Abstract
Craniofacial anomalies (CFA) are a diverse group of deformities, which affect the growth of the head and face. Dysregulation of cranial neural crest cell (NCC) migration, proliferation, differentiation, and/or cell fate specification have been reported to contribute to CFA. Understanding of the mechanisms through which cranial NCCs contribute for craniofacial development may lead to identifying meaningful clinical targets for the prevention and treatment of CFA. Isolation and culture of cranial NCCs in vitro facilitates screening and analyses of molecular cellular mechanisms of cranial NCCs implicated in craniofacial development. Here, we present a method for the isolation and culture of cranial NCCs harvested from the first branchial arch at early embryonic stages. Morphology of isolated cranial NCCs was similar to O9-1 cells, a cell line for neural crest stem cells. Moreover, cranial NCCs isolated from a transgenic mouse line with enhanced bone morphogenetic protein (BMP) signaling in NCCs showed an increase in their chondrogenic differentiation capacity, suggesting maintenance of their in vivo differentiation potentials observed in vitro. Taken together, our established method is useful to visualize cellular behaviors of cranial NCCs.
Keywords: NCCs, First branchial arch, Isolation, Primary culture, BMPs, Chondrogenic differentiation, Craniofacial development
Background
Cranial neural crest cells (NCCs) form the face and the anterior part of the head ( Jiang et al., 2002 ; Chai and Maxson, 2006; Mishina and Snider, 2014). Cranial NCCs are one of the subpopulations of NCCs that show unique characteristicis by differentiating into bones and cartilage, while other populations of NCCs do not have this differentiation potential (Stemple and Anderson, 1992; Jiang et al., 2000 ; Ishii et al., 2012 ). Cranial NCCs delaminate from the neural tube and migrate into branchial arches (BAs) (Santagati and Rijli, 2003; Kaucka et al., 2016 ; Yang et al., 2021 ). Subsequently, they give rise to neural tissues, including peripheral nerve systems but also connective tissues, such as bones, cartilage, and adipose tissues (Santagati and Rijli, 2003). Several signaling pathways are known to regulate differentiation, proliferation, migration, and cell specification for cranial NCCs, which have been reported to develop craniofacial anomalies (CFA), such as cleft palate and craniosynostosis ( Teng et al., 2008 ; Komatsu et al., 2013 ; He and Soriano, 2013). CFA are a diverse group of pathologic conditions caused by environmental and genetic reasons. Therefore, a deeper understanding of the molecular and cellular mechanisms underpinning cranial NCCs function may result in improved identification, prevention, and treatment of CFA in human patients.
In the 1980s, a method to generate chick-quail chimeras was developed, and cell lineage tracing experiments of neural crest cells during embryonic development became popular (Couly and Dourin, 1985). After establishment of Cre-loxP technologies, transgenic mouse lines expressing Cre recombinase driven by promoters of NCC marker genes, such as Wnt1 and Protein zero (P0) (Wnt1-Cre and P0-Cre lines), facilitated cell lineage tracing during neural crest migration and differentiation ( Danielian et al., 1998 ; Yamauchi et al., 1999 , Chen et al., 2017 ). Using the Cre-loxP technologies, genes of interest are specifically mutated in NCCs to introduce either gain-of-function (GOF) or loss-of-function (LOF) mutations, which significantly deepen our understanding of the mechanisms of craniofacial and trunk development mediated by NCCs. To further gain insight into mechanisms which control proliferation, cell fate specification, and differentiation of cranial NCCs, it is important to establish an in vitro culture system to maintain the character of cranial NCCs.
Bone morphogenetic proteins (BMPs) are a group of growth factors that bind to receptors on the plasma membrane that transduce a signal mediated by SMAD proteins. The levels of BMP-SMAD signaling are important to determine a range of cell behaviors including fate specification; therefore, developing an understanding of the fine-tuning mechanisms of BMP signaling in cranial NCCs is critical. We and others have reported that mutant mice with GOF or LOF of BMP type1A receptor (Bmpr1a) in NCCs display deformities in the craniofacial region ( Komatsu et al., 2013 ; Li et al., 2013 ; Gu et al., 2014 ). In addition to that, we have recently reported that when ACVR1 (caAcvr1), another BMP type 1 receptor, is constitutively activated in NCCs of transgenic mice (caAcvr1; P0-Cre mice, hereafter), these formed ectopic cartilage in their face ( Yang et al., 2021 ). Thus, we have proposed that enhanced BMP signaling pushes cranial NCCs to the chondrogenic fate at early embryonic stages.
We established the isolation and culture method for cranial NCCs from the first branchial arch (BA1) at an early embryonic stage, day 10.5 embryos (Figures 1,2). Isolated cranial NCCs from caAcvr1; P0-Cre mice showed greater chondrogenic differentiation capacity than cranial NCCs from control mice (Figure 3), suggesting that the isolated cranial NCCs from BA1 maintain the differentiation potential observed in vivo. This isolation method is an important tool to analyze molecular and cellular mechanisms of craniofacial development.
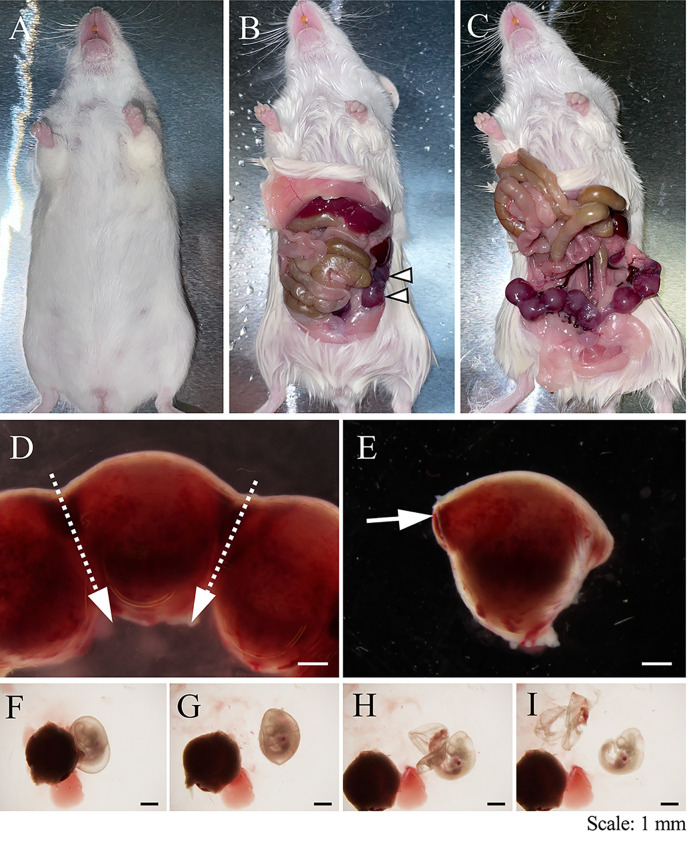
Figure 1. Harvest embryos from a pregnant mouse.
(A–C). A pregnant mouse was euthanized with CO2. The skin was cut with a pair of scissors, and a uterus with embryos (arrowheads) was exposed by pushing the digestive tract upward anteriorly. (D). The uterus was cut with a pair of scissors, by following the dotted arrows (top to bottom). (E). A tweezer was inserted as per the arrow, to separate the uterine wall from the embryo. (F–I). Embryos were carefully removed from the placenta, the yolk sac, and the amnion. Scale bars: 1 mm.
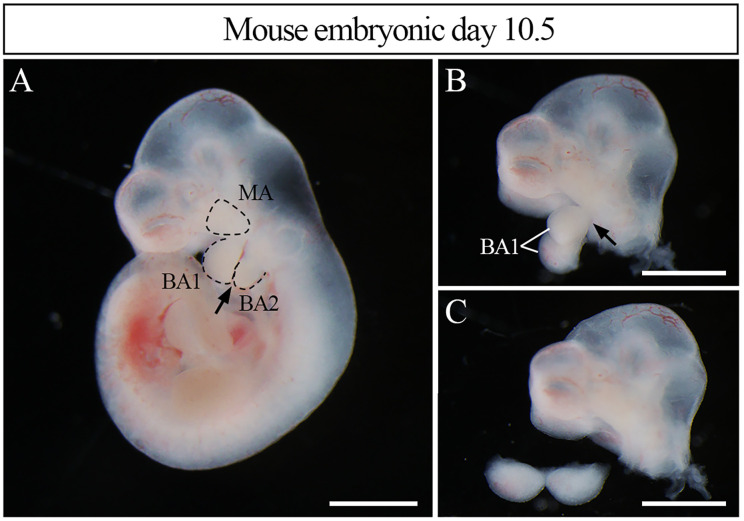
Figure 2. Harvest the first branchial arch from embryo.
(A). Embryos were divided into the head and the trunk. A tweezer was inserted between BA1 and BA2, as following the arrow. (B, C). The BA1 was divided from the MA with a tweezer, as following the arrow in B, and the isolated BA1 was transferred to a 1.5 ml tube. MA; maxillary arch, BA1; first branchial arch, BA2; second branchial arch. Scale bars: 1 mm.
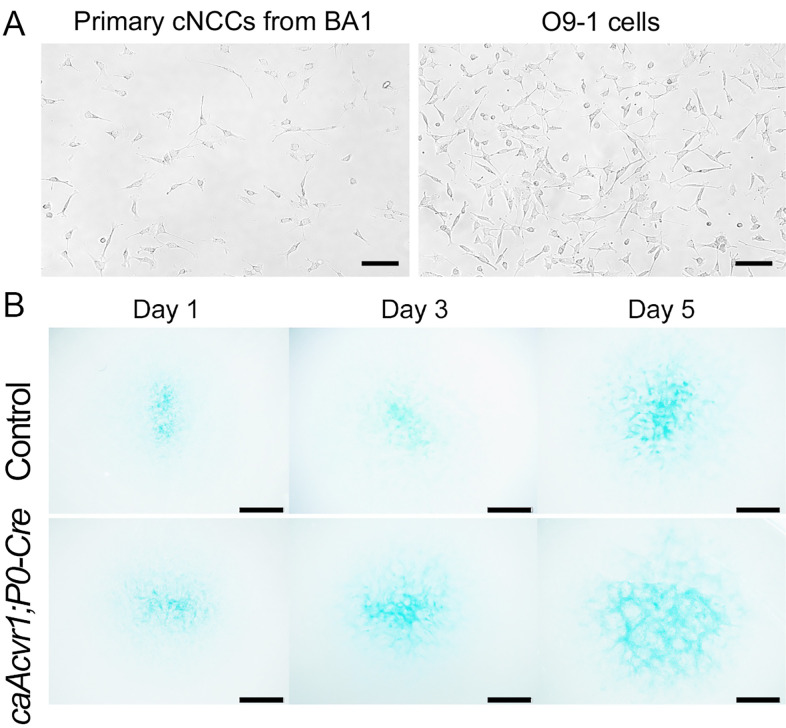
Figure 3. Observation of primary cranial NCCs in vitro.
(A). Harvested primary cranial NCCs from BA1 are shown. The morphology of the cranial NCCs from BA1 is close to O9-1 cells ( Ishii et al., 2012 ), a neural crest stem cell line. (B). Micromass culture for cranial NCCs from control and caAcvr1; P0-Cre mice embryos was performed. Cells were stained with Alcian blue. Scale bars (A) 100 µm and (B) 1 mm.
Materials and Reagents
1.5 mL tube
Wide bore filtered 1,000 µL pipette tips (Thermo Fisher Scientific, catalog number: 2079G)
35 mm tissue culture-treated dish (SPL Life Sciences, catalog number: 20060)
Sterile Cell Strainer, 40 µm Nylon Mesh (Thermo Fisher Scientific, catalog number: 22363547)
Phosphate buffered saline (PBS), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010023)
Cell-dissociation enzyme, TrypLE Express (Thermo Fisher Scientific, catalog number: 12605028)
Trypan Blue Solution, 0.4% (Thermo Fisher Scientific, catalog number: 15250061)
Advanced DMEM (Thermo Fisher Scientific, catalog number: 12491015)
Ham’s F-12 Nutrient Mix (Thermo Fisher Scientific, catalog number: 11765054)
Penicillin-Streptomycin, 100× (Thermo Fisher Scientific, catalog number: 15140122)
Fetal bovine serum (FBS)-US source, heat inactivated (Denville Scientific, catalog number: FB5001-H)
Fibronectin, human protein, plasma (Thermo Fisher Scientific, catalog number: 33016015)
Bouin’s fixative (Polysciences, catalog number: 16045)
Ammonium hydroxide solution, 28–30% (Sigma-Aldrich, catalog number: 221228)
Alcian blue, 8GX (Sigma-Aldrich, catalog number: A3157)
Acetic acid, glacial (Sigma-Aldrich, catalog number: 695092-2.5L)
Guanidine hydrochloride (Sigma-Aldrich, catalog number: G3272)
Fibronectin stock solution (1 mg/mL) (see Recipes)
100 µg/mL fibronectin in 10% FBS/Advanced DMEM (see Recipes)
10 µg/mL fibronectin in 10% FBS/Advanced DMEM (see Recipes)
4% FBS in Advanced DMEM/Ham’s F-12 (see Recipes)
Solution A for alcian blue staining (see Recipes)
Solution B for alcian blue staining (see Recipes)
1 mg/mL Alcian blue solution (see Recipes)
6M guanidine hydrochloride (see Recipes)
Equipment
Dumont #5 tweezer, tip size 0.10 × 0.06 mm (Roboz, catalog number: RS-5045), sterilized by autoclave
Dumont #7 tweezer, tip size 0.17 × 0.10 mm (Roboz, catalog number: RS-5047), sterilized by autoclave
Micro dissecting scissors, 3.5” straight (Roboz, catalog number: RS-5910), sterilized by autoclave
Stereo microscope, any brand (e.g., Leica M165-FC)
Centrifuge (Thermo Sorval Legend Micro 21)
Class II A/B3 Biological safety cabinet
CO2 incubator, NAPCO SERIES 8000DH
Varioskan Flash Spectral Scanning Multimode Reader (Thermo Scientific)
Procedure
-
Cell isolation
Pour 12 mL of PBS in a 10 cm dish on ice.
Pre-warm TrypLE to 37°C.
Euthanize a pregnant mouse (E10.5) via methods approved by the Institutional Animal Care and Use Committee (IACUC), such as inhalation of CO2 followed by thoracotomy.
Harvest the uterus and place it in ice-cold PBS (Figure 1).
Harvest embryos from the uterus and keep them in ice-cold PBS (Figure 1, Video 1).
Isolate the BA1 (Figure 2, Video 2), and place them into 1.5 mL tubes with 100 µL of ice-cold PBS (one BA1 to a tube).
Centrifuge at 100 × g and room temperature (RT) for 5 min.
Remove supernatant, then add pre-warmed 100 µL of TrypLE to the 1.5 mL tube.
Incubate at 37°C for 5–10 min. Gently pipette up and down with wide bore 1,000 µL tips.
Add 900 µL of 10% FBS in Advanced DMEM, then gently pipette 5–10 times.
Centrifuge at 100 × g and RT for 5 min, then discard supernatant.
Resuspend the cells with 500 µL of PBS.
Centrifuge at 100 × g and RT for 5 min, then discard supernatant.
Resuspend with 150 µL of PBS.
Filter the cells through a 40 µm cell strainer.
Take 10 µL of cell suspension into 1.5 mL tube, and mix with 10 µL of Trypan blue, to assess cell viability.
-
Micromass culture
Dilute the fibronectin stock solution with 10% FBS in Advanced DMEM, to prepare 100 µg/mL fibronectin solution.
Resuspend the cells with 100 µg/mL fibronectin solution (5 × 105 cells/100 µL).
Seed 20 µL of cells onto a 35 mm dish.
Incubate at 37°C for 1.5 h, to allow them to attach to well.
Add 1 mL of Advanced DMEM including 10 µg/mL Fibronectin and 10% FBS in Advanced DMEM.
Incubate at 37°C for 24 h.
Remove media, then add 2 mL of 4% FBS in Advanced DMEM/Ham's F-12.
Culture for 5 days.
Measure chondrogenic differentiation by Alcian blue staining (Figure 3 and part C), and set up a qRT-PCR to determine the levels of chondrogenic marker genes, such as Sox9 and type 2 collagen ( Yang et al., 2021 ).
-
Alcian Blue staining for micromass culture
Aspirate medium from micromass culture.
Fix with 1mL of Bouin’s fixative at RT for 15 min.
Wash with Solution A (see Recipes).
Wash with Solution B (see Recipes).
Stain with 2 mL of 1 mg/mL Alcian blue in 3% acetic acid (see Recipes) at RT for 1 h.
Wash dye off with double distilled water (ddH2O) four times.
Keep in 2 mL of ddH2O.
Take photos.
Wash dye off with ddH2O.
Extract the dye by incubating cultures with 1 mL of 6 M guanidine hydrochloride (see Recipes) at RT overnight.
Measure the optical density at OD595 using a spectrophotometer, such as the Thermo Scientific Varioskan Flash Spectral Scanning Multimode Reade.
Video 1. Harvest an E10.5 embryo from the uterus.

An embryo is located at the light red area of the isolated uterus (Figure 1E, upper part), and the placenta is located at the dark red area (Figure 1E, lower part). A tweezer will be carefully inserted from one of the cut sites of the separated uterus. The uterine wall will be carefully removed to harvest an embryo with yolk sac. Subsequently, the yolk sac and the amnion will be removed as shown in Figure 1H–I.
Video 2. Harvest the BA1 from an embryo.
The trunk of an embryo is gently pressed down with a tweezer, and another tweezer is inserted between the BA1 and the BA2. The head is then gently pressed down with a tweezer, and the BA1 is separated from the head.
Notes
To avoid cell death, embryos/cells should be kept on ice until they are seeded.
In case of cell viability less than 70–80%, consider removing dead cells by using a dead cell removal kit, such as Miltenyi dead cell removal kit (Miltenyi Biotech, Catalog number: 130-090-101).
When cells are seeded on 35 mm dishes, the drop should be kept maintaining a high-dense cell area. After culture for 1.5 h, cells should be attached on the 35 mm dish. Carefully add Advanced DMEM including Fibronectin.
The isolated cells can be used for micromass culture at the day of isolation. However, they can be passaged up to three times, with a total duration of culture of 10 days after isolation.
Recipes
-
Fibronectin stock solution (1 mg/mL)
1 mg fibronectin
1 mL of PBS
-
100 µg/mL fibronectin in 10% FBS/Advanced DMEM
60 µL of fibronectin stock solution
540 µL of 10% FBS/Advanced DMEM
-
10 µg/mL fibronectin in 10% FBS/Advanced DMEM
100 µL of fibronectin stock solution
1 mL of 10% FBS/Advanced DMEM
-
4% FBS in Advanced DMEM/Ham’s F-12
48 mL of Advanced DMEM
48 mL of Ham’s F-12
4 mL of FBS
-
Solution A for alcian blue staining
Add 0.5 mL of Ammonium hydroxide solution to 500 mL of 70% ethanol
-
Solution B for alcian blue staining
Add 25 mL of acetic acid, glacial, to 70% ethanol to make final volume 500 mL.
-
1 mg/mL Alcian blue solution
Add 0.6 mL of acetic acid, glacial, to ddH2O to make final volume 20 mL. Then add 20 mg of Alcian blue 8GX.
-
6 M guanidine hydrochloride
Dissolve 57.3 g of guanidine hydrochloride in 100 mL of ddH2O.
Acknowledgments
This work was supported by grant from NIH (R01DE020843 to Y.M. and R01DE025897 to Y.K.), International Fibrodysplasia Ossificans Progressiva Association (to Y.M.), and the grant-in-aid from the National Natural Science Foundation of China (31500788 to J.Y.). We thank Dr. W. Benton Swanson for critical reading. This protocol is derived from the original research paper, Yang et al., Augmented BMP signaling commits cranial neural crest cells to a chondrogenic fate by suppressing autophagic b-catenin degradation. Sci Signal 14(665): eaaz9368. (DOI: 10.1126/scisignal.aaz9368).
Competing interests
The authors declare no conflict of interest.
Ethics
All experimental procedures have been approved by the Institutional Animal Care and Use Committee at the University of Michigan (#PRO00009613) and the University of Texas (AWC-18-0137).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Chai Y. and Maxson R. E.,Jr (2006).Recent advances in craniofacial morphogenesis.Dev Dyn 235(9):2353-2375. [DOI] [PubMed] [Google Scholar]
- 2. Chen G.,Ishan M.,Yang J.,Kishigami S.,Fukuda T.,Scott G.,Ray M. K.,Sun C.,Chen S. Y.,Komatsu Y.,et al.(2017).Specific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos.Genesis 55(6): 10.1002/dvg.23034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Couly G. F. and Le Douarin N. M.(1985).Mapping of the early neural primordium in quail-chick chimeras. I. Developmental relationships between placodes, facial ectoderm, and prosencephalon.Dev Biol 110(2):422-439. [DOI] [PubMed] [Google Scholar]
- 4. Danielian P. S.,Muccino D.,Rowitch D. H.,Michael S. K. and McMahon A. P.(1998).Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase.Curr Biol 8(24):1323-1326. [DOI] [PubMed] [Google Scholar]
- 5. Gu S.,Wu W.,Liu C.,Yang L.,Sun C.,Ye W.,Li X.,Chen J.,Long F. and Chen Y.(2014).BMPRIA mediated signaling is essential for temporomandibular joint development in mice.PLoS One 9(8):e101000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He F. and Soriano P.(2013).A critical role for PDGFRα signaling in medial nasal process development.PLoS Genet 9(9):e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishii M.,Arias A. C.,Liu L.,Chen Y. B.,Bronner M. E. and Maxson R. E.(2012).A stable cranial neural crest cell line from mouse.Stem Cells Dev 21(17):3069-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaucka M.,Ivashkin E.,Gyllborg D.,Zikmund T.,Tesarova M.,Kaiser J.,Xie M.,Petersen J.,Pachnis V.,Nicolis S. K.,et al.(2016).Analysis of neural crest-derived clones reveals novel aspects of facial development.Sci Adv 2(8):e1600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang X.,Rowitch D. H.,Soriano P.,McMahon A. P. and Sucov H. M.(2000).Fate of the mammalian cardiac neural crest.Development 127(8):1607-1616. [DOI] [PubMed] [Google Scholar]
- 10. Jiang X.,Iseki S.,Maxson R. E.,Sucov H. M. and Morriss-Kay G. M.(2002).Tissue origins and interactions in the mammalian skull vault.Dev Biol 241(1):106-116. [DOI] [PubMed] [Google Scholar]
- 11. Komatsu Y.,Yu P. B.,Kamiya N.,Pan H.,Fukuda T.,Scott G. J.,Ray M. K.,Yamamura K. and Mishina Y.(2013).Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice.J Bone Miner Res 28(6):1422-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li L.,Wang Y.,Lin M.,Yuan G.,Yang G.,Zheng Y. and Chen Y.(2013).Augmented BMPRIA-mediated BMP signaling in cranial neural crest lineage leads to cleft palate formation and delayed tooth differentiation.PLoS One 8(6):e66107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teng L.,Mundell N. A.,Frist A. Y.,Wang Q. and Labosky P. A.(2008).Requirement for Foxd3 in the maintenance of neural crest progenitors.Development 135(9):1615-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mishina Y. and Snider T. N.(2014).Neural crest cell signaling pathways critical to cranial bone development and pathology.Exp Cell Res 325(2):138-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Santagati F. and Rijli F. M.(2003).Cranial neural crest and the building of the vertebrate head.Nat Rev Neurosci 4(10):806-818. [DOI] [PubMed] [Google Scholar]
- 16. Stemple D. L. and Anderson D. J.(1992).Isolation of a stem cell for neurons and glia from the mammalian neural crest.Cell 71(6):973-985. [DOI] [PubMed] [Google Scholar]
- 17. Yamauchi Y.,Abe K.,Mantani A.,Hitoshi Y.,Suzuki M.,Osuzu F.,Kuratani S. and Yamamura K.(1999).A novel transgenic technique that allows specific marking of the neuralcrest cell lineage in mice.Dev Biol 212(1):191-203. [DOI] [PubMed] [Google Scholar]
- 18. Yang J.,Kitami M.,Pan H.,Toda-Nakamura M.,Zhang H.,Liu F.,Zhu L.,Komatsu Y. and Mishina Y.(2021).Augmented BMP signaling commits cranial neural crest cells to a chondrogenic fate by suppressing autophagic b-catenin degradation.Sci Signal 14(665):eaaz9368. [DOI] [PMC free article] [PubMed] [Google Scholar]