Sigma–Aldrich). Cells were passaged every 6 days to obtain neurospheres originating from a single primary cell. For in vitro differentiation, cell suspensions were plated at a density of 2 × 105 cells/ml in 6-well plates or 10 mm dishes with DMEM/F-12 medium supplemented with 2% B27 and 2% fetal bovine serum (Gibco) and cultured for 7 days. For the mixed coculture experiments, the isolated exosomes were mixed with NSCs and processed in different ways after the cocultivation.
Primary neurons were isolated using standard methods, as previously described (47). Briefly, hippocampi were dissected from E14.5 embryos. The single-cell suspension was diluted in serum-free neurobasal medium (Gibco) containing 2% B27 supplement and 0.5 mM l-glutamine (Gibco) and then seeded onto precoated plates with poly-d-lysine. Half of the medium was replaced every 3 days.
Primary astrocytes were derived from cerebral cortices of 1-day-old neonatal rats as previously described (47). Briefly, dissociated cortical cells were suspended in DMEM/F-12 medium containing 10% fetal bovine serum and plated in flasks. After 3 to 4 days, the primary mixed cells were orbitally shaken to remove microglia and oligodendrocytes. Astrocytes were dissociated by trypsinization and then replated into flasks.
Cell infection
To construct overexpression vector, the linear sequence of circAcbd6 was inserted into the (poly A-MCS-UBI) RV-SV40-EGFP-IRES-puromycin and Osbpl2 were inserted into the Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin vector. The inhibition fragment of Osbpl2 used synthetic interference sequences and was cloned into hU6-MCS-CBh-gcGFP-IRES-puromycin vector, whereas the mock vector with no target gene sequence was used as a control (GeneChem). NSCs were plated at a density of 5 × 104 cells per well in 24-well plates and cultured overnight prior to transduction. LVs (LV-circAcbd6 and LV-Osbpl2) were added to the cultures at a multiplicity of infection of 10.
NSCs were transiently transfected using Lipofectamine3000 (Invitrogen) according to the manufacturer’s instructions. Vector controls and miRNA compounds (miR-320-5p/NC mimic or inhibitor) were purchased from RiboBio Co, Ltd.
TEM analysis
A 10 μl suspension of exosomes was diluted into a suitable concentration with PBS. Exosomes were fixed with glutaraldehyde at 4 °C overnight. After washing, exosomes were loaded onto formvar/carbon-coated grids, stained with aqueous phosphotungstic acid for 60 s, and imaged with a transmission electron microscope.
DLS analysis
The size of the vesicles was determined by a DLS technique using a ZetasizerNano ZS90 analysis system (Zetasizer, version 7.12; Malvern Instruments). A size distribution plot, for size distribution by intensity, with the x-axis showing the distribution of estimated particle diameter (nm) and the y-axis showing the relative percentage, was created.
Actinomycin D and RNase R treatment
To block transcription, actinomycin D (1 μg/ml; APExBIO Technology) or dimethyl sulfoxide (Sigma–Aldrich) as an NC was added into the cell culture medium. For RNase R treatment, 1 μg of total RNA was incubated for 30 min at 37 °C with or without 2 units/μg of RNase R and purified following the RNeasy MinElute cleaning Kit (QIAGEN).
RT–qPCR analysis
Total RNA from cells and tissues was isolated using TRIzol reagent (Vazyme Biotech) according to the manufacturer’s instructions. For mRNA expression analysis, 1 μg of RNA were used to synthesize cDNA using HiScript Q RT SuperMix for qPCR (+genomic DNA wiper) (Vazyme Biotech), and the SYBR green (Roche) method was performed in StepOnePlus real-time PCR system (Applied Biosystems) according to the manufacturer’s instructions. mRNA primers for PCR were designed and synthesized by Sangon Biotech and are listed in Table S1.
For miRNA expression analysis, the miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech) and the miRcute miRNA qPCR Detection Kit (SYBR Green; Tiangen Biotech) were used. circRNA and miRNA primers for PCR were designed and synthesized by RiboBio. The relative quantities of mRNA were calculated using 2−ΔCT method after normalization to the controls.
Western blot
Proteins were extracted by ultrasonication with radioimmunoprecipitation assay lysis buffer containing proteinase inhibitor. Equivalent amounts of protein lysates were loaded into each lane, separated by 10% SDS-PAGE, then transferred to 0.2 mm polyvinylidene fluoride membranes, and sealed with 5% skim milk for 2 h. After incubating with primary antibodies overnight at 4 °C, the membranes were incubated with horseradish peroxidase–linked secondary antibodies for 2 h. The immunoreactive bands were viewed by enhanced chemiluminescence reagents (Bio-Rad). Primary antibodies included anti-CD9 (1:1000 dilution; Abcam), anti-CD63 (1:1000 dilution; Abcam), anti-Tsg101 (1:1000 dilution; Abcam), anti-Tuj1 (1:1000 dilution; Millipore), anti-ChAT (1:1000 dilution; Abcam), and anti-β-actin (1:1000 dilution; Abcam).
Immunofluorescence assay
Cells or sections were fixed with 4% paraformaldehyde (PFA) for 30 min, permeabilized with and blocked with 10% normal goat containing 0.3% Triton X-100 and 1% bovine serum albumin for 2 h, and then incubated with primary antibody overnight at 4 °C. On the next day, cells were incubated with secondary antibodies at room temperature for 2 h. Cell nuclei were counterstained with Hoechst 33342 (1:1000 dilution; Pierce). Primary antibodies included anti-Tuj1 (1:1000 dilution; Millipore), anti-NeuN (1:200 dilution; Abcam), anti-ChAT (1:1000 dilution; Abcam), and anti-Osbpl2 (1:1000 dilution; LifeSpan BioSciences).
Immunohistochemistry
Cells were fixed with 4% PFA for 30 min and stained with Super-Sensitive Horseradish Peroxidase Immunohistochemistry Kit (Sangon Biotech) according to the manufacturer’s instructions. Briefly, cells were incubated with endogenous peroxidase blocking solution for 15 min. After rinsed in PBS, cells were incubated with blocking solution for 30 min at room temperature and incubated with rabbit anti-VAChT (1:100 dilution; Abcam) overnight at 4 °C. On the next day, cells were incubated with poly-horseradish peroxidase–conjugated anti-rabbit immunoglobulin G for 1 h and detected with diaminobenzidine.
Flow cytometry assay
Cells in a culture dish were trypsinized and collected, washed twice in PBS, and fixed in 1× Fix/Perm Buffer working solution at 4 °C for 40 min. After washing with 1× Perm/Wash Buffer, cell samples were mixed with 100 μl of 1× Perm/Wash Buffer and incubated with allophycocyanin-conjugated anti-Tuj1 antibody or allophycocyanin-conjugated IgG2A control (5 μl/test) (BD Biosciences) (Fig. S2) at 4 °C for 2 h. Cell suspension was subjected to flow cytometry.
FISH
After washing with PBS, the cells were fixed in 4% PFA for 10 min and then incubated in PBS with 0.3% Triton X-100 for 10 min. Before hybridization overnight at 4 °C, cells were incubated in the prehybridization solution for 30 min at 37 °C. On the next day, the cells were stained with Hoechst 33342 after washing with 4× saline sodium citrate (SSC) twice, 2× SSC once, 1× SSC once, and 1× PBS once at 42 °C.
ELISA
For ACh release detection, 5 × 105 cells were seeded in 6-well plates. The culture medium was collected after 7 days, and 10 μl of the culture medium was measured using commercially available ELISA kits (Lengton) according to the manufacturer’s protocol. The spectrophotometry of panels was read at 450 nm and calculated according to the standard curve.
Luciferase activity assay
The luciferase reporter vectors were constructed by GeneChem Company. miR-320-5p and circAcbd6/Osbpl2-Wild or circAcbd6/Osbpl2-Mut were cotransfected into 293T cells using the Dual-Luciferase Reporter Assay System (Promega). Dual-Glo Luciferase assay was applied to quantitate the relative luciferase activity at 72 h after transfection, which was computed by luciferase activity of firefly to Renilla.
Morris water maze test
To evaluate spatial memory and learning, rats were trained in the Morris water maze for 5 days prior to sacrifice. Rats were randomly divided into three groups: PBS group (cholinergic injury + PBS), LV–NC group (cholinergic injury + NC lentivirus injection), and LV-circAcbd6 group (cholinergic injury + circAcbd6-overexpressing lentivirus injection). In each trial, the rats were required to find the location of a hidden platform below the surface of the water and introduced from four different starting points. The time was limited to 120 s per trial, and the activities were recorded. If rats failed to escape on the platform within 120 s, they were guided to the platform. An autotracking system was used to measure escape latency (time to find the platform) and time in each quadrant. On the fifth day, the platform was removed, and the animals were allowed to swim in the maze for 120 s, and the number of platform crossings was measured.
Statistical analysis
Each experiment included at least three independent samples. All data are presented as mean ± SD and analyzed by GraphPad Prism 9.0 software (GraphPad Software, Inc). Comparison of differences between two groups was made by unpaired Student's two-tailed t test, and comparisons among multiple groups were made by one-way ANOVA. The results were considered statistically significant when ∗p < 0.05.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the Medical School of Nantong University.
Data availability
All data are included in this article.
Supporting information
This article contains supporting information (four supporting figures and one supporting table).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We gratefully acknowledge the Graduate Scientific Research Innovation Program of Jiangsu Province, the Priority Academic Program Development of Jiangsu Higher Education institutions for the research grants and the Science and Technology Program of Nantong City. We also acknowledge the editor and reviewers for their helpful comments on this article.
Contract grant sponsors: Graduate Scientific Research Innovation Program of Jiangsu Province (contract grant number: KYCX19 2066); a project funded by the Priority Academic Program Development of Jiangsu Higher Education institutions (contract grant number: 03081023); and The Science and Technology Program of Nantong City (contract grant numbers: JC2020024 and JC2021056).
Author contributions
W. L., X. Z., and G. J. conceptualization; B. S., X. C., H. H., J. Q., H. Z., and M. T. methodology; W. L. validation; W. L. formal analysis; W. L. investigation; B. S., X. C., H. H., J. Q., H. Z., and M. T. resources; B. S., X. C., H. H., J. Q., H. Z., and M. T. data curation; X. Z., G. J., and W. L. writing–original draft; X. Z. and G. J. writing–review & editing; X. Z. and G. J. project administration; X. Z., G. J., and W. L. funding acquisition.
Edited by Paul Fraser
Contributor Information
Xinhua Zhang, Email: zhangxinhua@ntu.edu.cn.
Guohua Jin, Email: jguohua@ntu.edu.cn.
Supporting information
References
- 1.Zhao X., Moore D.L. Neural stem cells: Developmental mechanisms and disease modeling. Cell Tissue Res. 2018;371:1–6. doi: 10.1007/s00441-017-2738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kempermann G., Song H., Gage F.H. Neurogenesis in the adult hippocampus. Cold Spring Harb. Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehouse P.J., Price D.L., Struble R.G., Clark A.W., Coyle J.T., Delon M.R. Alzheimer's disease and senile dementia: Loss of neurons in the basal forebrain. Science. 1982;215:1237–1239. doi: 10.1126/science.7058341. [DOI] [PubMed] [Google Scholar]
- 4.He H., Li W., Peng M., Qin J., Shi J., Li H., Tian M., Zhang X., Lv G., Jin G. MicroRNA expression profiles of neural stem cells following valproate inducement. J. Cell Biochem. 2018;119:6204–6215. doi: 10.1002/jcb.26831. [DOI] [PubMed] [Google Scholar]
- 5.Ruivo C.F., Adem B., Silva M., Melo S.A. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 6.Samanta S., Rajasingh S., Drosos N., Zhou Z., Dawn B., Rajasingh J. Exosomes: New molecular targets of diseases. Acta Pharmacol. Sin. 2018;39:501–513. doi: 10.1038/aps.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashed M.H., Bayraktar E., Helal G.K., Abd-Ellah M.F., Amero P., Chavez-Reyes A., Rodriguez-Aguayo C. Exosomes: From garbage bins to promising therapeutic targets. Int. J. Mol. Sci. 2017;18:538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y., Alexandrov P.N., Jaber V., Lukiw W.J. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes (Basel) 2016;7:116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Zhu D., Li H., Li H., Feng C., Zhang W. Characterization of circRNA-Associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol. Ther. 2017;25:2053–2061. doi: 10.1016/j.ymthe.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Ikonen S., Tapiola T., Tanila H., van Groen T. Fimbria-fornix lesion does not affect APP levels and amyloid deposition in the hippocampus of APP+PS1 double transgenic mice. Exp. Neurol. 2002;177:565–574. doi: 10.1006/exnr.2002.8015. [DOI] [PubMed] [Google Scholar]
- 13.van Groen T., Kadish I. Transgenic AD model mice, effects of potential anti-AD treatments on inflammation and pathology. Brain Res. Brain Res. Rev. 2005;48:370–378. doi: 10.1016/j.brainresrev.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 14.Zou L., Jin G., Zhang X., Qin J., Zhu H., Tian M., Tan X. Proliferation, migration, and neuronal differentiation of the endogenous neural progenitors in hippocampus after fimbria fornix transection. Int. J. Neurosci. 2010;120:192–200. doi: 10.3109/00207450903464579. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X., Jin G., Tian M., Qin J., Huang Z. The denervated hippocampus provides proper microenvironment for the survival and differentiation of neural progenitors. Neurosci. Lett. 2007;414:115–120. doi: 10.1016/j.neulet.2006.07.078. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Jin G., Wang L., Hu W., Tian M., Qin J., Huang H. Brn-4 is upregulated in the deafferented hippocampus and promotes neuronal differentiation of neural progenitors in vitro. Hippocampus. 2009;19:176–186. doi: 10.1002/hipo.20498. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Vincent H.A., Deutscher M.P. Insights into how RNase R degrades structured RNA: Analysis of the nuclease domain. J. Mol. Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M., Pan M., Ge L., Liu J., Deng J., Wang X., Li L., Wen J., Tan D., Zhang H., Hu X., Fu L., Xu Y., Li Z., Qiu X., et al. NeuroD1 overexpression in spinal neurons accelerates axonal regeneration after sciatic nerve injury. Exp. Neurol. 2020;327:113215. doi: 10.1016/j.expneurol.2020.113215. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez C., Diaz-Nido J., Avila J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 2000;61:133–168. doi: 10.1016/s0301-0082(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 21.Saxena S., Choudhury S., Mohan K.N. Reproducible differentiation and characterization of neurons from mouse embryonic stem cells. MethodsX. 2020;7:101073. doi: 10.1016/j.mex.2020.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haam J., Yakel J.L. Cholinergic modulation of the hippocampal region and memory function. J. Neurochem. 2017;142:111–121. doi: 10.1111/jnc.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hampel H., Mesulam M.M., Cuello A.C., Farlow M.R., Giacobini E., Grossberg G.T., Khachaturian A.S., Vergallo A., Cavedo E., Snyder P.J., Khachaturian Z.S. The cholinergic system in the pathophysiology and treatment of Alzheimer's disease. Brain. 2018;141:1917–1933. doi: 10.1093/brain/awy132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishino H., Hida H., Takei N., Kumazaki M., Nakajima K., Baba H. Mesencephalic neural stem (progenitor) cells develop to dopaminergic neurons more strongly in dopamine-depleted striatum than in intact striatum. Exp. Neurol. 2000;164:209–214. doi: 10.1006/exnr.2000.7426. [DOI] [PubMed] [Google Scholar]
- 25.Malm T., Loppi S., Kanninen K.M. Exosomes in Alzheimer's disease. Neurochem. Int. 2016;97:193–199. doi: 10.1016/j.neuint.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Weidle U.H., Birzele F., Kollmorgen G., Ruger R. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics. 2017;14:1–15. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou R., Chen K.K., Zhang J., Xiao B., Huang Z., Ju C., Sun J., Zhang F., Lv X.B., Huang G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol. Cancer. 2018;17:75. doi: 10.1186/s12943-018-0823-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudhof T.C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 29.Faergeman N.J., Wadum M., Feddersen S., Burton M., Kragelund B.B., Knudsen J. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol. Cell Biochem. 2007;299:55–65. doi: 10.1007/s11010-005-9040-3. [DOI] [PubMed] [Google Scholar]
- 30.Fan J., Liu J., Culty M., Papadopoulos V. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): An emerging signaling molecule. Prog. Lipid Res. 2010;49:218–234. doi: 10.1016/j.plipres.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao S., Chye M.L. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 2009;47:479–484. doi: 10.1016/j.plaphy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Xiao S., Chye M.L. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 2011;50:141–151. doi: 10.1016/j.plipres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Soupene E., Serikov V., Kuypers F.A. Characterization of an acyl-coenzyme A binding protein predominantly expressed in human primitive progenitor cells. J. Lipid Res. 2008;49:1103–1112. doi: 10.1194/jlr.M800007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soupene E., Kuypers F.A. ACBD6 protein controls acyl chain availability and specificity of the N-myristoylation modification of proteins. J. Lipid Res. 2019;60:624–635. doi: 10.1194/jlr.M091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang Z., Tan T., Zhang X., Wan J., Zhou Y., Jiang G., Yang D., Guo X., Liu T. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019;10:932. doi: 10.1038/s41419-019-2161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang L., Wang J., Yuan Y., Zhang Y., Liu H., Wu C., Yan Y. MicRNA-320 facilitates the brain parenchyma injury via regulating IGF-1 during cerebral I/R injury in mice. Biomed. Pharmacother. 2018;102:86–93. doi: 10.1016/j.biopha.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 37.Shen W., Lu Y., Hu J., Le H., Yu W., Xu W., Yu W., Zheng J. Mechanism of miR-320 in regulating biological characteristics of ischemic cerebral neuron by mediating Nox2/ROS pathway. J. Mol. Neurosci. 2020;70:449–457. doi: 10.1007/s12031-019-01434-5. [DOI] [PubMed] [Google Scholar]
- 38.Escajadillo T., Wang H., Li L., Li D., Sewer M.B. Oxysterol-related-binding-protein related Protein-2 (ORP2) regulates cortisol biosynthesis and cholesterol homeostasis. Mol. Cell Endocrinol. 2016;427:73–85. doi: 10.1016/j.mce.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kentala H., Koponen A., Kivelä A.M., Andrews R., Li C., Zhou Y., Olkkonen V.M. Analysis of ORP2-knockout hepatocytes uncovers a novel function in actin cytoskeletal regulation. FASEB J. 2018;32:1281–1295. doi: 10.1096/fj.201700604R. [DOI] [PubMed] [Google Scholar]
- 40.Kentala H., Koponen A., Vihinen H., Pirhonen J., Liebisch G., Pataj Z., Kivelä A., Li S., Karhinen L., Jääskeläinen E., Andrews R., Meriläinen L., Matysik S., Ikonen E., Zhou Y., et al. OSBP-related protein-2 (ORP2): A novel Akt effector that controls cellular energy metabolism. Cell Mol. Life Sci. 2018;75:4041–4057. doi: 10.1007/s00018-018-2850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koponen A., Arora A., Takahashi K., Kentala H., Kivelä A.M., Jääskeläinen E., Peränen J., Somerharju P., Ikonen E., Viitala T., Olkkonen V.M. ORP2 interacts with phosphoinositides and controls the subcellular distribution of cholesterol. Biochimie. 2019;158:90–101. doi: 10.1016/j.biochi.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Wang H., Lin C., Yao J., Shi H., Zhang C., Wei Q., Lu Y., Chen Z., Xing G., Cao X. Deletion of OSBPL2 in auditory cells increases cholesterol biosynthesis and drives reactive oxygen species production by inhibiting AMPK activity. Cell Death Dis. 2019;10:627. doi: 10.1038/s41419-019-1858-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao J., Zeng H., Zhang M., Wei Q., Wang Y., Yang H., Lu Y., Li R., Xiong Q., Zhang L., Chen Z., Xing G., Cao X., Dai Y. OSBPL2-disrupted pigs recapitulate dual features of human hearing loss and hypercholesterolaemia. J. Genet. Genomics. 2019;46:379–387. doi: 10.1016/j.jgg.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Hefti F. Nerve growth factor promotes survival of septal cholinergic neurons after fimbrial transections. J. Neurosci. 1986;6:2155–2162. doi: 10.1523/JNEUROSCI.06-08-02155.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H., Jin T., Cheng X., Qin J., Zhang L., He H., Xue J., Jin G. GAS5 which is regulated by Lhx8 promotes the recovery of learning and memory in rats with cholinergic nerve injury. Life Sci. 2020;260:118388. doi: 10.1016/j.lfs.2020.118388. [DOI] [PubMed] [Google Scholar]
- 46.Linqing Z., Guohua J., Haoming L., Xuelei T., Jianbing Q., Meiling T. Runx1t1 regulates the neuronal differentiation of radial glial cells from the rat hippocampus. Stem Cells Transl. Med. 2015;4:110–116. doi: 10.5966/sctm.2014-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing C., Wang X., Cheng C., Montaner J., Mandeville E., Leung W., van Leyen K., Lok J., Wang X., Lo E.H. Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke. 2014;45:2085–2092. doi: 10.1161/STROKEAHA.114.005733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this article.