Abstract
Arp2/3 complex is an actin filament nucleation and branching machinery conserved in all eukaryotes from yeast to human. Arp2/3 complex branched networks generate pushing forces that drive cellular processes ranging from membrane remodeling to cell and organelle motility. Several molecules regulate these processes by directly inhibiting or activating Arp2/3 complex and by stabilizing or disassembling branched networks. Here, we review recent advances in our understanding of Arp2/3 complex regulation, including high-resolution cryo-electron microscopy structures that illuminate the mechanisms of Arp2/3 complex activation and branch formation, and novel cellular pathways of branch formation, stabilization, and debranching. We also identify major gaps in our understanding of Arp2/3 complex inhibition and branch stabilization and disassembly.
Keywords: Arp2/3 complex, nucleation-promoting factors, inhibitors, branched network stabilization and disassembly, mechanosensation
Arp2/3 complex and nucleation-promoting factors (NPFs)
The actin cytoskeleton is a dynamic system of hundreds of proteins that helps maintain cell shape and polarity and provides forces and directionality for a myriad of motile functions in cells. At the core of this system is actin, the most abundant protein in the cytosol of eukaryotic cells. Actin is in constant flux between monomeric and filamentous forms to produce networks with distinct architectures. One such architecture is that consisting of branched actin networks (also called dendritic networks), resulting from the filament nucleation and branching activities of Arp2/3 complex. Activated by proteins known as nucleation-promoting factors (NPFs), Arp2/3 complex binds to the side of a pre-existing (mother) filament and nucleates the formation of a new (branch) filament that grows at a ~70° angle relative to the mother filament.
The 7-subunit Arp2/3 complex comprises two actin-related proteins, Arp2 and Arp3, and five scaffolding subunits, ArpC1 to ArpC5. The Arps (Arp2 and Arp3) act as a pseudo-actin dimer during nucleation, whereas the scaffolding subunits hold the Arps together and mediate key interactions with the mother filament in the branch [1]. In the basal state, Arp2/3 complex is inactive, with the Arps splayed apart [2]. During activation, the NPFs perform three main functions: a) they trigger a conformational change that repositions the Arps into a filament-like (short-pitch) conformation [3], b) they recruit actin subunits to the barbed end of the Arps to form the initial polymerization nucleus [4–6], and c) they promote binding of the complex to the side of the mother filament [7]. Wiskott-Aldrich Syndrome Protein (WASP)-family NPFs, also called class-I NPFs, are generally unrelated, but all share C-terminal proline (Pro)-rich and WH2/Central/Acidic (WCA) domains (Fig. 1a). Recent work shows that the Pro-rich regions play an essential role by mediating the recruitment of profilin-actin to feed the growth of newly nucleated filaments via NPFs clustered on membranes (reviewed in [8]). The WCA region is necessary and sufficient to activate Arp2/3 complex in vitro and comprises one to three WASP-homology 2 (WH2 or W) domains that bind actin [4], and Central (C) and Acidic (A) domains that bind to two distinct sites on Arp2/3 complex [9, 10].
Figure 1. Activators, inhibitors, and other regulators of Arp2/3 complex.
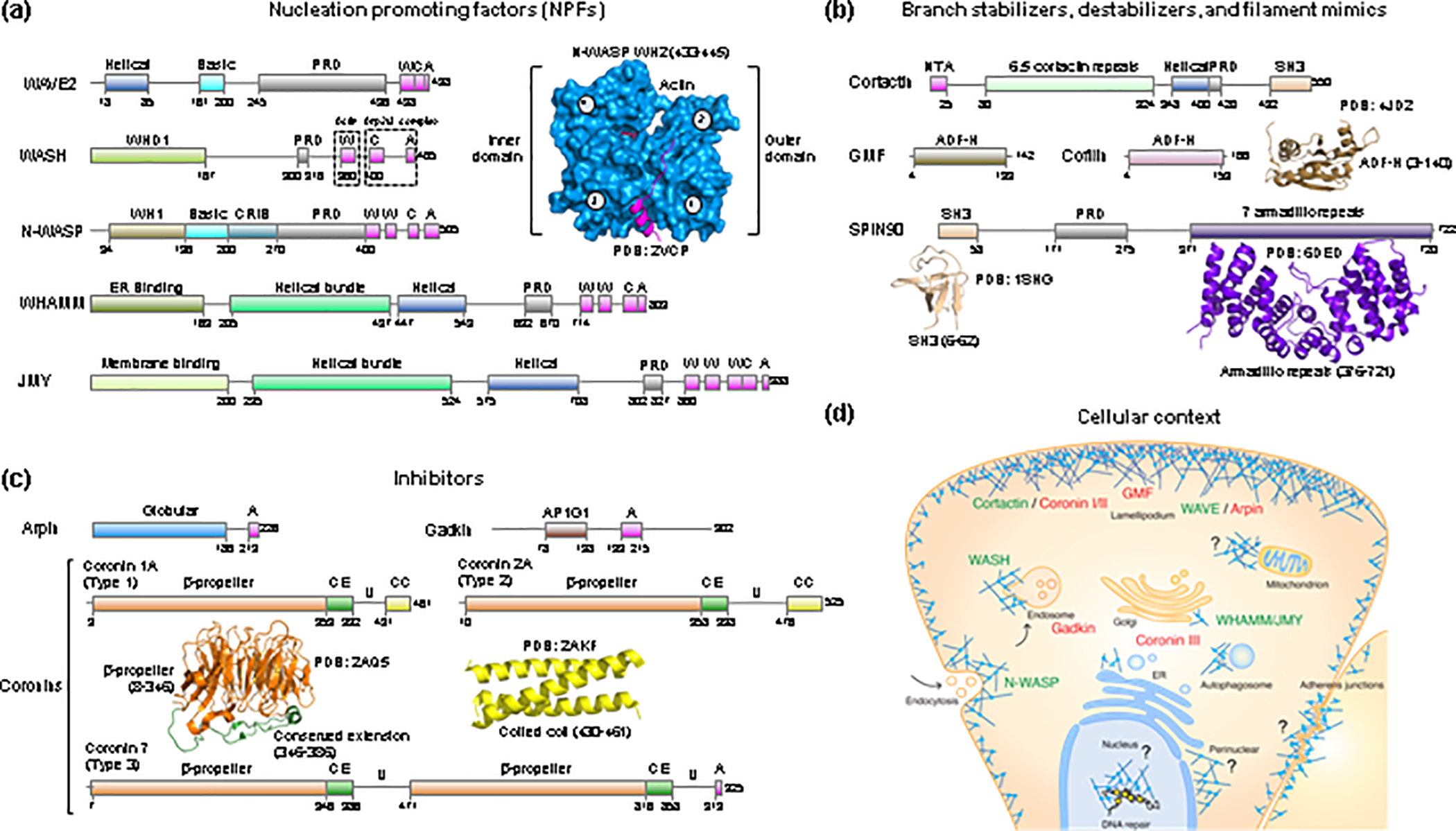
(a – c) Domain diagrams of human NPFs (a), branch stabilizers and destabilizers and mother filament mimics (b), and Arp2/3 complex inhibitors (c). WASP-homology 2 (WH2), central (C) and acidic (A) domains are shown in magenta. Proline rich domains (PRDs) are shown in gray. Other domains specific to each proteins include: WH1, WASP-homology domain 1; CRIB, Cdc42/Rac interactive binding; AP1G1, adaptor protein-1 gamma-1 interacting domain; NTA, N-terminal acidic domain; SH3, Src homology 3 domain; ADF-H, actin-depolymerizing factor homology; CE, conserved extension; U, Unique region; CC, coiled-coil. The UniProt accession codes of the human proteins shown are: WAVE2 (Q9Y6W5); WASH (A8K0Z3); N-WASP (O00401); WHAMM (Q8TF30); JMY (Q8N9B5); Cortactin (Q14247); GMF (O60234); Cofilin (P23528); SPIN90 (Q9NZQ3); Arpin (Q7Z6K5); Gadkin (Q63HQ0); Coronin 1A (P31146); Coronin 2A (Q92828); Coronin 7 (P57737). Ribbon diagrams of determined structures of domains of these proteins are shown along with the PDB accession codes. In the structure of the WH2 domain bound to actin, the outer (subdomains 1 and 2) and inner (subdomains 3 and 4) domains of actin are indicated. (d) Cellular context of NPFs and branch stabilizers (green text) and inhibitors and branch destabilizers (red text). Question marks indicate Arp2/3 complex regulators yet to be identified in connection with specific functions or subcellular locations.
The variable N-terminal domains of NPFs are typically involved in localization and integration of upstream signaling, giving rise to a certain division of roles among NPFs, which tend to fulfil specialized functions at specific subcellular locations (Fig. 1d). The NPF literature is vast and not a focus of this review (reviewed in [11]). Only some of their better-documented or recently-established functions are listed here. Thus, N-WASP is primarily implicated in endocytosis, whereas WAVE (isoforms 1–3) forms part of a pentameric complex called the WAVE regulatory complex (WRC) that is implicated in the formation of cell protrusions and cell migration (reviewed in [12]). WASH, which forms part of a similar pentameric complex, together with WHAMM and JMY are primarily implicated in remodeling of intracellular membranes, including endosomes, ER, Golgi, and autophagosomes [13–15]. However, such a division of roles among NPFs is an oversimplification. For example, N-WASP can substitute for WAVE in the generation of membrane protrusions [16].
Branched actin networks generate pushing forces at diverse subcellular locations
Branched actin networks are present along the cell cortex and near most membranous organelles in the cell (Fig. 1d). These networks exert forces during processes such as cell motility, vesicular trafficking, and membrane scission. At the cortex, the pushing forces of branched actin networks are counterbalanced by pulling forces from myosin motors on linear actin filaments. This balance of forces is particularly evident at adherens junctions in epithelial cells, where Arp2/3 complex-dependent protrusive forces push the membranes of neighboring cells to maintain cadherin-mediated cell-cell adhesions [17]. If the Arp2/3 complex is inhibited, myosin pulls the membranes of neighboring cells away from one another, resulting in the formation of membrane bulges [18]. Imbalances in this “tug-of-war” between protrusive and contractile forces results in defects in tissue mechanics and morphology, as observed during epithelial folding in development [19] and the sprouting of blood vessels [20]. The forces exerted by branched networks are also used to counteract membrane tension and separate transport intermediates from the plasma membrane during clathrin-mediated endocytosis [21] and from endosomes along the endocytic pathway [22]. Actin pushing forces with the help of BAR domain proteins constrict the neck of clathrin-coated endocytic vesicles [23, 24] and elongate endocytic invaginations [25, 26].
Lately, branched actin networks have been shown to push and squeeze the nucleus to help cells migrate through constricted spaces [27] and to direct the movement of nuclei from the center to the periphery of skeletal muscle myofibers during myogenesis [28]. Arp2/3 complex also has functions inside the nucleus, where it was recently shown to facilitate DNA repair and promote homology-directed recombination by moving and clustering double-strand DNA breaks [29, 30]. In symmetrically dividing cells, a meshwork of actin filaments maintains the uniform distribution of mitochondria around the mitotic spindle and shuffles mitochondria to ensure their equal and random inheritance by daughter cells [31]. Mitochondria were propelled through the cell by Arp2/3 complex-dependent actin comet tails, similar to those associated with the movement of intracellular pathogens.
Molecular mechanism of branched network formation and stabilization
The role of NPFs in Arp2/3 complex activation was discovered in 1998 [32, 33] and for the following 10 years, it was thought that activation resulted from NPF binding to a single site on Arp2/3 complex. A breakthrough occurred in 2008, when it was discovered that activation proceeded through binding of the Central-Acidic (CA) region of NPFs to two distinct sites on Arp2/3 complex [9]. Several studies subsequently confirmed this finding, and models of CA-bound Arp2/3 complex were proposed [5, 6, 34, 35]. While these models all captured the generally accepted idea that one NPF binds to Arp2-ArpC1 whereas the other binds to Arp3, they differed in important ways; they disagreed about the precise interactions of each NPFs, their order of binding and role in activation, and the importance of actin binding to NPFs for activation.
Recent cryo-electron microscopy (cryo-EM) structures of Arp2/3 complex substantially advance our understanding of the mechanisms of nucleation and branch formation [1, 10, 36, 37]. The first of these structures was that of human Arp2/3 complex with the CA region of N-WASP bound determined at 3.8-Å resolution [10]. This structure showed the precise binding paths of NPFs on Arp2-ArpC1 and Arp3 (Fig. 2a). The ability to recombinantly express human Arp2/3 complex using a novel insect cell expression system further allowed for mutagenesis and crosslinking studies that resulted in several new findings: a) actin binding to the W domains of NPFs favors the transition of the Arps toward a filament-like short-pitch conformation, b) actin-NPF binding to Arp2-ArpC1 precedes binding to Arp3 and is sufficient to shift the equilibrium toward the short-pitch conformation, with no additional gain from binding to Arp3, c) while the short-pitch transition is part of the activation pathway, it is insufficient to fully activate mammalian Arp2/3 complex, which also requires NPF-mediated delivery of actin to the barbed end of both Arps. As we discuss below, the latter finding is crucial to understand the difference between actual NPFs that contain monomeric actin-binding sites and other molecules that regulate Arp2/3 complex. The scaled-up expression of human Arp2/3 complex in Sf9 cells also enabled the determination of the 4.2-Å resolution cryo-EM structures of the most and least active isoform variants of Arp2/3 complex, containing respectively subunits ArpC1B and ArpC5L vs. ArpC1A and ArpC5 [37]. This study, however, did not reveal substantial differences in structure between variants, suggesting that their different activities may result from different energetic barriers for activation. For instance, it has long been known that Saccharomyces cerevisiae Arp2/3 complex has substantial background activity in the absence of NPFs, consistent with a lower activation barrier [38].
Figure 2. Cryo-EM structures of Arp2/3 complex with bound NPFs, at the pointed end and in the branch.
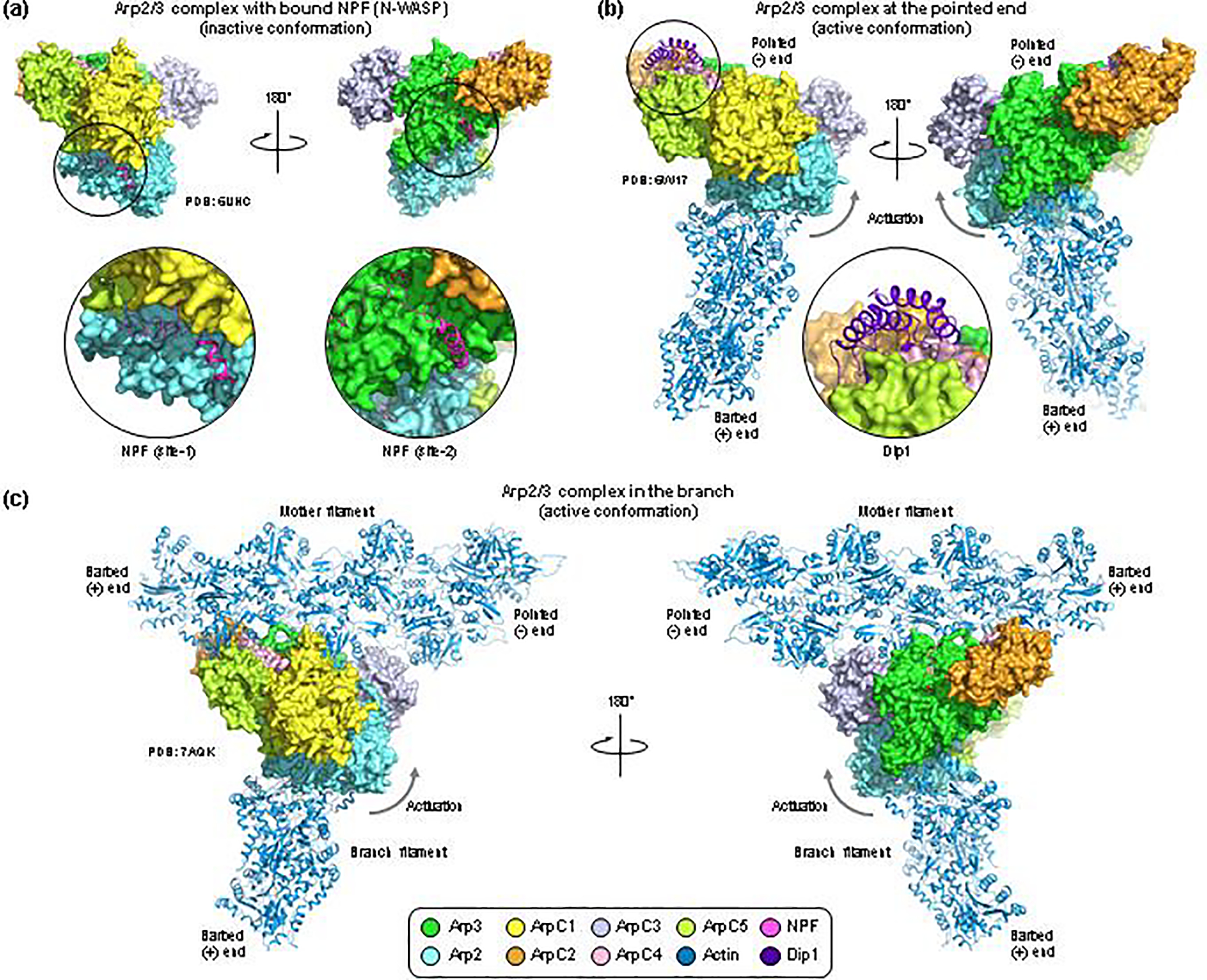
(a) Cryo-EM structure of human Arp2/3 complex with bound NPF N-WASP. The coloring scheme is given at the bottom of the figure, and the PDB accession codes are listed for each structure. The CA region of N-WASP binds to two sites on the complex (close-up views). In site-1, the C helix binds in the hydrophobic cleft at the barbed end of Arp2 whereas the A domain interacts with ArpC1. In site-2, both the C helix and the A domain bind to Arp3, with the C helix interacting in the hydrophobic cleft of Arp3. Albeit NPFs drive the equilibrium toward activation, the conformation of Arp2/3 complex in this structure is inactive. (b) Cryo-EM structure of yeast Arp2/3 complex at the pointed end of the actin filament, also showing a fragment of the armadillo repeat of Dip1 (residues G235-E366, close-up view). The two orientations shows are the same as in part a. The conformation of Arp2/3 complex in this structure is active, with the Arp2 subcomplex (Arp2-ArpC1-ArpC4-ArpC5) rotated up ~19° relative to the Arp3 subcomplex (Arp3-ArpC2-ArpC3), as indicated by the arrow. (c) Cryo-ET structure of Arp2/3 complex in the branch. This structure contains only main-chain atoms (i.e. it lacks side chains). To produce a comparable view to those shown in parts a and b, an all-atom model of Arp2/3 complex was generated by superimposing individual subunits from a high-resolution crystal structure onto the main-chain backbone of these subunits in the cryo-ET structure. The orientation is the same as in parts a and b, and the conformation of Arp2/3 complex is active, and similar to that at the pointed end (part b). A notable difference is subunit ArpC3, which in this structure moves substantially to make contacts with Arp2, Arp3 and the mother filament.
An essential component of the Arp2/3 complex nucleation reaction is the mother filament [33]. While bona fide WASP-family NPFs all contain actin monomer and profilin-actin binding sites (Fig. 1a) that play essential roles in nucleation [10] and elongation [8], respectively, other Arp2/3 complex regulators either bind to the mother filament or bypass the need for the mother filament during nucleation. These include cortactin [39] and WISH/DIP/SPIN90 (WDS)-family proteins [40]. These molecules are sometimes referred to as class-II NPFs, although their function does not appear to be Arp2/3 complex activation. Indeed, on their own cortactin and WDS-family proteins activate mammalian Arp2/3 complex only weakly [39, 40], requiring high, micromolar concentrations, whereas nanomolar concentrations of WASP-family NPFs produce very high polymerization rates [33]. It is also generally accepted that cortactin’s major biochemical activity is the stabilization of Arp2/3 complex branch junctions [39], and unlike WASP-family NPFs that dissociate after nucleation [41], cortactin binds stably to these junctions and may also accelerate the release of NPFs [42]. Moreover, contrary to WASP-family NPFs that tend to have specialized roles and localization, cortactin is found rather ubiquitously in association with branched networks (reviewed in [43]). These properties allow cortactin to fulfil countless roles in cells by promoting the formation and stabilization of actin branched networks.
Although it has been proposed that WDS-family proteins (Fig. 1) act as bona fide NPFs, capable of activating Arp2/3 complex while avoiding the need for the mother filament, actin monomer binding and delivery to the Arps, and profilin-actin delivery to the newly formed branch [40], their actual role and mechanism in branched network formation is only beginning to emerge. Like cortactin [40], but unlike NPFs [41], WDS-family proteins remain bound to Arp2/3 complex at the pointed end of newly formed filaments, making them single-turnover regulators [44]. Recent evidence offers a more nuanced role for these proteins, showing that yeast Dip1 works synergistically with Wsp1 (yeast WASP) to activate Arp2/3 complex during endocytosis, without the need for preexisting filaments [45]. Of note, Wsp1 was required for both initiation and propagation of endocytic actin networks, consistent with its role as the NPF in this process, whereas Dip1 may have served as a substitute for mother filaments to kick-start endocytosis. Another recent study in mammalian cells showed that SPIN90 (mammalian ortholog of Dip1) synergizes with both Arp2/3 complex and the formin mDia1 to nucleate fast-growing filaments leading to an increase in the density of linear actin arrays at the cell cortex [46]. A recent 3.9-Å resolution cryo-EM structure of Dip1-bound yeast Arp2/3 complex at the pointed end of the filament revealed the active conformation of Arp2/3 complex (Fig. 2b) [36]. The transition toward the activated stated was ascribed to the presence of Dip1 in the reconstruction, but this conclusion is questionable. First, Arp2/3 complex has been long known to also act as a pointed end-capping protein [47], and a previous EM reconstruction already showed Arp2/3 complex at the pointed end in the absence of NPFs [48]. Although the lower resolution of this study precluded a precise description of the structure of Arp2/3 complex at the pointed end, the authors concluded that the Arps were realigned into a filament-like conformation, consistent with the addition of actin subunits at the barbed end of the Arps. Second, the Dip1-bound structure was determined in the presence of 1 μM Wsp1 WCA, presumably used to shift the equilibrium toward the active conformation, although this was not discussed. Third, Dip1, a globular protein consisting of 374 amino acids, was mostly disordered in the structure (only 132 amino acids were observed, G235-E366), suggesting that it is weakly bound to Arp2/3 complex and thus unlikely to be the source of the conformational change. Fourth, a previous crystal structure showed Arp2/3 complex in the inactive conformation when bound to SPIN90 [49], suggesting that on their own WDS-family proteins are unable to trigger the activating transition. The cryo-EM structure is nonetheless informative in that it reveals the structural changes that take place during Arp2/3 complex binding to the pointed end. The Arps are realigned into a filament-like conformation, as previously suggested [48], and their outer and inner domains (referring to the position of these domains in the filament, Fig. 1a) are rotated with respect to one another to produce a flatter conformation analogous to that of actin subunits in the filament.
A recent breakthrough was the determination of the structure of the Arp2/3 complex branch junction in cells using cryo-electron tomography (cryo-ET) [1]. While the technical difficulties of this study limited the resolution to 9-Å, the main-chain backbone of all the subunits of Arp2/3 complex, eight subunits of the mother filament and three subunits of the branch filament were unambiguously defined. The transition of the Arps toward the short pitch conformation is brought about by a ~19° rotation around an axis that coincides with that of the coiled coil formed by ArpC2 and ArpC4, similar to what was observed when Arp2/3 complex was bound at the pointed end [36]. This rotation divides Arp2/3 complex into two subcomplexes, one comprising Arp2 (Arp2-ArpC1-ArpC4-ArpC5) and one comprising Arp3 (Arp3-ArpC2-ArpC3). The rotation creates binding sites for the first two subunits of the branch filament at the barbed ends of the Arps. On the opposite side, five subunits of Arp2/3 complex (Arp3, ArpC1, ArpC3, ArpC2, ArpC4) contact five subunits of the mother filament, without this resulting in any substantial change in the structure of the mother filament. As previously suggested [36, 48], the conformation of Arp2/3 complex is very similar in the branch and at the pointed end, with some differences. Notably, ArpC3 is positioned differently in the branch, allowing it to make contacts with Arp3, Arp2 and the mother filament that help stabilize the short-pitch conformation and the branch junction. Although the contact surface of Dip1 on Arp2/3 complex overlaps only with one of the five subunits of the mother filament that interact with Arp2/3 complex (compare Figs. 2 b and c), it is now easy to see how Dip1 can compete with and possibly substitute for the mother filament in certain cellular contexts.
Another twist on the source of mother filaments at cellular locations where these may be lacking is a recent study that established dynactin as a seed for mother filament formation and Arp2/3 complex activation on endosomes [50]. The dynactin complex, better known for its role as a general scaffold of the dynein motor, is built around an actin-like minifilament comprising eight actin-related protein-1 (Arp1) subunits and one β-actin subunit, and capped at the barbed end by the capping protein (CP) αβ heterodimer and at the pointed end by four subunits (Arp11, p62, p25 and p27) [51]. The pentameric WASH complex, the NPF present on endosomes, has evolved the necessary attributes to convert dynactin into a seed for the mother filament while simultaneously activating Arp2/3 complex. One of the subunits of the WASH complex, FAM21, contains a capping protein interaction (CPI) motif that binds CPαβ and uncaps dynactin. Once uncapped the dynactin minifilament elongates from the barbed end through the addition of actin subunits to form the mother filament. Then, another subunit of the WASH complex (WASH) recruits and activates Arp2/3 complex on the side of the dynactin-formed filament. This study adds meaning to intriguing observations, such as the role of the actin minifilament in dynactin, the role of some of the subunits of the WASH complex and the role of the CPI motif in FAM21.
Mechanosensation, reinforcement and disassembly of branched actin networks
In recent years, it has become increasingly clear that the actin cytoskeleton not only generates forces but also senses extracellular forces [52–54], which in turn triggers actin network remodeling and several cellular and tissue processes [55–57]. Branched actin networks placed under mechanical load in vitro are strengthened through a force feedback mechanism that increases their density, pushing power, and mechanical efficiency [52]. For actin filaments buckling under load, Arp2/3 complex branching occurs preferentially on the convex than the concave side of the bend, which may be one way in which the density of branched networks is biased by force [58]. Increased debranching has also been observed under load [59], indicating that both branching and debranching can be affected by force. Similar observations have been made in cells, where dendritic actin networks adapt to mechanical load by transitioning toward a configuration dominated by filaments growing perpendicularly to the plasma membrane [55]. In another example in vivo, mechanical load was shown to trigger the formation of Rac1- and Arp2/3 complex-dependent cell protrusions to perform the phagocytic clearance of apoptotic cells by the surface epithelium of zebrafish and mouse embryos [57]. Cell cycle progression through the G1/S transition also depends on mechanical inputs, such as substratum rigidity and epithelial cell density [56]. These inputs are mechanotransduced via the Rac1-WAVE-Arp2/3 complex pathway and depend specifically on Arp2/3 complex subunit ArpC1B, which is the predominant isoform in branch junctions at the cell cortex. Indeed, three of the Arp2/3 complex subunits (Arp3, ArpC1, and ArpC5) have different isoforms, resulting in Arp2/3 complexes with different properties. Thus, it was recently established that ArpC1B-containing Arp2/3 complexes at the cortex display higher nucleation and branching activities and are more resistant to coronin-mediated branch disassembly than complexes containing ArpC1A [56, 60].
The positive feedback loop for branched network assembly and reinforcement is likely stopped by Arp2/3 complex inhibitors such as Arpin at the cell cortex and other inhibitors at other subcellular locations [61, 62]. Branched networks then need to be disassembled, which primarily involves the coordinated action of cofilin-family members, with the nucleotide state on actin and Arp2/3 complex acting as a molecular clock to signal disassembly (Figs. 1b and 3). A characteristic feature of members of the cofilin family is to bind ADP-actin monomers and filaments with higher affinity than ATP-actin monomers and filaments. This property allows cofilin to specifically target older, ADP-actin segments of the filament for disassembly (Fig. 3). Like with most actin-binding proteins, however, properly aligning biochemical and cell biological data requires that proteins be analyzed at physiologically meaningful concentrations. Case in point, cofilin; at saturating (equimolar) occupancy, cofilin stabilizes the actin filament by forming additional contacts between subunits [63]. At low concentrations, however, clusters of just two to three cofilin molecules efficiently sever filaments [64] by inducing structural perturbations at the boundary between cofilin-bound and unbound subunits [63]. Recent studies reveal that cofilin-mediated depolymerization can be further accelerated by novel mechanisms, including functional synergy between cofilin and cyclase-associated-protein [65, 66] and actin oxidation by MICAL [67, 68]. MICAL also oxidizes Arp2/3 complex subunit Arp3B, accelerating the disassembly of branched networks enriched with this Arp3 isoform [69].
Figure 3. Model of nucleation, stabilization, destabilization, and inhibition of branched actin networks.
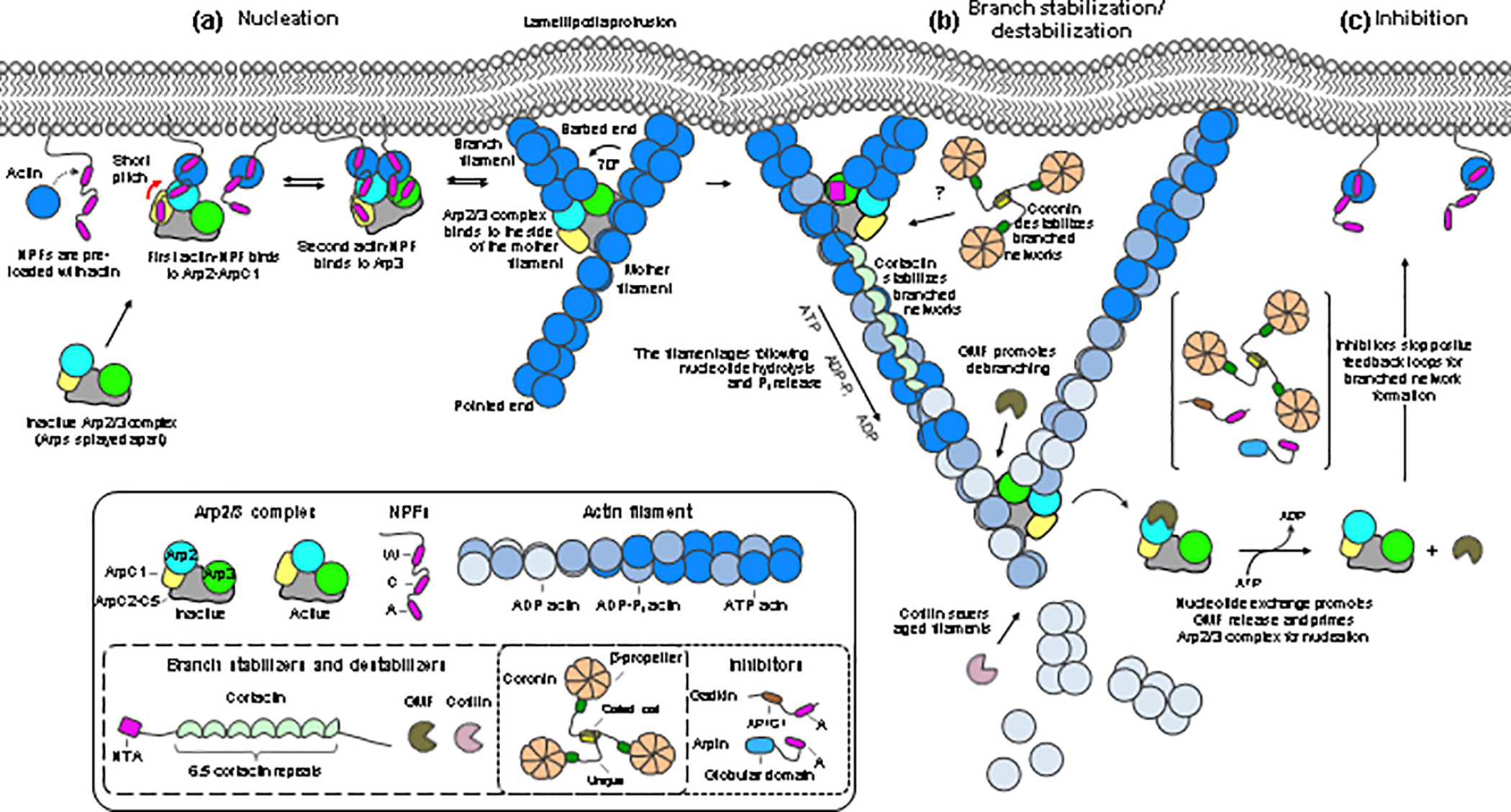
Proteins are colored and labeled according to figure 1. Boxed insets show cartoon diagrams of the proteins and domains. (a) Nucleation is a multi-step process. NPFs clustered at the membrane, with actin monomers pre-bound to their WH2 domains, recruit Arp2/3 complex through site-1 (Arp2-ArpC1). This shifts the equilibrium toward the activated conformation (indicated by a red arrow), allowing for NPF binding to site-2 (Arp3). The NPFs deliver actin subunits at the barbed ends of both Arps. The complex then binds to the side of a pre-existing filament (mother filament), which stabilizes the active conformation, prompts the release of NPFs, and allows the branch to grow at a ~ 70° angle relative to the mother filament. Growth of the branch and mother filaments is accelerated by profilin-actin delivery through the Pro-rich regions of NPFs clustered at the membrane. (b) Proteins like cortactin stabilize the branched network. Network reinforcement depends on positive feedback loops that converge on signaling pathways, such as the Rac1-WAVE-Arp2/3 complex pathway. Older networks, containing ADP-bound actin and Arp2/3 complex, are disassembled by members of the cofilin family, a process that can be accelerated by other factors (see text). (c) Arp2/3 complex inhibitors such as Arpin stop positive feedback loops for network reinforcement.
Although glial maturation factor (GMF) has been described as an inhibitor of Arp2/3 complex [70, 71], this view is controversial. GMF belongs to the cofilin family of proteins. Like cofilin, GMF binds with higher affinity to ADP-Arp2/3 complex than ATP-Arp2/3 complex [72]. GMF’s low affinity for ATP-Arp2/3 complex prevents competition with NPFs during nucleation, whereas its preference for ADP-Arp2/3 complex is consistent with its role in branch disassembly. Indeed, a new study reveals that older ADP-Arp2/3 complex branches are more sensitive to mechanical force and GMF-mediated debranching [59]. There is also disagreement about the mechanism of interaction of GMF with Arp2/3 complex. Both isothermal titration calorimetry [72] and x-ray crystallography [73] reveal a single binding site for GMF on Arp2/3 complex. The crystal structure shows GMF bound to the hydrophobic cleft at the barbed end of Arp2, which is strikingly similar to the interactions of other cofilin-family members with actin [68]. However, a secondary binding site on Arp3 has been proposed based on low-resolution EM of negatively stained samples [71], and therefore the precise mechanism of GMF-mediated branch disassembly remains incompletely characterized.
Arp2/3 complex inhibition
Like with most cellular processes, Arp2/3 complex inhibition is as important as activation. Several inhibitors of Arp2/3 complex have been proposed, including coronins [74], Arpin [61], GMF [70], gadkin [75], and protein interacting with C kinase (PICK1) [76]. However, while our understanding of nucleation mechanisms has advanced substantially, inhibitory mechanisms are poorly understood, and some of the existing evidence is controversial.
The first proposed and most extensively studied Arp2/3 complex inhibitor is coronin [74], a protein found at sites of dynamic actin assembly in eukaryotes from yeast to mammals [77, 78]. Budding yeast has a single coronin (Crn1), whereas mammals have seven coronins subdivided into three types: type-1 (coronins 1A, 1B, 1C and 6), type-2 (coronins 2A and 2B); and type-3 (coronin 7) (Fig. 1c). All coronins have a 7-bladed WD40 β-propeller domain, immediately followed by a 40-amino acid conserved sequence that packs tightly against the β-propeller and is required for its structural stability [79]. All type 1 and 2 coronins have a C-terminal coiled-coil domain that mediates trimerization [80–82], but are distinguished from one another by the so-called “Unique” region that separates the β-propeller and coiled-coil domains and varies significantly in length and sequence. Coronin 7 is substantially different in that it has two β-propeller domains and a C-terminal CA region similar to that of NPFs but lacks the coiled-coil domain.
While coronins differ enough in sequence as to anticipate functional and mechanistic differences among them [78], most studies have focused on type-1 mammalian coronins and yeast Crn1, which more closely resemble one another. Some functions, however, are likely conserved among all coronins, such as F-actin binding mediated by the most highly conserved β-propeller domain and shown for both yeast [83] and type-1 mammalian coronins [84]. Coronins bind with higher affinity to ATP/ADP•Pi- than ADP-actin filaments [84]. The β-propeller domain has been also implicated in plasma membrane binding [81].
Direct binding and inhibition of Arp2/3 complex has been reported for yeast [74, 82] and mammalian [85, 86] coronins, although the role and mechanism of inhibition is debated. Studies on Crn1, including low-resolution negative stain EM, support a model in which the trimeric coiled coil domain unravels and Crn1 binds as a monomer via its C-terminal sequence to subunit ArpC2 of Arp2/3 complex [71, 74]. Another study, however, found that low concentrations of Crn1 do not inhibit but activate Arp2/3 complex [82]. These authors identified an Arp2/3 complex-binding site within a CA sequence, analogous to that of NPFs, located within the Unique region of yeast Crn1. They mapped the binding site of this sequence to subunit Arp2 and showed that Crn1 can activate Arp2/3 complex synergistically with NPFs. They also observed Arp2/3 complex inhibition, but only at high Crn1 concentrations and saturating binding to the mother filament, suggesting that inhibition results from competition with Arp2/3 complex for binding to the mother filament. While this model is appealing, particularly because it does not invoke the structurally unlikely event of coiled coil unraveling, it cannot be extended to mammalian coronins, which lack a CA sequence within their Unique region. Moreover, this mechanism of inhibition is indirect, analogous to competitive inhibition of Arp2/3 complex by tropomyosin [87], and does not qualify coronin as a direct inhibitor of Arp2/3 complex. So, the question remains, are mammalian coronins actual inhibitors of Arp2/3 complex?
Most of the data available is consistent with a role for type-1 coronins as branched network destabilizers (Fig. 3). Thus, type-1 coronins destabilize actin filament branches in vitro and in cells by inducing dissociation of Arp2/3 complex from the sides of actin filaments and by competing with branch stabilization by cortactin [88]. Type-1 coronins specifically promote disassembly of Arp2/3 complex networks containing subunit isoforms ArpC1A and ArpC5, whereas cortactin preferentially stabilizes networks containing isoforms ArpC1B and ArpC5L [60]. Both yeast [89, 90] and mammalian type-1 [91, 92] coronins have been also shown to synergize with cofilin and actin-interacting protein 1 (Aip1) to promote actin network disassembly. Mammalian coronin 7 remains virtually uncharacterized. A recent study of the C. elegans coronin 7 ortholog POD-1 suggests that it induces filament debranching and plays a role in cell migration and the establishment of cell polarity [93].
Arpin (Figs. 1 and 3) was discovered through a bioinformatics search for proteins containing a C-terminal A domain analogous to that of NPFs, but experiments in vitro showed that contrary to NPFs Arpin inhibited Arp2/3 complex [61]. Arpin depletion in cells and in vivo resulted in more persistent lamellipodial protrusions, whereas injection of purified Arpin destabilized lamellipodia protrusions [61]. Lamellipodial branched networks drive directional and persistent cell migration through a Rac1-WAVE-Arp2/3 complex-dependent positive feedback loop. Arpin also functions under Rac1 control, such that it does not appear to inhibit Arp2/3 complex globally but rather acts locally to terminate the Rac1-WAVE-Arp2/3 complex positive feedback loop [61]. It has now become apparent that the relative levels of Arpin and WAVE critically control the growth and invasion of tumor cells and act as a prognosis factor for multiple types of cancers [94, 95]. While Arpin is thought to function through direct competition with WAVE [61], the exact mechanism of inhibition is unknown. A low-resolution negative stain EM study suggested that two Arpin molecules bind through their A domains to Arp2/3 complex, one to each of the Arps [71]. However, the A domain accounts only partially for the inhibitory capacity of full-length Arpin, suggesting that other regions of the protein participate in binding and inhibition of Arp2/3 complex [61].
Gadkin is another proposed inhibitor that contains an A domain (Figs. 1 and 3), which mediates binding to Arp2/3 complex and competition with NPFs [75]. Yet, in vitro gadkin does not alter the nucleation activity of Arp2/3 complex. In cells, gadkin depletion leads to partial redistribution of Arp2/3 complex to the plasma membrane and increased cell migration. Thus, gadkin appears to function as a negative regulator of Arp2/3 complex by sequestering the complex at intracellular sites without directly altering its activity [75]. Other inhibitors of Arp2/3 complex have been proposed, including PICK1 [76] and WD repeat-containing protein 63 (WDR63) [96], but remain mostly uncharacterized. Particularly, PICK1 does not bind or inhibit Arp2/3 complex in vitro [97].
Concluding remarks
Recent advances in the cryo-EM field have led to substantial progress in our understanding of the mechanisms of Arp2/3 complex nucleation and branch formation. We anticipate this trend will continue in the near future, since many Arp2/3 complex interactors exist, but their molecular mechanisms remain poorly understood. This concerns specifically the mechanisms of Arp2/3 complex inhibition by Arpin, branch stabilization by cortactin and branch destabilization by GMF and coronins, all areas of intense interest that should become major topics of structural studies in the immediate future. Cryo-ET in cells is a still developing method that offers great potential, with the promise to observe some of these regulatory pathways in action at high resolution directly inside cells. Functional studies are also leading to new understanding of the many ways in which the Arp2/3 complex and branched networks control cellular processes from the nucleus to the cell cortex as well as undesirable activities such as pathogen and cancer cell motility. We are also just beginning to understand how the cell uses positive feedback loops centered on Arp2/3 complex to control processes such as cell-cell communication, mechanosensation and cancer cell invasion. Owing to the combined power of novel gene editing, structural, and cell visualization methods the research in the cytoskeleton field and particularly Arp2/3 complex is undergoing a renaissance.
Outstanding questions box.
What are the NPFs responsible for Arp2/3 complex recruitment and activation for novel functions such as nuclear deformation, DNA repair and mitochondria reshuffling?
Are there other inhibitors of Arp2/3 complex in cells? Does a precise NPF-inhibitor correlation exist?
What is the cellular and mechanistic role of SPIN90, the mammalian ortholog of Dip1? Does it synergize with mammalian NPFs for branch network assembly like its yeast ortholog Dip1?
What is the precise role of NPFs vs. the mother filament in the conformational change that leads to Arp2/3 complex activation?
Are mammalian coronins Arp2/3 complex inhibitors, activators, branch stabilizers or destabilizers? Do they bind Arp2/3 complex and how?
How is the relatively weak inhibitory activity of Arpin in vitro reconciled with its strong inhibitory phenotype in cells? Are there factors in cells that enhance Arpin’s inhibitory capacity? Is Arpin part of an inhibitory complex? Is Arpin’s activity modulated through post-translational modification?
How is the Arp2/3 complex branch stabilized by cortactin?
Highlights.
Positive feedback loops allow Arp2/3 complex networks to generate persistent pushing forces at membranes in a myriad of processes ranging from cell-cell communication to cell migration and mechanosensation
The mother filament plays an essential role in Arp2/3 complex activation and novel sources or substitutes of mother filaments are emerging, including dynactin and WDS-family proteins
Mammalian Arp2/3 complex activation requires binding of two NPFs along Arp2-ArpC1 and Arp3 and NPF-mediated delivery of actin at the barbed end of both Arps
The transition toward a filament-like conformation is brought about by a ~19° rotation of two subcomplexes relative to one another, one including Arp2 (Arp2-ArpC1-ArpC4-ArpC5) and one including Arp3 (Arp3-ArpC2-ArpC3)
There is an acute need to realign the biochemical/structural and cell biological data in the Arp2/3 complex literature, with unresolved inconsistencies persisting in several areas, such as inhibition and branch disassembly
Acknowledgements
Supported by National Institutes of Health grants R01 GM073791 and RM1 GM136511 to R.D. and Agence Nationale de la Recherche grant ANR-20-CE13-0016-01 and Institut National du Cancer grant INCA_6521 to A.M.G.
Footnotes
Declaration of Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fassler F et al. (2020) Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nat Commun 11 (1), 6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson RC et al. (2001) Crystal structure of Arp2/3 complex. Science 294 (5547), 1679–84. [DOI] [PubMed] [Google Scholar]
- 3.Espinoza-Sanchez S et al. (2018) Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments. Proc Natl Acad Sci U S A 115 (37), E8642–E8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chereau D et al. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proceedings of the National Academy of Sciences of the United States of America 102 (46), 16644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padrick SB et al. (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proceedings of the National Academy of Sciences of the United States of America 108 (33), E472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boczkowska M et al. (2014) Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun 5, 3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith BA et al. (2013) Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proceedings of the National Academy of Sciences of the United States of America 110 (4), 1285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullins RD et al. (2018) From solution to surface to filament: actin flux into branched networks. Biophys Rev 10 (6), 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padrick SB et al. (2008) Hierarchical regulation of WASP/WAVE proteins. Mol Cell 32 (3), 426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmet A et al. (2020) Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci Adv 6 (23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molinie N and Gautreau A (2018) The Arp2/3 Regulatory System and Its Deregulation in Cancer. Physiol Rev 98 (1), 215–238. [DOI] [PubMed] [Google Scholar]
- 12.Rottner K et al. (2021) WAVE regulatory complex. Curr Biol 31 (10), R512–R517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fokin AI and Gautreau AM (2021) Assembly and Activity of the WASH Molecular Machine: Distinctive Features at the Crossroads of the Actin and Microtubule Cytoskeletons. Front Cell Dev Biol 9, 658865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kast DJ et al. (2015) WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr Biol 25 (13), 1791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutts AS and La Thangue NB (2015) Actin nucleation by WH2 domains at the autophagosome. Nat Commun 6, 7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H et al. (2013) Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol 23 (2), 107–17. [DOI] [PubMed] [Google Scholar]
- 17.Efimova N and Svitkina TM (2018) Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J Cell Biol 217 (5), 1827–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JXH et al. (2020) Actin protrusions push at apical junctions to maintain E-cadherin adhesion. Proc Natl Acad Sci U S A 117 (1), 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin E et al. (2021) Arp2/3-dependent mechanical control of morphogenetic robustness in an inherently challenging environment. Dev Cell 56 (5), 687–701 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueiredo AM et al. (2021) Endothelial cell invasion is controlled by dactylopodia. Proc Natl Acad Sci U S A 118 (18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulant S et al. (2011) Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nature cell biology 13 (9), 1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautreau A et al. (2014) Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb Perspect Biol 6 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins A et al. (2011) Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol 21 (14), 1167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suetsugu S and Gautreau A (2012) Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends in cell biology 22 (3), 141–50. [DOI] [PubMed] [Google Scholar]
- 25.Kaksonen M and Roux A (2018) Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19 (5), 313–326. [DOI] [PubMed] [Google Scholar]
- 26.Akamatsu M et al. (2020) Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiam HR et al. (2016) Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun 7, 10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman W et al. (2017) Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat Cell Biol 19 (10), 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrank BR et al. (2018) Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559 (7712), 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caridi CP et al. (2018) Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559 (7712), 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore AS et al. (2021) Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591 (7851), 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch MD et al. (1998) Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science 281 (5373), 105–8. [DOI] [PubMed] [Google Scholar]
- 33.Machesky LM et al. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A 96 (7), 3739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ti SC et al. (2011) Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proceedings of the National Academy of Sciences of the United States of America 108 (33), E463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luan Q et al. (2018) Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc Natl Acad Sci U S A 115 (7), E1409–E1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaaban M et al. (2020) Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat Struct Mol Biol 27 (11), 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Loeffelholz O et al. (2020) Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms. Biol Open 9 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen KK and Rubenstein PA (2005) Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J Biol Chem 280 (25), 24168–74. [DOI] [PubMed] [Google Scholar]
- 39.Weaver AM et al. (2001) Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol 11 (5), 370–4. [DOI] [PubMed] [Google Scholar]
- 40.Wagner AR et al. (2013) Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol 23 (20), 1990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith BA et al. (2013) Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife 2, e01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helgeson LA and Nolen BJ (2013) Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. Elife 2, e00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnoor M et al. (2018) Cortactin: Cell Functions of A Multifaceted Actin-Binding Protein. Trends Cell Biol 28 (2), 79–98. [DOI] [PubMed] [Google Scholar]
- 44.Balzer CJ et al. (2019) Single-Turnover Activation of Arp2/3 Complex by Dip1 May Balance Nucleation of Linear versus Branched Actin Filaments. Curr Biol 29 (19), 3331–3338 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balzer CJ et al. (2020) Synergy between Wsp1 and Dip1 may initiate assembly of endocytic actin networks. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao L et al. (2020) SPIN90 associates with mDia1 and the Arp2/3 complex to regulate cortical actin organization. Nat Cell Biol 22 (7), 803–814. [DOI] [PubMed] [Google Scholar]
- 47.Mullins RD et al. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A 95 (11), 6181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volkmann N et al. (2014) Three-dimensional reconstructions of actin filaments capped by Arp2/3 complex. Eur J Cell Biol 93 (5–6), 179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luan Q et al. (2018) Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. EMBO J 37 (22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fokin AI et al. (2021) The Arp1/11 minifilament of dynactin primes the endosomal Arp2/3 complex. Sci Adv 7 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urnavicius L et al. (2015) The structure of the dynactin complex and its interaction with dynein. Science 347 (6229), 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bieling P et al. (2016) Force Feedback Controls Motor Activity and Mechanical Properties of Self-Assembling Branched Actin Networks. Cell 164 (1–2), 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oakes PW et al. (2018) Lamellipodium is a myosin-independent mechanosensor. Proc Natl Acad Sci U S A 115 (11), 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papalazarou V and Machesky LM (2021) The cell pushes back: The Arp2/3 complex is a key orchestrator of cellular responses to environmental forces. Curr Opin Cell Biol 68, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller J et al. (2017) Load Adaptation of Lamellipodial Actin Networks. Cell 171 (1), 188–200 e16. [DOI] [PubMed] [Google Scholar]
- 56.Molinie N et al. (2019) Cortical branched actin determines cell cycle progression. Cell Res 29 (6), 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoijman E et al. (2021) Cooperative epithelial phagocytosis enables error correction in the early embryo. Nature 590 (7847), 618–623. [DOI] [PubMed] [Google Scholar]
- 58.Risca VI et al. (2012) Actin filament curvature biases branching direction. Proc Natl Acad Sci U S A 109 (8), 2913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pandit NG et al. (2020) Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches. Proc Natl Acad Sci U S A 117 (24), 13519–13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abella JV et al. (2016) Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol 18 (1), 76–86. [DOI] [PubMed] [Google Scholar]
- 61.Dang I et al. (2013) Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503 (7475), 281–4. [DOI] [PubMed] [Google Scholar]
- 62.Chanez-Paredes S et al. (2019) Cellular and pathophysiological consequences of Arp2/3 complex inhibition: role of inhibitory proteins and pharmacological compounds. Cell Mol Life Sci 76 (17), 3349–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huehn AR et al. (2020) Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments. Proc Natl Acad Sci U S A 117 (3), 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bibeau JP et al. (2021) Clusters of a Few Bound Cofilins Sever Actin Filaments. J Mol Biol 433 (7), 166833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotila T et al. (2019) Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat Commun 10 (1), 5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shekhar S et al. (2019) Synergy between Cyclase-associated protein and Cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat Commun 10 (1), 5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wioland H et al. (2021) Actin filament oxidation by MICAL1 suppresses protections from cofilin-induced disassembly. EMBO Rep 22 (2), e50965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grintsevich EE et al. (2017) Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun 8 (1), 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galloni C et al. (2021) MICAL2 enhances branched actin network disassembly by oxidizing Arp3B-containing Arp2/3 complexes. J Cell Biol 220 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandhi M et al. (2010) GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Current biology : CB 20 (9), 861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sokolova OS et al. (2017) Structural Basis of Arp2/3 Complex Inhibition by GMF, Coronin, and Arpin. J Mol Biol 429 (2), 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boczkowska M et al. (2013) Glia maturation factor (GMF) interacts with Arp2/3 complex in a nucleotide state-dependent manner. J Biol Chem 288 (36), 25683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luan Q and Nolen BJ (2013) Structural basis for regulation of Arp2/3 complex by GMF. Nature Structural & Molecular Biology 20 (9), 1062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humphries CL et al. (2002) Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol 159 (6), 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maritzen T et al. (2012) Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc Natl Acad Sci U S A 109 (26), 10382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rocca DL et al. (2008) Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nature cell biology 10 (3), 259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goode BL et al. (1999) Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol 144 (1), 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan KT et al. (2011) Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol 21 (8), 481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Appleton BA et al. (2006) The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure 14 (1), 87–96. [DOI] [PubMed] [Google Scholar]
- 80.Kammerer RA et al. (2005) A conserved trimerization motif controls the topology of short coiled coils. Proc Natl Acad Sci U S A 102 (39), 13891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gatfield J et al. (2005) Association of the leukocyte plasma membrane with the actin cytoskeleton through coiled coil-mediated trimeric coronin 1 molecules. Mol Biol Cell 16 (6), 2786–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu SL et al. (2011) Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem 286 (19), 17039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gandhi M et al. (2010) Functional surfaces on the actin-binding protein coronin revealed by systematic mutagenesis. J Biol Chem 285 (45), 34899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai L et al. (2007) F-actin binding is essential for coronin 1B function in vivo. J Cell Sci 120 (Pt 10), 1779–90. [DOI] [PubMed] [Google Scholar]
- 85.Cai L et al. (2007) Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128 (5), 915–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan M et al. (2007) Coronin function is required for chemotaxis and phagocytosis in human neutrophils. J Immunol 178 (9), 5769–78. [DOI] [PubMed] [Google Scholar]
- 87.Blanchoin L et al. (2001) Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol 11 (16), 1300–4. [DOI] [PubMed] [Google Scholar]
- 88.Cai L et al. (2008) Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134 (5), 828–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gandhi M et al. (2009) Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell 34 (3), 364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin MC et al. (2010) Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci 123 (Pt 8), 1329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brieher WM et al. (2006) Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol 175 (2), 315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jansen S et al. (2015) Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat Commun 6, 7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xie C et al. (2021) Actin filament debranching regulates cell polarity during cell migration and asymmetric cell division. Proc Natl Acad Sci U S A 118 (37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lomakina ME et al. (2016) Arpin downregulation in breast cancer is associated with poor prognosis. Br J Cancer 114 (5), 545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang SR et al. (2019) Arpin downregulation is associated with poor prognosis in pancreatic ductal adenocarcinoma. Eur J Surg Oncol 45 (5), 769–775. [DOI] [PubMed] [Google Scholar]
- 96.Zhao K et al. (2020) WDR63 inhibits Arp2/3-dependent actin polymerization and mediates the function of p53 in suppressing metastasis. EMBO Rep 21 (4), e49269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Madasu Y et al. (2015) PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner. Mol Biol Cell 26 (7), 1308–22. [DOI] [PMC free article] [PubMed] [Google Scholar]


