Abstract
A rare African ancestry–specific germline deletion variant in HOXB13 (X285K, rs77179853) was recently reported in Martinican men with early-onset prostate cancer. Given the role of HOXB13 germline variation in prostate cancer, we investigated the association between HOXB13 X285K and prostate cancer risk in a large sample of 22 361 African ancestry men, including 11 688 prostate cancer cases. The risk allele was present only in men of West African ancestry, with an allele frequency in men that ranged from 0.40% in Ghana and 0.31% in Nigeria to 0% in Uganda and South Africa, with a range of frequencies in men with admixed African ancestry from North America and Europe (0–0.26%). HOXB13 X285K was associated with 2.4-fold increased odds of prostate cancer (95% confidence interval [CI] =1.5–3.9, p = 2 × 10–4), with greater risk observed for more aggressive and advanced disease (Gleason ≥8: odds ratio [OR] = 4.7, 95% CI = 2.3–9.5, p = 2 × 10–5; stage T3/T4: OR = 4.5, 95% CI = 2.0–10.0, p = 2 × 10–4; metastatic disease: OR = 5.1, 95% CI = 1.9–13.7, p = 0.001). We estimated that the allele arose in West Africa 1500–4600 yr ago. Further analysis is needed to understand how the HOXB13 X285K variant impacts the HOXB13 protein and function in the prostate. Understanding who carries this mutation may inform prostate cancer screening in men of West African ancestry.
Keywords: African ancestry, Allelic age, Genetics, Health disparities, HOXB13, Prostate cancer, Rare genetic variants
Patient summary:
A rare African ancestry–specific germline deletion in HOXB13, found only in men of West African ancestry, was reported to be associated with an increased risk of prostate cancer. Understanding of who carries this mutation may help inform screening for prostate cancer in men of West African ancestry.
A rare African ancestry–specific germline deletion in HOXB13 (X285K) was associated with an increased risk of overall and advanced prostate cancer and was found only in men of West African ancestry. Men carrying this variant may benefit from earlier prostate cancer screening.
The nonsynonymous rare germline HOXB13 G84E variant (rs138213197) is a major risk factor for prostate cancer, accounting for ~5% of hereditary prostate cancer in men of European ancestry [1,2]. Rare prostate cancer HOXB13 risk variants have also been observed in other populations, including missense variants G132E in Japanese men (rs1286034091; allele frequency = 0.04%) [3] and G135E in Chinese men (rs769634543; allele frequency = 0.004%) [4]. Recently, a rare African ancestry–specific germline deletion variant in HOXB13 (rs77179853, allele frequency = 0.2%), which removes the stop codon (X285K) and elongates the HOXB13 protein, was observed in three Martinican men (French West Indies) with early-onset prostate cancer (allele frequency = 3.2%) [5]. Given the critical role of HOXB13 germline variation in prostate cancer, we investigated the association between the HOXB13 X285K variant and prostate cancer risk in a large sample of men of African ancestry.
This investigation included 11 688 prostate cancer cases and 10 673 controls from the African Ancestry Prostate Cancer (AAPC) Consortium, ELLIPSE/PRACTICAL OncoArray Consortium, California/Uganda Prostate Cancer Study, Ghana Prostate Study, and Men of African Descent and Carcinoma of the Prostate (MADCaP) Network (Supplementary Tables 1 and 2). The HOXB13 X285K variant was not included on genome-wide association studies (GWAS) arrays used in the African ancestry prostate cancer studies and was imputed separately using the Trans-Omics for Precision Medicine (TOPMed) r2 and 1000 Genomes Project (1KGP) phase 3 reference panels; the variant was observed in approximately 126 of 97 256 TOPMed participants and three of 2504 1KGP participants (Supplementary material). We imputed 101 carriers among 22 361 men in the African ancestry studies when using the TOPMed panel (imputation info score range across studies: 0.92–0.97) versus 60 when using 1KGP (imputation info score range across studies: 0.68–0.82; Supplementary Table 3). The carrier concordance between imputation panels was 0% (Supplementary Fig. 1). Confirmatory genotyping of 82 TOPMed imputed carriers, 42 1KGP imputed carriers, and 1431 imputed noncarriers confirmed 81 of 82 TOPMed but none of the 1KGP imputed genotypes (Supplementary Fig. 1 and Supplementary material). Other pathogenic and deleterious variants in HOXB13 were observed in our African ancestry populations (Supplementary Table 4), but were extremely rare and not able to be imputed with high confidence and tested in the current study.
HOXB13 X285K was present only in men of West African ancestry (Fig. 1 and Supplementary Fig. 2), with an allele frequency ranging from 0% in men from Uganda and South Africa to 0.31% in controls from Nigeria and 0.40% in controls from Ghana (Supplementary Table 5). Allele frequencies ranged from 0% to 0.26% in African ancestry controls from North America, the UK, and France (Supplementary Table 5), likely due to the high degree of European admixture in these populations. Of the 22 361 men, those with greater West African ancestry were found to have larger risk allele frequencies, ranging from 0.05% in cases with 0–20% West African ancestry to 0.90% in cases with 80–100% West African ancestry (Fisher’s exact test p = 2 × 10–4; Supplementary Fig. 3 and Supplementary material).
Fig. 1 –
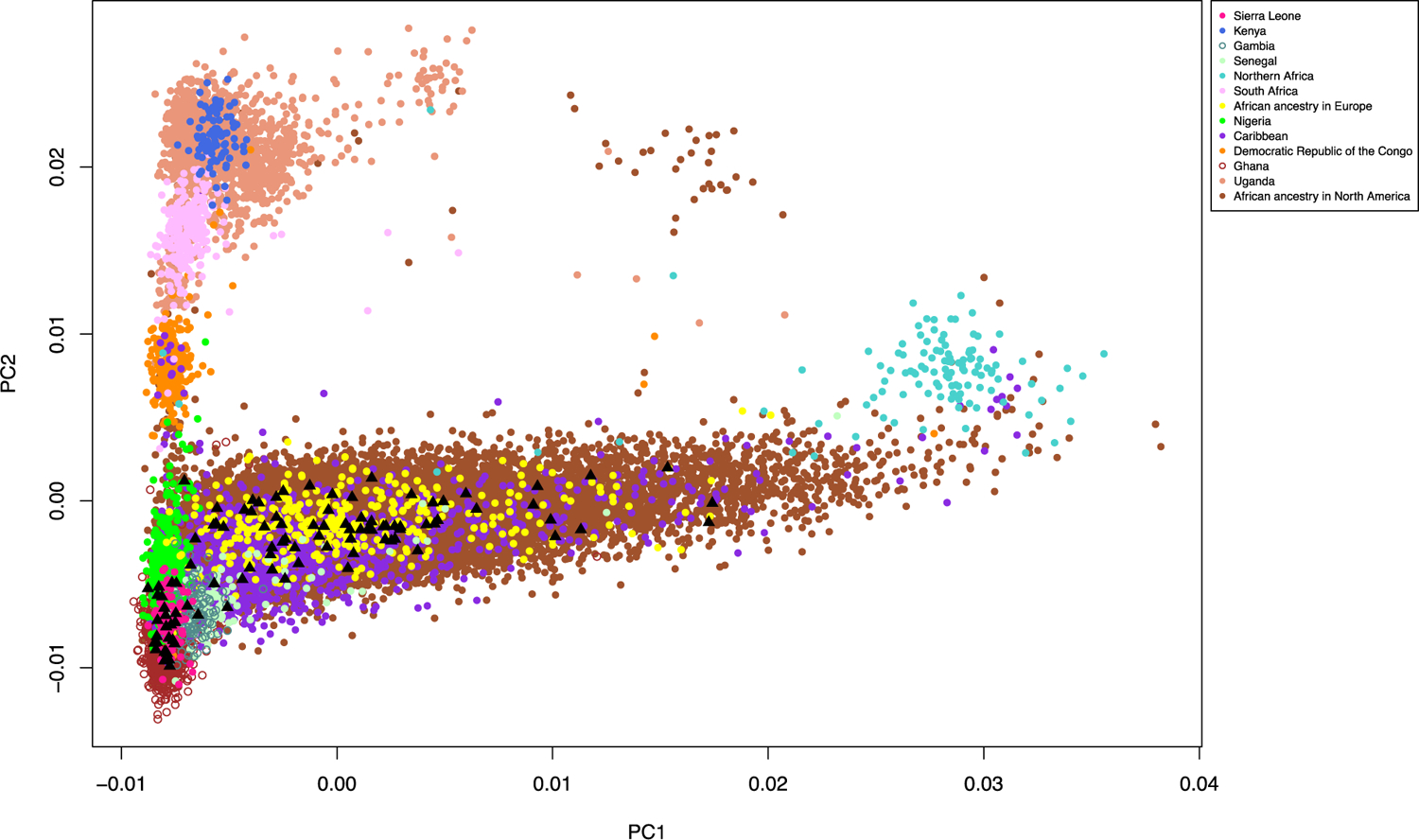
Distribution of HOXB13 rs77179853 by genetic ancestry comparing principal components 1 and 2 calculated in our sample of 22 361 men of African ancestry. Men carrying the rs77179853 delA risk allele are highlighted by black triangles.
In studies where the variant was observed (10 477 cases and 9688 controls; Supplementary Table 2), the HOXB13 X285K variant was significantly associated with 2.4-fold increased odds of prostate cancer (95% confidence interval [CI] = 1.5–3.9, p = 2 × 10–4; allele frequency in cases = 0.35% and controls = 0.14%; Table 1 and Supplementary material). The allele frequency was more common in cases with higher Gleason scores (0.29% in men with Gleason ≤6 tumors [odds ratio {OR} = 2.6, 95% CI = 1.3–5.2, p = 0.01], 0.36% in men with Gleason 7 tumors [OR = 2.3, 95% CI = 1.2–4.2, p = 0.01], and 0.84% in men with Gleason ≥8 tumors [OR = 4.7, 95% CI = 2.3–9.5, p = 2 × 10–5]), in cases diagnosed with higher-stage disease (0.34% in men with stage T1/T2 disease [OR = 2.3, 95% CI = 1.3–4.1, p = 0.01] and 0.73% in men with stage T3/T4 disease [OR = 4.5, 95% CI = 2.0–10.0, p = 2 × 10–4]), and in cases with metastatic (or prostate-specific antigen ≥100 ng/ml) disease (1.08% [OR = 5.1, 95% CI = 1.9–13.7, p = 0.001]; Table 1 and Supplementary Table 6).
Table 1 –
Association of HOXB13 germline variant rs77179853 with prostate cancer risk and disease aggressivenessa
| Group | n | Carriers, n | Risk allele frequency (%) | Carrier frequency (%) | OR (95% CI) | p value |
|---|---|---|---|---|---|---|
| Overall prostate cancer | ||||||
| Controls (reference) | 9688 | 28 | 0.14 | 0.29 | – | – |
| Cases | 10 477 | 73 | 0.35 | 0.70 | 2.42 (1.52–3.87) | 2 × 10−4 |
| Men of African ancestry from North America | ||||||
| Controls (reference) | 8766 | 21 | 0.12 | 0.24 | – | – |
| Cases | 9192 | 44 | 0.24 | 0.48 | 1.98 (1.16–3.38) | 0.01 |
| Men of African ancestry from West African countries (Ghana, Nigeria, and Senegal) | ||||||
| Controls (reference) | 922 | 7 | 0.38 | 0.76 | – | – |
| Cases | 920 | 22 | 1.20 | 2.39 | 3.99 (1.46–10.9) | 0.01 |
| Disease aggressiveness | ||||||
| Controls (reference) | 9180 | 28 | 0.15 | 0.31 | – | – |
| Gleason ≤6 tumors | 3319 | 19 | 0.29 | 0.57 | 2.58 (1.28–5.18) | 0.01 |
| Gleason 7 tumors | 3056 | 22 | 0.36 | 0.72 | 2.28 (1.22–4.24) | 0.01 |
| Gleason ≥8 tumorsb | 1126 | 19 | 0.84 | 1.69 | 4.65 (2.28–9.47) | 2 × 10–5 |
| Controls (reference) | 8918 | 28 | 0.16 | 0.31 | – | – |
| Stage T1/T2 | 4784 | 33 | 0.34 | 0.69 | 2.30 (1.28–4.14) | 0.01 |
| Stage T3/T4 | 959 | 14 | 0.73 | 1.46 | 4.48 (2.01–9.98) | 2 × 10–4 |
| Controls (reference) | 6526 | 20 | 0.15 | 0.31 | – | – |
| Metastatic or PSA ≥100 ng/ml | 511 | 11 | 1.08 | 2.15 | 5.08 (1.88–13.7) | 0.001 |
| Controls (reference) | 9688 | 28 | 0.14 | 0.29 | – | – |
| Cases with low-risk diseasec,d | 2795 | 12 | 0.21 | 0.43 | 1.84 (0.87–3.88) | 0.11 |
| Cases with intermediate-risk diseasec | 2721 | 12 | 0.22 | 0.44 | 1.51 (0.73–3.15) | 0.27 |
| Cases with high-risk disease | 3082 | 33 | 0.54 | 1.07 | 3.09 (1.75–5.45) | 1 × 10–4 |
CI = confidence interval; OR = odds ratio; PSA = prostate-specific antigen; RAF = risk allele frequency.
Analyses were limited to studies that carried the variant (see Supplementary Table 2).
Compared with 8329 controls (25 carriers, RAF = 0.15%) as the CA UG study did not have Gleason ≥8 tumor case carriers.
Compared with 9400 controls (27 carriers, RAF = 0.14%) as MADCaP did not have low- or intermediate-risk case carriers.
Low risk disease: Gleason <7, stage T1/T2, and PSA <10 ng/ml; intermediate-risk disease: Gleason = 7, stage T1/T2, and PSA = 10–20 ng/ml; high-risk disease: Gleason 8–10, stage T3/T4, PSA >20 ng/ml, metastatic disease, or died of prostate cancer.
The absolute risk of prostate cancer was 15.9% (95% CI = 15.9–16.0%) in noncarriers and 32.9% (95% CI = 22.0–44.6%) in carriers by age 85 yr (Supplementary Fig. 4). We did not observe associations between the variant and age at diagnosis (Supplementary Tables 7 and 8), family history of prostate cancer (Supplementary Table 9), or prostate-specific antigen levels (Supplementary Table 10).
We estimated that the HOXB13 X285K variant arose approximately 1500–4600 yr ago (refer to Supplementary Fig. 5 and Supplementary material for details on allelic age estimates based on two complementary approaches) and likely occurred after the Bantu migration from Western to Southern and Eastern Africa [6], which may explain why it is found only in men of West African ancestry. These findings, together with the established prostate cancer susceptibility G84E founder mutation that is more prevalent in Scandinavian populations [7] and the East Asian–specific G132E and G135E mutations [3,4], underscore the importance of ancestry-specific germline prostate cancer risk variants in the HOXB13 gene.
The HOXB13 X285K variant adds to growing evidence of regional differences in Africa for prostate cancer risk variants [8,9] and provides the first evidence of a genetic factor that is limited to specific African ancestry populations, although studies in other populations are needed to better understand the distribution of this variant in Africa. This investigation also demonstrates the importance and necessity of building diverse reference panels to facilitate the discovery of rare ancestry-specific risk variants and the need for larger sequencing studies in prostate cancer. At an exome-wide significance threshold of p < 5 × 10–7, 18 000 cases and 18 000 controls would be needed to detect an OR of 2.4 for an allele frequency of 0.14% (eg, HOXB13 X285K) with 90% power. The X285K stop codon is predicted to result in a 34% elongation of the HOXB13 protein, extending it by 96 amino acids [10]. Further studies are needed to understand how the HOXB13 X285K variant impacts the function of this homeobox transcription factor in the prostate. Understanding who carries this mutation may help inform screening for prostate cancer in men of West African ancestry.
Supplementary Material
Acknowledgments:
A full listing of acknowledgments is detailed in the Supplementary material.
Financial disclosures:
Burcu F. Darst certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor:
This work was supported by the National Cancer Institute at the National Institutes of Health, grants U19 CA148537, U19 CA214253, R01 CA165862, and K99CA246063 and the Prostate Cancer Foundation, grant 20CHAS03. Dr. Burcu F. Darst was supported in part by an award from the Achievement Rewards for College Scientists Foundation Los Angeles Founder Chapter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ewing CM, Ray AM, Lange EM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med 2012;366:141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xu J, Lange EM, Lu L, Zheng SL, et al. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium for Prostate Cancer Genetics (ICPCG). Hum Genet 2013;132:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Momozawa Y, Iwasaki Y, Hirata M, et al. Germline pathogenic variants in 7636 Japanese patients with prostate cancer and 12 366 controls. J Natl Cancer Inst 2020;112:369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin X, Qu L, Chen Z, et al. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate 2013;73:169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Marlin R, Creoff M, Merle S, et al. Mutation HOXB13 c.853delT in Martinican prostate cancer patients. Prostate 2020;80:463–70. [DOI] [PubMed] [Google Scholar]
- [6].Choudhury A, Aron S, Botigue LR, et al. High-depth African genomes inform human migration and health. Nature 2020;586:741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Karlsson R, Aly M, Clements M, et al. A population-based assessment of germline HOXB13 G84E mutation and prostate cancer risk. Eur Urol 2014;65:169–76. [DOI] [PubMed] [Google Scholar]
- [8].Lachance J, Berens AJ, Hansen MEB, Teng AK, Tishkoff SA, Rebbeck TR. Genetic hitchhiking and population bottlenecks contribute to prostate cancer disparities in men of African descent. Cancer Res 2018;78:2432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harlemon M, Ajayi O, Kachambwa P, et al. A custom genotyping array reveals population-level heterogeneity for the genetic risks of prostate cancer and other cancers in Africa. Cancer Res 2020;80:2956–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Akbari MR, Trachtenberg J, Lee J, et al. Association between germline HOXB13 G84E mutation and risk of prostate cancer. J Natl Cancer Inst 2012;104:1260–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


