Highlights
-
•
30 ccRCC patients were used, to follow the epigenetic changes (γH2A.X, H3K4me3 and H3K9me3) and the alterations in the level of RNA polymerase II (RNAPII).
-
•
The variabilities between the tumorous and non-tumorous parts of the tissue were measured by image analysis in which we monitored 30 cells from different positions of either the tumorous or the non-tumorous part of the tissue sections.
-
•
These markers were classified to predict patient outcomes based on their individual cellular background. These results also support that detection of any alteration in the level of H3K4me3, H3K9me3, and γH2AX can account valuable information for presuming the progression of ccRCC and the clinical benefits to select the most efficient personalized therapy.
Keywords: ccRCC, epigenetics, H3K4me3 and H3K9me3, RNAPII, Renal cell carcinoma, γH2A.X
Abstract
Patients diagnosed with clear cell renal cell carcinoma (ccRCC) have poor prognosis for recurrence and approximately 30–40% of them will later develop metastases. For this reason, the appropriate diagnosis and the more detailed molecular characterisation of the primary tumour, including its susceptibility to metastasis, are crucial to select the proper adjuvant therapy by which the most prosperous outcome can be achieved. Nowadays, clinicopathological variables are used for classification of the tumours. Apart from these, molecular biomarkers are also necessary to improve risk classification, which would be the most beneficial amongst modern adjuvant therapies. As a potential molecular biomarker, to follow the transcriptional kinetics in ccRCC patients (n=30), we analysed epigenetic changes (γH2A.X, H3K4me3, and H3K9me3) and the alterations in the level of RNA polymerase II (RNAPII) by immunohistochemical staining on dissected tissue sections. The variabilities between the tumorous and non-tumorous parts of the tissue were detected using quantitative image analysis by monitoring 30 cells from different positions of either the tumorous or the non-tumorous part of the tissue sections. Data obtained from the analyses were used to identify potential prognostic features and to associate them with the progression. These markers might have a value to predict patient outcomes based on their individual cellular background. These results also support that detection of any alteration in the level of H3K4me3, H3K9me3, and γH2A.X can account for valuable information for presuming the progression of ccRCC and the clinical benefits to select the most efficient personalised therapy.
Introduction
Among the various types of renal cell carcinoma (RCC), clear cell renal cell carcinoma (ccRCC) is the most frequently occurred subtype, accounting for approximately 70–80% of all cases, and it has the sixth highest mortality rate in Europe and US [1]. According to the WHO database, RCC is considered one of the most common epithelial cancers worldwide, with 430,000 newly diagnosed and 180,000 death cases in 2020. The mortality rate and severeness of the disease are highly related to late-stage diagnoses as only 10% of the patients developed typical symptoms, like haematuria, backache, or abdominal mass [2]. The current WHO classification distinguishes 14 RCC subsets, including clear cell RCC, papillary RCC, and chromophobe RCC, as the most frequent subtypes based on morphological and histopathological characteristics [3], [4], [5]. Due to the lack of molecular biomarkers, RCC classification is difficult, and the adequate identification of stages requires reliable tools. Additionally, it is also important to develop proper tools for predicting clinical outcomes to improve the standard treatment guidelines for RCC patients. In modern diagnostics, novel methods include prognostic modelling which relies on DNA-, RNA-, and protein-based biomarkers, to predict the possible behaviour of the tumour [6,7]. For patients diagnosed with high-risk tumour, adjuvant therapeutic inventions are crucial, while for patients diagnosed with low-risk cancer, overtreatment can be dangerous [8], [9], [10]. Applying large-scale genomic and proteomic analyses, the establishment of biomarkers with diagnostic quality is feasible, and alterations associated with RCC can be identified [11]. These techniques also have the potential to reveal novel therapeutic targets which can be implicated into unprecedented diagnostic and therapeutic strategies. However, to describe the overall performance of the tumour progression, an individual biomarker does not provide valuable data in terms of sensitivity and specificity [12], [13], [14].
In the progression of ccRCC, mutations of the Von Hippel-Lindau Tumor Supressor (VHL) tumour suppressor gene are one of the main drivers [15,16]. Additionally, mutations of the Polybromo 1 (PBRM1), BRCA1 Associated Protein 1 (BAP1), and SET domain containing 2 (SETD2) are also found to be associated with ccRCC but with a lower frequency [17,18]. This genetic heterogeneity observed in ccRCC leads to difficulties in achieving a prosperous treatment and promotes recurrence. These mutations in PBRM1, SETD2, and BAP1 genes are strongly associated with epigenetic changes since the proteins produced from these genes are responsible for modulating the chromatin structural changes [18,19]. The PBRM1 gene encodes the bromodomain-containing protein BAF180, which is a subunit of the Polybromo- and BRG1-associated factors containing complex (PBAF) SWItching defective/Sucrose Non-Fermenting (SWI/SNF) chromatin remodelling complex. BAP1 encodes the histone deubiquitinating enzyme BRCA1-Associated Protein 1, while SETD2 encodes a methyltransferase that is specifically trimethylates at lysine 36 of histone H3 (H3K36me3) [20]. The loss of these genes leads to abnormal epigenetic alterations, providing new opportunities for the diagnostic application of these proteins and for developing novel therapeutic strategies in ccRCC. Some drugs, targeting the epigenetic system, are currently under investigation; however, strategies that combine therapies targeting the epigenetic machinery with conventional therapies are still in progress [21], [22], [23].
Here, we provide an overview of the epigenetic landscape of 30 patients diagnosed with ccRCC by using immunohistochemical profiling and automated quantification of the γH2A.X, H3K4me3, H3K9me3, and RNAPII level in tumorous and non-tumorous parts of the tissue. In this report, we highlight that the pattern of these marks is dependent on the tumour stages and grades. Therefore, our results establish the prognostic value of γH2A.X, H3K4me3, H3K9me3, and RNAPII patterns in ccRCC.
Materials and methods
Selection of the cohort
A total of 30 patients, who underwent radical or partial nephrectomy at the University of Szeged, were selected for further investigation. This study was conducted with the permission of the Regional and Institutional Human Medical Biological Research Ethics Committee, University of Szeged (No. 188/2019-SZTE). All patients gave his/her written informed consent for the publication of all data and images. A histopathologist evaluated all the cases for the histological subtype, tumour stage, and ISUP grade. Haematoxylin and eosin staining was performed on each section to confirm the tumour content of the renal tissues. After surgery, tissue samples were dissected from both the tumorous and normal parts of the tissues, then they were flash frozen, and deposited in the Biobank of Pathology, University of Szeged. All the samples were stored at −80°C till further processing.
Preparations of the normal and tumorous tissues
For immunofluorescence staining, the tissues were embedded in Shandon Cryomatrix gel (Thermo Fisher Scientific), and 5 µm sections were prepared on Superfrost Ultra Plus slides by cryostat (Cryostar NX50).
Immunostaining of frozen tissues
The tissues were fixed for 10 minutes using acetone, and then washed 3 times with PBS solution. Subsequently, the tissues were permeabilized with 0.3% Triton-X-100/PBS for 20 minutes at 25°C. Alternatively, for permeabilization 0.3% Na-DOC/0.3% Triton-X-100/PBS was used for 20 minutes at 25°C. Then sections were blocked with 5% BSA/0.3% Triton X-100/PBS for 1 hour. Samples were incubated with primary antibodies diluted in 1% BSA/PBST: anti-γH2A.X (Millipore 05-636) in 1:100, anti-H3K9me3 (Abcam ab8898) in 1:100, anti-H3K4me3 (Abcam ab8580) in 1:300, and RNAPII (1BP7G5 IGBMC) in 1:250 dilution. After washing steps, the following secondary antibodies were used: GAR Alexa 555 (Invitrogen, A21429) in 1:100, and GAM Alexa 488 (Molecular Probes, A11029) in 1:500 dilution. Finally, cells were mounted with DAPI-containing ProLong Gold antifade reagent (Life technologies). Samples were visualised with Olympus FluoView FV10i confocal microscopy. The same exposition time was used for every image capturing. Images were quantified with ImageJ software.
Image analysis of the confocal pictures
For ImageJ analysis, 10-10 cells from 3 different parts of each tissue section were selected. The selection was established on the haematoxylin and eosin staining-based separation of the tumorous and non-tumorous tissue section which was defined by expert pathologists. During the analysis, the size of the nuclei of each cell and their mean grey/intensity values were recorded and added to the database. The CTCF values (Corrected Cell Fluorescence) were calculated accordingly: integrated density–(nuclear area * average background intensity). The calculated values were visualized by box plots in Sigma Plot 12.5, and the significance of each sample was calculated by independent samples t-test in IBM SPSS Statistics 27.0.
Haematoxylin and eosin staining
Tissues were stained with haematoxylin dye, supplemented with aluminium potassium sulphate dodecahydrate (Sigma-Aldrich). Following washing with distilled water, sections were stained with 1 g/l eosin-B solution (Sigma-Aldrich). Then samples were washed twice with distilled water. Next, the tissues were washed with isopropyl-alcohol and distilled water, then the sections were mounted. Finally, the slides were scanned with 3DHISTECH Pannoramic Midi digital slide scanner for monitoring histoarchitectural changes.
CNV (copy number variation) measurement
CNV values were determined by RT-qPCR reactions performed on Thermo Scientific PIKO 96-well Thermal Cycler (Thermo Fischer Scientific) platform using SYBR Green chemistry (Promega, GoTaq qPCR Master Mix) as it is described in Ujfaludi et al. 2022 (manuscript accepted in POR).
Statistical analysis for correlation study
Pearson correlation was executed to verify the strength of a linear association between the examined markers. For this, first mean value was calculated from the CTCF values of each specimen, then the mean value of the tumorous part was divided with the mean value of the non-tumorous part of each tissue section in case of each marker. Subsequently, using IBM SPSS Statistics 27.0, these values were applied for determination of the Pearson correlation coefficient (referred to as ‘r’) between the examined markers.
Results
Selection and classification of the cohort
The age distribution of the 30 candidates selected for the current study was between 39 to 78 years, with an average age of 62. 14 tissue sections were derived from females, and 16 were dissected from male patients, representing an almost 1:1 gender ratio. According to the haematoxylin-eosin staining of the dissected tissue, patients were categorized based on either tumour stage (stadium pT1-3) or grade (low [ISUP grade 1+2] or high [ISUP grade 3+4]) by an expert pathologist. In 3 patients, metastases were also detected in the bone marrow or in the adrenal gland. The patients selected for this study were also characterised in order to test their von Hippel-Lindau Tumor Supressor (VHL), Polybromo 1 (PBRM1), BRCA1-Associated Protein 1 (BAP1), and SET domain containing 2 (SETD2) status. We detected pathological mutation of the VHL gene in approximately one third of the samples. Although in most cases, the copy number variation of these genes was close to the control, in some cases they were found to be haploid: (I) PBRM1 in three, (II) SETD2 in four, and (III-IV) BAP1 and VHL in five patients. The detailed clinical and genetic parameters are represented in Table 1. All the 30 patients were further analysed for H3K4me3, H3K9me3, H2AXS139P (referred to as γH2A.X), and RNA polymerase II (RNAPII) levels.
Table 1.
The components of the cohort
| Code | Sample ID number | Gender | Age | Tumour size (mm) | Stadium | Metastasis | Grade | PBRM1 (CNV) | BAP1 (CNV) | SETD2 (CNV) | VHL (CNV) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | T1207 | female | 62 | 75 | pT3a | adrenal gland | High | N | N | N | N |
| #2 | T1257 | male | 50 | 52 | pT1b | not detected | Low | L | N | N | N |
| #3 | T1337 | female | 61 | 30 | pT1a | not detected | Low | N | N | N | N |
| #4 | T1434 | female | 60 | 125 | pT3a | not detected | High | N | N | N | L |
| #5 | T1441 | male | 66 | 62 | pT3a | not detected | High | H | N | H | L |
| #6 | T1447 | female | 59 | 45 | pT1b | not detected | Low | N | N | N | H |
| #7 | T1453 | male | 78 | 60 | pT3a | not detected | Low | N | N | N | N |
| #8 | T1526 | male | 73 | 44 | pT1b | not detected | High | N | N | N | N |
| #9 | T1528 | male | 59 | 48 | pT3a | not detected | Low | N | N | N | N |
| #10 | T1531 | female | 66 | 95 | pT3a | not detected | Low | N | N | N | N |
| #11 | T1533 | male | 67 | 49 | pT3a | not detected | High | N | N | N | N |
| #12 | T1538 | female | 40 | 42 | pT3a | not detected | Low | H | N | N | N |
| #13 | T1622 | female | 62 | 112 | pT3a | not detected | High | N | H | N | N |
| #14 | T1625 | male | 62 | 39 | pT3a | not detected | Low | N | N | N | N |
| #15 | T1652 | female | 71 | 125 | pT3a | adrenal gland | High | N | H | H | H |
| #16 | T1725 | female | 40 | 51 | pT1b | not detected | Low | N | N | N | N |
| #17 | T1731 | female | 68 | 25 | pT1a | not detected | Low | N | N | N | N |
| #18 | T1742 | female | 55 | 32 | pT1a | not detected | Low | N | H | N | N |
| #19 | T1761 | male | 77 | 34 | pT1a | not detected | High | N | N | N | N |
| #20 | T747 | male | 68 | 90 | pT3a | not detected | High | L | N | N | N |
| #21 | T788 | female | 68 | 72 | pT3a | bone marrow | High | N | H | H | H |
| #22 | T790 | male | 61 | 65 | pT3a | not detected | High | N | N | N | N |
| #23 | T795 | male | 57 | 50 | pT3a | not detected | High | L | N | N | N |
| #24 | T803 | female | 51 | 35 | pT3a | not detected | Low | L | N | N | N |
| #25 | T808 | female | 63 | 33 | pT1a | not detected | High | N | H | N | H |
| #26 | T815 | male | 68 | 40 | pT1a | not detected | High | N | N | N | N |
| #27 | T823 | male | 55 | 30 | pT1a | not detected | Low | N | N | N | N |
| #28 | T827 | male | 78 | 56 | pT1b | not detected | High | N | N | N | N |
| #29 | T888 | male | 39 | 72 | pT2a | not detected | High | N | H | N | N |
| #30 | T904 | male | 78 | 39 | pT1a | not detected | High | N | N | N | N |
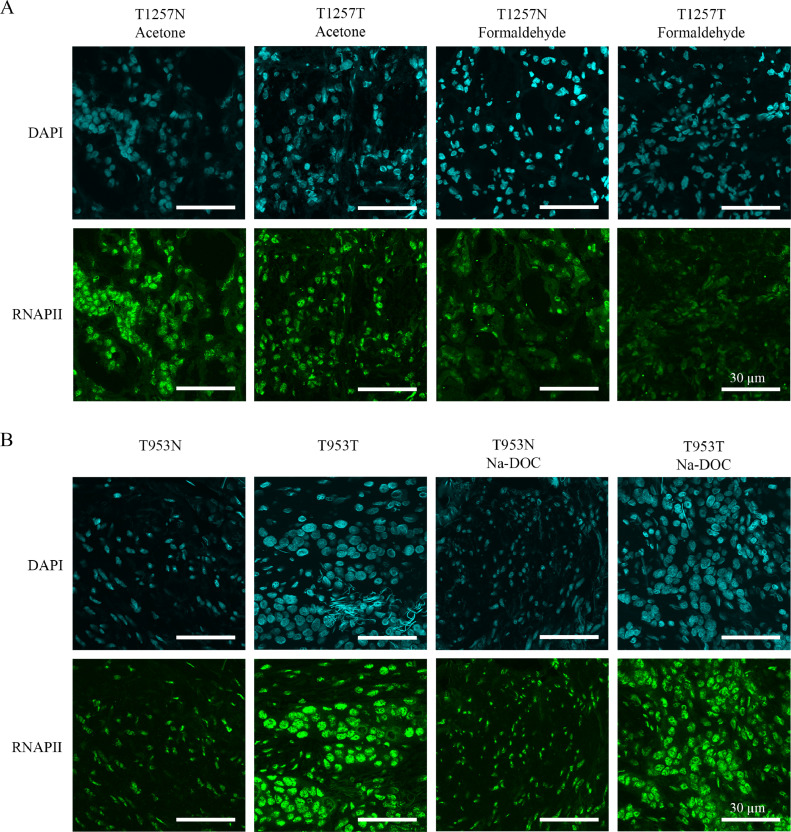
First, the reliability and specificity of the antibodies against H3K4me3, H3K9me3, γH2A.X, and RNAPII on frozen material were optimised by setting the immunostaining procedure on slices derived from ccRCC and the surrounding non-tumorous parts of the tissue (Fig. 1). As expected, strong nuclear expression of the RNAPII protein was observed in all the conditions we tested, underlying the specificity of the antibody used for this study [24]. Intriguingly, the widely used formaldehyde fixation resulted in both nuclear and cytoplasmic RNAPII positivity in the non-tumorous parts and loss of signal in the ccRCC regions (Fig. 1A). To achieve comparable staining efficiency from the different tissue parts, we tried acetone fixation which appeared to be applicable for further investigations (Fig. 1A). Moreover, we also tested different permeabilization methods, but it did not affect the staining efficiency (Fig. 1B).
Fig. 1.
Adjustment of fixation and permeabilization steps of immunohistochemistry method on normal and tumorous tissue section parts from patients with clear cell renal cell carcinoma (ccRCC). A) Fixation with acetone or formaldehyde, B) Permeabilization with solely Triton X-100 or the combinatorial usage of Triton X-100 and Na-DOC. DAPI (blue) was used to visualize the nuclei. Scale bars represent 30 µm.
Global levels of H3K4me3, RNAPII, and H3K9me3 show altered patterns in ccRCC
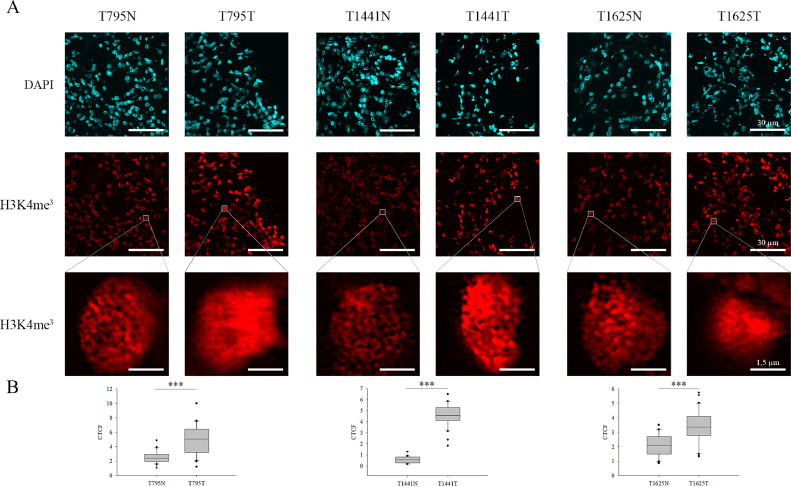
To reveal whether H3K4me3 level, a widely applied euchromatic mark, correlates with the progression of ccRCC, immunohistochemistry staining was performed on the dissected tissues from all the 30 patients who were classified according to stages (1–4) and grades (low and high). To perform comparable immunohistochemical detection on non-tumorous and ccRCC parts, following microtome dissection of the tissues, the segments obtained from either the tumour mass or the surrounding normal part of the tissue were placed on the same microscopic slide as it is shown in Supplementary Fig. 1. Subsequently, in case of each patient, the staining protocol was performed simultaneously on the non-tumorous and tumorous tissue parts. Regardless of tumour grades, the H3K4me3 level significantly increased in 50% of the cohort, while we detected less H3K4me3 in 5 tumorous specimens (Fig. 2 and Supplementary Fig. 2). Although the H3K4me3 level generally altered in 63% of the samples, we did not find any correlation with cancer progression considering the patients’ tumour stages and grades (Table 1). We also monitored the H3K4me3 status and the cancer survival rate, and we found that the alteration of this PTM resulted in better survivability (Supplementary Fig. 3). Based on these, we believe this marker presumably has a prognostic value which has to be further validated on an extended clinical cohort.
Fig. 2.
Immunofluorescent spectra of H3K4me3 detected in normal and ccRCC tumorous tissue parts. A) N represents non-tumorous tissue parts, while T corresponds to ccRCC sections. Representative images of H3K4me3 staining in peritumoral (left) and tumour tissues (right) from ccRCC patients. For the better visualization of the changes in H3K4me3 level, one cell is represented in a magnified resolution in each specimen. Scale bars represent 30 µm and 1.5 µm, respectively. B) The observed alterations in the quantification of relative fluorescence intensity (CTCF) of H3K4me3 level between the tumorous and non-tumorous tissue parts obtained from automated image acquisition (n=30) were illustrated in box plots and evaluated based on independent samples t-test, ***P < 0.001.
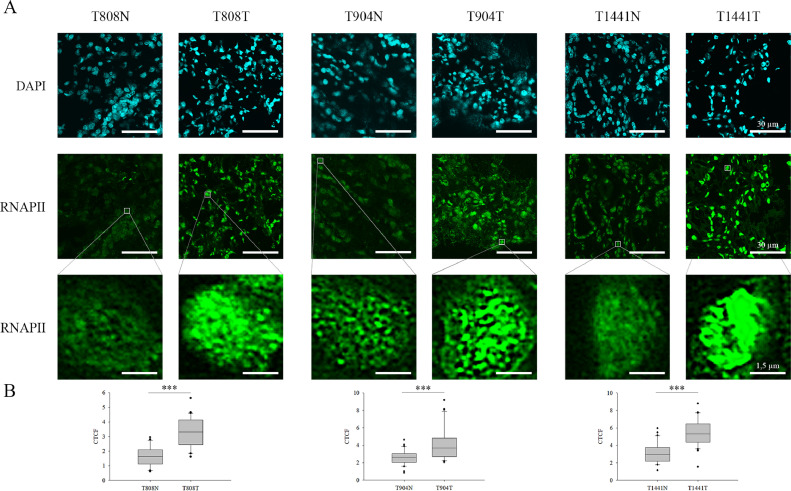
To study whether the alteration in the euchromatic H3K4me3 level correlates with the transcriptional profile changes of these patients, we performed co-immunofluorescence staining with H3K4me3 and RNAPII antibodies. Henceforth, the RNAPII levels of the tumorous and adjacent normal tissue parts were analysed in all the examined ccRCC patients simultaneously with H3K4me3. These results revealed a significantly strong correlation between the alteration of H3K4me3 and RNAPII levels, which presumes a transcriptomic profile change in these tumours (Fig. 3 and Supplementary Fig. 4). In 14 samples increased levels, while in 3 specimens a decreased level of RNAPII was detected in the tumorous part compared to the corresponding adjacent normal tissue part. The elevated level of RNAPII mainly concerns high grade ccRCC in these patients (10 versus 4 samples).
Fig. 3.
Immunofluorescent spectra of RNAPII detected in normal and ccRCC tumorous tissue parts. A) N represents non-tumorous tissue parts, while T corresponds to ccRCC sections. Representative images of RNAPII staining in peritumoral (left) and tumour tissues (right) from ccRCC patients. For the better visualization of the changes in RNAPII level, one cell is represented in a magnified resolution in each specimen. Scale bars represent 30 µm and 1.5 µm, respectively. B) The observed alterations in the quantification of relative fluorescence intensity (CTCF) of RNAPII level between the tumorous and non-tumorous tissue parts obtained from automated image acquisition (n=30) were illustrated in box plots and evaluated based on independent samples t-test, ***P < 0.001.
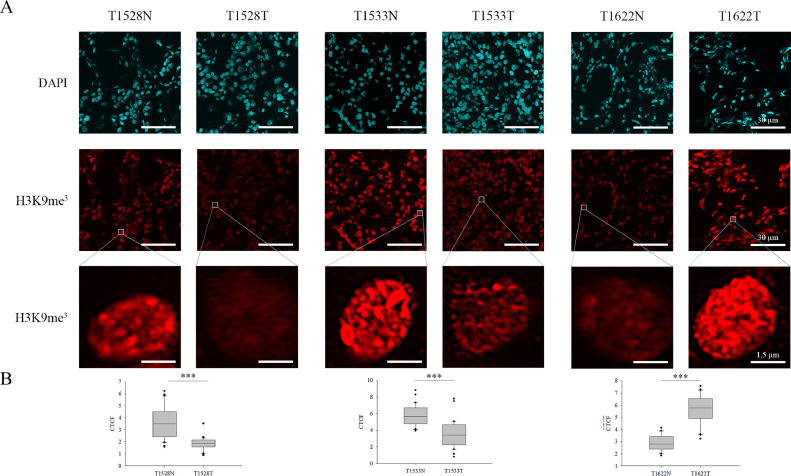
We hypothesized that the increased RNAPII binding and the elevated level of H3K4me3 might contribute to an imbalance in the euchromatic–heterochromatic milieu of these cells. To decipher this issue, H3K9me3 heterochromatic mark was selected and detected by the same immunostaining procedure described before. We observed altered H3K9me3 level in 20 patients: decreased in 11 cases, while increased in 9 samples (Fig. 4 and Supplementary Fig. 5).
Fig. 4.
Immunofluorescent spectra of H3K9me3 detected in normal and ccRCC tumorous tissue parts. A) N represents non-tumorous tissue parts, while T corresponds to ccRCC sections. Representative images of H3K9me3 staining in peritumoral (left) and tumour tissues (right) from ccRCC patients. For the better visualization of the changes in H3K9me3 level, one cell is represented in a magnified resolution in each specimen. Scale bars represent 30 µm and 1.5 µm, respectively. B) The observed alterations in the quantification of relative fluorescence intensity (CTCF) of H3K9me3 level between the tumorous and non-tumorous tissue parts obtained from automated image acquisition (n=30) were illustrated in box plots and evaluated based on independent samples t-test, ***P < 0.001.
Higher rate of DNA damage is detected in ccRCC
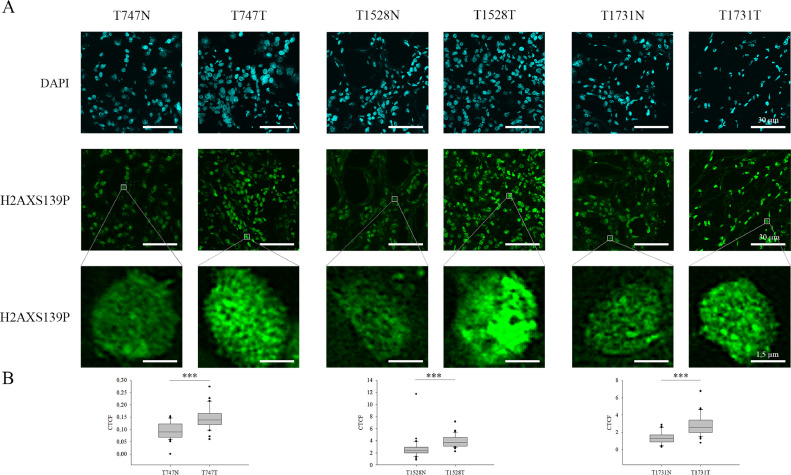
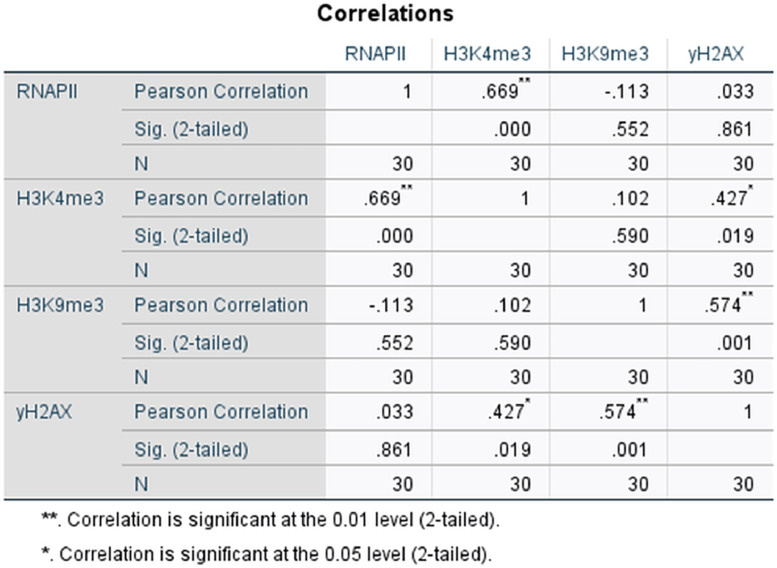
As mentioned previously, ccRCC is characterised as an aggressive tumour type, in which high genomic mutation frequency is peculiar [25]. It also affects certain tumour suppressor genes, including VHL and TP53, which play a significant role in DNA repair [26]. However, the severity of DNA damage and the maintenance of genome stability in ccRCC have yet to be characterised. Since the metastatic capability of the tumour is notably influenced by the type of DNA damage and mutation frequency, we explored the possible alterations in the ccRCC-related DNA repair process. To follow the DNA repair foci formation in primary ccRCC tumours, we applied immunostaining with an antibody against one of the initial DDR factors, γH2A.X (Fig. 5 and Supplementary Fig. 6). We observed increased γH2A.X levels in 50% of the specimens irrespectively of tumour grading, which underlined our hypothesis that persistent DNA damage occurs in these cells, which is presumably one of the driving forces of the metastatic progression in ccRCC cells. Additionally, in ccRCC patients, we revealed significant correlations between γH2A.X and H3K4me3 or H3K9me3, which support the assumption that chromatin structural changes, occurred in the tumorous regions, associate with persistent DNA damage which is more prevalent in tumorous regions than in the adjacent non-tumorous parts (Fig. 6).
Fig. 5.
Immunofluorescent spectrum of γH2A.X (H2AXS139P) detected in normal and ccRCC tumorous tissue parts. A) N represents non-tumorous tissue parts, while T corresponds to ccRCC sections. Representative images of γH2A.X staining in peritumoral (left) and tumour tissues (right) from ccRCC patients. For the better visualization of the changes in γH2A.X level, one cell is represented in a magnified resolution in each specimen. Scale bars represent 30 µm and 1.5 µm, respectively. B) The observed alterations in the quantification of relative fluorescence intensity (CTCF) of γH2A.X level between the tumorous and non-tumorous tissue parts obtained from automated image acquisition (n=30) were illustrated in box plots and evaluated based on independent samples t-test, ***P < 0.001.
Fig. 6.
Bivariate Pearson correlation calculated between H3K4me3, RNAPII, H3K9me3 and γH2A.X is represented, *P < 0.05 and **P < 0.01.
Discussion
ccRCC is considered a highly invasive tumour type, associated with poor prognosis and high chance of recurrence [27]. To diagnose these patients, a classical pathological scoring system is still widely used to predict the prognosis of ccRCC; however, it mainly relies on anatomical information and has no biological characteristics [28]. Additionally, mutations have been recently used to characterise the status of VHL, PBRM1, SETD2, and BAP1 genes [17,19]. On the other hand, due to the alteration in histone post-translational modifications (PTMs), chromatin structural changes have been associated with carcinogenesis and turned to be potential key regulators of cancer-related pathways [29,30]. Dysregulation in histone PTMs can be originated from the malfunction of either PTM writers or erasers and also from the genome-wide transcriptional reprogramming occurring during tumorigenesis. Additionally, DNA methylation can be used to characterise epigenetic changes in ccRCC, and the methylome dataset can be applied to predispose prognostic signatures [31]. This emphasizes the biological and diagnostic relevance of the epigenetic marks used to characterise ccRCC tumours regarding their invasiveness and response to the applied therapy. In this study, we characterised tumorous and adjacent normal tissues dissected from 30 patients diagnosed with ccRCC, and we validated the signatures of H3K4me3, and RNAPII associated with actively transcribed genomic regions. Although previous studies described an increased level of H3K4me3 during ccRCC progression, they found a decrease in the level of this PTM at later stages of ccRCC [32]. In contrast to this finding, we detected a synergistic elevation both in H3K4me3 and RNAPII level which confirms the reliability of our data. Our findings are further supported by the fact that extensive transcriptional reprogramming and overactivation occur during tumour progression in various types of cancer. The present study also establishes a strong correlation between H3K4me3 and RNAPII marks. Finally, the sample processing and automated analysis we applied in our experimental setup also minimise the appearance of experimental bias. Our data is in accordance with a previous study that established a correlation between the level of H3K4me3 and later tumour stages, indicating the prognostic implication of this marker [33]. In accordance with this, our study also supports the idea that global histone modifications, including H3K4me3, are also altered during ccRCC progression.
As it was mentioned before, the altered DNA methylation pattern is also a hallmark of cancerous malformations, i.e. DNA hypermethylation contributes to heterochromatinization which leads to the appearance of H3K9me3 mark at the given genomic locus, which then recruits Heterochromatic Protein 1 (HP1), resulting in constitutive heterochromatin formation [34]. We also found that the alteration in the global level of H3K9me3 corresponding with changes in the level of H3K4me3 and RNAPII was correlated with the presence of ccRCC. This finding is in accordance with already published data which demonstrated that patients diagnosed with ccRCC exhibited low level of heterochromatin-related H3K9 mono- and di-methylations as well as increased level of euchromatin-related H3K9ac and H3K18ac [35], [36], [37]. Taken together, these data reveal that the global level of different histone modifications is altered in unfavourably localized ccRCC. The correlation between histone methylation and acetylation presumes epigenetic confusion in ccRCC cells due to aberrant activity and/or expression of histone-modifying enzymes. These findings are further supported by our previous study, in which we demonstrated that the PBRM1, BAP1, and SETD2 expression levels were completely altered in patients developing ccRCC (accepted manuscript at POR), and it was also published that Jumonji histone demethylases are also dysregulated in ccRCC patients [38]. Despite the correlation between histone methylation and acetylation, these modifications may have different relevance to ccRCC stages [39]. Thus, epigenetic alterations may represent a novel therapeutic potential: Histone Deacetylase (HDAC) inhibitors show an antineoplastic effect on ccRCC patients as valproic acid inhibits the growth of RCC tumours and vorinostat increases the anticancer activity of the novel therapeutic drugs [40]. Additionally, the globally altered histone methylation levels detected in ccRCC can be restored with histone demethylase inhibitors, such as Lysine-specific histone demethylase 1 (LSD1) [41]. We believe that our study has the potential in combining clinicopathological variables with methylation signatures, by which the current risk classification of non-metastatic ccRCC can be improved.
Finally, in ccRCC tumour-derived specimens, we observed increased γH2A.X level, which is the hallmark of persistence DNA damage [42,43]. In correlation with the perpetual presence of γH2A.X, in ccRCC patients considerable number of DNA damage or insufficient DNA repair takes place. In agreement with this, previous studies revealed that during ccRCC progression, the DNA repair mechanism is also malfunctional [44]. First, the application of PARP and ATR inhibitors on PBRM1-deficient cells, is known to be synthetically lethal [45]. In the absence of exogenous DNA damage, PBRM1-defective ccRCC-derived cells exhibited elevated levels of replication stress and micronuclei formation. It unveils that the loss of PBRM1 during ccRCC progression is strongly associated with genome instability [45]. Our findings also supported this idea as insufficient DNA repair occurs in these cells, which opens the possibility of using PARPi or other chemotherapeutic drugs targeting DNA repair in ccRCC-related tumour therapy. Second, a large number of differentially expressed DNA repair genes were in silico identified, which reflect on the patient outcomes in the development of ccRCC [46]. These data also support our findings that the improper function of DNA repair genes is probably the consequence of epigenetic changes which contribute to an aggravated mutagenic rate in ccRCC and largely affect the tumour progression. Hence, these markers can be used to predict the clinical benefits and efficacy of the applied therapy.
Funding
This research was funded by the National Research, Development and Innovation Office grant GINOP-2.2.1-15-2017-00052 and NKFI-FK 132080. T.P. was funded by National Research, Development and Innovation Office grant GINOP-2.2.1-15-2017-00052, the János Bolyai Research Scholarship of the Hungarian Academy of Sciences BO/27/20, UNKP-20-5-SZTE-265, by NKFI-FK 132080, UNKP-21-5-SZTE-563 and EMBO short-term fellowship 8513. L.K. was funded by the University of Szeged, Faculty of Medicine Research Fund-Hetényi Géza Grant (Grant No. 5S 340 A202) and the New National Excellence Programme (Grant No. UNKP-21-4-SZTE-131).
Author contribution
Study design, N.Ö., B.N.B., H.M., L.K., and T.P.; investigation, N.Ö., Z.U.; data collections, N.Ö., B.N.B., H.M., Z.U., G.P.B., S.B., F.S., L.K., T.P.; data analysis, N.Ö., B.N.B., U.Z., T.P.; writing—original draft preparation, N.Ö., T.P.; writing—review and editing, N.Ö., B.N.B., H.M., Z.U., G.P.B., S.B., F.S., L.K., T.P.; visualization, N.Ö., B.N.B., Z.U.; funding acquisition, B.N.B., L.K., T.P. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank Ivett Csikós for her expert technical help.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101420.
Contributor Information
Levente Kuthi, Email: kuthi.levente@med.u-szeged.hu.
Tibor Pankotai, Email: pankotai.tibor@szte.hu.
Appendix. Supplementary materials
References
- 1.Bratu O., Mischianu D., Marcu D., Spinu D., Iorga L., Cherciu A., Balescu I., Bacalbasa N., Diaconu C., Savu C., Savu C., Anghel R. Renal tumor biomarkers (Review) Exp. Ther. Med. 2021;22:1297. doi: 10.3892/etm.2021.10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorstenson A., Bergman M., Scherman-Plogell A.H., Hosseinnia S., Ljungberg B., Adolfsson J., Lundstam S. Tumour characteristics and surgical treatment of renal cell carcinoma in Sweden 2005-2010: a population-based study from the national Swedish kidney cancer register. Scand. J. Urol. 2014;48:231–238. doi: 10.3109/21681805.2013.864698. [DOI] [PubMed] [Google Scholar]
- 3.Prasad S.R., Humphrey P.A., Catena J.R., Narra V.R., Srigley J.R., Cortez A.D., Dalrymple N.C., Chintapalli K.N. Common and uncommon histologic subtypes of renal cell carcinoma: imaging spectrum with pathologic correlation. Radiographics. 2006;26:1795–1806. doi: 10.1148/rg.266065010. discussion 1806-1710. [DOI] [PubMed] [Google Scholar]
- 4.Srigley J.R., Delahunt B. Uncommon and recently described renal carcinomas. Mod. Pathol. 2009;22(2):S2–S23. doi: 10.1038/modpathol.2009.70. Suppl. [DOI] [PubMed] [Google Scholar]
- 5.Srigley J.R., Delahunt B., Eble J.N., Egevad L., Epstein J.I., Grignon D., Hes O., Moch H., Montironi R., Tickoo S.K., Zhou M., Argani P., Panel I.R.T. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am. J. Surg. Pathol. 2013;37:1469–1489. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 6.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farber N.J., Kim C.J., Modi P.K., Hon J.D., Sadimin E.T., Singer E.A. Renal cell carcinoma: the search for a reliable biomarker. Transl. Cancer Res. 2017;6:620–632. doi: 10.21037/tcr.2017.05.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharyya G.S., Doval D.C., Desai C.J., Chaturvedi H., Sharma S., Somashekhar S.P. Overview of breast cancer and implications of overtreatment of early-stage breast cancer: an indian perspective. JCO Glob. Oncol. 2020;6:789–798. doi: 10.1200/GO.20.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tacconi E.M.C., Tuthill M., Protheroe A. Review of adjuvant therapies in renal cell carcinoma: evidence to date. Onco. Targets Ther. 2020;13:12301–12316. doi: 10.2147/OTT.S174149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekar T., Klaassen Z., Goldberg H., Sayyid R.K., Kulkarni G.S., Fleshner N.E. High competing risks minimize real-world utility of adjuvant targeted therapy in renal cell carcinoma: a population-based analysis. Oncotarget. 2018;9:16731–16743. doi: 10.18632/oncotarget.24675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q.K., Pavlovich C.P., Zhang H., Kinsinger C.R., Chan D.W. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): A critical step towards the personalized care of renal cancers. Semin. Cancer Biol. 2019;55:8–15. doi: 10.1016/j.semcancer.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrnes S.A., Weigl B.H. Selecting analytical biomarkers for diagnostic applications: a first principles approach. Expert Rev. Mol. Diagn. 2018;18:19–26. doi: 10.1080/14737159.2018.1412258. [DOI] [PubMed] [Google Scholar]
- 14.Moch H., Srigley J., Delahunt B., Montironi R., Egevad L., Tan P.H. Biomarkers in renal cancer. Virchows. Arch. 2014;464:359–365. doi: 10.1007/s00428-014-1546-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim E., Zschiedrich S. Renal Cell Carcinoma in von Hippel-Lindau Disease-From Tumor Genetics to Novel Therapeutic Strategies. Front. Pediatr. 2018;6:16. doi: 10.3389/fped.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H., Shim B.Y., Lee S.J., Lee J.Y., Lee H.J., Kim I.H. Loss of Von Hippel-Lindau (VHL) tumor suppressor gene function: VHL-HIF pathway and advances in treatments for metastatic Renal Cell Carcinoma (RCC) Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22189795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bihr S., Ohashi R., Moore A.L., Ruschoff J.H., Beisel C., Hermanns T., Mischo A., Corro C., Beyer J., Beerenwinkel N., Moch H., Schraml P. Expression and mutation patterns of PBRM1, BAP1 and SETD2 mirror specific evolutionary subtypes in clear cell renal cell carcinoma. Neoplasia. 2019;21:247–256. doi: 10.1016/j.neo.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piva F., Santoni M., Matrana M.R., Satti S., Giulietti M., Occhipinti G., Massari F., Cheng L., Lopez-Beltran A., Scarpelli M., Principato G., Cascinu S., Montironi R. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies. Expert Rev. Mol. Diagn. 2015;15:1201–1210. doi: 10.1586/14737159.2015.1068122. [DOI] [PubMed] [Google Scholar]
- 19.Brugarolas J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J. 2013;19:324–332. doi: 10.1097/PPO.0b013e3182a102d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Cubas A.A., Rathmell W.K. Epigenetic modifiers: activities in renal cell carcinoma. Nat. Rev. Urol. 2018;15:599–614. doi: 10.1038/s41585-018-0052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly T.K., De Carvalho D.D., Jones P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angulo J.C., Manini C., Lopez J.I., Pueyo A., Colas B., Ropero S. The role of epigenetics in the progression of clear cell renal cell carcinoma and the basis for future epigenetic treatments. Cancers (Basel) 2021;13 doi: 10.3390/cancers13092071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borsos B.N., Huliak I., Majoros H., Ujfaludi Z., Gyenis A., Pukler P., Boros I.M., Pankotai T. Human p53 interacts with the elongating RNAPII complex and is required for the release of actinomycin D induced transcription blockage. Sci. Rep. 2017;7:40960. doi: 10.1038/srep40960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S., Li X. A genomic instability-derived risk index predicts clinical outcome and immunotherapy response for clear cell renal cell carcinoma. Bioengineered. 2021;12:1642–1662. doi: 10.1080/21655979.2021.1922330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson C.M., Ohh M. The multifaceted von Hippel-Lindau tumour suppressor protein. FEBS Lett. 2014;588:2704–2711. doi: 10.1016/j.febslet.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh J.J., Purdue M.P., Signoretti S., Swanton C., Albiges L., Schmidinger M., Heng D.Y., Larkin J., Ficarra V. Renal cell carcinoma. Nat. Rev. Dis. Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delahunt B., McKenney J.K., Lohse C.M., Leibovich B.C., Thompson R.H., Boorjian S.A., Cheville J.C. A novel grading system for clear cell renal cell carcinoma incorporating tumor necrosis. Am. J. Surg. Pathol. 2013;37:311–322. doi: 10.1097/PAS.0b013e318270f71c. [DOI] [PubMed] [Google Scholar]
- 29.Audia J.E., Campbell R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pankotai T., Komonyi O., Bodai L., Ujfaludi Z., Muratoglu S., Ciurciu A., Tora L., Szabad J., Boros I. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 2005;25:8215–8227. doi: 10.1128/MCB.25.18.8215-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasseigne B.N., Brooks J.D. The role of DNA methylation in renal cell carcinoma. Mol Diagn Ther. 2018;22:431–442. doi: 10.1007/s40291-018-0337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A., Kumari N., Sharma U., Ram S., Singh S.K., Kakkar N., Kaushal K., Prasad R. Reduction in H3K4me patterns due to aberrant expression of methyltransferases and demethylases in renal cell carcinoma: prognostic and therapeutic implications. Sci. Rep. 2019;9:8189. doi: 10.1038/s41598-019-44733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Shen L., Chen K.N. Association between H3K4 methylation and cancer prognosis: a meta-analysis. Thorac. Cancer. 2018;9:794–799. doi: 10.1111/1759-7714.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monaghan L., Massett M.E., Bunschoten R.P., Hoose A., Pirvan P.A., Liskamp R.M.J., Jorgensen H.G., Huang X. The emerging role of H3K9me3 as a potential therapeutic target in acute myeloid leukemia. Front. Oncol. 2019;9:705. doi: 10.3389/fonc.2019.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minardi D., Lucarini G., Filosa A., Milanese G., Zizzi A., Primio R.D., Montironi R., Muzzonigro G. Prognostic role of global DNA-methylation and histone acetylation in pT1a clear cell renal carcinoma in partial nephrectomy specimens. J. Cell. Mol. Med. 2009;13:2115–2121. doi: 10.1111/j.1582-4934.2008.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosashvilli D., Kahl P., Mertens C., Holzapfel S., Rogenhofer S., Hauser S., Buttner R., Von Ruecker A., Muller S.C., Ellinger J. Global histone acetylation levels: prognostic relevance in patients with renal cell carcinoma. Cancer Sci. 2010;101:2664–2669. doi: 10.1111/j.1349-7006.2010.01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocco E., Leo M., Canzonetta C., Di Vito S., Mai A., Rotili D., Di Napoli A., Vecchione A., De Nunzio C., Filetici P., Stoppacciaro A. KAT3B-p300 and H3AcK18/H3AcK14 levels are prognostic markers for kidney ccRCC tumor aggressiveness and target of KAT inhibitor CPTH2. Clin Epigenetics. 2018;10:44. doi: 10.1186/s13148-018-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramadoss S., Chen X., Wang C.Y. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramakrishnan S., Ellis L., Pili R. Histone modifications: implications in renal cell carcinoma. Epigenomics. 2013;5:453–462. doi: 10.2217/epi.13.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakrishnan S., Pili R. Histone deacetylase inhibitors and epigenetic modifications as a novel strategy in renal cell carcinoma. Cancer J. 2013;19:333–340. doi: 10.1097/PPO.0b013e3182a09e07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu L., Wang J., Kong W., Huang J., Dong B., Huang Y., Xue W., Zhang J. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharm Sin B. 2019;9:324–334. doi: 10.1016/j.apsb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji J., Zhang Y., Redon C.E., Reinhold W.C., Chen A.P., Fogli L.K., Holbeck S.L., Parchment R.E., Hollingshead M., Tomaszewski J.E., Dudon Q., Pommier Y., Doroshow J.H., Bonner W.M. Phosphorylated fraction of H2AX as a measurement for DNA damage in cancer cells and potential applications of a novel assay. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pankotai T., Bonhomme C., Chen D., Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 44.Guo E., Wu C., Ming J., Zhang W., Zhang L., Hu G. The clinical significance of DNA damage repair signatures in clear cell renal cell carcinoma. Front Genet. 2020;11 doi: 10.3389/fgene.2020.593039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chabanon R.M., Morel D., Eychenne T., Colmet-Daage L., Bajrami I., Dorvault N., Garrido M., Meisenberg C., Lamb A., Ngo C., Hopkins S.R., Roumeliotis T.I., Jouny S., Henon C., Kawai-Kawachi A., Astier C., Konde A., Del Nery E., Massard C., Pettitt S.J., Margueron R., Choudhary J.S., Almouzni G., Soria J.C., Deutsch E., Downs J.A., Lord C.J., Postel-Vinay S. PBRM1 deficiency confers synthetic lethality to DNA repair inhibitors in cancer. Cancer Res. 2021;81:2888–2902. doi: 10.1158/0008-5472.CAN-21-0628. [DOI] [PubMed] [Google Scholar]
- 46.Clark D.J., Dhanasekaran S.M., Petralia F., Pan J., Song X., Hu Y., da Veiga Leprevost F., Reva B., Lih T.M., Chang H.Y., Ma W., Huang C., Ricketts C.J., Chen L., Krek A., Li Y., Rykunov D., Li Q.K., Chen L.S., Ozbek U., Vasaikar S., Wu Y., Yoo S., Chowdhury S., Wyczalkowski M.A., Ji J., Schnaubelt M., Kong A., Sethuraman S., Avtonomov D.M., Ao M., Colaprico A., Cao S., Cho K.C., Kalayci S., Ma S., Liu W., Ruggles K., Calinawan A., Gumus Z.H., Geiszler D., Kawaler E., Teo G.C., Wen B., Zhang Y., Keegan S., Li K., Chen F., Edwards N., Pierorazio P.M., Chen X.S., Pavlovich C.P., Hakimi A.A., Brominski G., Hsieh J.J., Antczak A., Omelchenko T., Lubinski J., Wiznerowicz M., Linehan W.M., Kinsinger C.R., Thiagarajan M., Boja E.S., Mesri M., Hiltke T., Robles A.I., Rodriguez H., Qian J., Fenyo D., Zhang B., Ding L., Schadt E., Chinnaiyan A.M., Zhang Z., Omenn G.S., Cieslik M., Chan D.W., Nesvizhskii A.I., Wang P., Zhang H. C. Clinical proteomic tumor analysis, integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell, 2019;179:964–983. doi: 10.1016/j.cell.2019.10.007. e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.