Abstract
Noncanonical amino acid mutagenesis has emerged as a powerful tool for the study of protein structure and function. While triplet nonsense codons, especially the amber codon, have been widely employed, quadruplet codons have attracted attention for the potential of creating additional blank codons for noncanonical amino acids mutagenesis. In this review, we discuss methodologies and applications of quadruplet codon decoding in genetic code expansion both in vitro and in vivo.
Keywords: Quadruplet codon, +1 frameshift, four-base codon, non-canonical amino acids, genetic code
1. Introduction
The canonical genetic code consists of 64 triplet codons that encode the common twenty proteinogenic amino acids, with the rare exception of selenocysteine and pyrrololysine, and three termination signals. An undesirable shift in the triplet reading frame results in nonsense translation products that are usually truncated or degraded prematurely in the cell. Therefore, reading frame shift only occurs at a low rate, less than 10−5 per codon.1, 2 Although triplet codons are the predominant form of genetic code in nature, “programmed frameshift” exists and is essential for the translation of mRNAs that contain an extra base (+1 frameshift), one less base (−1 frameshift), or other variations with respect to a hypothetical “canonical gene”.3–5 Such programmed frameshifts give rise to the idea of more complex encryption of information than the standard triplet form. Indeed, it is frequently used by viruses to regulate the production of key enzymes (e.g., gag-pol frameshift in HIV and other retroviruses) by switching between alternative reading frames. Such frameshift also increases the amount of genetic information that can be encoded by the small genome size of viruses. This review focuses on quadruplet codon decoding (an apparent +1 frameshift event) in cellular genetic code expansion.
While ribosome is a well-refined machine, certain mutations to itself and/or other translation factors (e.g., tRNA) could “break” the machine’s triplet reading frame. For instance, tRNA mutants that contain extended anticodon loops (8-base instead of normal 7-base loop) with an apparent quadruplet anticodon could induce +1 frameshift.6–8 These naturally occurring programmed +1 frameshift events are generally promoted by the pausing of elongating ribosome,5 which are facilitated by the local secondary structure of mRNA, the interaction between an internal Shine-Dalgarno (SD) sequence upstream of the shift site and the anti-Shine-Dalgarno (aSD) sequence at the 3’ end of 16S rRNA, and the presence of adjacent rare codons. Two working models were proposed for the programmed +1 frameshift: (1) The yardstick model (Figure 1A) states that the anticodon of tRNA interacts with all four bases of a quadruplet codon in the A site of the ribosome, which leads to subsequent quadruplet translocation.8–16 In this case, the anticodon of tRNA measures and determines the codon size during the mRNA translation; (2) The slippery model (Figure 1B) entails that tRNA makes a normal three-base codon-anticodon interaction in the A site of ribosome and translocation is triplet. An mRNA-anticodon realignment subsequently occurs in the P site with a slip of the mRNA by one base, which leads to the alternate reading frame.17–21
Figure 1. Proposed models for +1 frameshift (quadruplet codon decoding) with tRNAs containing extended anticodon loop.
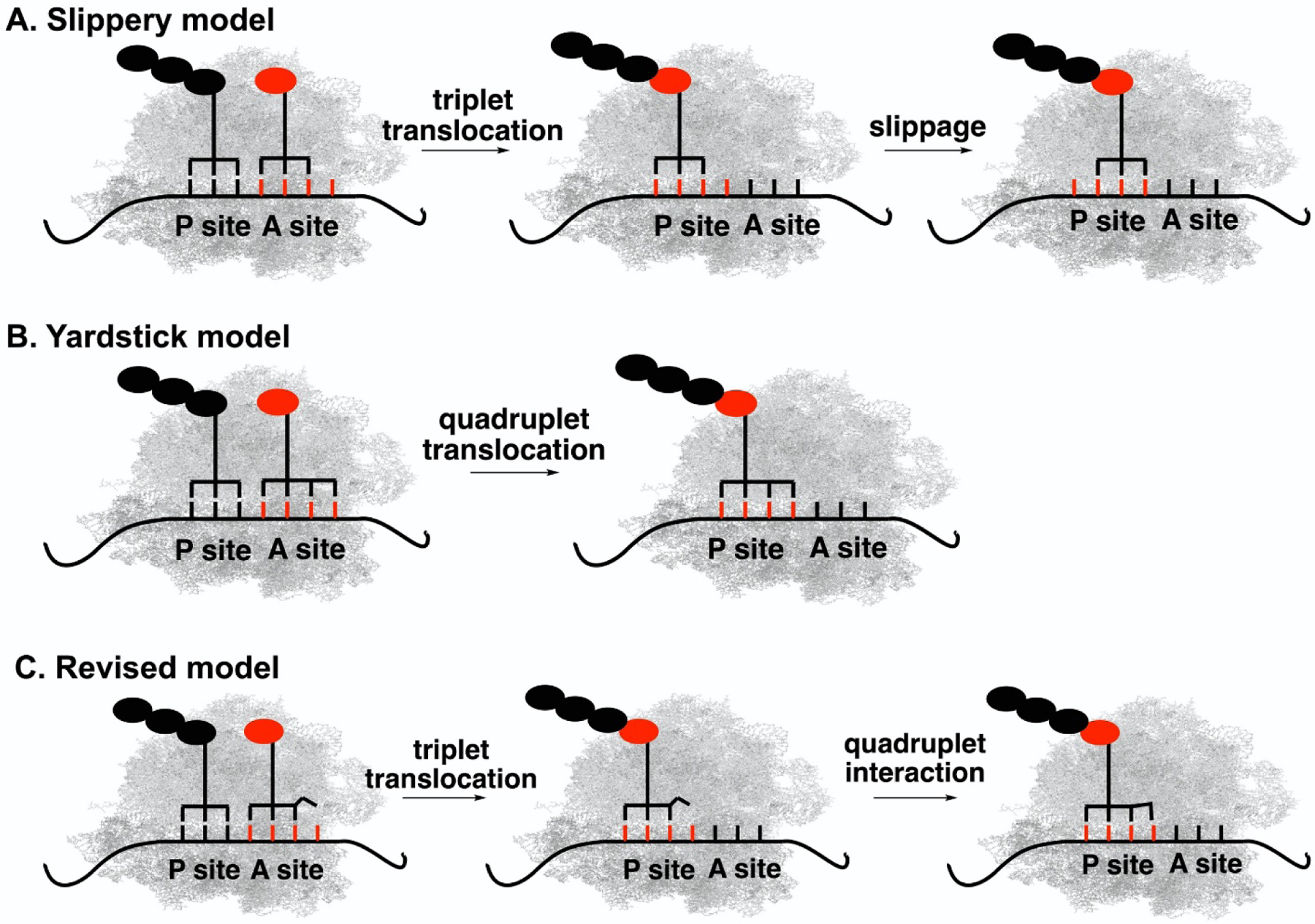
(A) The Slippery model features a normal triplet translocation followed by a slippage in the P site. (B) The Yardstick model features quadruplet anticodon-codon interaction in the A site followed by quadruplet translocation. (C) The revised model features a triplet translocation followed by a quadruplet anticodon-codon interaction in the P site.
One motivation of studying quadruplet codon decoding is to significantly expand the genetic code and use this decoding mechanism as an enabling tool for biochemical studies or biomedical applications. In recent years, the amber nonsense codon has been extensively used to encode noncanonical amino acids (ncAAs) with defined chemical properties into proteins in live cells.22–28 This general approach is proven to be a useful tool for the evolution, manipulation, and study of protein structure and function. A major focus of this research field is on how to further expand the cellular genetic code so that multiple ncAAs can be simultaneously encoded in a single cell. Multiple ncAA incorporation is of significant importance for a broader application of ncAA mutagenesis in biological experiments, such as fluorescence resonance energy transfer (FRET) and nuclear magnetic resonance (NMR) studies, or even the synthesis of completely unnatural biopolymers as new biomaterials. However, the number of available nonsense triplet codons severely limits the ultimate number of different ncAAs that can be simultaneously encoded by any organism. To unleash the full potential of ncAA mutagenesis in biological studies, more blank codons (codon that does not encode natural proteinogenic amino acids) are needed. One approach is to reprogram codon usage in the host cell. Since there are 64 triplet codons and only twenty proteinogenic amino acids, it is possible to reduce codon degeneracy and ‘free up’ some codons for ncAA mutagenesis. Indeed, the Church group was able to remove amber codon, two arginine codons, two leucine codons, and two serine codons from the E. coli genome.29, 30 However, due to resource requirements and limitations in current genome-editing technologies, this approach is currently limited to bacteria hosts and is clearly not readily applicable to more complex eukaryotic systems. As a second approach, the quadruplet codon decoding serves as an excellent choice. In theory, a quadruplet codon table would provide a total of 256 “blank” codons that can be used to further expand the genetic code. Even with potential cross talk among them, significant number of quadruplet codons can be used simultaneously to encode multiple different ncAAs. In this review, we will focus on ncAA mutagenesis in response to quadruplet codons, approaches to improve efficiency of quadruplet codon decoding, and applications of quadruplet codon decoding.
2. ncAA mutagenesis in response to quadruplet codons.
Quadruplet codons can be decoded by naturally occurring tRNA mutants with natural amino acids (also known as +1 frameshift).7, 8, 11, 31–36 These tRNA mutants usually contain an extra nucleotide in the anticodon loop (8-bases instead of 7-bases). Following this rationale, ncAA mutagenesis has been achieved using tRNA mutants containing expanded anticodon loop. The incorporation of ncAAs in response to quadruplet codons has been achieved in cell-free systems using in vitro protein translation methods, in live cells with either chemically or biologically aminoacylated tRNAs,37–40 and in animal.41
Cell-free systems
In the cell-free system, tRNA is either aminoacylated prior or after its addition to the translation mix. It offers a convenient and flexible means for ncAA mutagenesis. In comparison to cell-based system, the cell-free system has no concerns over potential cytotoxicity due to undesirable frameshifts induced by quadruplet decoding tRNAs.
In 1993, the Hardesty group synthesized E. coli alanyl-tRNA with different anticodons and showed that these aminoacylated tRNAs could be used to decode either triplet or quadruplet codons for in vitro protein engineering.42 In this study, the CAC anticodon of alanyl-tRNA was replaced with CUA, CCU, ACCU, or CCUA in order to decode UAG, AGG, AGGU, and UAGG codons, respectively. It was found that AGGU codon was decoded more efficiently than UAGG codon. In 1996, the Sisido group modified a yeast phenylalanyl-tRNA to contain apparent NCCU (N = A, G, C, U) anticodons.43 These tRNAs were synthesized by ligating aminoacyl dinucleotides to tRNAs that lack the 3’-CA. Decoding AGGN codons with nitrophenylalanine and protein synthesis were conducted in E. coli S-30 extract. All four AGGN codons could be effectively decoded, and expression levels of mutant proteins were comparable to that of the wild-type protein. Later, the Sisido lab also made efforts for the decoding of various quadruplet codons for the incorporation of one or more ncAAs into proteins using cell-free translation system.44–47 In their 1999 work, two different ncAAs were incorporated into streptavidin by combining the use of CGGG and AGGU codons. It was observed that the decoding efficiency of the AGGU codon was lower than that of the CGGG codon. The yield of the ncAA-containing streptavidin mutant was estimated to be 9% of that of the wild-type protein. In addition to the Sisido group, the Hecht and Hohsaka labs also demonstrated that quadruplet codon decoding using cell-free translation system could be a useful methodology for applications in FRET analysis48, 49 and saturation mutagenesis.50 In addition to E. coli extract, quadruplet codon decoding has been achieved in cell-free system using insect and mammalian cell extracts.51–53
Quadruplet codon decoding in live cells with chemically aminoacylated tRNAs
The Hecht and Schultz groups developed and optimized a method for the synthesis of aminoacylated tRNAs through the combination of chemically synthesis of dinucleotide and subsequent enzymatic ligation to truncated tRNAs lacking the 3′-terminal dinucleotide.54–56 These aminoacylated tRNAs can then be microinjected into cells, such as Xenopus oocytes for ncAA mutagenesis. The advantage of this approach is that it has less restriction on the structure of ncAAs. However, it is limited to cells that can be microinjected. In addition, the chemically aminoacylated tRNAs can potentially be hydrolyzed and cannot be re-acylated inside cells, and therefore, the yields of protein are generally low.
In 2004, the Voss group successfully incorporated ncAAs that contained spin or a fluorescence probes in response to either an amber nonsense or a GGGU quadruplet codon in Xenopus oocytes.57 It was observed that the efficiency of quadruplet codon decoding was significantly lowered in comparison to that of amber suppression. It is likely due to a competition from the endogenous glycyl-tRNAs bearing a CCC anticodon. It was proposed that a GCGU codon may be more suitable for ncAA mutagenesis since GCG has the lowest usage (~0.05) in genes found in oocytes. In 2006, the Dougherty group also showed that yeast phenylalanine frameshift suppressor tRNAs could be used to decode CGGG and GGGU codons for ncAA incorporation in Xenopus oocytes.37 Again, the quadruplet decoding tRNAs are less efficient than that of an amber suppressor tRNA. To demonstrate the utility of quadruplet codon decoding, the site-specific incorporation of two and three ncAAs simultaneously into a neuroreceptor was achieved by combining nonsense and frameshift suppression.
Quadruplet codon decoding in live cells with endogenously aminoacylated tRNAs
Besides chemical aminoacylation, tRNAs can be aminoacylated by aminoacyl-tRNA synthetases (aaRSs) in live cells. This is the basis of a general approach that has been developed for the genetic incorporation of ncAAs in live cells.22–28 In this system, orthogonal aaRS/tRNA pairs, which do not cross-react with any of the endogenous tRNAs and aaRSs of the host, are engineered to recognize a blank codon (codon that does not encode natural proteinogenic amino acids, e.g., amber UAG codon). The orthogonal aaRS is then modified to charge its cognate tRNA with only the desired ncAA.
The quadruplet codon decoding studies in live cells with endogenously aminoacylated tRNAs were first conducted with the incorporation of natural amino acids. In 2000, the Atkins group showed that a tRNA mutant with an expanded anticodon loop was able to decode UAGA codon.15 This tRNA was derived from an amber suppressor allele of the E. coli leuX (supP) gene and contained an extra U at position 33.5 of its anticodon loop. With a partial inactivation of release factor 1, the decoding efficiency could reach 13 to 26 %. With a further modification of mRNA, two tandem UAGA codons could be decoded with an overall efficiency of 10%.34 In 2001, the Schultz group identified E. coli seryl-tRNA mutants, with alterations in their anticodon loops, that could decode various quadruplet codons.35 Among them, AGGA, UAGA, CCCU, and CUAG codons could be very efficiently decoded without any cross-reactivity. Interestingly, the most efficient tRNA mutants contained anticodons that had complementary Watson-Crick pairing with quadruplet codons. In the same year, the O’Connor group also showed that E. coli glutaminylt-RNA and lysyl-tRNA with an expanded anticodon loop could decode CAAA codons in E. coli.58 The glutaminyl-tRNA mutant is identical to the sufG class of tRNAs that induce +1 frameshift in Salmonella enterica serovar Typhimurium and E. coli. Although only natural amino acids, leucine serine, glutamine, or lysine, were incorporated in these works, they represent an excellent and successful first step towards ncAA mutagenesis in response to quadruplet codons in live cells.
In 2004, the Schultz group demonstrated the genetic incorporation of a ncAA, L-homoglutamine, in response to an AGGA codon in E. coli.38 This was achieved by using an engineered lysyl-tRNA and synthetase pair from Pyrococcus horikoshii. Their data showed that suppression of AGGA codon did not significantly affect protein yields or cell growth rates. Indeed, AGG is a rare codon in E. coli. It is also likely that the lysyl-tRNA mutant cannot efficiently suppress endogenous AGGA codons in essential genes. Together with an evolved O-methyl-L-tyrosine-specific synthetase and tRNA pair from Methanococcus jannaschii, two different ncAAs were efficiently incorporated into distinct sites of a single protein. In 2010, the Chin group demonstrated that a M. jannaschii-derived tyrosyl-tRNA and tyrosyl-tRNA synthetase pair could decode AGGA codon in E. coli.39 Since the anticodon of M. jannaschii tyrosyl-tRNA is recognized by its cognate tyrosyl-tRNA synthetase, mutations were introduced in order to accommodate the artificially installed quadruplet UCCU anticodon. In addition, to improve quadruplet codon decoding efficiency and to minimize the impact of AGGA decoding to the host, the decoding center of an orthogonal ribosome59 was modified. In combination with an amber suppressor tRNA, two different ncAAs could now be simultaneously incorporated into a single protein in response to an amber and an AGGA codon. In the same year, the Liu group showed that a pyrrolysyl-tRNA mutant that contained a UCUA anticodon could be used to decode a UAGA codon in E. coli.60 In this work, however, the Liu group mostly focused on the genetic incorporation of two different ncAAs in response to two nonsense codons, UAA and UAG. In 2012, the Schultz group engineered a prolyl-tRNA/prolyl-tRNA synthase pair to decode AGGG and CUAG codons in E. coli.61 This was achieved by engineering the anticodon-binding pocket of Pyrococcus horikoshii prolyl-tRNA synthase to recognize Archaeoglobus fulgidus prolyl-tRNAs bearing an expanded anticodon loop.
In 2013, the Guo group identified pyrrolysyl-tRNA mutants that were able to decode AGGA codon for the incorporation of ncAAs into proteins in E. coli.62 Significant level of AGGA decoding was observed by simply replacing the CUA anticodon of pyrrolysyl-tRNA with an UCCU anticodon. This is because that the anticodon of pyrrolysyl-tRNA is not used as a recognition element by its cognate pyrrolysyl-tRNA synthetase.63–65 As a step forward, the Guo group showed that the AGGA-decoding efficiency could be enhanced by modifying the anticodon stem and loop of the tRNA. In 2014, the Schultz group also showed that a UAGA codon can be efficiently decoded by a M. jannaschii-derived tyrosyl-tRNA and tyrosyl-tRNA synthetase pair.40 In this work, the anticodon-binding pocket of the tyrosyl-tRNA synthetase was not altered. Significant level of UAGA decoding was only observed in a genomically recoded E. coli C321,29 which lacks any in frame UAG codons and release factor 1. However, in a later work, the Yoo group showed that M. jannaschii-derived tyrosyl-tRNA synthetase could charge its cognate tRNA bearing an UCCU anticodon without any additional engineering of the anticodon-binding pocket.66 It is possible that the UCCU anticodon is better recognized than the UAGS anticodon by the M. jannaschii-derived tyrosyl-tRNA synthetase. In 2018, the Chin group engineered mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs for the genetic incorporation of ncAAs in response to the amber nonsense codon and a quadruplet AGGA codon.67
In addition to combining one amber and one quadruplet codon, the use of multiple quadruplet codons simultaneously for ncAA mutagenesis have also been achieved. In 2014, the Chin group generated a set of tRNA mutants that could decode various quadruplet codons. While this work still focused on using both amber and quadruplet-decoding tRNAs to encode ncAAs E. coli, they indeed showed that two distinct quadruplet codons, AGGA and AGUA, could be used together to encode two different ncAAs.68 In 2019, the Guo group demonstrated that two mutually orthogonal quadruplet codons, AGGA and UAGA, could be used to simultaneously encode two different ncAAs within a single protein in bacterial cells by using a tyrosyl-tRNA and a pyrrolysyl-tRNA mutant.69 In this work, the anticodon-binding pocket of the tyrosyl-tRNA synthetase was altered in order to improve UAGA-decoding efficiency. In 2020, the Chin group took another step to use engineered orthogonal pyrrolysyl–tRNA synthetase/tRNA pairs for the genetic incorporation of three distinct ncAAs in response to UAG, AGUA, and AGGA codons.70 Later, in 2021, the same group showed for the first time that four distinct ncAAs could be simultaneously encoded by CUAG, UAGA, AGGA, and AGUA codons in E. coli.71
In addition to bacteria, the decoding of quadruplet codons has also been achieved in mammalian cells. In 2006, Taki, Sisido, and coworkers were able to decode UAGN codons with tyrosine in CHO cells by engineering human tyrosyl-tRNA.16 This was the first time that the genetic code of a mammalian cell was expanded by using quadruplet codons with endogenously aminoacylated tRNAs. However, only natural amino acid incorporation was reported in this work. In 2013, the Guo group showed that an engineered pyrrolysyl-tRNA that contains a UCCU anticodon could be used to efficiently decode a AGGA codon with a ncAA in 293T cells.62 This was achieved by an effort to improve AGGA-decoding efficiency with tRNA engineering. Later, in 2018, the same group demonstrated that a pyrrolysyl-tRNA mutant with a UCUA anticodon was able to decode a UAGA codon with a ncAA in mammalian cells.72 As a proof of concept study to demonstrate the utility of quadruplet codon decoding, they constructed an HIV-1 mutant that lacked any in-frame UAGA codons and showed that this HIV-1 mutant can be precisely activated by the decoding of a genomically embedded UAGA codon with a ncAA. In 2021, the Tsai group demonstrated that quadruplet codon-encoded ncAAs can be used to construct logic gates in mammalian cells.73 Albeit with recent developments, in comparison to the bacterial system, quadruplet codon decoding is largely unexplored in mammalian cells. This is mainly due to the lack of a facile engineering platform to evolve quadruplet codon decoding tRNAs for ncAA mutagenesis in mammalian cells.
Quadruplet codon decoding in an animal
While the ncAA mutagenesis in response to amber nonsense codon has been successfully applied to animals, it was only recently that quadruplet codon decoding was achieved in an animal. In 2021, the Greiss group used quadruplet codons to encode ncAAs into proteins in Caenorhabditis elegans.41 This was achieved by engineering hybrid pyrrolysyl-tRNA mutants that can efficiently decode quadruplet codons in C. elegans. These hybrid tRNAs contain anticodon loops optimized for quadruplet codon decoding62, 74 and tRNA scaffolds optimized for interaction with the eukaryotic translational machinery.75 It is intriguing that such modular assembly yielded functional and efficient tRNA mutants. Such hybrid tRNA approach can likely serve as a general method to generate tRNA mutants with desirable properties. As a demonstration of utility, photocaged ncAAs were incorporated into proteins in response to UAGA codons. In one application, photocaged lysine was site-specifically incorporated into Cre recombinase and was used for optical control of gene expression in C. elegans. In a second application, photocaged cysteine was successfully incorporated into Caspase-3 and was used for optical control of apoptosis induction in C. elegans.
3. Improving efficiency of quadruplet codon decoding
While the use of quadruplet codons for ncAA mutagenesis is intriguing and promising, such approach is hampered by low decoding efficiency. This is because the translation machinery maintains a reading frame that consists of triplet codons, which are the predominant form of genetic code in nature. Quadruplet codon-decoding tRNAs also face a significant competition with endogenous, triplet-recognizing tRNAs of the host. Efforts have been made to improve quadruplet codon decoding efficiency by focusing on multiple components of the translation machinery.
Tuning the interaction between tRNA and aminoacyl-tRNA synthetase
The aminoacylation of tRNAs by aminoacyl-tRNA synthetases is an early and essential step in protein synthesis. Many aminoacyl-tRNA synthetases recognize anticodons of their cognate tRNAs in order to maintain a high fidelity of protein translation. Since quadruplet codon decoding tRNAs usually contain an expanded anticodon loop, the recognition of tRNA mutants by aminoacyl-tRNA synthetases can be weakened, which would lead to a slower aminoacylation reaction and result in an overall lower quadruplet codon decoding efficiency. This issue was first recognized by the Chin group. In an effort to decode an AGGA codon by a M. jannaschii-derived tyrosyl-tRNA bearing an expanded anticodon loop, the cognate tyrosyl-tRNA synthetase was modified at the region that recognizes the UCCU anticodon.39 This led to a more efficient tRNA/synthetase pair and enabled the genetic incorporation of two ncAAs in response to UAG and AGGA codons, respectively. In another work, the Guo group sought to decode an UAGA codon by using a M. jannaschii-derived tyrosyl-tRNA.69 They fine-tuned the interaction between aminoacyl-tRNA synthetase and tRNA by modifying the anticodon-binding pocket of aminoacyl-tRNA synthetase. Many functional and enhanced aminoacyl-tRNA synthetase mutants were obtained. It indicates that a large number of solutions exist to better recognize an expanded anticodon loop of tRNA by its cognate aminoacyl-tRNA synthetase. The use of engineered aminoacyl-tRNA synthetases afforded up to 21-fold improvement in quadruplet codon decoding efficiency.
Tuning the interaction between tRNA and ribosome
A proper interaction between ribosome and tRNA is essential for a codon-dependent binding of correct aminoacylated tRNAs. Since ribosome was evolved to maintain a triplet reading frame, it is necessary to manipulate interaction between ribosome and tRNA to allow more efficient quadruplet codon decoding. One can engineer either ribosome or tRNA in order to establish a favorable interaction between them for quadruplet codon decoding.
The Chin group pioneered the engineering of ribosome to accommodate quadruplet codon decoding tRNAs that possess extended anticodon loops.39 Since the small subunit of ribosome mediates the correct interactions between the anticodons of tRNAs and codons on mRNA, the Chin group focused on the modification of decoding centre using saturation mutagenesis. By using a growth-based selection scheme, ribosome mutants that led to higher efficiency in quadruplet codon decoding were obtained. Interestingly, only two or three mutations were found in each hit although the ribosome mutant libraries covered 127 nucleotides in the decoding centre. Indeed, one of the best hits, ribo-Q1, contained only two mutations, A1196G and A1197G. It is possible that many mutations that benefits quadruplet codon decoding interfere with the overall maintenance of triplet reading frame of the reporter protein, and therefore are not emerged from a cell growth-based selection. Although it only contains two mutations, ribo-Q1 functioned very well and allowed both triplet and quadruplet codon decoding events to happen efficiently. It was also observed that the quadruplet codon decoding fidelity of ribo-Q1 could rival the triplet codon decoding fidelity of the wild-type ribosome. By using both an AGGA-decoding tRNA and an amber suppressor tRNA, the Chin group demonstrated that two different ncAAs could be simultaneously incorporated into a single protein. It should be noted that ribo-Q1 was derived from an orthogonal ribosome,59 which recognizes an unique ribosomal binding site. Since an orthogonal ribosome is not responsible for the synthesis of the proteome, it would mitigate a potential interference of the quadruplet codon decoding system to the host.
The other approach for tuning the interaction between ribosome and tRNA is to engineer tRNA. In comparison to ribosome engineering, tRNA engineering is technically less demanding. In 2001, the Schultz group engineered E. coli seryl-tRNA mutants with alterations in their anticodon loops.35 The wild-type anticodon loop of seryl-tRNA, which contains seven nucleotides, were replaced with eight or nine random nucleotides. The tRNA libraries were cross tested against a set of randomized quadruplet codons (NNNN; N = G, A, U, C). A number of tRNA mutants were identified for efficient decoding of various quadruplet codons, including AGGA, UAGA, CCCU, and CUAG. In 2013, the Guo group conducted a similar combinatorial approach in order to identify pyrrolysyl-tRNA mutants that could decode an AGGA codon. According to structural data,63–65 it is unlikely that the anticodon of pyrrolysyl-tRNA is used as a recognition element by its cognate pyrrolysyl-tRNA synthetase. This would allow a thorough tRNA engineering without interfering with the essential interaction between tRNA and aminoacyl-tRNA synthetase. In addition to the anticodon loop, the anticodon stem was also randomized and included in the library construction. The mutations in anticodon stem may facilitate a required communication from the detection of a desirable codon-anticodon interaction to the dissociation of EF-Tu. Indeed, beneficial mutations were found in both anticodon loop and stem. Interestingly, all clones contained a non-Watson-Crick base pairing and two strong G/C base pair in the anticodon stem. These mutants displayed several folds improvement in AGGA decoding as well as maintained a comparable fidelity to the wild-type parent. Furthermore, it was demonstrated that the evolved AGGA-decoding tRNA mutants were able to decode the same quadruplet codon in mammalian cells. In 2014, the Chin group further engineered pyrrolysyl-tRNAs to efficiently decode several different quadruplet codons, such as AGGA, UAGA, and AGUA.68 Their strategy also included mutagenesis of both anticodon loop and stem. In combination with ribo-Q1,39 a previously engineered orthogonal ribosome for quadruplet codon decoding, a significant increase in ncAA mutagenesis efficiency was obtained in response to quadruplet codons. In 2016, the Guo group conducted a systematic evolution and study of UAGN decoding tRNAs in E. coli. Through randomizing bases in anticodon stem and loop, tRNA mutants with significantly improved UAGN decoding efficiency were identified. The best UAGA-, UAGG-, and UAGU-decoding tRNA mutant displayed approximately 55-, 40-, and 9-fold improvement over their parents.
Host engineering
With recent breakthrough in E. coli genome engineering, one is now able to truly expand the cellular genetic code in E. coli C321.ΔA29 host using UAGN (N = A, G, U, C) codons with an increased decoding efficiency (no competition from release factor 1 for translation stop) and a reduced fitness penalty (no proteome-wide incorporation of ncAAs due to the lack of any in frame UAG codons). In 2014, the Schultz group showed that a M. jannaschii-derived tRNA could more efficiently decode a UAGA codon in this recoded C321.ΔA strain.40 In 2016, the Guo group also performed a comprehensive study of UAGN decoding in C321.74 Again, a significant improvement in UAGN decoding was observed when C321.ΔA was used. These findings indicate that elimination of competing triplet recognition in E. coli can lead to enhanced suppression efficiency of quadruplet codons.
4. Applications of the quadruplet codon decoding system
In theory, a quadruplet codon table would provide a total of 256 “blank” codons that can be used to greatly expand the genetic code. Therefore, the use of quadruplet codons would significantly augment current efforts in applying ncAA mutagenesis in biological studies and biomedical applications. In this section, we will discuss three major applications of quadruplet codon decoding, including the genetic incorporation of multiple ncAAs for biochemical studies, the construction of logic gates for synthetic biological applications, and the mechanistic studies of reading frame in translation.
Genetic incorporation of multiple ncAAs
Multiple ncAA incorporation is of significant importance for a broader application of ncAA mutagenesis in biological experiments, such as fluorescence resonance energy transfer (FRET) and nuclear magnetic resonance (NMR) studies, or even the synthesis of completely unnatural biopolymers as new biomaterials. Indeed, one major focus of using quadruplet codons in genetic code expansion is to encode two or more ncAAs for biological studies. The Chin group is a major player in this field. In two separate reports, the Chin group successfully labeled a single protein with two ncAAs in response to a UAG and an AGGA codon.39, 68 These two ncAAs contained orthogonal chemical handles that allowed further modifications of proteins in a site-specific manner. In one study, the two ncAAs contained an azide and an alkyne functional group, respectively.39 These two ncAAs enabled the cyclization of a protein through the formation of a triazole cross-link, realized by a Huisgen [2+3] cycloaddition reaction. In the other study, one ncAA contained a tetrazine moiety and the other had a strained alkene.68 Two fluorophores were then attached to the protein with a BODIPY-FL-tetrazine conjugate (reacts with the strained alkene) and a BODIPY-TMR-X-bicyclononyne conjugate (reacts with the tetrazine). In order to avoid cross reaction between BODIPY-FL-tetrazine and BODIPY-TMR-X-bicyclononyne, the protein was first labeled with one reagent, purified from excess reagent, and then labeled with the second reagent. Nevertheless, such site-specific introduction of a FRET pair into a single protein provides a general method for the study of protein dynamics. While the genetic incorporation of two or three ncAAs can be achieved by using only triplet codons,60, 76, 77 the availability of usable quadruplet codons provides alternative solutions.
Construction of logic gates
While ncAA mutagenesis has been widely applied to a range of different applications, the use of ncAA as a cue to dictate the behavior of an organism through amber suppression-based translational control was only reported recently. This was pioneered by the Guo group. In their work, an amber suppression-based and ncAA-mediated nonnatural genetic switch was devised to enable the development of novel live-attenuated HIV-1 vaccines.78, 79 This system entails the manipulation of essential HIV-1 protein biosynthesis through amber codon suppression that is precisely controlled by three mutually dependent exogenous regulatory components, including a unique amber suppressor tRNA (component 1) that can decode an amber codon and a special aminoacyl-tRNA synthetase (component 2) that charges the suppressor tRNA with an ncAA (component 3). All three components are required for the replication and spread of HIV-1 within the host. Since the viral replications can be tightly controlled at any time by supplying or withdrawing ncAA as the key control component, this approach greatly improves the safety of live-attenuated HIV-1 vaccines that have the best efficacy. Furthermore, the temporal control of HIV-1 replication can mimic prime and boost vaccination for the elicitation of protective immune responses as needed. If a booster vaccination is required to mount a recall immune response, virus replication can be transiently turned on and then off again at a later time. This approach can be used to improve the safety of other live-attenuated or replication-competent vector-based vaccines, and to generate vaccines against other viral and bacterial pathogens. Similar amber suppression-based and ncAA-mediated controls were later reported for potential applications in biocontainment80–83 and influenza vaccine production.84
Although amber suppression-based control worked well in many cases, some viruses have high mutation rate and may be able to escape from the control by mutating an installed amber codon back to a sense codon. One approach to mitigate this problem is to introduce multiple amber codons (e.g., three or more) into the essential genes of a virus and/or insert amber codons into a region with lower mutation rate. As an alternative approach, the Guo group reported in 2018 a novel strategy that used a quadruplet codon, instead of a triplet amber codon, as part of the control element.72 In this case, simple substitution of nucleotides in the quadruplet codon does not allow a virus to replicate since it causes +1 frameshift and does not lead to the expression of functional proteins. Although nucleotide insertion/deletion at or close to the quadruplet codon may lead to nonessential amino acid changes and allow the expression of functional proteins, such events are much less common than nucleotide substitution. In their work, the Guo group constructed a HIV-1 mutant that lacked any in-frame amber nonsense codons and demonstrated that it could be precisely activated by the decoding of a UAGA codon with a ncAA. Such conditionally activatable HIV-1 mutant can likely facilitate both fundamental investigations of HIV-1 as well as vaccine developments.
In 2021, the Tsai group demonstrated that quadruplet codon-encoded ncAAs could act as biologically inert switches for the construction of logic gates in mammalian cells.73 In this work, they screened eleven quadruplet codon-decoding pyrrolysyl-tRNA mutants from literature reports and found that all variants were able to decode CUAG or AGGA codons in mammalian cells. By combining an amber suppressor tyrosyl-tRNA and an AGGA-decoding pyrrolysyl-tRNA, an AND logic gate was constructed by using a split GFP system. This AND gate used two ncAAs, BocK (encoded by an AGGA codon) and AzF (encoded by a UAG codon) as its inputs and GFP fluroescence as its output. In the presence of both BocK and AzF, the UAG and AGGA codons embeded in genes encoding the two GFP fragments were decoded, which led to the assembly of a functional GFP. While this is only a simple demonstration, with the availability of genetic incoporation of up to four ncAAs,71 more complex logic gates could be generated in the future to performe desirable tasks.
Mechanistic studies
While the major goal is to further expand the genetic code, the development of artificial quadruplet codon decoding system also provides an excellent platform to better understand the underlying mechanism of naturally occurring +1 frameshift. Indeed, the Guo group has been working on this front. During a system evolution of UAGN codon decoding tRNAs, they found that all highly efficient tRNA hits contained a U at position 33.5 (The extra base in the anticodon loop is numbered as 33.5) no matter which UAGD (D = A, G, and U) codons they decode.74 In addition, some tRNA hits were able to decode all three UAGD codons with good efficiency. It suggests that a complete quadruplet codon-anticodon base pairing is apparently not required for UAGD decoding and is more consistent with the slippery model of +1 frameshift.17–19 On the other hand, a much higher decoding efficiency was observed for UAGA codon by the evolved tRNA variants that have potential to form a four-nucleotide Watson-Crick base-pairing with the UAGA codon, which cannot be explained by the slippery model. In addition, the evolved tRNA variants did not have an alternative cognate or near-cognate site for re-pairing in the P site, which is a hallmark of the slippery model. Even re-pairing would happen, the efficiency of UAGG decoding would be higher than that of UAGA decoding since the re-pairing of the triplet anticodon of tRNA is energetically more favored for UAGG. In contrast, higher decoding efficiency was observed for the UAGA codon in comparison to the UAGG codon.
The Guo group proposed a revised slippery model for UAGN decoding (Figure 1C).74 In this model, the tRNA rearrangement in the P site does not involve a slippage event. Instead, it is likely that the anticodon engages an interaction with all four bases of a quadruplet codon in the P site and facilitates a +1 frameshifting. More specifically, the extra U at position 33.5 of tRNA partially occupies the fourth base of the UAGN codon. This creates a steric hindrance with the incoming tRNA in the A site and induces +1 frameshift by preventing the incoming tRNA to interact with the fourth base of the UAGN sequence. Indeed, the highest decoding efficiency was observed for the UAGA codon that forms a full four-nucleotide Watson-Crick base-pairing with the UCUA anticodon. The wobble interaction between U33.5 of tRNA and G of UAGG codon is also favorable and led to the second highest decoding efficiency. On the other hand, the U33.5-U interaction in UAGU decoding and the U33.5-C interaction in UAGC decoding are less favorable and led to the lowest decoding efficiency. In fact, no hit was obtained that could efficiently decode a UAGC codon. While this new model may not be suitable to explain all +1 frameshifting cases, it provides a new aspect on the +1 frameshifting involving UAGN codons.
5. Conclusion and future perspective
Quadruplet codon decoding system has been extensively used in ncAA mutagenesis. It holds a high potential to further expand the genetic code and to implement novel design principles for synthetic biology applications. In principle, there are 256 quadruplet codons for ncAA mutagenesis. However, studies by the Guo group indicate that significant cross-decoding activity exist for similar quadruplet codons, especially those that share the same sequence in the first three bases. For instance, a tRNA with an apparent UCUA anticodon was able to decode not only UAGA codon, but also UAGG and UAGU codons. It is likely that the first three bases of a quadruplet codon are the major recognition element by the decoding tRNA and ribosome. Therefore, this would significantly reduce the table size of quadruplet codons that can be used for ncAA mutagenesis with good fidelity. Nevertheless, one can expect that the 64 non-cross-decoding triplet codons would lead to 64 non-cross-decoding quadruplet codons, which is still a large number and is sufficient for the current efforts in genetic code expansion. In addition, it would be interesting to develop strategies so that the fourth base pairing is essential for the efficient decoding of a quadruplet codon. This would make a 320-codon (64 triplet codons plus 256 quadruplet codons) genetic code possible. While the availability of more quadruplet codons is intriguing, they do not represent the current bottleneck for ncAA mutagenesis and genetic code expansion. There are now more usable quadruplet codons than the number of orthogonal tRNA/aminoacyl-tRNA synthetase pairs. Among all available quadruplet codons, UAGA and AGGA are the two most explored ones. Since AGG is a rare codon in E. coli, this improves AGGA decoding efficiency since it faces less competition from endogenous, AGG-recognizing tRNAs. Amber UAG codon are rare codons in both E. coli and mammalian cells. The deletion of release factor I can also improve the efficiency of UAGN codon decoding. As a general principle, any rare codon-containing quadruplet codons are good choice for ncAA mutagenesis. It should be noted that different hosts have different preference for codon usage. Therefore, it is necessary to examine the codon usage data of the target host before selecting suitable quadruplet codons. Since nonsense codons are usually least used ones in any organisms, UAGN, UAAN, and UGAN codons are generally good choices.
The efficiency of quadruplet codon decoding is essential for its application in ncAA mutagenesis. By engineering ribosome, tRNA, aminoacyl-tRNA synthetase, and host, one can significantly improve quadruplet codon decoding efficiency. In the future, further improving quadruplet codon decoding efficiency could potentially be achieved by modifying other components of the protein translation machinery that are critical in maintaining decoding accuracy. For example, the mRNA itself may also play an active role in maintaining reading frame beyond simply providing a template to base pair with tRNAs. Indeed, efficient +1 frameshifts in nature usually require certain recoding signals that are embedded at the close proximity of frameshift sites.5 Therefore, it would be possible to identify recoding signals and use them to improve quadruplet codon decoding efficiency.
As an important note, the current efforts in improving quadruplet codon decoding efficiency focus on bacterial host, such as E. coli. This is partly due to the availability of a facile platform for an easy selection of more efficient quadruplet codon decoding system. Indeed, it is still challenging to engineer mammalian ribosomes or host cells. The Guo group showed that some tRNA mutants that were evolved in E. coli could efficiently decode quadruplet codons in mammalian cells.62 The anticodon loop of a tRNA mutant evolved by the Guo group was also used successfully as part of a hybrid tRNA by the Greiss group in their C. elegans work.41 It indicates that tRNAs (or at least anticodon loops) evolved in E. coli may be generally transferrable to eukaryotic cells. However, it would still be ideal to perform such engineering efforts directly in mammalian cells, or at least in eukaryotic cells, such as yeast.
While amber codon is widely used to encode ncAAs, concerns remain that the suppression of endogenous amber stop codons could cause undesirable interference to cells’ physiology. One approach to overcome this problem is to conduct massive genome recoding to replace all amber codons.29 However, this approach is resource-demanding and is currently only doable in bacteria. Furthermore, the elimination of any abundant sense codons in either bacteria or mammalian cells would require tremendous efforts. As an alternate approach, quadruplet codon decoding can partially mitigate the suppression of endogenous stop codons. For instance, an UAGA codon-decoding tRNA displayed low activity on UAGU and UAGC codons, and therefore would not have a significant readthrough of endogenous amber codons that are followed by a nucleotide U or C. In comparison to a complete genome recoding, quadruplet codon decoding approach can be readily applicable to many cell types. However, this approach still cannot completely overcome the concerns of either reading through endogenous stops codons when any stop codon-containing quadruplet codons are used or causing frameshifts at other positions when any sense codon-containing quadruplet codons are used. The higher the quadruplet codon decoding efficiency would lead to more read throughs or frameshifts. In this regard, an orthogonal ribosome can be useful to reduce these undesirable events. However, orthogonal ribosome is still not available in mammalian system for live cell studies. Efforts in this direction would be useful. In addition, a recoding signal-dependent quadruplet codon decoding approach may be employed to further reduce the decoding of undesirable quadruplet codons in the host genome. In this case, significant level of quadruplet codon decoding can only happen in the presence of a recoding signal. Since such recoding signals are rarely found within the host genome, the chance of undesirable decoding events can be reduced.
Overall, ncAA mutagenesis in response to quadruplet codons is still at an early stage of development. A mechanistic understanding of quadruplet codon decoding (+1 frameshift) can provide useful guidance to further improve decoding efficiency and fidelity. In addition to introducing multiple ncAAs into a single protein and functioning in logic gates, novel applications using quadruplet codon decoding system are desirable to move this field forward.
Introduction to quadruplet codon decoding (+1 frameshift)
Noncanonical amino acid mutagenesis in response to quadruplet codons
Improving efficiency of quadruplet codon decoding
Applications of the quadruplet codon decoding system
Conclusion and future perspective
Acknowledgments
This work was supported by National Science Foundation (grant 1553041 to J.G.), National Institute of Health (grant 1R01GM138623 to J.G. and W.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare no competing interests.
References
- 1.Kurland CG (1992) Translational accuracy and the fitness of bacteria. Annu. Rev. Genet 26, 29–50. [DOI] [PubMed] [Google Scholar]
- 2.Kurland CG (1979) Reading frame errors on ribosomes, In Nonsense mutations and tRNA suppressors, pp 97–108, Academic, London. [Google Scholar]
- 3.Atkins JF, Weiss RB, Thompson S, and Gesteland RF (1991) Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu. Rev. Genet 25, 201–228. [DOI] [PubMed] [Google Scholar]
- 4.Farabaugh PJ (1996) Programmed translational frameshifting. Microbiol. Rev 60, 103–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins JF, and Bjoerk GR (2009) A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev 73, 178–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riddle DL, and Roth JR (1970) Suppressors of frameshift mutations in Salmonella typhimurium. J. Mol. Biol 54, 131–144. [DOI] [PubMed] [Google Scholar]
- 7.Yourno J, and Kohno T (1972) Externally suppressible proline quadruplet ccc U. Science 175, 650–652. [DOI] [PubMed] [Google Scholar]
- 8.Riddle DL, and Carbon J (1973) Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol 242, 230–234. [DOI] [PubMed] [Google Scholar]
- 9.Curran JF, and Yarus M (1987) Reading frame selection and transfer RNA anticodon loop stacking. Science 238, 1545–1550. [DOI] [PubMed] [Google Scholar]
- 10.Phelps SS, Gaudin C, Yoshizawa S, Benitez C, Fourmy D, and Joseph S (2006) Translocation of a tRNA with an extended anticodon through the ribosome. J. Mol. Biol 360, 610–622. [DOI] [PubMed] [Google Scholar]
- 11.Walker SE, and Fredrick K (2006) Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol 360, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taira H, Hohsaka T, and Sisido M (2006) In vitro selection of tRNAs for efficient four-base decoding to incorporate non-natural amino acids into proteins in an Escherichia coli cell-free translation system. Nucleic Acids Res. 34, 1653–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernhardt HS, and Tate WP (2008) Evidence from glycine transfer RNA of a frozen accident at the dawn of the genetic code. Biol. Direct 3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill CW, Combriato G, Steinhart W, Riddle DL, and Carbon J (1973) Nucleotide sequence of the GGG-specific glycine transfer ribonucleic acid of Escherichia coli and of Salmonella typhimurium. J. Biol. Chem 248, 4252–4262. [PubMed] [Google Scholar]
- 15.Moore B, Persson BC, Nelson CC, Gesteland RF, and Atkins JF (2000) Quadruplet codons: implications for code expansion and the specification of translation step size. J. Mol. Biol 298, 195–209. [DOI] [PubMed] [Google Scholar]
- 16.Taki M, Matsushita J, and Sisido M (2006) Expanding the genetic code in a mammalian cell line by the introduction of four-base codon/anticodon pairs. ChemBioChem 7, 425–428. [DOI] [PubMed] [Google Scholar]
- 17.Qian Q, Li J-N, Zhao H, Hagervall TG, Farabaugh PJ, and Bjork GR (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1, 471–482. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen S (1984) Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 3, 2895–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaber RF, and Culbertson MR (1984) Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol. Cell. Biol 4, 2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steitz TA (2008) A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol 9, 242–253. [DOI] [PubMed] [Google Scholar]
- 21.Herr AJ, Nelson CC, Wills NM, Gestland RF, and Atkins JF (2001) Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol 309, 1029–1048. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, and Schultz PG (2009) Synthesis at the interface of chemistry and biology. J. Am. Chem. Soc 131, 12497–12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, and Yokoyama S (2008) Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun 371, 818–822. [DOI] [PubMed] [Google Scholar]
- 24.Dumas A, Lercher L, Spicer CD, and Davis BG (2015) Designing logical codon reassignment - Expanding the chemistry in biology. Chemical Science 6, 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young DD, and Schultz PG (2018) Playing with the molecules of life. ACS Chem. Biol 13, 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukai T, Lajoie MJ, Englert M, and Soll D (2017) Rewriting the genetic code. Annu. Rev. Microbiol 71, 557–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin JW (2017) Expanding and reprogramming the genetic code. Nature 550, 53–60. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Xie J, and Schultz PG (2006) Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct 35, 225–249. [DOI] [PubMed] [Google Scholar]
- 29.Lajoie MJ, Rovner AJ, Goodman DB, Aerni H-R, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, and Isaacs FJ (2013) Genomically recoded organisms expand biological functions. Science 342, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, Shrock E, Pruitt BW, Conway N, Goodman DB, Gardner CL, Tyree G, Gonzales A, Wanner BL, Norville JE, Lajoie MJ, and Church GM (2016) Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822. [DOI] [PubMed] [Google Scholar]
- 31.Bossi L, and Smith DM (1984) Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc. Natl. Acad. Sci. U. S. A 81, 6105–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossi L, Kohno T, and Roth JR (1983) Genetic characterization of the sufj frameshift suppressor in Salmonella typhimurium. Genetics 103, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bossi L, and Roth JR (1981) Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell 25, 489–496. [DOI] [PubMed] [Google Scholar]
- 34.Moore B, Nelson CC, Persson BC, Gesteland RF, and Atkins JF (2000) Decoding of tandem quadruplets by adjacent tRNAs with eight-base anticodon loops. Nucleic Acids Res 28, 3615–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magliery TJ, Anderson JC, and Schultz PG (2001) Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol 307, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson JC, Magliery TJ, and Schultz PG (2002) Exploring the limits of codon and anticodon size. Chem Biol 9, 237–244. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez EA, Lester HA, and Dougherty DA (2006) In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc. Natl. Acad. Sci. U. S. A 103, 8650–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, and Schultz PG (2004) An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A 101, 7566–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann H, Wang K, Davis L, Garcia-Alai M, and Chin JW (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee A, Lajoie MJ, Xiao H, Church GM, and Schultz PG (2014) A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem 15, 1782–1786. [DOI] [PubMed] [Google Scholar]
- 41.Xi Z, Davis L, Baxter K, Tynan A, Goutou A, and Greiss S (2021) Using a Quadruplet Codon to Expand the Genetic Code of an Animal. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma C, Kudlicki W, Odom OW, Kramer G, and Hardesty B (1993) In vitro protein engineering using synthetic tRNA(Ala) with different anticodons. Biochemistry 32, 7939–7945. [DOI] [PubMed] [Google Scholar]
- 43.Hohsaka T, Ashizuka Y, Murakami H, and Sisido M (1996) Incorporation of Nonnatural Amino Acids into Streptavidin through In Vitro Frame-Shift Suppression. Journal of the American Chemical Society 118, 9778–9779. [Google Scholar]
- 44.Murakami H, Hohsaka T, Ashizuka Y, and Sisido M (1998) Site-Directed Incorporation of p-Nitrophenylalanine into Streptavidin and Site-to-Site Photoinduced Electron Transfer from a Pyrenyl Group to a Nitrophenyl Group on the Protein Framework. Journal of the American Chemical Society 120, 7520–7529. [Google Scholar]
- 45.Hohsaka T, Ashizuka Y, Sasaki H, Murakami H, and Sisido M (1999) Incorporation of Two Different Nonnatural Amino Acids Independently into a Single Protein through Extension of the Genetic Code. J. Am. Chem. Soc 121, 12194–12195. [Google Scholar]
- 46.Hohsaka T, Ashizuka Y, Taira H, Murakami H, and Sisido M (2001) Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry 40, 11060–11064. [DOI] [PubMed] [Google Scholar]
- 47.Ohtsuki T, Manabe T, and Sisido M (2005) Multiple incorporation of non-natural amino acids into a single protein using tRNAs with non-standard structures. FEBS Lett. 579, 6769–6774. [DOI] [PubMed] [Google Scholar]
- 48.Anderson RD III, Zhou J, and Hecht SM (2002) Fluorescence Resonance Energy Transfer between Unnatural Amino Acids in a Structurally Modified Dihydrofolate Reductase. J. Am. Chem. Soc 124, 9674–9675. [DOI] [PubMed] [Google Scholar]
- 49.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, and Hohsaka T (2006) FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat. Methods 3, 923–929. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe T, Muranaka N, and Hohsaka T (2008) Four-base codon-mediated saturation mutagenesis in a cell-free translation system. J. Biosci. Bioeng 105, 211–215. [DOI] [PubMed] [Google Scholar]
- 51.Taira H, Fukushima M, Hohsaka T, and Sisido M (2005) Four-base codon-mediated incorporation of nonnatural amino acids into proteins in a eukaryotic cell-free translation system. J. Biosci. Bioeng 99, 473–476. [DOI] [PubMed] [Google Scholar]
- 52.Taki M, Tokuda Y, Ohtsuki T, and Sisido M (2006) Design of carrier tRNAs and selection of four-base codons for efficient incorporation of various nonnatural amino acids into proteins in Spodoptera frugiperda 21 (Sf21) insect cell-free translation system. J. Biosci. Bioeng 102, 511–517. [DOI] [PubMed] [Google Scholar]
- 53.Tokuda Y, Taki M, and Sisido M (2006) Efficient incorporation of a nonnatural amino acid into a protein in an insect cell-free translation system. Nucleic Acids Symp. Ser, 277–278. [DOI] [PubMed] [Google Scholar]
- 54.Heckler TG, Chang LH, Zama Y, Naka T, Chorghade MS, and Hecht SM (1984) T4 RNA ligase mediated preparation of novel “chemically misacylated” tRNAPhes. Biochemistry 23, 1468–1473. [DOI] [PubMed] [Google Scholar]
- 55.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, and Schultz PG (1989) The use of 5’-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res. 17, 9649–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson SA, Ellman JA, and Schultz PG (1991) A general and efficient route for chemical aminoacylation of transfer RNAs. J. Am. Chem. Soc 113, 2722–2729. [Google Scholar]
- 57.Shafer AM, Kálai T, Bin Liu SQ, Hideg K, and Voss JC (2004) Site-Specific Insertion of Spin-Labeled l-Amino Acids in Xenopus Oocytes. Biochemistry 43, 8470–8482. [DOI] [PubMed] [Google Scholar]
- 58.O’Connor M (2002) Insertions in the anticodon loop of tRNAGln1 (sufG) and tRNALys promote quadruplet decoding of CAAA. Nucleic Acids Res. 30, 1985–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rackham O, and Chin JW (2005) A network of orthogonal ribosome·mRNA pairs. Nat. Chem. Biol 1, 159–166. [DOI] [PubMed] [Google Scholar]
- 60.Wan W, Huang Y, Wang Z, Russell WK, Pai P-J, Russell DH, and Liu WR (2010) A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew. Chem., Int. Ed 49, 3211–3214. [DOI] [PubMed] [Google Scholar]
- 61.Chatterjee A, Xiao H, and Schultz PG (2012) Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 109, 14841–14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu W, Schultz PG, and Guo J (2013) An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem Biol 8, 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, and Söll D (2007) Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. U. S. A 104, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, and Yokoyama S (2008) Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J. Mol. Biol 378, 634–652. [DOI] [PubMed] [Google Scholar]
- 65.Nozawa K, O’Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Söll D, and Nureki O (2009) Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature 457, 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee BS, Kim S, Ko BJ, and Yoo TH (2017) An efficient system for incorporation of unnatural amino acids in response to the four-base codon AGGA in Escherichia coli. Biochim. Biophys. Acta, Gen. Subj 1861, 3016–3023. [DOI] [PubMed] [Google Scholar]
- 67.Willis JCW, and Chin JW (2018) Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem 10, 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, Mehl RA, and Chin JW (2014) Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem 6, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hankore ED, Zhang L, Chen Y, Liu K, Niu W, and Guo J (2019) Genetic incorporation of noncanonical amino acids using two mutually orthogonal quadruplet codons. ACS Synth. Biol 8, 1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunkelmann DL, Willis JCW, Beattie AT, and Chin JW (2020) Engineered triply orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids. Nat. Chem 12, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunkelmann DL, Oehm SB, Beattie AT, and Chin JW (2021) A 68-codon genetic code to incorporate four distinct non-canonical amino acids enabled by automated orthogonal mRNA design. Nat. Chem, Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Wan Y, Wang N, Yuan Z, Niu W, Li Q, and Guo J (2018) Controlling the Replication of a Genomically Recoded HIV-1 with a Functional Quadruplet Codon in Mammalian Cells. ACS Synth. Biol 7, 1612–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mills EM, Barlow VL, Jones AT, and Tsai Y-H (2021) Development of mammalian cell logic gates controlled by unnatural amino acids. Cell Reports Methods, 100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang N, Shang X, Cerny R, Niu W, and Guo J (2016) Systematic Evolution and Study of UAGN Decoding tRNAs in a Genomically Recoded Bacteria. Sci Rep 6, 21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serfling R, Lorenz C, Etzel M, Schicht G, Boettke T, Moerl M, and Coin I (2018) Designer tRNAs for efficient incorporation of non-canonical amino acids by the pyrrolysine system in mammalian cells. Nucleic Acids Res. 46, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Italia JS, Addy PS, Erickson SB, Peeler JC, Weerapana E, and Chatterjee A (2019) Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J. Am. Chem. Soc 141, 6204–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tharp JM, Vargas-Rodriguez O, Schepartz A, and Soll D (2021) Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons. ACS Chem. Biol 16, 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang N, Li Y, Niu W, Sun M, Cerny R, Li Q, and Guo J (2014) Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew. Chem., Int. Ed 53, 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan Z, Wang N, Kang G, Niu W, Li Q, and Guo J (2017) Controlling multicycle replication of live-attenuated HIV-1 using an unnatural genetic switch. ACS Synth. Biol 6, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, and Church GM (2015) Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, and Isaacs FJ (2015) Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gan F, Liu R, Wang F, and Schultz PG (2018) Functional replacement of histidine in proteins to generate noncanonical amino acid dependent organisms. Journal of the American Chemical Society 140, 3829–3832. [DOI] [PubMed] [Google Scholar]
- 83.Koh M, Yao A, Gleason PR, Mills JH, and Schultz PG (2019) A general strategy for engineering noncanonical amino acid dependent bacterial growth. Journal of the American Chemical Society 141, 16213–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Si L, Xu H, Zhou X, Zhang Z, Tian Z, Wang Y, Wu Y, Zhang B, Niu Z, Zhang C, Fu G, Xiao S, Xia Q, Zhang L, and Zhou D (2016) Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 354, 1170–1173. [DOI] [PubMed] [Google Scholar]


