Abstract
Artificial intelligence (AI)-based image analysis is increasingly being used for preclinical safety-assessment studies in the pharmaceutical industry. In this paper, we present an AI-based solution for preclinical toxicology studies. We trained a set of algorithms to learn and quantify multiple typical histopathological findings in whole slide images (WSIs) of the livers of young Sprague Dawley rats by using a U-Net-based deep learning network. The trained algorithms were validated using 255 liver WSIs to detect, classify, and quantify seven types of histopathological findings (including vacuolation, bile duct hyperplasia, and single-cell necrosis) in the liver. The algorithms showed consistently good performance in detecting abnormal areas. Approximately 75% of all specimens could be classified as true positive or true negative. In general, findings with clear boundaries with the surrounding normal structures, such as vacuolation and single-cell necrosis, were accurately detected with high statistical scores. The results of quantitative analyses and classification of the diagnosis based on the threshold values between “no findings” and “abnormal findings” correlated well with diagnoses made by professional pathologists. However, the scores for findings ambiguous boundaries, such as hepatocellular hypertrophy, were poor. These results suggest that deep learning-based algorithms can detect, classify, and quantify multiple findings simultaneously on rat liver WSIs. Thus, it can be a useful supportive tool for a histopathological evaluation, especially for primary screening in rat toxicity studies.
Keywords: digital pathology, machine learning, pharmaceutical development, toxicity study, hepatotoxicity, lesion detection and quantification
Introduction
Toxicity studies, including the histopathological examination of tissues from experimental animals, play an important role in the risk assessment of chemicals. The reliability of evaluation results is dependent on the level of experience, expertise, and diagnostic skill of the pathologist. In addition, because most areas of the specimen are within normal limits and without any abnormal features, the manual reading of histopathological slides is time consuming and labor intensive. Moreover, hundreds of glass slides must be evaluated in a single toxicity study. Accurately detecting and evaluating a few abnormal findings from vast areas of normal histology without committing errors of omission is difficult. Inter-pathologist variability is inevitable between individual pathologists or study sites owing to the inherent qualitative/semi-quantitative nature of histopathological evaluations. Although a pathology peer review can reduce bias, such reviews can be augmented through quantitative results of specific histopathological findings. Moreover, highlighting and quantifying abnormal areas in a pathological image, an easy comparison using the reference values, summary tables, and graphs makes the evaluation evidence-based and adds to the level of confidence during the evaluation process. This is also useful in explaining the findings to researchers and team members who are unfamiliar with pathology.
In recent years, the digitization of glass slides has enabled the application of digital pathology in diverse areas such as human surgical pathology, cancer diagnosis, and preclinical toxicity studies. The use of machine learning and deep learning networks for recognizing and quantifying histopathological features has recently been increasingly applied in these areas1, 2, 3, 4. Particularly in the field of human clinical medicine, several cancer tissues can be diagnosed with high accuracy using an AI-based pathological image analysis5, 6. In addition, diagnostic techniques used to detect subtle changes in digital images acquired through endoscopy and computed tomography have been established7, 8. In the field of human hepatology, AI-based diagnostic imaging technology has been used in a variety of tests, such as the diagnosis and prediction of hepatobiliary cancers, screening for nonalcoholic fatty liver disease, and accurate hepatic steatosis quantification of liver biopsies9, 10, 11.
Such techniques are gradually being used in preclinical toxicity studies for pharmaceuticals. However, developing AI-based pathological image-analysis methods for preclinical toxicity studies using laboratory animals has proved challenging12. The diversity of histology due to the wide variety of animal species used in toxicity studies (such as rats, mice, dogs, monkeys, and mini pigs) and the large number of organs and tissues to be evaluated in a single study are major hurdles in training algorithms. Some AI-based image-analysis algorithms for laboratory animals using histopathological digital images have recently been reported, such as for detecting and quantifying testicular stage classification in rats13, of rodent cardiomyopathy14 and hypertrophy and vacuolation of rat liver15, 16. These reports showed that, for abnormal findings, the algorithms could detect and quantify only a single type of finding in each case, and none could immediately detect or classify multiple types of findings simultaneously on a WSI.
In this study, we trained a U-Net-based deep learning network17 to segment and quantify seven typical histopathological findings in rat livers. Our algorithm can detect, classify, and quantify multiple findings simultaneously using a WSI. To train the algorithm, digitized liver WSIs scanned from glass slide specimens of young Sprague Dawley (SD) rats (male, 8 weeks old) treated with various compounds during toxicity studies were used. We optimized the model by testing and retraining using 92 WSIs in the training dataset and 59 WSIs in the test dataset. After training, the algorithm was validated on 255 liver WSIs to detect, classify, and quantify seven types of histopathological findings: vacuolation of hepatocytes (spontaneous and drug-induced), single-cell necrosis, bile duct hyperplasia, hepatocellular hypertrophy, microgranuloma, and extramedullary hematopoiesis in the liver. The classification and quantification performance of the model for the histopathological findings was evaluated. The quantitative values computed by the algorithm were then compared with the information on the histopathological grade labels, as diagnosed by in-house board-certified pathologists (diplomates of the Japanese Society of Toxicologic Pathology). Subsequently, thresholds were calculated to discriminate between “no findings (within normal limits)” and “abnormal findings” based on the quantitative results.
Materials and Methods
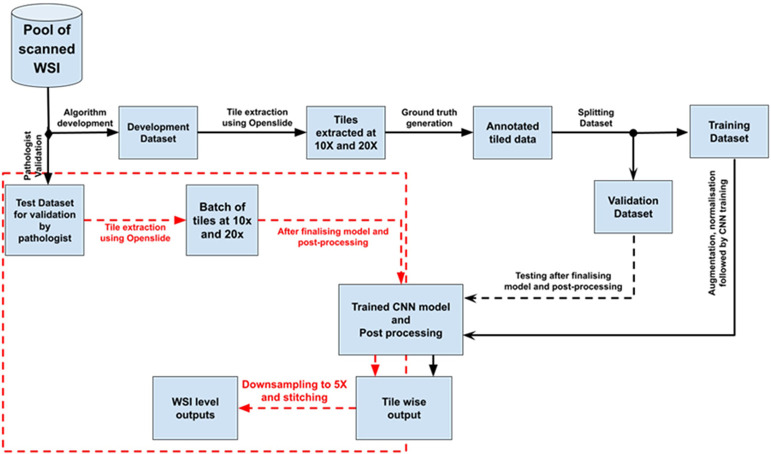
For training the deep learning-based analysis model, we selected seven histopathological findings that have been observed in toxicity studies in SD rat livers: vacuolation of hepatocytes (spontaneous and drug-induced), single cell necrosis, bile duct hyperplasia, hepatocellular hypertrophy, microgranuloma, and extramedullary hematopoiesis (hereafter referred to as lesions). Vacuolation is often observed in toxicity studies as a drug-induced lesion; however, because some degree of vacuolation is also spontaneously observed in normal rats, we distinguished spontaneous vacuolation from drug-induced vacuolation in this study. Whole slide images (WSIs) of hematoxylin and eosin (HE) stained specimens of rat liver were used for the development of the algorithm. A subset of the total data was used for training the algorithm, and the remaining data were used for testing, finetuning, and validation of the model to establish the performance metrics. Progressive improvements of the algorithm were introduced until acceptable degrees of accuracy and precision were achieved between the algorithm and the pathologist. Figure 1 summarizes the algorithm development process.
Fig. 1.
Workflow of algorithm development. Black dotted lines are used to connect the process steps after the model development is completed. Red dotted lines are used to highlight the process steps connected with a validation by the pathologists.
• Animals
All SD rats were housed individually in a climate-controlled room with a temperature of 23 ± 1 °C, humidity of 55 ± 15%, and a 12 h lighting cycle. A pelleted diet (CR-LPF, Oriental Yeast Co., Ltd., Tokyo, Japan) was provided ad libitum. All animal protocols used in this study were in compliance with our laboratory guidelines for animal experimentation and were approved by the Institutional Animal Care and Use Committee of the Central Pharmaceutical Research Institute, Japan Tobacco, Inc. Before necropsy, the animals were fasted overnight on the last day of the dosing period. The animals were euthanized by exsanguination from the abdominal aorta under isoflurane anesthesia and examined in detail for gross lesions. The livers were collected, fixed in 10% neutral buffered formalin, and prepared for histopathological examination by embedding in paraffin wax, sectioning, and staining with HE.
• Generation of WSIs
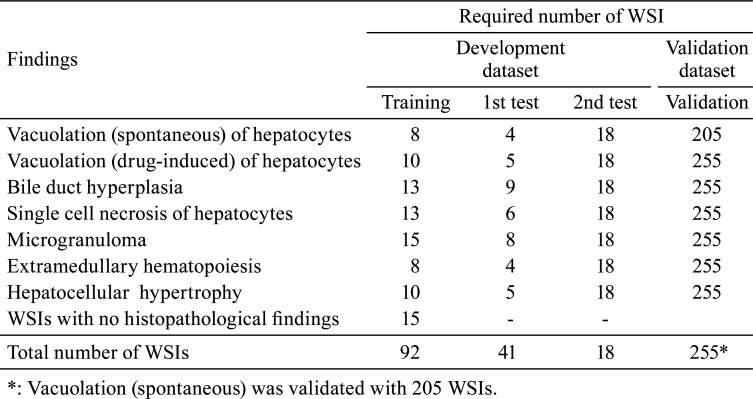
We prepared the WSIs for the training data for model construction, as described in Table 1. A total of 406 HE-stained glass slides of pathological liver specimens from 8 week-old male SD rats, which were treated with several compounds in toxicity studies conducted at Japan Tobacco, Inc., were scanned using a NanoZoomer S360 (Hamamatsu Photonics K.K., Shizuoka, Japan) at 20× magnification and converted into WSI.
Table 1. Number of WSIs Used for Training and Validation of the Algorithm.
• Datasets
Collecting extensive and high-quality data is essential for the development of a deep-learning-based algorithm. For this study, we used 406 WSIs for the algorithm development, including training and validation. The total dataset, made up of 406 WSIs, was divided into two mutually exclusive groups, namely, a development dataset containing 151 WSIs for training, testing, and finetuning of the models and a validation dataset of 255 WSIs for validation by the pathologists. A total of 92 WSIs from the development dataset were used to train the deep learning models for the seven lesions. Data from 15 WSIs without histopathological findings and 77 WSIs with histopathological findings were used. The models were tested and progressively finetuned based on two rounds of feedback from the pathologists on two different test datasets comprising 41 and 18 WSIs. Table 1 summarizes the data distributions.
• Tile extraction
The digital images were read as tiles of chosen sizes and magnifications using the OpenSlide software library, which is a vendor-neutral software for digital pathology. We extracted 512 × 512 × 3 dimensional colored tiles at magnifications of 10× and 20×.
• Ground truth generation
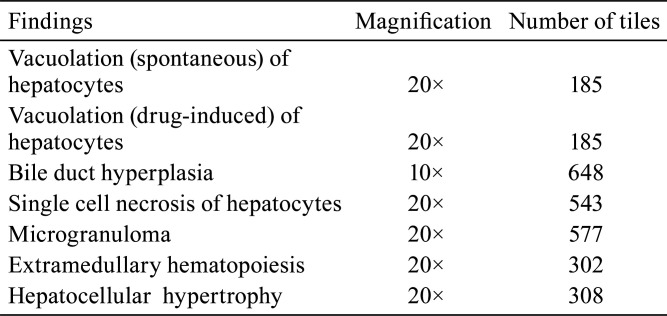
Ground truth annotations for the seven lesions were generated by data-marking experts under the guidance of pathologists, who further verified the annotated data after marking. Annotated tiles at appropriate magnifications (10× or 20×) were then used to train the models to detect individual lesions. Table 2 lists the magnification and number of tiles used to develop the algorithms for each lesion.
Table 2. Magnification and Number of Tiles Used to Develop the Algorithm for Each Lesion.
• Data preparation
The training dataset for a lesion consisted of tiles with annotated foci of the lesion (hereafter referred to as positive tiles) along with some (8–10%) tiles without any focus (hereafter referred to as negative tiles). The latter were used to define the contextual information for the background of a lesion.
The rationale behind adding the negative samples is to help the models better learn the contrast between the positive and negative classes, thereby improving the decision boundary and in turn resulting in a smaller number of false cases (particularly false positives). The performance of the model depends upon the optimum ratio of the positive and negative samples. However, there is no general rule regarding this optimization. In practice, the optimum ratio is determined empirically for a given model based on the availability of positive data. In our study, 8–10% of the negative data was observed to be the ideal ratio.
• Data augmentation
Both color and geometric augmentations were applied to the training set. Color augmentation was conducted by varying the saturation and hue of the colored tiles. Rotating a tile at multiple angles of 90° and flipping a tile horizontally and vertically are examples of geometric augmentations.
• Algorithm development
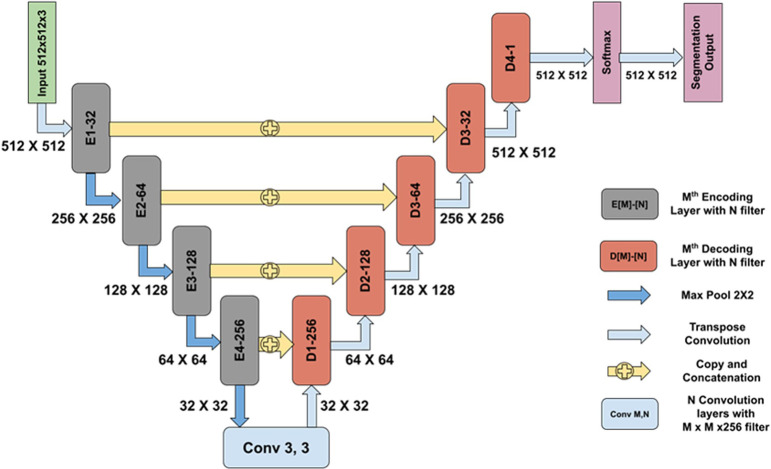
For each lesion, except for the hepatocellular hypertrophy, an accurate binary segmentation was achieved using a deep learning model based on a customized U-Net architecture13 as shown in Fig. 2. The annotated tiles from the training dataset were used to train the models. The models were then tested, and were gradually altered and improved to ensure that the algorithm and pathologists achieved agreement.
Fig. 2.
Network architecture used for segmentation of various parameters. An encoder–decoder convolutional neural network (CNN) was trained to segment the lesions using annotated training datasets. The network architecture is similar to the U-Net architecture, which has skip connections from the encoding layers to the decoding layers. This helps in eliminating the vanishing gradient problem and thereby simplifies the optimization during the backpropagation of the gradients. In the architecture, E [M]-[N] denotes the Mth encoding layer with N convolutional filters incorporating an inception-like module, whereas D [M]-[N] denotes the Mth decoding layer with N convolutional filters followed by a transposed convolution. Inception modules possess the ability to extract features at multiple scales by employing convolution filters of varying sizes. After each encoding block, the feature map is downsampled by a factor of 2 using a max pool operation on a 2 × 2 receptive field with a stride of 2. This reduces the number of learnable parameters, which in turn reduces the computational cost. At each decoder block (D[M]-[N]), outputs from both the previous decoding layer and the encoding layer are concatenated; they are further convolved with convolution filters followed by a transposed convolution of 2 × 2 with a stride of 2. Unlike the plane upsampling layer, a transposed convolution has learnable parameters that help in the better reconstruction of a segmentation map. Each decoding block increases the feature map size by a factor of 2. A softmax layer is used after the last decoding block to obtain a probability map that determines the probability of each pixel belonging to each class. An argmax operation is performed in the last layer to obtain the segmentation output.
A customized U-Net architecture was utilized to separate the hepatic nuclei to locate hypertrophic lesions. In addition, the nuclei per unit area and nucleo–cytoplasmic ratio for areas around the central vein and portal triads in the WSIs of liver tissue were calculated. Hypertrophic hepatocytes can be recognized by comparing the mean values of these quantitative measurements obtained from a series of control images with those obtained from the test image.
• Training, testing, and finetuning
The deep learning model was trained on a GeForce GTX TITAN X with 12 GB of memory (NVIDIA Co., Santa Clara, CA, USA). For each lesion, separate training was conducted using the convolutional neural network (CNN) architecture, as previously mentioned. Approximately 20% of the training tiles (along with annotations) were used for model validation (differing from the pathologist validation). Before feeding to the CNN, all training and validation tiles were normalized using the channel-wise (red, green, and blue) mean and standard deviation. This helped achieve a faster convergence and invariance of the training data against possible local variations. The training was conducted in batches of 12 tiles. The focal Tversky Loss function18 was used to reduce the impact of class imbalance. An Adam optimizer19 was used with a learning rate scheduler with an initial value of 1e−3 and a stepwise reduction by a factor of 10 after every 50 epochs. Necessary postprocessing using morphological operations was applied to the segmented outputs.
The initial models were tested on test dataset-1, which consisted of 36 WSIs after training. The training labels were adjusted in response to the pathologist’s feedback on individual lesions. The revised labels were then used to retrain the model. To establish agreement between the algorithm and the pathologists, the models were finetuned based on a second round of testing and pre-validation of all lesions on a separate test dataset-2 with 18 WSIs.
• Testing and validation
The trained models for each lesion were tested on 255 WSIs of the validation dataset. The testing was conducted on the tiles extracted from the dataset at the same magnification as that of the training. The bile duct hyperplasia model was tested on the 10× magnified tiles, whereas the models for vacuolation of hepatocytes (spontaneous and drug-induced), single cell necrosis of hepatocytes, hepatocellular hypertrophy, microgranuloma, and extramedullary hematopoiesis were tested on the 20× magnified tiles. The lesions were quantified in terms of the count or percentage area for the given tissue sections in each WSI, as described in Table 3 below.
Table 3. Measurement Parameters for Each Finding.
• Validation of the algorithm
From the analysis of the 255 WSIs of the liver (validation set) by the trained algorithm, 2 categories of information were gathered. The first shows abnormal results (at the image level) and offers a diagnosis based on the WSIs discovered by the algorithm, whereas the second includes a quantification for each of the findings. First, a group of pathologists double-checked the annotated data to ensure that the true lesion locations were marked. Then, histopathological data (“no findings (−)” or “abnormal findings (+)”) diagnosed by the pathologists were concatenated with the quantitative values obtained from the algorithm for each specimen. Histopathological data were obtained as follows. A pathologist first observed the HE-stained specimens and provided draft data. A peer review pathologist double-checked both the data and specimens. After the peer review, five of the pathologists, including the original and peer-review pathologists, discussed the validity of the draft data using the specimens and finalized the results.
The most reliable thresholds were calculated for each finding based on a receiver operating characteristic (ROC) curve using JMP software (version 13.0.0, SAS Institute, Inc., Cary, NC, USA). The ROC curves were drawn by plotting (1 − Specificity) on the x-axis and Recall on the y-axis for all possible thresholds. The best threshold value was calculated by maximizing Youden’s index (Recall + Specificity − 1) in the ROC curve. The discriminative performance was evaluated based on the area under the ROC curve (AUC-ROC).
Based on the threshold value from the ROC curve, binary diagnostic results by the pathologists were classified into four classes: true positive, false positive, false negative, or true negative for each finding. The following values were calculated.
• Comparison of quantitative values between the “no findings (−)” and “abnormal finding (+)” groups
The quantitative values of each finding by the algorithm were classified into two groups: those with no findings (−) or those with abnormal findings (+), based on the diagnosis by pathologists. For all the learned findings, it was confirmed that the populations were equally distributed through an F-test, and thus a comparison of the mean values between the two groups was conducted using an unpaired t-test (Student’s t-test).
Results
The analysis of WSIs by the algorithm generated two types of data: a) image dataset annotating the areas of each histopathological finding detected on WSI and b) a dataset quantifying the lesion areas.
Image data annotation
Figures 3–8 show the results of the detection and annotation of the abnormal areas of each histopathological finding by the algorithm.
Fig. 3.
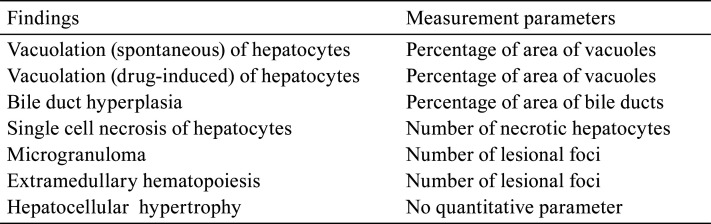
A: (Original WSI): Vacuolation (drug-induced) at the periportal area to the midzonal and normal bile ducts within the Glisson’s sheath were observed. B: (Annotation by the algorithm): The abnormal area (vacuolation) in Fig. 3A was annotated (filled) with yellow. (Bar=200 μm). C: Higher magnification of the dashed area in Fig. 3A. D: Higher magnification of the dashed area in Fig. 3B. The abnormal area (vacuolation) in Fig. 3C is annotated (filled) with yellow (Bar=100 μm).
• Vacuolation of hepatocytes
In this study, spontaneous vacuolation and drug-induced vacuolation were separately trained and evaluated. Spontaneous vacuolation refers to vacuoles observed in the periportal area of the liver of the control group. Specimens with few or no vacuolated areas in the hepatocytes were classified as “no findings” of spontaneous vacuolation. By contrast, if vacuolated areas were observed to be larger than the areas of spontaneous vacuolation, these specimens were classified as drug-induced vacuolation. Therefore, the “no finding” group for drug-induced vacuolation consisted of specimens with no vacuoles and specimens with spontaneous vacuolation. Drug-induced small vacuoles in the hepatocytes in the periportal area to the midzonal region were observed (Fig. 3A and 3C), and vacuoles were detected and annotated (filled) as vacuolation in yellow by the algorithm (Fig. 3B and 3D). The normal bile ducts within the Glisson’s sheath were annotated with black lines (Fig. 3B). Drug-induced vacuolation or spontaneous vacuolation was classified based on the threshold (6.23%).
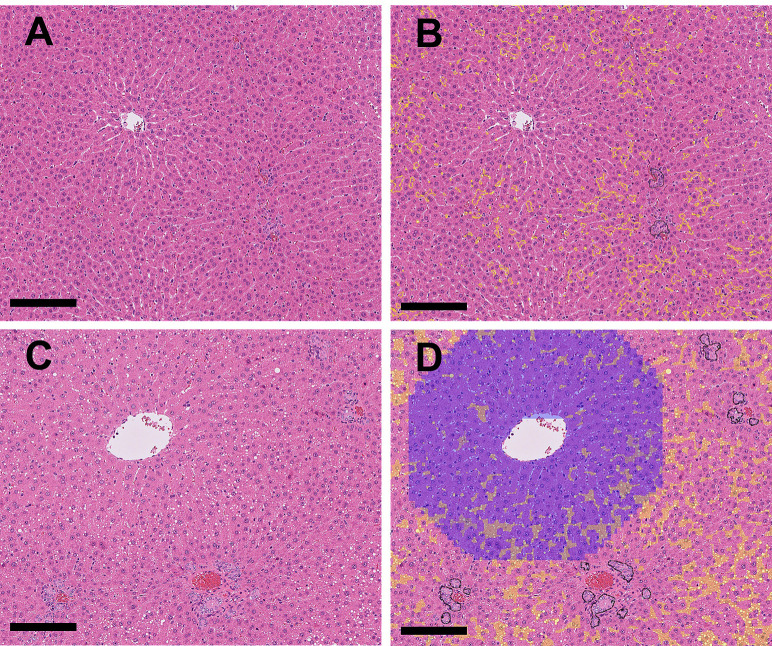
• Bile duct hyperplasia
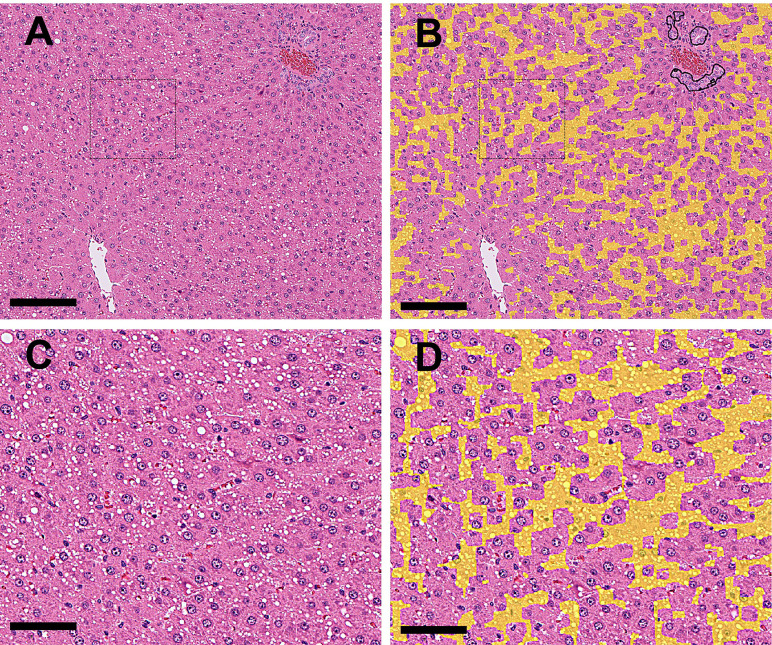
The bile ducts proliferated in the periportal area (Fig. 4A and 4C), and the structure of the bile ducts was detected and annotated with black lines by the algorithm (Fig. 4B and 4D). Normal bile ducts observed in the control group or drug-induced bile duct hyperplasia were determined based on whether the percentage of bile duct positive areas (black annotated areas) on the WSI exceeded the threshold. If the percentage of such areas exceeded the threshold, the lesion was considered to be drug-induced hyperplasia.
Fig. 4.
A: (Original WSI): Bile duct hyperplasia (drug-induced) at the periportal areas was observed. B: (Annotation by the algorithm): The abnormal areas in Fig. 4A (bile duct hyperplasia and vacuolation) were annotated with black and yellow, respectively (Bar=500 μm). C: Higher magnification of the dashed area in Fig. 4A, showing bile duct hyperplasia and vacuolation of hepatocytes in the periportal area. D: Higher magnification of the dashed area in Fig. 4B, annotating the abnormal areas (bile duct hyperplasia and vacuolation) in Fig. 4C with black and yellow, respectively (Bar=100 μm).
• Single cell necrosis of hepatocytes
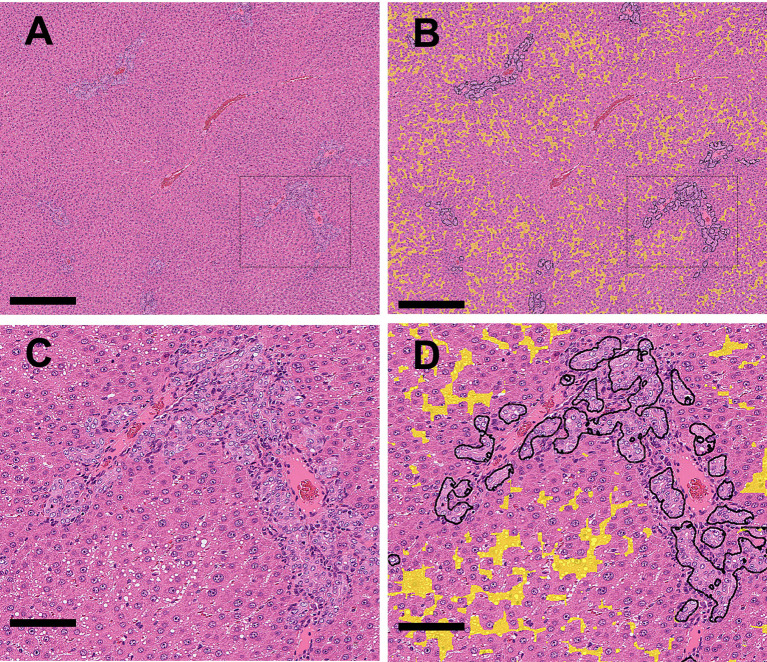
Single-cell necrosis of hepatocytes is annotated with light blue lines. Slightly vacuolated hepatocytes were observed in the same area and were also detected and annotated as vacuolated using yellow lines (Fig. 5A and 5B).
Fig. 5.
A: (Original WSI): Single cell necrosis (arrowheads) and slightly vacuolated hepatocytes were found at the periportal area (drug-induced). B: (Annotation by the algorithm): Abnormal areas (single cell necrosis and vacuolation) in Fig. 5A were annotated with light blue and yellow, respectively (Bar=100 μm).
• Hepatocellular hypertrophy
In the case of hepatocellular hypertrophy, no quantitative values were generated by this model because the algorithm detects this finding based on a variety of parameters, not just a single parameter. Therefore, qualitative data (normal or abnormal) and annotation results were generated. Hepatocellular hypertrophy was not observed in the non-treated liver and was not annotated as “abnormal findings” (Fig. 6A and 6B). By contrast, in the treated liver, drug-induced hepatocellular hypertrophy was observed in the central area and annotated with blue (Fig. 6C and 6D).
Fig. 6.
A: (Original WSI): Non-treated liver (control animal). B: (Annotation by the algorithm): The areas of vacuolation (spontaneous) and bile ducts in Fig. 6A were annotated with yellow and black, respectively. (Bar=200 μm). C: Hepatocellular hypertrophy (drug-induced) is observed in the central area. D: The abnormal area (hypertrophy) in Fig. 6C is annotated with blue.
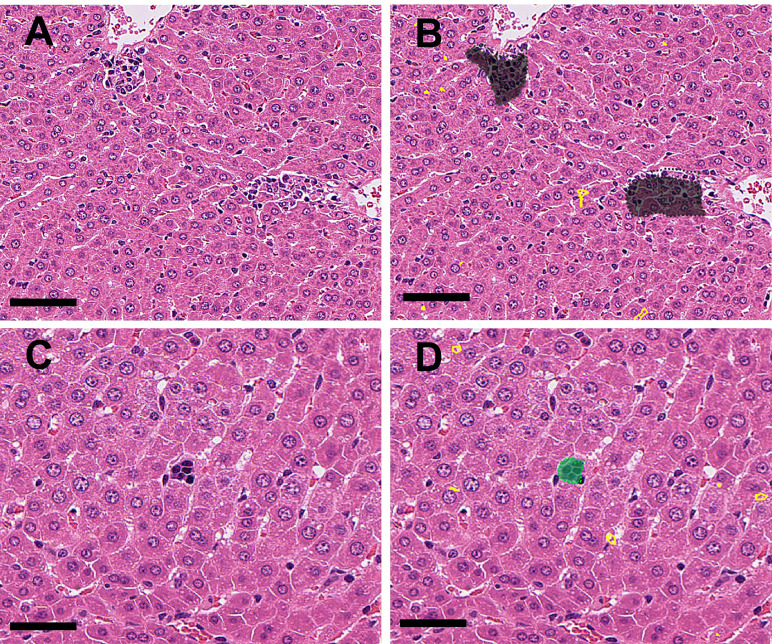
• Microgranuloma and extramedullary hematopoiesis
Microgranuloma (Fig. 7A) and extramedullary hematopoiesis (erythrocytic islands, Fig. 7C), which were also observed as spontaneous lesions in the control group, were detected and annotated with gray and green, respectively (Fig. 7B and 7D).
Fig. 7.
A: (Original WSI): Microgranuloma (spontaneous) near central veins were observed. B: (Annotation by the algorithm): The abnormal area (microgranuloma) in Fig. 7A was annotated with gray (Bar=100 μm). C: An erythroblastic island (spontaneous) was observed in the sinusoids. D: The abnormal area (extramedullary hematopoiesis) in Fig. 7C is annotated as green (Bar=50 μm).
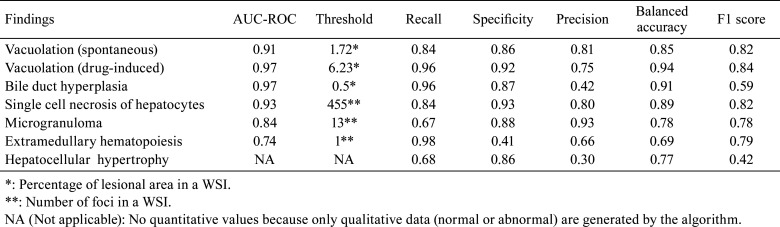
Discriminate performance
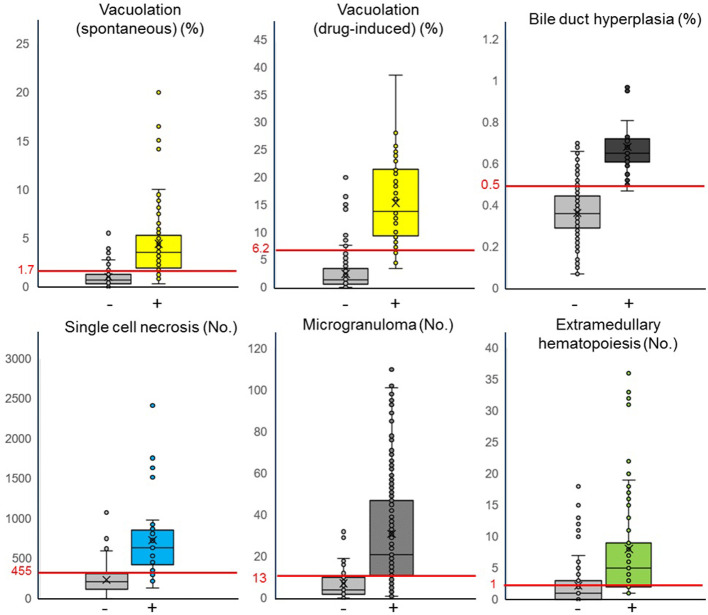
The algorithm quantifies the areas or numbers of annotated lesions for each finding described above. The best threshold values for the quantitative value from the algorithm were calculated through an ROC analysis with the pathologist’s qualitative diagnosis of “no findings” or “abnormal findings” (Table 4). Based on the best threshold values, each quantitative value of the findings below or above the thresholds was judged as “no findings” or “abnormal findings”, respectively (Fig. 8). Figure 8 shows box-and-whisker diagrams based on each quantitative value and the binary classification diagnosed by the pathologists; in addition, the threshold calculated by the ROC curve was found to separate the binary classes well regarding the five findings (two types of vacuolation, bile duct hyperplasia, single cell necrosis, and microgranuloma). This indicates that approximately 75% of the total sample could be classified as a true positive or true negative. By contrast, for extramedullary hematopoiesis, the binary class was not separated well by the threshold, leading to a possible over-detection.
Table 4. Statistical Parameters Derived as Indices for Performance of Lesion Detection for Each Finding.
Fig. 8.
Comparison of the quantitative values between binary classifications by the pathologists. The horizontal axis shows binary classification judged as “no findings (−)” or “abnormal findings (+)” by pathologists, and the vertical axis shows the quantitative values calculated by the algorithm. Here, [%] indicates the ratio and [No.] indicates the number of annotated areas of abnormal findings in the WSI. The red line crosses the vertical axis and its numerical value indicates the threshold value of the finding calculated from the ROC curve. In the “no findings (−)” group, plotted samples above the threshold value indicate false positives, and plotted samples below the threshold value indicate true negatives. By contrast, in the “abnormal finding (+)” group, plotted samples above the threshold value indicate true positives, and plotted samples below the threshold value indicate false negatives. Vacuolation (spontaneous): n=120 (−), 85 (+). Vacuolation (drug-induced): n=205 (−), 50 (+). Bile duct hyperplasia: n=222 (−), 23 (+). Single cell necrosis: n=192 (−), 63 (+). Microgranuloma: n=76 (−), 179 (+). Extramedullary hematopoiesis: n=118 (−), 137 (+). As for the five findings other than extramedullary hematopoiesis, the thresholds bisected the body of the box, indicating that approximately 75% of the total sample could be classified as a true positive or true negative. However, for extramedullary hematopoiesis, the thresholds intersect the body of the box in both groups.
The performance of the algorithm for histopathological findings was examined statistically using the measures indicated in Table 4. Based on the trend of the results of the five assessment indices, the detection performance of the algorithm for each lesion was divided into four groups.
1) A high recall, specificity, precision, balanced accuracy, and F1-score group for vacuolation (spontaneous), vacuolation (drug-induced), and single-cell necrosis had few false positives and false negatives.
2) A high specificity and precision and low recall group for microgranuloma showed a number of false negatives.
3) A high recall and low precision group for bile duct hyperplasia and extramedullary hematopoiesis showed numerous false positives.
4) A high specificity and low recall and precision group for hepatocellular hypertrophy showed numerous false positives and false negatives.
Discussion
In this paper, we present a U-Net-based deep learning algorithm for the classification and quantification of seven histopathological findings in the livers of SD rats. The algorithm detects and quantifies different histopathological findings simultaneously in WSIs. These features are difficult to analyze using conventional image-analysis models.
Table 4 shows that six findings, except for hepatocellular hypertrophy, indicated a high AUC on the ROC curve and the F-score, which is a comprehensive evaluation index of accuracy and comprehensiveness. Figure 8 shows that the bodies of the box for the “no findings” group and “abnormal findings” group were almost neatly divided into two parts at the threshold, indicating that approximately 75% of the total sample could be classified as a true positive or true negative. Although several false positives were reported for some findings, indicating a low precision, the number of false positives was high presumably because we tried to minimize the number of oversights during training. Because the primary purpose of this algorithm is to screen for abnormal findings, the advantage of fewer false negatives was given priority over the disadvantage of more false positives.
In the detection of abnormal areas, findings with clear boundaries with the surrounding normal structures, such as vacuolation and single-cell necrosis, were accurately detected and the statistical scores were high; however, ambiguous boundaries such as hepatocellular hypertrophy were not accurately detected and false positives were frequently found.
Vacuolation and single cell necrosis were well classified and quantified by the algorithm and correlated well with the diagnosis of the pathologist. For vacuolation, we set the quantitative threshold values for the following groups: the no-vacuolation group with almost no vacuolated areas, the vacuolation group with vacuolated areas up to the level of the control group (spontaneous vacuolation), and the vacuolation group with vacuolated areas larger than the areas of spontaneous lesions in the control group (drug-induced vacuolation). The performance of the annotation was high, and vacuoles with a diameter of approximately 2 μm (approximately one-third of the diameter of the nucleus) can be accurately detected even in the 20× scanned WSIs. As a drawback, in addition to the vacuole area, the cytoplasm area of hepatocytes with vacuoles of a certain extent was also detected and quantified, which was higher than the actual vacuolated area. This may be attributed to the insufficient resolution of the scanned image (20× magnification). If scanning at 40× magnification or higher becomes the mainstream in the future, the detection performance will be improved. At present, the algorithm can adequately detect differences among the groups with no findings, spontaneous vacuolation, and drug-induced vacuolation, which is sufficient for toxicity screening studies. In this study, we did not consider the lack of grading of the lesions within the drug-induced group because the first purpose of this study was to detect and classify the abnormalities. To further subdivide the histopathological grade or classify drug-induced “macrovesicular” or “microvesicular” lesions based on the quantitative values, it is necessary to train the algorithm using specimens with more variation in the degree of vacuolation.
For single-cell necrosis, the algorithm can accurately detect areas with lesions in the WSIs. Because a certain number of necrotic cells are usually observed even in the control group (group of “no findings”), setting a threshold and quantifying necrotic cells as above and below the threshold, is an extremely useful way to objectively classify areas “within normal limits” and those showing “drug-induced single cell necrosis”. As the drawback of this algorithm, normal hepatocytes in the control group are occasionally detected as single cell necrosis. The detection algorithm for this finding is based on indicators such as irregularly shaped nuclei (pyknotic nuclei), more eosinophilic cytoplasm, and a change in cytoplasmic morphology from polygonal to round. However, normal hepatocytes may be judged as necrotic cells depending on the variation of the cut surface derived from the specimen preparation process (artifact). In addition, the basic premise of the application of this algorithm is to prevent an overlooking of abnormal findings (false negatives) as much as possible, while allowing the detection of a certain number of false positives. Because this model must be operated in anticipation of a certain number of false positives, it is beneficial to set a threshold to reduce the subsequent noise.
With microgranuloma, the majority of lesions in the WSIs were accurately detected by the algorithm, although the detection accuracy was not equal to that for vacuolation and single-cell necrosis. The specimens were occasionally classified as false negatives based on the threshold. One-fourth of the specimens were classified as false negative because there was a relatively large difference in the cut-off criteria used by pathologists. In the future, this problem can be solved using graded data that correct the differences in criteria among the pathologists. In this study, because the number of microgranuloma rarely increases with drug treatment in rat toxicity studies, as based on our experience, and specimens with drug-induced lesions are sparingly available, only spontaneous microgranuloma were used for training and validating the algorithm. However, we were able to collect values for the background levels of spontaneous microgranuloma. Therefore, in the pathological evaluation, specimens that showed quantitative values exceeding the background level of spontaneous microgranuloma should be carefully examined by pathologists.
Regarding bile duct hyperplasia, the algorithm could detect and classify a wide range of bile duct morphologies from normal to abnormally proliferated bile ducts. In addition, false detection of other structures, such as veins and arteries, as bile ducts is extremely rare. However, large bile duct structures are occasionally found in a specimen, and may be reflected in a higher area percentage of bile ducts. As a result, the number of specimens classified as false positives may occasionally be higher. The noise in this problem can be minimized through the careful examination of each group for dose dependency by the pathologists and setting study-specific threshold values based on the control specimens. In the future, it will be necessary to include training for variations in the grade of lesions of bile duct hyperplasia as well as vacuolation to enable an accurate quantitative grading.
For extramedullary hematopoiesis, the algorithm could detect erythroblast islands on the specimen, with occasional false-positive detection of mononuclear cell infiltration and microgranuloma. Further training is needed to improve the accuracy of this parameter because the livers of 8 week-old male rats show a small number of extramedullary hematopoiesis foci, and even a few false positives can greatly affect the accuracy of the diagnosis.
Typical areas of hepatocellular hypertrophy can be detected. However, even in the control group, normal areas that appeared to be hypertrophic were occasionally judged as abnormal (false positive) depending on artificial variations in the specimen preparation, such as a poor fixation, deformation during fixation, and section extension by hot water. Conversely, the true hypertrophic areas were occasionally overlooked. Overall, the accuracy is insufficient. We need to investigate the parameters necessary for the algorithm to determine normal/abnormal (such as the area ratio of hepatocytes at the periportal and central regions) cases and conduct training to increase the accuracy of these parameters. Certain algorithms have been reported to achieve a high accuracy in detecting hepatocellular hypertrophy12; therefore, we would like to improve our algorithm further by using a variety of samples in the future.
Previous toxicity assessment algorithms in nonclinical studies using AI image-analysis technology focused on detecting and quantifying only a single type of finding in each case. Focusing on and detecting only one specific finding is an extremely useful approach. However, the previous algorithms could neither detect nor classify multiple types of findings simultaneously on a WSI. By contrast, our algorithm can detect, classify, and quantify multiple changes simultaneously on a WSI and supports a wide variety of typical findings for detection. We believe that our algorithm is more useful than others for screening a wide variety of findings in the early stages of drug development.
Importantly, our algorithm can generate accurate analysis results by using WSIs scanned at 20×; therefore, it significantly reduces the data size and duration of digital scanning by 25% compared to WSIs scanned at 40×, which is commonly used in this field.
It is challenging to speed up the selection of candidate compounds for drug development, particularly in the early stages of development. In preclinical toxicity studies during the early stage of drug development, this deep-learning-based algorithm can be a useful tool to optimize the time needed for data generation to evaluate the development feasibility after necropsy. The algorithm can reduce the time and effort pathologists expend in screening normal areas in images, as well as prevent fatigue-induced errors of omission owing to cumbersome work processes. In addition, such solutions can set an objective and quantifiable threshold between cases “within normal limits” and “findings present” to improve the consistency among pathologists.
In actual situations, liver specimens can be scanned at 20× magnification and stored on a server during working hours, with the algorithm batch analysis of the images taking place during non-working hours. The results will then be ready the next working day. The pathologist can then review the analyzed WSIs, and if necessary, cross-check them with glass slides of the histopathology. This allows pathologists to quickly screen areas that the algorithm judges as “non-abnormalities”, and to focus more time on the areas that the algorithm annotates as “abnormal findings”. Therefore, this can be a useful and supportive tool for a histopathological evaluation, particularly for the primary early screening in rat toxicity studies when the speed of a go or no-go decision is critical. This is expected to greatly improve the work efficiency and prevent errors owing to an oversight.
In conclusion, we trained efficient algorithms for the classification and quantification of seven specific histopathological findings in the liver of young SD rats, which, with the exception of hepatocellular hypertrophy, exhibited a high correlation with the diagnoses of pathologists. We believe that this algorithm will contribute to the improvement of the objectivity, accuracy, and reproducibility of histopathological evaluations in toxicity studies. Moreover, the quantification of morphological changes adds a novel dimension to this evaluation method. In addition to the seven findings described above, as well as findings that require further improvement, we are training algorithms to classify and quantify additional findings in the liver that are often encountered in toxicity studies, such as degenerative lesions, in addition to lesions occurring in other organs such as the kidney.
Regarding the detection of drug-induced findings other than the seven findings examined in this study, an unsupervised approach can be adopted to detect them as anomalies/outliers in otherwise normal/known data. The current algorithm for identifying the seven lesions described in this study is based on a supervised approach. However, the learning in terms of normal histology and the specific abnormalities combined with the training data provided us with a strong footing to develop an anomaly detection algorithm based on unsupervised methods. Although this function was not well developed during this study because such findings are rare in young SD rat specimens, we would like to verify and develop this function further in the future. The goal is to develop a versatile and innovative tool for a faster and more efficient pathological evaluation, which will play an auxiliary role for pathologists in future toxicity studies. We will continue to expand this application to other organs to ultimately develop a solution that will accelerate the speed of pharmaceutical drug development.
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest directly relevant to the content of this article.
Acknowledgments
The authors are grateful to the following people for their support of the present study and helpful discussions: Dr. Uttara Joshi of AIRA Matrix and Mr. Yusuke Mashimo, Ms. Mayuko Konoike, and our colleagues from the pathology group of Japan Tobacco, Inc.
References
- 1.Kuklyte J, Fitzgerald J, Nelissen S, Wei H, Whelan A, Power A, Ahmad A, Miarka M, Gregson M, Maxwell M, Raji R, Lenihan J, Finn-Moloney E, Rafferty M, Cary M, Barale-Thomas E, and O’Shea D. Evaluation of the use of single- and multi-magnification convolutional neural networks for the determination and quantitation of lesions in nonclinical pathology studies. Toxicol Pathol. 49: 815–842. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horai Y, Akatsuka A, Mizukawa M, Nishina H, Nishikawa S, Ono Y, Takemoto K, and Mochida H. Current status and prospects for quantitative analysis of digital image of pathological specimen using image processing software including artificial intelligence. Translat Regulat Sci. 2: 72–79. 2020. [Google Scholar]
- 3.Kwak JT, Hewitt SM, Kajdacsy-Balla AA, Sinha S, and Bhargava R. Automated prostate tissue referencing for cancer detection and diagnosis. BMC Bioinformatics. 17: 227. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atupelage C, Nagahashi H, Kimura F, Yamaguchi M, Tokiya A, Hashiguchi A, and Sakamoto M. Computational hepatocellular carcinoma tumor grading based on cell nuclei classification. J Med Imaging (Bellingham). 1: 034501. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, and Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 21: 249–257. 2018. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto Y, Offord CP, Kimura G, Kuribayashi S, Takeda H, Tsuchiya S, Shimojo H, Kanno H, Bozic I, Nowak MA, Bajzer Ž, and Dingli D. Tumour and immune cell dynamics explain the PSA bounce after prostate cancer brachytherapy. Br J Cancer. 115: 195–202. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namikawa K, Hirasawa T, Yoshio T, Fujisaki J, Ozawa T, Ishihara S, Aoki T, Yamada A, Koike K, Suzuki H, and Tada T. Utilizing artificial intelligence in endoscopy: a clinician’s guide. Expert Rev Gastroenterol Hepatol. 14: 689–706. 2020. [DOI] [PubMed] [Google Scholar]
- 8.Sharma P, Suehling M, Flohr T, and Comaniciu D. Artificial intelligence in diagnostic imaging: status quo, challenges, and future opportunities. J Thorac Imaging. 35(Suppl 1): S11–S16. 2020. [DOI] [PubMed] [Google Scholar]
- 9.Calderaro J, and Kather JN. Artificial intelligence-based pathology for gastrointestinal and hepatobiliary cancers. Gut. 70: 1183–1193. 2021. [DOI] [PubMed] [Google Scholar]
- 10.Okanoue T, Shima T, Mitsumoto Y, Umemura A, Yamaguchi K, Itoh Y, Yoneda M, Nakajima A, Mizukoshi E, Kaneko S, and Harada K. Artificial intelligence/neural network system for the screening of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatol Res. 51: 554–569. 2021. [DOI] [PubMed] [Google Scholar]
- 11.Roy M, Wang F, Vo H, Teng D, Teodoro G, Farris AB, Castillo-Leon E, Vos MB, and Kong J. Deep-learning-based accurate hepatic steatosis quantification for histological assessment of liver biopsies. Lab Invest. 100: 1367–1383. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa T, Horai Y, Asaoka Y, Sakurai T, Kikuchi S, Yamaoka M, and Tanaka M. Current status of pathological image analysis technology in pharmaceutical companies: a questionnaire survey of the Japan Pharmaceutical Manufacturers Association. J Toxicol Pathol. 33: 131–139. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creasy DM, Panchal ST, Garg R, and Samanta P. Deep learning-based spermatogenic staging assessment for hematoxylin and eosin-stained sections of rat testes. Toxicol Pathol. 49: 872–887. 2021. [DOI] [PubMed] [Google Scholar]
- 14.Tokarz DA, Steinbach TJ, Lokhande A, Srivastava G, Ugalmugle R, Co CA, Shockley KR, Singletary E, Cesta MF, Thomas HC, Chen VS, Hobbie K, and Crabbs TA. Using artificial intelligence to detect, classify, and objectively score severity of rodent cardiomyopathy. Toxicol Pathol. 49: 888–896. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pischon H, Mason D, Lawrenz B, Blanck O, Frisk AL, Schorsch F, and Bertani V. Artificial intelligence in toxicologic pathology: quantitative evaluation of compound-induced hepatocellular hypertrophy in rats. Toxicol Pathol. 49: 928–937. 2021. [DOI] [PubMed] [Google Scholar]
- 16.Ramot Y, Zandani G, Madar Z, Deshmukh S, and Nyska A. Utilization of a deep learning algorithm for microscope-based fatty vacuole quantification in a fatty liver model in mice. Toxicol Pathol. 48: 702–707. 2020. [DOI] [PubMed] [Google Scholar]
- 17.Ronneberger O, Fischer P, and Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer. 2015.
- 18.Abraham N, and Khan NM. A novel focal Tversky loss function with improved attention U-Net for lesion segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Venice. 683–687. 2019.
- 19.Kingma D, and Ba J. Adam: A Method for Stochastic Optimization. International Conference on Learning Representations. In: 3rd International Conference for Learning Representations. San Diego. 2015.