Abstract
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative, facultative intracellular bacterium, which causes gastrointestinal disorders in humans, and systemic, typhoid fever-like infections in mice. Our current knowledge regarding the involvement of cellular and humoral immunity in the defense from S. Typhimurium infections is largely based on animal models with attenuated strains. Cells of the innate immune system act as one of the first barriers in the defense from bacteria. We established a robust experimental model for the characterization of these cell types and their response during host-pathogen interactions. Therefore, this protocol focuses on the characterization of macrophages, monocytes, and neutrophils in the spleens of infected animals by employing multi-color flow cytometry.
Keywords: Macrophages, Intracellular bacteria, Gram-negative bacteria, Infection control
Background
Salmonella enterica is a highly iron-dependent siderophilic intracellular Gram-negative pathogen, which can cause local intestinal disease, or severe systemic infection and septicemia. Hence, it was included in the WHO list of the most serious infectious disease threats to human health (Keestra-Gounder et al., 2015; Bumann and Schothorst, 2017). Salmonella invades and multiplies within mononuclear phagocytic cells in the liver, spleen, lymph nodes, and Peyer’s patches, in so-called Salmonella-containing vacuoles (SCV) (Steele-Mortimer, 2008). However, Salmonella exhibits different mechanisms to evade antimicrobial activities, for example by inhibiting phagolysosomal fusion (Baumler and Fang, 2013; Bhutta et al., 2018).
Several immune cells are important in the defense against bacterial infections. While some act as the first barrier, by taking up and killing bacteria, others are responsible for long-term immunity or for coordinating efficient anti-microbial immune responses. Macrophages, dendritic cells (DCs), and neutrophils represent one of the first lines of defense against invading pathogens like Salmonella, by recognizing specific pathogen-associated molecular patterns (PAMPs), and danger-associated-molecular patterns (DAMPs) (de Jong et al., 2012). Recognition of Gram-negative bacteria in macrophages involves the binding of LPS to a receptor complex, leading to the secretion of pro- and anti-inflammatory cytokines (Nagai et al., 2002; Bode et al., 2012). Macrophages can polarize dependending on local factors within the microenvironment, microbial stimuli, and cytokines secreted by other cells. Pro-inflammatory macrophages are essential components of anti-microbial host defense mechanisms, and can be characterized by the synthesis of inducible nitric oxide synthase (iNOS). Anti-inflammatory macrophages dampen the inflammatory state, by producing anti-inflammatory cytokines and Arginase-1 (ARG1). ARG1 cleaves L-arginine, the substrate of iNOS, and thereby impairs the control of various intracellular pathogens (Mosser and Edwards, 2008; Murray, 2017).
Several studies have shown that Salmonella is also phagocytosed by dendritic cells, which serve as an important link between innate and adaptive immunity (Steinman and Hemmi, 2006). Upon phagocytic internalization of bacteria, DCs mature and migrate to defined lymphoid tissues. Thereby, they increase the expression of the major histocompatibility complex II (MHCII) to present specific antigens to T cells, initiating adaptive immune response.
Due to the widespread development of multidrug resistances against antibiotics, current antibiotic treatments of invasive salmonellosis is often not successful. In addition, it is uncertain whether standard antibiotics can penetrate into the Salmonella-containing vacuole. Thus, it is necessary to better understand the host-pathogen interplay in salmonellosis, to develop novel effective antimicrobial therapies targeting intracellular pathogens (Lahiri et al., 2010;Navarre et al., 2010;Mastroeni and Grant, 2011).
Materials and Reagents
10 cm bacteriological Petri dish (Falcon, catalog number: 351029)
Cell strainer 100 µm (Falcon, catalog number: 352360)
5 ml syringe (BD, DiscarditTM II 309050)
Sterile wedge shaped spreader (Microspec, catalog number: 211738)
Syringes for intraperitoneal injection (30G × ½’’ 0.3 mm × 12 mm, Braun, Omnican F 9161502S)
1.5 mL Eppendorf Tubes (Eppendorf, catalog number: T9661)
96 well round-bottom plates (BRAND, catalog number: 781600)
Cryovials 2 mL round bottom (Simport, catalog number: T311-3)
Salmonella enterica serovar Typhimurium ATCC14028 (ATCC)
LB Broth Lennox (Roth, catalog number: X964.2)
Phosphate buffer saline (PBS) (Lonza, catalog number: 17-515 F)
Agar-Agar Kobe I (Roth, catalog number: 5210.3)
Paraformaldehyde (Sigma-Aldrich, Formalin-solution, neutral buffered 10%, HT501128)
APC anti-mouse CD11b (BioLegend, catalog number: 101212)
FITC anti-mouse CD45 (BioLegend, catalog number: 103108)
BV421 rat anti-mouse F4/80 (BD Biosciences, catalog number: 565411)
BV510 anti-mouse Ly6C (BioLegend, catalog number: 128033)
PerCP-eFluorTM 710 anti-mouse Ly6G (Invitrogen, catalog number: 46-9668-82)
PerCP/Cyanine5.5 anti-mouse MHCII (BioLegend, catalog number: 116416)
BV421 anti-mouse CD11c (BioLegend, catalog number: 117330)
PE-Cyanine7 anti-mouse iNOS (Invitrogen, catalog number: 25-5920-80)
anti-mouse ARG1 (Invitrogen, catalog number: 17-3697-82)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Fetal Bovine Serum heat inactivated (FBS) (PAN Biotech, catalog number: P30-3031)
Ethylenedinitrilotetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: 254134)
Potassium hydrogen carbonate (KHCO3) (Sigma-Aldrich, catalog number: 237205)
Ethylenediaminetetraacetic acid, disodium salt (Na2EDTA) (Sigma-Aldrich, catalog number: E5134)
Triton X-100 (Roth, catalog number: 3051.3)
LB-Agar plates (see Recipes)
ACK lysis buffer (see Recipes)
FACS buffer (see Recipes)
Triton buffer (see Recipes)
LB agar plates (see Recipes)
LB medium (see Recipes)
4% PFA (see Recipes)
LB-medium with 30% glycerol (see Recipes)
Equipment
Incubator 37°C (ELOS Breed, catalog number: B110N)
Shaking incubator (VWR, GFL, catalog number: 3031)
Photometer (Eppendorf, BioPhotometer D30, catalog number: 6133000001)
Centrifuge (Hettich Micro 200R, catalog number: Z652113)
CytoFLEX S V4-B4-R2-I2 Flow Cytometer (13 detectors, 4 lasers, Beckman Coulter, catalog number: C01161)
CASY TT counting system (OMNI Life Science, catalog number: TT-20A-2571)
Software
FlowJo v10.7.0 (BD Biosciences, https://www.flowjo.com/solutions/flowjo)
GraphPad Prism V8.4.1 (https://www.graphpad.com/scientific-software/prism)
Procedure
-
Grow Salmonella enterica serovar Typhimurium ATCC14028 in an Erlenmeyer flask, until they reach the logarithmic growth phase
Open the original vial and rehydrate the entire pellet in 1 mL of LB-medium.
Mix well by pipetting up and down.
-
Pipette 10 µL in 10 mL of LB-medium as pre-culture.
Note: Centrifuge the remaining 950 µL at 1920 × g at room temperature (RT) for 5 min, resuspend the pellet in 1 mL of LB-medium containing 30% Glycerol, aliquot as 10 × 100 µL in cryovials, and freeze the aliquots at -80°C. This will be the stock for the pre-culture in further experiments.
Incubate the pre-culture in a rotating incubator at 200 rpm at 37°C overnight.
Next day, pipette 50 µL of this overnight pre-culture in 10 mL of LB-medium in an Erlenmeyer flask.
Incubate in an orbital shaker at 200 rpm at 37°C for 1–2 h, until an OD600 of 0.5 (measured in a photometer using LB-medium as blank).
-
Counting of viable Salmonella enterica serovar Typhimurium using for example a CASY TT counting system
Use the 45 µm capillary.
Measure the background, by placing a new Casy cup with fresh Casy ton buffer under the measuring unit.
Select program for background measurement (Table 1).
Measure background. This should be below 30 counts and 1 µm size. Otherwise, wash the system.
Prepare a new Casy cup with 10 mL of Casy ton buffer, and add 5 µL of S. typhimurium solution.
Shake gently.
Place sample under the measuring unit.
Select program for measuring between 1–3 µm (Table 1).
Measure.
Click next to get the number of viable counts per milliliter = viable S.tm/mL.
After the measurement is completed, remove the sample cup, and add a fresh Casy cup with 10 mL Casy ton buffer.
Perform Casy Clean up to five times.
Select Program for Washing (Table 1).
After washing is completed, check the background.
If the background is below 30, the Casy counting system can be turned off; otherwise, continue washing.
-
Intraperitoneal injection of mice
Using Omnican F syringes (30G × ½’’ (0.3 mm × 12 mm), intraperitoneally (i.p.) inject mice with 1000 bacteria in 200 µL of PBS, according to the measurement in the Casy counting system.
Negative control mice are injected with 200 µL of PBS.
A video demonstrating the procedure can be found at: Intraperitonelal Injection in the Mouse: https://researchanimaltraining.com/articles/intraperitoneal-injection-in-the-mouse/.
Notes:
The injection of mice is performed in a bio-safety level 2 animal facility, where mice are housed in individually ventilated cages (IVC). After infection, all cages, litter, and used material must be autoclaved.
Weight and body surface temperature are important markers for monitoring the mice during the infection. Therefore, mice should be familiarized to daily weighing and temperature measurements 3–5 days prior to the experiment. This will avoid stress-induced weight loss and changes in body temperature.
All substances used should be warmed to RT. The injection of cold substances influences the wellbeing of the mice, and can lead to a sudden drop in body temperature.
-
Monitoring of mice during the infection
Note: Mice on a C57BL/6 background express a non-functional NRAMP1 (natural resistance associated macrophage protein 1) protein. NRAMP is a phagolysosomal membrane transporter for iron, protons, and divalent cations, and is associated with host resistance to various intracellular pathogens. Non-functional NRAMP leads to intracellular persistence of S. typhimurium within macrophages, and early death after 5 days at the latest. Therefore, it is crucial to monitor the mice in the most careful way.
-
Body weight:
The body weight should be measured at the same time each day, to avoid behavioral changes due to food intake. It is important to use a lockable box and a precision scale, which can be disinfected.
-
Body temperature:
The body temperature is measured with an infrared thermometer at the same time each day.
Pick the mouse up by the middle of the tail and expose its abdomen. Allow the mouse to hold on to the grid of a cage with its forepaws, which allows the mouse to stretch its upper body and expose the abdomen. Hold the trigger to measure the temperature, taking care to aways measure at the same location of the abdomen.
-
General appearance of the mice:
The general appearance of the mice should be documented twice a day, for example according to the M-CASS scoring sheet in Lilley et al. (2015), and your experimental animal license. Severe pain, suffering, and distress must be prevented.
-
-
Isolation of splenocytes
After 72 h of infection, isolate the spleens, and press the organs through a 100 µm cell strainer with the plunger of a 5 mL syringe, onto a Petri dish with 5 mL of cold PBS.
Centrifuge at 300 × g and 4°C for 5 min, and wash once with 5 mL of PBS (300 × g at 4°C for 5 min), discard the supernatant.
Subject the cell pellet to erythrocyte lysis, by resuspending in 2 mL of ACK lysis buffer.
Incubate at RT for 2 min.
Wash once with 5 mL of FACS buffer (300 × g at 4°C for 5 min).
-
Flow Cytometry stain of innate immune cells
All procedures are performed in 1.5 mL Eppendorf tubes.
-
Extracellular stain (Table 2)
Prepare an appropriate mix of antibodies (1:200) in 100 µL of FACS buffer per sample (Table 2).
Resuspend the cell pellet in this mix.
Incubate in the dark at 4°C for 15 min.
Wash once with 1,000 µL of FACS buffer (300 × g at 4°C for 5 min).
Resuspend the pellets in 4% paraformaldehyde (PFA) to fix the cells for intracellular staining.
Transfer into 96-well round-bottom plates.
Analyze directly in a flow cytometer.
-
Intracellular stain (Table 3)
Prepare extracellular stain as described in steps 1a–d.
Resuspend the pellet in 4% PFA.
Incubate at 4°C for 15 min in the dark.
Centrifuge and resolve the pellet in Triton buffer.
Mix well and incubate at room temperature for 15 min in the dark.
Prepare appropriate mix of antibodies (1:100) in 50 µL of Triton buffer per sample (Table 3).
Resuspend the cell pellet in this mix.
Incubate at 4°C for 45 min.
Wash once with 1,000 µL Triton buffer (300 × g at 4°C for 5 min).
Resuspend the pellets in 4% PFA.
Transfer into 96-well round-bottom plates.
Analyze directly in a flow cytometer.
-
Table 1. Programs CASY TT Counting system.
| Background Measurement | Measurement of S. Typhimurium |
Washing Program |
|
|---|---|---|---|
| Capillary | 45 µm X-Axis: 5 µm | 45 µm X-Axis: 3 µm | 45 µm X-Axis: 5 µm |
| Sample Volume | 200 µL Cycles: 1 | 200 µL Cycles: 3 | 200 µL Cycles: 10 |
| Dilution | 1.00 × 100 | 2.00 × 103 | 1.00 × 100 |
| Y-Axis | Auto | Auto | Auto |
| Eval.Cursor | 1.00–4.89 µm | 0.75–2.93 µm | 0.00–5.00 µm |
| Norm. Cursor | 0.5–4.89 µm | 0.3–2.93 µm | 0.00–5.00 µm |
| %Calculation | %ViaDebris: On | %Via Debris: On | %Via Debris: On |
| Aggregation Correction | Auto | Auto | Auto |
| Interface | Par P.Feed: On | Par P.Feed: On | Par P.Feed: On |
| Print Mode | Manual Graphic: On | Manual Graphic: On | Manual Graphic: On |
Table 2. Antibodies for flow cytometry – extracellular stain.
| Antibody | Clone | Company | Catalog number | expressed on |
|---|---|---|---|---|
| CD11b APC | M1/70 | BioLegend | 101212 | neutrophils, monocytes |
| CD45 FITC | 30-F11 | BioLegend | 103108 | leukocytes |
| F4/80 BV421 | T45-2342 | BD Biosciences | 565411 | macrophages |
| Ly6C BV510 | HK1.4 | BioLegend | 128033 | monocytes |
| Ly6G PerCP-eFluorTM710 | 1A8-Ly6g | Invitrogen | 46-9668-82 | neutrophils |
| MHCII PerCP/Cyanine5.5 | AF6-120.1 | BioLegend | 116416 | dendritic cells |
| CD11c BV421 | N418 | BioLegend | 117330 | dendritic cells |
Table 3. Antibodies for flow cytometry – intracellular stain.
| Antibody | Clone | Company | Catalog number | expressed on |
|---|---|---|---|---|
| iNOS PE-Cyanine7 | CXNFT | Invitrogen | 25-5920-80 | pro-inflammatory macrophages |
| ARG1 APC | A1exF5 | Invitrogen | 17-3697-82 | anti-inflammatory macrophages |
Data analysis
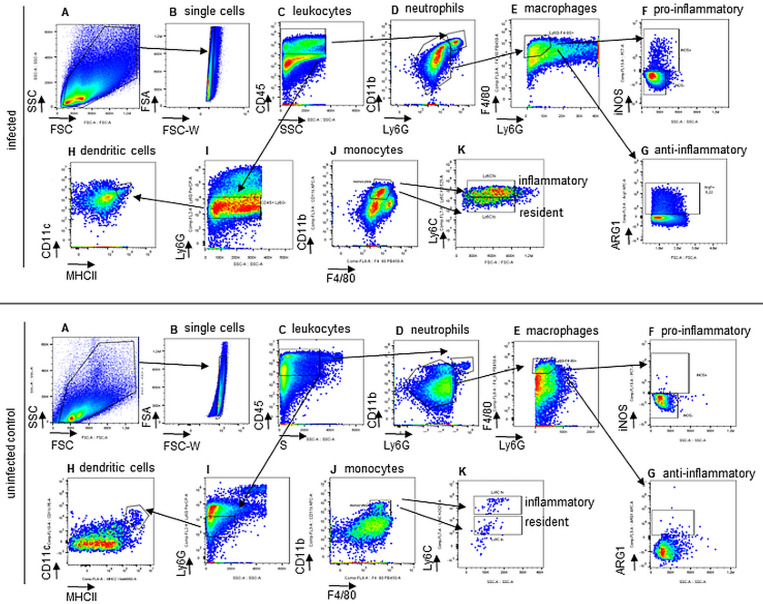
FlowJo software can be used to analyze data. Plot a linear FSC (forward scatter) versus SSC (side scatter) dot plot, and create a gate to select all cells. Using these cells, select CD45+ cells (pan-leucocytes), and follow the gating strategy described in Figure 1.
Figure 1. Gating strategy for the analysis of innate immune cells (infected mouse - upper panel, and uninfected mouse – lower panel).
(A) forward scatter (FSC) against side scatter (SSC), (B) exclusion of doublets, (C) leukocytes: CD45+, (D) neutrophils: CD45+Ly6G+CD11b+, (E) macrophages: CD45+CD11b-Ly6G-F4/80+, (F) proinflammatory macrophages express iNOS, (G) antiinflammatory macrophages express Arginase, (H) dendritic cells: CD45+Ly6G-CD11c+MHCII+, (I) + (J) monocytes: CD45+Ly6G-F4/80+CD11b+, (K) inflammatory and resident monocytes: Ly6Chigh or Ly6Clow, respectively.
Statistical analysis and graphs are performed and generated in GraphPad Prism. When data are normally distributed (assess by using histograms and statistical tools, for example the Shapiro-Wilk test), analyze by the unpaired Student’s t-test for samples with equal variances, or by two-way ANOVA (F test). When data are not normally distributed, the Mann-Whitney U, or an ANOVA on ranks tests can be performed.
Recipes
-
FACS buffer
PBS with 0.5% heat-inactivated FBS
2 mM EDTA
-
ACK (Ammonium-Chloride-Potassium) lysis buffer
150 mM NH4Cl
10 mM KHCO3
0.1 mM Na2EDTA
pH 7.2–7.4
-
Triton buffer
PBS with 0.05% Triton X-100
-
LB agar plates
2% LB-Broth
1.5% Agar-Agar
Autoclave (121°C for 20 min), cool down, and pipette 15 mL in 10 cm bacteriological Petri dishes.
Store at 4°C.
-
LB medium
2% LB-Broth
Autoclave (121°C for 20 min).
-
4% PFA
Add 40 µL of PFA to 960 µL of PBS.
-
LB-medium with 30% glycerol
Add 300 µL of glycerol to 700 µL of LB-medium.
Acknowledgments
Author G.W. is supported by grants from the Christian Doppler Society and an ERA-NET grant by the FWF (EPICROSS, I-3321), N.B. was supported by the FWF doctoral college project W1253 HOROS. This protocol was adapted and modified after Nairz et al. (2009).
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Ethics
Animal experiments were approved by the Austrian Federal Ministry of Science and Research according to the directive 2010/63/EU.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Baumler A. and Fang F. C.(2013).Host specificity of bacterial pathogens.Cold Spring Harb Perspect Med 3(12):a010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhutta Z. A.,Gaffey M. F.,Crump J. A.,Steele D.,Breiman R. F.,Mintz E. D.,Black R. E.,Luby S. P. and Levine M. M.(2018).Typhoid Fever: Way Forward.Am J Trop Med Hyg 99(3_Suppl): 89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lahiri A.,Lahiri A.,Iyer N.,Das P. and Chakravortty D.(2010).Visiting the cell biology of Salmonella infection.Microbes Infect 12(11):809-818. [DOI] [PubMed] [Google Scholar]
- 4. Lilley E.,Armstrong R.,Clark N.,Gray P.,Hawkins P.,Mason K.,Lopez-Salesansky N.,Stark A. K.,Jackson S. K.,Thiemermann C.,et al.(2015).Refinement of animal models of sepsis and septic shock.Shock 43(4):304-316. [DOI] [PubMed] [Google Scholar]
- 5. Mastroeni P. and Grant A. J.(2011).Spread of Salmonella enterica in the body during systemic infection: unravelling host and pathogen determinants.Expert Rev Mol Med 13:e12. [DOI] [PubMed] [Google Scholar]
- 6. Navarre W. W.,Zou S. B.,Roy H.,Xie J. L.,Savchenko A.,Singer A.,Edvokimova E.,Prost L. R.,Kumar R.,Ibba M.,et al.(2010).PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica.Mol Cell 39(2):209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bode J. G.,Ehlting C. and Haussinger D.(2012).The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis.Cell Signal 24(6):1185-1194. [DOI] [PubMed] [Google Scholar]
- 8. Bumann D. and Schothorst J.(2017).Intracellular Salmonella metabolism.Cell Microbiol 19(10). [DOI] [PubMed] [Google Scholar]
- 9. de Jong H. K.,Parry C. M.,van der Poll T. and Wiersinga W. J.(2012).Host-pathogen interaction in invasive Salmonellosis.PLoS Pathog 8(10):e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keestra-Gounder A. M.,Tsolis R. M. and Baumler A. J.(2015).Now you see me, now you don't: the interaction of Salmonella with innate immune receptors.Nat Rev Microbiol 13(4):206-216. [DOI] [PubMed] [Google Scholar]
- 11. Mosser D. M. and Edwards J. P.(2008).Exploring the full spectrum of macrophage activation.Nat Rev Immunol 8(12):958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murray P. J.(2017).Macrophage Polarization.Annu Rev Physiol 79:541-566. [DOI] [PubMed] [Google Scholar]
- 13. Nagai Y.,Akashi S.,Nagafuku M.,Ogata M.,Iwakura Y.,Akira S.,Kitamura T.,Kosugi A.,Kimoto M. and Miyake K.(2002).Essential role of MD-2 in LPS responsiveness and TLR4 distribution.Nat Immunol 3(7):667-672. [DOI] [PubMed] [Google Scholar]
- 14. Nairz M.,Theurl I.,Schroll A.,Theurl M.,Fritsche G.,Lindner E.,Seifert M.,Crouch M. L.,Hantke K.,Akira S.,et al.(2009).Absence of functional Hfe protects mice from invasive Salmonella enterica serovar Typhimurium infection via induction of lipocalin-2.Blood 114(17):3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steele-Mortimer O.(2008).The Salmonella-containing vacuole: moving with the times.Curr Opin Microbiol 11(1):38-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steinman R. M. and Hemmi H.(2006).Dendritic cells: translating innate to adaptive immunity.Curr Top Microbiol Immunol 311:17-58. [DOI] [PubMed] [Google Scholar]