Summary
Spindle- or lemon-shaped viruses infect archaea in diverse environments. Due to the highly pleomorphic nature of these virions, which can be found with cylindrical tails emanating from the spindle-shaped body, structural studies of these capsids have been challenging. We have determined the atomic structure of the capsid of Sulfolobus monocaudavirus 1, a virus that infects hosts living in nearly boiling acid. A highly hydrophobic protein, likely integrated into the host membrane before the virions assemble, forms seven strands that slide past each other in both the tails and the spindle body. We observe the discrete steps that occur as the tail tubes expand, and these are due to highly conserved quasi-equivalent interactions with neighboring subunits maintained despite significant diameter changes. Our results show how helical assemblies can vary their diameters, becoming nearly spherical to package a larger genome, and suggest how all spindle-shaped viruses have evolved from archaeal rod-like viruses.
In Brief
Structural analysis of an archaea-specific virus reveals principles for how filamentous assemblies of hydrophobic proteins can readily accommodate changes in morphology relevant for virus evolution and potentially for contexts as diverse as eukaryotic membrane dynamics and intermediate filaments.
Graphical Abstract

Introduction
Capsids are a hallmark of viruses, which distinguishes them from all other types of mobile genetic elements (Forterre et al., 2014; Raoult and Forterre, 2008). Over billions of years, viruses have ‘invented’ capsids on multiple independent occasions from non-homologous and structurally unrelated proteins (Krupovic et al., 2019). Nevertheless, the majority of known viruses package their genomes into icosahedral or filamentous helical protein capsids (also called nucleocapsids if surrounded by an additional layer, e.g., lipid membrane) (Krupovic and Koonin, 2017; Sevvana et al., 2021). Viruses infecting archaea notoriously deviate from this general paradigm by producing virions with unique, odd-shaped morphologies, which are not observed among bacterial or eukaryotic viruses (Dellas et al., 2014; Prangishvili et al., 2017). Such archaea-specific virion architectures resemble droplets, champagne bottles or spindles (Baquero et al., 2020). Viruses with spindle-shaped (or lemon-shaped) virions are particularly common in diverse extreme and moderate environments and infect a wide range of archaeal lineages from the phyla Crenarchaeota, Euryarchaeota and Thaumarchaeota as well as Asgardarchaeota, a group of archaea widely considered to represent the closest archaeal relatives of eukaryotes (Medvedeva et al., 2021). Because of the broad distribution in Archaea, it has been suggested that spindle-shaped viruses were associated with the last archaeal common ancestor, and possibly, even the last universal cellular ancestor (LUCA) (Krupovic et al., 2020).
Based on virion characteristics and genomic relationships, most spindle-shaped viruses fall into two groups (Krupovic et al., 2014). One of the groups comprises smaller spindle-shaped viruses classified into families Fuselloviridae (e.g., Sulfolobus spindle-shaped virus 1 [SSV1]), Halspiviridae (e.g., haloarchaeal virus His1) and Thaspiviridae (e.g., Nitrosopumilus spindle-shaped virus 1 [NSV1]) as well as several unclassified viruses. These viruses encode homologous major capsid proteins (MCP) containing two hydrophobic, potentially membrane-spanning domains and are extruded from the host cell through a budding-like mechanism without causing cell lysis (Kim et al., 2019; Quemin et al., 2016). Attempts to determine the structures of SSV1 (Stedman et al., 2015) and His1 (Hong et al., 2015) by cryo-EM did not yield atomic models and provided little insight into virion organization. The second group includes members of the family Bicaudaviridae, which have larger virions and genomes. A characteristic feature of this virus assemblage is the presence of ‘tails’ emanating from one or both pointed ends of the spindle. Notably, Acidianus two-tailed virus (ATV) and Sulfolobus monocaudavirus 1 (SMV1) have been reported to develop tails extracellularly, with the only requirement for this transformation being the incubation at temperatures close to those of the natural habitats (75–90°C) (Haring et al., 2005; Uldahl et al., 2016). It has been suggested that virus-encoded MoxR-type AAA+ ATPase and a von Willebrand domain A-containing cochaperone play an active role in this process by hydrolyzing ATP stored within the virions (Scheele et al., 2011). However, the exact mechanism underlying this virion morphogenesis outside of the host cells has not been elucidated. Other bicaudaviruses, namely, Acidianus tailed spindle virus (ATSV) and Sulfolobus tengchongensis spindle-shaped viruses 1 and 2 (STSV1 and STSV2, respectively) are released from the cells with tails extending from one of the virion poles and do not undergo further transformation (Erdmann et al., 2014; Hochstein et al., 2016; Xiang et al., 2005). Remarkably, upon infection, STSV2 and SMV1 block normal cell division, transforming the host cell into a giant virion-producing factory, which is up to 20 times larger compared to non-infected cells (Liu et al., 2021). The soluble structural protein repeatedly identified as the major component of bicaudavirus virions (Erdmann et al., 2014; Hochstein et al., 2016; Prangishvili et al., 2006; Xiang et al., 2005) is unrelated to that of the smaller spindle-shaped viruses from the first group, leading to the suggestion that the two groups of viruses are evolutionarily distinct (Krupovic et al., 2014). The protein has been crystallized for ATV and ATSV and shows a four-helix bundle fold (Goulet et al., 2010; Hochstein et al., 2018).
Filamentous and spherical capsids (displaying helical and icosahedral symmetries, respectively) embody the simplest virion designs dictated by the principle of ‘genetic economy’ inherent to viral genomes, whereby relatively large capsids are built from a small number of distinct protein subunits interacting with each other in a way that is identical throughout the capsid (Crick and Watson, 1956). For icosahedral viruses, Caspar and Klug proposed that interactions between the capsomers are quasi-equivalent (rather than identical) and developed the quasi-equivalence theory which allows predicting the arrangement of capsomers in larger icosahedral capsids (Caspar and Klug, 1962). How complex capsids, such as those of spindle-shaped viruses, are organized and how they have evolved remains unclear. While two theoretical models have been postulated for the geometric basis of the spindle shape in archaeal viruses (Perotti et al., 2016; Perotti et al., 2019), we show that they bear no relation to the actual structure of these viruses. Similarly, we can show that the crystal structures for the putative MCPs in both ATV (Goulet et al., 2010) and ATSV (Hochstein et al., 2018) were not of the MCPs, but unrelated viral proteins. Since spindle-shaped viruses appear to have evolved from archaeal rod-like viruses, our results explain why spindle-shaped viruses have thus far only been observed to infect archaea.
Results
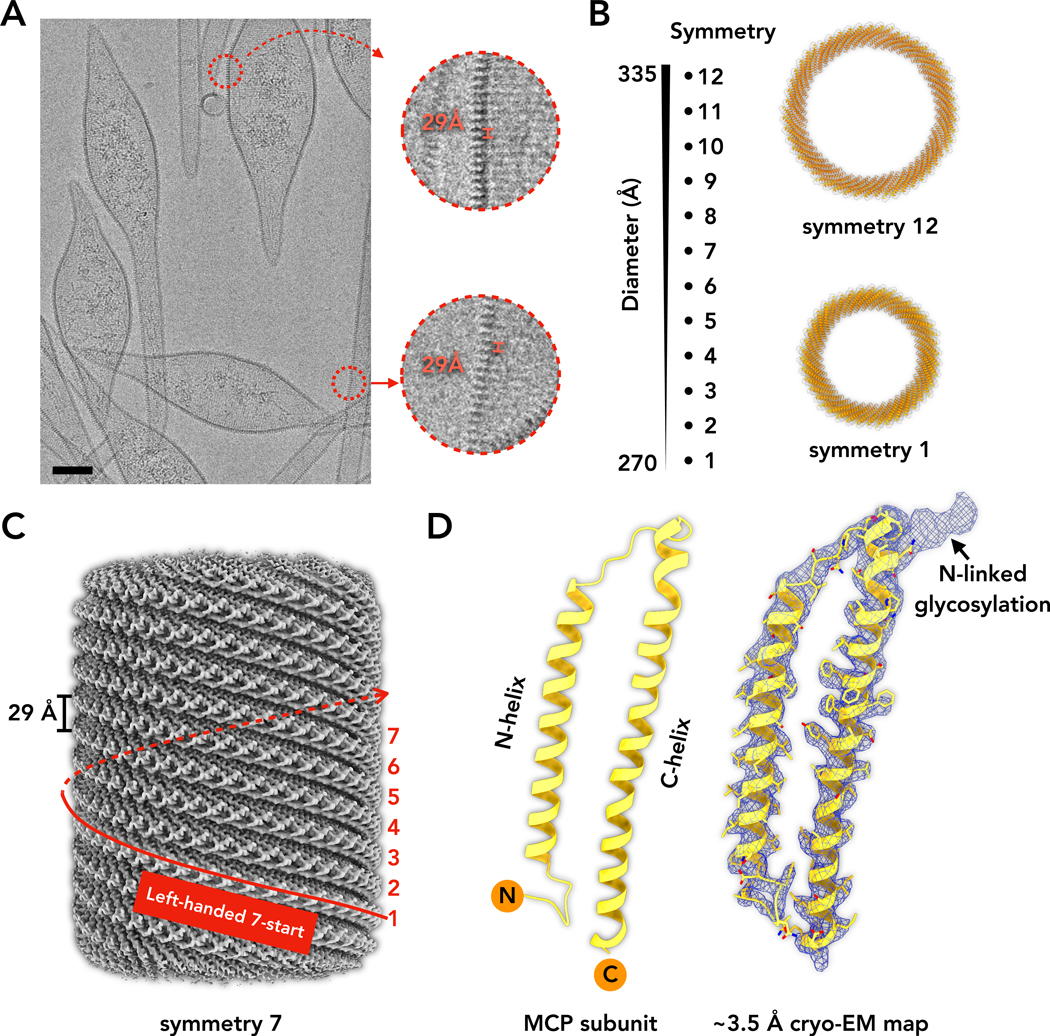
Cryo-EM images of SMV1 (Figure 1A) show that the spindle-shaped body of the virion is continuous with the tubular tails extending out from the body. As observed for ATSV (Hochstein et al., 2018), the periodicity in the spindle body is the same as that in the tails, strongly suggesting a seamless transition between the tails and the spindles. While this periodicity was described as ~40 Å for ATSV (Hochstein et al., 2018), we found a 29 Å periodicity in SMV1 (Figure 1A). The presence or absence of tails can be modulated by prolonged incubation at high temperatures (Haring et al., 2005; Uldahl et al., 2016). However, the heat-treated SMV1 population remained very heterogeneous with tails displaying variable lengths and diameters. We noticed that increase in pH from 6 to 10 results in nearly uniform transformation of spindle-shaped virions into tubular structures and is accompanied by the release of material present inside of the viral particle (Figure S1A). Fractionation of the SMV1 particles on 5–20% sucrose gradient followed by analysis of the different gradient fractions showed that the viral genome was released in the form of amorphous aggregates along with proteins present in the lumen of the viral particles, whereas tubular structures were largely devoid of DNA (Figure S1B). Thus, given that the tubular tails must have helical symmetry, and understanding the tails would reveal the structure of the more complicated spindle body, most of the structural studies in this paper were performed on the capsids which were transformed into a tubular state (see Methods). Nevertheless, extensive analysis of millions of segments revealed a nearly continuous variation in the diameter of the tail tubes. Progress was only made by starting from a dataset containing six million particles extracted from 16,401 micrographs, allowing us to use multiple 2D classification cycles to find nearly homogeneous subsets of constant diameter. This classification approach showed that rather than being continuously variable, there were discrete changes in diameter. We were then able to generate three-dimensional reconstructions at a near-atomic level of resolution for 12 discrete diameters, ranging from 270 to 335 Å (Figure 1B), each having a different helical symmetry, but each with seven strands (Table 1; Figure S2A). The α-helices in the map had a clear hand and showed unambiguously that the seven strands were left-handed, as had been determined at low-resolution by cryoelectron tomography and sub-tomogram averaging. The resolution of the best volume (Table 1, symmetry 7, ~3.5 Å, Figure 1C) would be more than adequate to build a full-atomic model ab initio when given the protein sequence.
Figure 1. Cryo-EM of bicaudavirus SMV1.
A, Representative cryo-EM of the Sulfolobus monocaudavirus 1 (SMV1). A 29 Å periodicity can be seen in both spindle-shaped bodies and tails. Scale bar, 50 nm.
B, Cryo-EM reconstructions of the SMV1 tails made from 12 different diameters. The top view models of the largest (symmetry 12) and smallest (symmetry 1) diameters are shown.
C, A surface of the 3.5 Å resolution reconstruction of symmetry 7. The left-handed 7-start protofilaments and 29 Å spacing between them are labeled.
D, Ribbon model of a single MCP protein (left). The map quality of the subunit is shown on the right. The arrow points to the N-linked glycosylation associated with Asn53.
Table 1.
Helical symmetry and model statistics for SMV1 and ATV
| SMV1 | ATV | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | --- |
| Helical symmetry | |||||||||||||
| Point group | C7 | C1 | C1 | C1 | C1 | C1 | C1 | C7 | C1 | C1 | C1 | C1 | C1 |
| Helical rise (Å) | 5.15 | 0.72 | 0.70 | 0.68 | 0.67 | 0.66 | 0.64 | 4.38 | 0.61 | 0.60 | 0.59 | 0.58 | 1.08 |
| Helical twist (°) | −9.0 | 50.2 | −155.5 | −104.1 | 101.7 | 153.1 | −52.6 | −7.7 | 50.4 | −155.3 | −103.9 | 101.8 | −156.0 |
|
| |||||||||||||
| Map resolution (Å) | |||||||||||||
| Model:map FSC (Å, 0.5) | 5.1 | 4.8 | 4.3 | 4.1 | 4.1 | 3.9 | 3.7 | 3.8 | 3.7 | 3.8 | 4.0 | 4.3 | --- |
| Map:map FSC (Å, 0.143) | 4.4 | 4.3 | 3.8 | 3.8 | 3.8 | 3.8 | 3.7 | 3.7 | 3.4 | 3.5 | 3.6 | 4.0 | 6.3 |
|
| |||||||||||||
| Refinement and Model validation | |||||||||||||
| Bond lengths RMSD (Å) | 0.004 | 0.004 | 0.006 | 0.005 | 0.006 | 0.004 | 0.003 | 0.004 | 0.006 | 0.007 | 0.005 | 0.004 | --- |
| Bond angles RMSD (°) | 0.658 | 0.640 | 0.770 | 0.841 | 0.671 | 0.699 | 0.635 | 0.703 | 0.681 | 0.735 | 0.651 | 0.658 | --- |
| Clashscore | 12.4 | 9.9 | 12.0 | 9.3 | 10.0 | 8.4 | 9.0 | 10.9 | 9.2 | 11.0 | 8.7 | 7.4 | --- |
| Ramachandran Favored (%) | 96.7 | 95.7 | 94.6 | 95.7 | 94.6 | 95.7 | 93.4 | 95.0 | 93.4 | 94.6 | 94.6 | 95.7 | --- |
| Ramachandran Outlier (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 | --- |
| RSCC | 0.86 | 0.87 | 0.87 | 0.87 | 0.88 | 0.87 | 0.87 | 0.88 | 0.87 | 0.87 | 0.86 | 0.87 | --- |
|
| |||||||||||||
| Deposition ID | |||||||||||||
| PDB (model) | 7RO2 | 7RO3 | 7RO4 | 7RO5 | 7RO6 | 7ROB | 7ROC | 7ROD | 7ROE | 7ROG | 7ROH | 7ROI | --- |
| EMDB (map) | 24585 | 24586 | 24587 | 24588 | 24589 | 24590 | 24591 | 24592 | 24593 | 24594 | 24595 | 24597 | 24596 |
To determine the identity of the MCP, SMV1 proteins were separated by SDS-PAGE and the major protein band of ~14 kDa was subjected to trypsin digestion followed by mass spectrometry analysis (Figure S3). The dominant protein in the band, gp11 (W0UU99), was the homolog of the MCPs previously identified in all other bicaudaviruses (Erdmann et al., 2014; Hochstein et al., 2016; Prangishvili et al., 2006; Xiang et al., 2005). However, it was impossible to thread the sequence of this putative MCP (gp11) through the density map, trying both possible N-to-C orientations. The absence of density in the map for a bulky side chain might be explained by disorder of that residue, but no threading could account for the pattern of bulky side chains seen in the map. Further, while the atomic structures of the ATV (PDB ID: 3FAJ) and ATSV (PDB ID: 5EQW) putative MCPs each displayed a compact four-helix bundle with a short fifth helix projecting out, the SMV1 capsid protein was clearly composed of two long helices connected by a short turn. We therefore searched for other possible candidates. A Cα trace of the density map suggested that the MCP is ~90 amino acids long. Assuming that there might be disordered residues at either the N- or C-terminus that were not present in the density map, we focused on SMV1 proteins containing between 90 and 150 residues. There were 17 such candidates (out of 96 proteins), but none could be successfully threaded through the map.
We next performed the N-terminal sequencing of the proteins in the major gel band. Two different five-residue N-terminal peptides were identified: one from a minor species, VEDYF, corresponding to the N-terminus of gp11 (the putative MCP that did not fit the map), and one from the major species, VTFGT, that did not correspond to the N-terminus from any of the 96 ORFs in the SMV1 genome. A search of the ORFs, found this pentapeptide uniquely in SMV1 gp03 (W0UUV5), a hypothetical 157-residue sequence annotated as a membrane protein. Removing 59 N-terminal residues would place the pentapeptide at the N-terminus of a processed protein, and the resulting 98-residue protein fit the density map perfectly (Figure 1D). The 98-residue protein contains no useful tryptic digestion sites (one at the C-terminus would create a three-residue peptide), explaining why peptides from this protein were never detected after tryptic digestion and mass spectrometry. The processing is likely performed by a cellular protease, because SMV1 and other bicaudaviruses do not encode identifiable proteases. Notably, however, gp03 does not contain a canonical signal sequence or recognizable signal peptidase cleavage site (Figure S4A). The fit revealed that additional density facing the outside of the capsid, near the turn between the two helices, was most likely due to heavy N-linked glycosylation of Asn53. Since the predicted MW for the 98-residue MCP is 10.1kDa, we suggest that extensive glycosylation of this single residue can explain why the protein runs in a gel with an apparent MW of ~14kDa. The glycosylation on the outside of the virion may play a role in protecting the capsid against very aggressive conditions, as was shown for extensively glycosylated pili of hyperthermophilic and acidophilic archaea (Wang et al., 2020; Wang et al., 2019). To further validate this, we used trifluoromethanesulfonic acid (TFMS), which removes both N- and O-linked glycans from proteins (Sojar and Bahl, 1987). Conditions were found under which a significant shift to lower mass could be obtained for a silver-stained band that ran at ~13 kDa before acid treatment and at ~11 kDa after acid treatment. These two bands were excised from the gel and analysed by mass spectrometry, which found that the gp03 can only be detected in the lower band. Interestingly, the protein gp11 was detected in both bands (Figure S3B-D). Thus, we designate SMV1 gp03 as the actual MCP. Given the abundance of gp11 in the virions of all known bicaudaviruses and lack of any discerneable contribution to capsid formation, we hypothesize that this protein functions as a nucleocapsid protein which compacts and, potentially, protects the viral DNA. Notably, in ATV, gp42, one of the two ATV-encoded homologs of SMV1 gp11, has been shown to act as a strong inhibitor of the host RNA polymerase (Sheppard et al., 2016), consistent with the possibility that this soluble protein is injected into the cell upon viral genome delivery.
Kinship among all spindle-shaped viruses
We have imaged ATV by cryo-EM (Figure 2A) and carried out a similar analysis. As with SMV1, ATV is formed from 7-start helical strands, and the subunit contains two α-helices linked by a short turn. The repeat of the 7-start helices in ATV is 32 Å compared with the 29 Å in SMV1. Although the resolution we have obtained for ATV (~6 Å) is much worse than for SMV1, there is no ambiguity in building a homology model for the ATV gp59 (YP_319890) based upon the SMV1 results, and this model fits well into the reconstructed density (Figure 2C). The crystal structure for the putative MCP in ATV, gp62 (PDB id: 3FAJ)(Goulet et al., 2010), cannot be fitted into the density, and clearly is the structure of a different protein. Thus, we designate gp59 as the actual MCP of ATV.
Figure 2. Cryo-EM of the Acidianus two-tailed virus (ATV).
A, Representative cryo-EM of ATV. A 32 Å periodicity can be seen in both spindle-shaped bodies and tails, indicated by black arrows. Scale bar, 50 nm.
B, Surface of the cryo-EM reconstruction of the ATV tails for one diameter, with the 7-start protofilaments labeled.
C, The homology model of ATV fit into the cryo-EM map in side and top views.
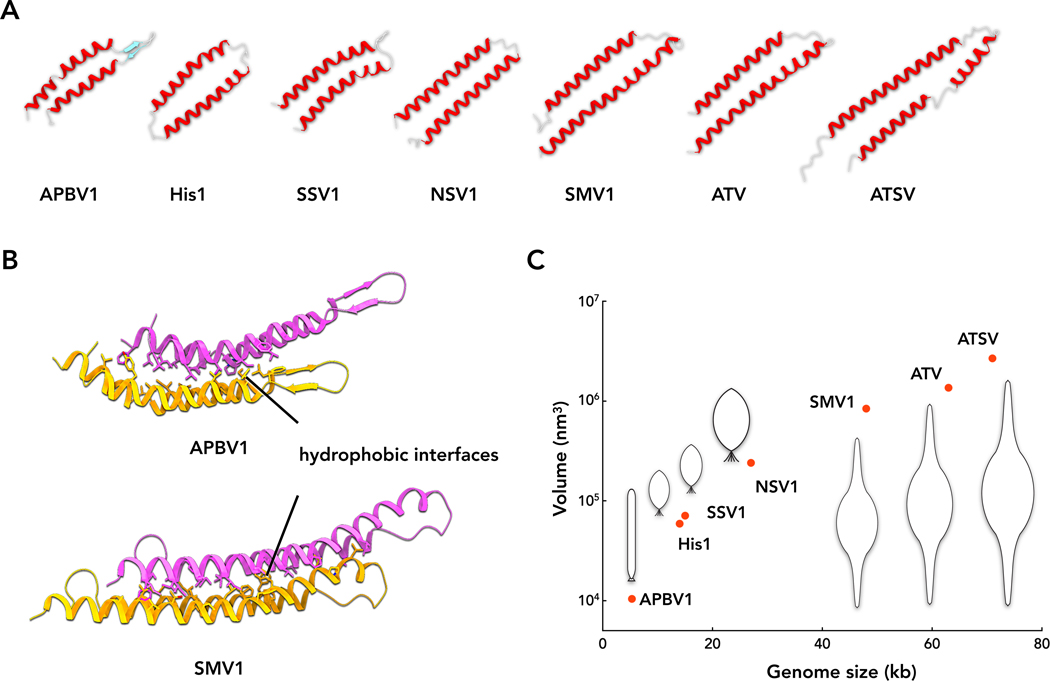
Homologs of SMV1 gp03 and ATV gp59 are conserved in all bicaudaviruses, including STSV1 and STSV2 (Figure S4B). Based upon the results for SMV1 and ATV, everything suggests that the actual MCP of ATSV is ATSV_F163, rather than ATSV_D135 for which an x-ray crystal structure was determined (Hochstein et al., 2018). The N-glycosylation sequon (NxS/T, where x is any amino acid) located at the beginning of the second α-helix is conserved in all homologs (Figure S4B), suggesting that glycosylation is important for virion assembly and/or stability. Features of the bicaudavirus MCPs are remarkably similar to those of the MCPs of group I spindle-shaped viruses, such as fusellovirus SSV1, halspivirus His1 and thaspivirus NSV1 (Figure S4C). Notably, the MCP of SSV1 is encoded as a pre-protein which is proteolytically processed at the N-terminus (Iverson et al., 2017; Quemin et al., 2015), as in the case of SMV1. Furthermore, MCPs of fuselloviruses also contain the conserved N-glycosylation sites at the position equivalent to that in bicaudaviruses and SSV1 MCP has been indeed shown to be glycosylated (Quemin et al., 2015). Finally, fuselloviruses, halspiviruses and thaspiviruses were all shown to exist in tubular form, either upon virion egress or following genome ejection (Hong et al., 2015; Kim et al., 2019; Quemin et al., 2016). In light of this evidence, combined with the results of bipartite network analysis showing that bicaudaviruses and group I spindle-shaped viruses share several signature genes (Iranzo et al., 2016), we suggest that all archaeal spindle-shaped viruses have evolved from a common ancestor and use similar principles of virion organization (Figure 3C). However, details in both the protein subunits and the virion morphology differ, as would be expected from evolutionary divergence. For instance, viruses STSV1 and STSV2 have only one, relatively short tail and do not undergo morphological changes outside of the cell (Erdmann et al., 2014; Xiang et al., 2005); ATSV has a very long and narrow tail of uniform length (Hochstein et al., 2018); whereas ATV and SMV1 analyzed in the present paper can “grow” tails from both pointed ends of the spindle (Haring et al., 2005; Uldahl et al., 2016). Strikingly, this emerging virus lineage might extend beyond spindle-shaped viruses to also include rod-shaped viruses of the family Clavaviridae (Mochizuki et al., 2010). In particular, a high resolution experimentally-determined structure exists for a rod-like clavavirus APBV1 virion (Ptchelkine et al., 2017), which is constructed from highly hydrophobic subunits with a helical-hairpin structure and an N-glycosylation site at the same position as in SMV1 and other bicaudaviruses (Figure 3B). Although MCPs of APBV1 and SMV1 do not display significant sequence similarity, which is not unexpected due to high sequence divergence and sparse sampling of archaeal viruses, the interactions between the MCP subunits in the two groups of viruses are very similar, full of hydrophobic interactions (Figure 3B).
Figure 3. Spindle-shaped viruses evolved to carry larger genomes.
A, Single subunit structures of seven MCPs. The structures of the APBV1 and SMV1 MCPs have been determined by high-resolution cryo-EM (PDB 5OXE and 7ROC). The other five MCP structures are homology models based on the SMV1 structure (see Methods).
B, Comparison of hydrophobic interactions of APBV1 and SMV1.
C, The estimated internal volumes of seven viruses plotted against genome size.
How the virions expand
The subunit model from “symmetry 7” of SMV1 (Figure 1D) fits the other 11 volumes with no modifications or perturbations. In addition, changes in the local packing of the subunit were almost infinitesimally small in the diameter range from 270 to 335 Å (Figure 4, Supplemental Movies 1 and 2), showing how the diameter could change significantly while at the same time preserving a highly conserved local environment for each subunit. The RMSD between two adjacent subunits in the same strand comparing the smallest and the largest diameter was only 0.3 Å (Figure 4A), while the RMSD between subunits in adjacent strands was only 0.4 Å (Figure 4B) when comparing the smallest and largest diameter.
Figure 4. The local contacts between capsid subunits are conserved as the diameter changes.
A, Interactions along the 7-start protofilament. One protofilament from the tube is highlighted on the left. Two subunits in one protofilament from symmetry 1 and 12 are aligned (center) and compared (right).
B, Interactions between adjacent 7-start protofilaments. Two protofilaments are highlighted on the left. The contacts between three subunits, two on the bottom strand (Sa and Sb) and one on the top (S’a) are compared in two views (center and right).
C, The rise per subunit along a strand for 12 different symmetries is plotted versus the diameter of the tubes (orange spheres), showing a nearly linear relationship. The blue dashed line shows the extrapolation to both smaller and larger diameters.
Helical symmetry can be defined as a rise per subunit, a rotation per subunit, and a possible Cn rotational symmetry. Two of the 12 structures had a C7 rotational symmetry (Table 1, symmetries 1 and 8), so the rise per subunit in each strand is the same as the helical rise. For the other symmetries in Figure 4C we have multiplied the helical rise by seven to yield the rise per subunit within a strand. The determined diameters for these 12 points fall into a small range, considering that the spindle bodies can have a maximum diameter of ~1,200 Å. Because the 29 Å periodicity remains constant in both spindle body and tail, and this comes from the repeat of the 7-start helices which have a fixed 203 Å pitch (7 × 29 Å) in both the tails and the spindle body, the number of subunits per strand in one turn will be u, which is 203 Å divided by the rise per subunit along a strand. We have found for the 12 symmetries reconstructed that u is incremented by seven for each larger diameter (which is an increment of one per 29 Å), explaining why the changes in diameter, D, are discrete, and not continuous. Let p be the path length along a 7-start strand per 203 Å turn, then:
We have shown that the local packing does not change as the diameter changes (Figure 4, Supplemental Movies 1,2), so the spacing between subunits along a strand, q, can be taken as a constant as the diameter changes, and p = uq. From the observed tubes, q=18.8 Å. Given this, the rise per subunit and the diameter can be easily calculated for each successive value of u. Therefore, we can extrapolate the results from the tubes to the larger diameters that would be found in the spindles (Figure 4C, Supplemental Movie 3).
How can the subunit structure explain the remarkable ability of these strands to slide past each other in discrete steps? An analysis of the protein sequence (Figure 5A) shows that most of the residues are quite hydrophobic. A plot of transmembrane probabilities for the sequence indicates a strong prediction for two transmembrane helices (Figure 5B). This can be seen in terms of the actual structure, where there is a hydrophobic region in the center of the subunit that is ~35 Å thick, with polar regions below and above it (Figure 5C). Therefore, these capsid proteins would not be soluble as monomers and must be integral to the host membrane before the virions are formed. When packed into virions (Figure 5D), the hydrophobic regions are completely buried and the polar regions are facing either the outside or the lumen. Notably, the interface between neighboring 7-start strands is almost completely hydrophobic, with minimal interactions between the hydrophilic regions. This will allow these strands to slide by each other while still excluding solvent (Supplemental Movies 4,5). Consistent with this, treatment of the virions with detergent, such as N-lauryl sarcosine, leads to rupture of the virions (Figure S5A), much as such treatment would destabilize a membrane.
Figure 5. The SMV1 MCP has clustered lipophilicity.
A, Amino acid composition of the SMV1 MCP.
B, TMHMM prediction of transmembrane probabilities for the SMV1 MCP sequence.
C, The spatial distribution of hydrophobic and hydrophilic residues in a single MCP.
D, Different views of the SMV1 tubes colored by lipophilicity.
The extensive hydrophobicity of the capsid and likely insertion of the protein subunits into the membrane prior to virion assembly motivated us to look for possible lipids associated with the virions (Figure S5B). The distribution of lipids in the SMV1 virions was not the same as that found in the host, suggesting that these lipids were selectively incorporated rather than being contaminants from cellular debris. In bacterial mating pili, the subunits also have a helical-hairpin structure (Costa et al., 2016; Zheng et al., 2020) and these subunits exist as integral membrane proteins before polymerization and after depolymerization. A single lipid molecule has been observed tightly bound to each subunit in the three published structures. However, we found no ordered lipid molecules in the SMV1 tube reconstructions. We can exclude the possibility that disordered lipid molecules (that we do not visualize) are present between the hydrophobic helices as the space between these helices is too narrow to accommodate a lipid. Since the tube segments used for three-dimensional reconstruction were carefully screened to only include the most ordered and regular ones, we are thus left with the possibility that the lipids may be found in the many dislocations or irregular regions in the virions, in particular, where additional subunits are added to helical turns as the diameter becomes larger. Alternatively, lipids could be associated with the specialized structures that are present at the termini of SMV1 virions.
Discussion
We have shown how a highly hydrophobic subunit can assemble into tubes with a variable diameter, and how these tubes can seamlessly expand and contract to form a spindle-shaped body for a virion that maintains the same quasi-equivalent contacts between subunits that are present in the tubes. The hydrophobic effect is, by definition, non-specific (Hillyer and Gibb, 2016; Newberry and Raines, 2019), and the hydrophobic surfaces of the helices (Figure 5) allow them to slide past each other (Supplemental Movies 4,5) while still maintaining the integrity of the capsid and excluding very acidic solvent in the environment from the interior of the virion. Our expectation is that the interior of the virions is close to the same near-neutral pH as the cytoplasm of the host (Baker-Austin and Dopson, 2007) and that is why the capsid must be impermeable to acidic solvent. While the hydrophobic effect alone is non-specific, specificity in the intersubunit contacts is provided by the helix-helix contacts that exist between neighboring subunits which dictate that discrete changes in diameter are required to maintain these quasi-equivalent contacts (Supplemental Movies 4,5). The notion of quasi-equivalence was first introduced by Caspar and Klug to explain how icosahedral viruses could be assembled from many copies of a single protein (Caspar and Klug, 1962). If one only had 60 copies of such a capsid protein a perfect icosahedron could be assembled so that every subunit was in an equivalent environment. It was also known that it was impossible to place more than 60 copies of a protein on the surface of a sphere such that all would be in strictly equivalent environments. But a capsid built from only 60 protein subunits would be quite small, and it had been observed that larger icosahedral viruses contained many more than 60 copies of the capsid protein. The solution was found in the possibility of quasi-equivalence (Caspar and Klug, 1962), wherein multiples of 60 protein subunits could be assembled into a spherical shell maintaining not identical, but quasi-equivalent contacts. This has now been amply confirmed with high resolution structures for many icosahedral viruses (Damodaran et al., 2002; Johnson and Olson, 2021; Johnson and Speir, 1997), although it has also been shown that certain icosahedral viruses can violate quasi-equivalence (Rayment et al., 1982; Stehle et al., 1994). However, the extension of quasi-equivalence theory to the radial expansion of helical viruses has not been envisioned.
Here we show that the quasi-equivalence principle explaining the ability of icosahedral viruses to expand the number of subunits needed to encapsulate larger genomes has a striking parallel in helical viruses. In APBV1 or other rod-like viruses, there will be a stoichiometric ratio between the number of capsid proteins and the number of bases or base pairs encapsidated. As pointed out by Caspar and Klug (Caspar and Klug, 1962), the length of tobacco mosaic virus virions is simply determined by the length of the single-stranded RNA genome given the stoichiometric ratio of three bases per protein subunit. In filamentous bacteriophage, the packaging of DNA involves charged residues in the lumen of the phage, and thus modifications of these residues can change the length of the virions while the length of the genome is unaltered (Hay and Lithgow, 2019). Thus, there is a linear relation between the length of the virion and the size of the genome, even though the actual stoichiometry can be modified. In contrast, for spherical viruses the number of capsid subunits will be proportional to the surface area, which is the radius squared, while the size of the genome encapsidated will scale as the radius cubed. Thus, the ratio of base pairs per capsid subunit will scale as the radius, becoming more efficient in genome packaging the larger the capsid.
With hydrophobic subunits having the ability to slide past each other, it is easy to imagine how a rod-like virus such as APBV1 could evolve into a spindle-shaped virus (Figure 3C) while still maintaining all of the local interactions that hold the capsid together. We suggest that the minimum energy conformation of the spindle-shaped capsids is actually a tube, as evidenced by the fact that when the genome is released, the SMV1 spindles relax into tubes (Figure S1). The same spindle-to-tube transformation has been observed upon genome ejection for a group I spindle-shaped virus His1 (Hong et al., 2015). It is therefore the pressure of the genome that expands the capsid that would normally be rod-like into a spindle shape. Thus, the unusual spindle morphology results from the radial expansion of a rod.
Archaeal and bacterial gas vesicles (Li and Cannon, 1998; Pfeifer et al., 2002; Strunk et al., 2011) have some remarkable similarity in morphology to spindle-shaped viruses. The gas vesicles are made of a single major structural protein of ~8 kDa and have a hydrophobic interior surface. However, whereas the capsid proteins contain two hydrophobic α-helices, those of gas vesicles from both bacteria and archaea have a α-β-β-α structure (Knitsch et al., 2017; Strunk et al., 2011), and the β-strands are predicted to mediate subunit-subunit interactions. Finally, unlike the capsid proteins which are predicted to contain two transmembrane domains, the proteins of gas vesicles are predicted to have none. Thus, we conclude that the superficial similarity between spindle-shaped viruses and gas vesicles is a result of convergent evolution. However, given the hydrophobic nature of both structures, we predict that changes in diameter described for both are mediated by these hydrophobic interactions.
There has been great interest in understanding the role of BAR domain proteins (Frost et al., 2008), ESCRT-III (Endosomal Sorting Complexes Required for Transport) (Pfitzner et al., 2021) and dynamin family proteins (Antonny et al., 2016) in membrane remodeling. All of these proteins form helical tubes of variable diameters that have been shown to constrict as part of their role in processes such as membrane fission and vesiculation. Structural studies of these assemblies have been extremely difficult due to the variability of tube diameters, and we still do not have any models at an atomic level of detail for the elementary steps in constriction. Similarly, keratin filaments, assembled from α-helical coiled-coils, have also been revealed to form variable diameter tubes in situ (Weber et al., 2021). While we do not suggest any homology between these proteins and the SMV1/ATV capsid proteins, we suggest that the general principle underlying the sliding of the strands of SMV1 proteins past each other while maintaining the structural integrity of the capsid might extend to other filamentous assemblies with variable diameters. The great advances in cryo-EM that have allowed us to determine in atomic detail the structure of SMV1 capsid will likely allow a similar level of understanding for other assemblies in the near future.
Limitations of the Study
We have determined a high resolution structure for only a single spindle-shaped virus, SMV1, and determined a lower resolution structure for a second spindle-shaped virus, ATV. These results have been combined with an existing high resolution structure for the clavavirus APBV1 (Ptchelkine et al., 2017) and homology models of the major capsid proteins from other spindle-shaped viruses to show the likely common ancestry and suggest how the spindle-shaped viruses have evolved from rod-like viruses such as APBV1. Future high resolution studies of other rod-like and spindle-shaped viruses will greatly extend our understanding of this evolution. Just as knowing the morphology and genetics of apes and humans suggests common ancestry, determining the intermediates that existed in this evolutionary process gives one a much more profound understanding of biological history.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Edward H. Egelman, egelman@virginia.edu
Materials Avalability
This study did not generate any new unique reagents.
Data and Code Availability
The SMV1 maps were deposited in the Electron Microscopy Data Bank (EMDB) with entry codes EMD-24585, EMD-24586, EMD-24587, EMD-24588, EMD-24589, EMD-24590, EMD-24591, EMD-24592, EMD-24593, EMD-24594, EMD-24595, and EMD-24597 and the respective atomic models were deposited in the Protein Data Bank (PDB) with entry codes 7RO2, 7RO3, 7RO4, 7RO5, 7RO6, 7ROB, 7ROC, 7ROD, 7ROE, 7ROG, 7ROH and 7ROI. The ATV map was deposited in the EMDB with entry code EMD-24596.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Archaeal strains and growth conditions
Sulfolobus monocaudavirus 1, SMV1, was propagated in Sulfolobus islandicus CRISPR deletion mutant delta C1C2 (Gudbergsdottir et al., 2011). The host cell culture was grown in Sulfolobus medium supplemented with 0.2% tryptone, 0.1% yeast extract, 0.2% sucrose and 0.002% uracil (Zillig et al., 1993). The culture was started from −80°C stock. The cells were grown in 50 ml of medium at 76°C with shaking. After 24 h of incubation, the cell culture was diluted 20 times in pre-warmed (76°C) medium and the growth was continued until it reached an OD600 of ~0.2. Then the cells were infected with SMV1 stock and incubation continued at 76°C with agitation for 24h. 24 h post infection the cells were removed by centrifugation in Sorval 3000 rotor, 7000 rpm, 20 min, 15°C, virus-containing supernatant was collected and virions precipitated with PEG600 (10.5% w/v) and NaCl (5.8% (w/v) for 2h at room temperature. The PEG-precipitate was then pelleted in a Sorval 3000 rotor, 9000 rpm, 30 min, 15°C and the resulting pellet was resuspended in one tenth of the original volume in 20 mM Tris-acetate (pH6) buffer.
METHODS DETAILS
Virus production and treatments
To purify the virus, solid CsCl was added to the virus suspension to a final concentration of 45% (w/v). The samples were run in a Beckman SW41 rotor, 38,000 rpm, at least 16 h, 15°C. After the run, the opalescent band corresponding to SMV1 virions was collected. The purified SMV1 was stored in CsCl at 4°C until used.
For SMV1 virion dissociation, CsCl purified virions were diluted 50 times in 20 mM Tris-acetate (pH6) buffer and pelleted down by centrifugation in a Beckman 45Ti rotor, 35,000 rpm, 2 h, 15°C. The resultant pellet was re-suspended in 20mM Tris-acetate (pH6) buffer and used for dissociation. The SMV1 virions were dissociated by addition of 0.1% (v/v) NaOH and incubation at 37°C for 1 h. Then non-treated (control) and NaOH-treated virions were loaded on top of the linear 5–20 % sucrose gradients and run in a Beckman SW41 rotor, 24,000 rpm, 20 min, 15°C. After the run, 12 fractions from the top of each tube were collected and analyzed in 4–12% Bis-Tris acrylamide gels. After the run the gels were stained for DNA with ethidium bromide and for proteins with InstantBlue. The contents of the collected fractions were also visualized by negative staining TEM. For that, 10 μl of the fraction of interest was adsorbed onto copper grids with carbon-coated Formvar films and negatively stained with 2.0% (w/v) uranyl acetate for 20 s. The samples were observed under a FEI Tecnai BioTwin 120 microscope operated at 120 kV.
Cryo-electron microscopy and image processing
The SMV1 sample was applied to glow-discharged lacey carbon grids and vitrified using a Vitrobot Mark IV (Thermo Fisher). Grids were imaged at on a Titan Krios (300 keV, Thermo Fisher) with a K3 camera (Gatan). 16,401 micrographs were collected under electron counting mode at 1.1 Å per pixel, using a defocus range of 1–2 μm with ~50 electrons/Å2 distributed into 40 fractions. Motion correction and CTF estimation were done in cryoSPARC (Punjani et al., 2017; Rohou and Grigorieff, 2015; Zheng et al., 2017). A total of seven million particles were auto-picked by “Filament Tracer” with a shift of 13 pixel, and non-virion bad particles were removed by 2D classification. About six million SMV1 particles remained having a range of diameters. From this six million particle dataset, 12 relatively homogeneous subsets were sorted out by iterative 2D classifications. The possible helical symmetries were calculated from an averaged power spectrum for each subset, generated from the raw particles. For each subset, the actual helical symmetry was determined in cryoSPARC by trial and error, until the hand of α-helices and amino acid side chains were seen (Egelman, 2000; Punjani et al., 2020). The resolution of each reconstruction was estimated by both Map:Map FSC and Model:Map FSC. The final volumes were then sharpened with a negative B-factor automatically estimated in cryoSPARC, and the statistics are listed in Table 1.
For ATV the same approach was used, with 2.8M particle images collected using a shift of 9 pixels between adjacent boxes.
Model building of SMV1 MCP
The density corresponding to a single SMV1 MCP was segmented from the experimental cryo-EM density using Chimera (Pettersen et al., 2004). Possible MCP sequences were threaded through the density using DeepTracer (Pfab et al., 2021). Only one sequence, W0UUV5, also detected by N-terminal sequencing, could be threaded through the map, matching the bulky side chain densities and the glycosylation site. This model was adjusted manually in Coot (Emsley and Cowtan, 2004) and real-space refined in PHENIX (Afonine et al., 2018). Using the determined helical symmetry, a filament model was generated in Chimera and refined against the full cryo-EM map using PHENIX real-space refinement. MolProbity was used to evaluate the quality of the filament model (Williams et al., 2018). The refinement statistics are shown in Table 1.
Cryo-electron tomography
For cryo-electron tomography, CsCl-purified virions were pelleted by ultracentifugation (Beckman Type 50.2 Ti rotor, 32000 rpm, 2h, 15 °C), resuspended in water and incubated at 90 °C for one week. Heat-treated SMV1 particles were mixed with CsCl-purified rudivirus SIRV2 as a control, since the helical hand had been determined previously (DiMaio et al., 2015). Data were collected on a Titan Krios microscope (TFS) operating at 300kV in EFTEM mode using a bioquatum/K3 (Gatan) energy filter and camera with a 20 eV slit. Tilt series were recorded at a magnification of 26kx corresponding to 3.4 Å/pixel. A tilt range of +/−60° was set with a 3° tilt increment and a total dose of 120 e-/Å2 with a dose symmetric tilting scheme using the TFS tomography software with defocus values between −4 and −6 micron defocus.
A total of 935 segments were manually picked along the long tails of SMV1 from 8× binned tomograms by “tomopick” in the tomography package i3 (Winkler, 2007). Sub-tomograms of SMV1 were first extracted from the 8× binned tomograms, and then aligned in the i3 software package. After initial alignment, sub-tomograms were extracted from unbinned tomograms for further refinement. Multivariate statistical analysis (MSA) implemented in the I3 package was then used for 3D classification and sub-tomogram averaging. As a control, a total of 200 segments were manually picked from the tubes of SMV2 from the same 8× binned tomograms. The similar procedure was used to generate a 3D averaged structure of the SIRV2 tube without applying any helical symmetry. The averaged structure of SMV1 is a left-handed helix with ~29 Å periodicity, while the averaged structure of SIRV2 is a right-handed helix with ~43 Å periodicity.
Homology modeling of ATV, SSV1, His1 and ATSV MCP
The MCP homologs in bicaudaviruses were identified by PSI-BLAST (Altschul et al., 1997) searches (E-value cutoff of 0.05) queried with the sequence of SMV1 MCP (YP_009008070) against the NCBI protein database restricted to members of the family Bicaudaviridae (taxid:423358). The sequences were aligned using PROMALS3D (Pei and Grishin, 2014). MCPs of SSV1 and His1 were determined experimentally (Pietila et al., 2013; Quemin et al., 2015). The MCP structures of ATV, SSV1, His1 and ATSV were first predicted by AlphaFold2 (Jumper et al., 2021). Then the predicted structures were flexibly aligned to SMV1 by FATCAT (Li et al., 2020). For ATV, the predicted MCP structure was then docked into the 6 Å resolution cryo-EM map and real-space refined in PHENIX (Afonine et al., 2018) with helical symmetry imposed.
N-terminal sequencing of SMV1 by Edman Degradation
The analysis was performed on an ABI Procise 494 sequencer. The sample was placed on a PVDF membrane for the Edman degradation with a cyclic procedure where residues were cleaved off one at a time and identified by chromatography. There were three steps in each cycle. In step 1 the PITC reagent was coupled to the N-terminal amino group under alkaline conditions. In step 2 the N-terminal residue was cleaved in acidic media. In step 3, the PITC-coupled residue was transferred to a flask, converted to a PTH-residue and identified by HPLC chromatography. The cycle was then started again for the identification of the next N-terminal residue.
SMV1 deglycosylation reaction and tandem mass spectrometry
Briefly, 50 μl of concentrated SMV1 were lyophilized. Then, 150 μl of TFMS was added to the tube and the mixture was incubated at 4 °C for 24 h. Following this, 150 μl of 60% pyridine solution, cooled to ~15 °C with a methanol dry ice bath, was added to the reaction tube, neutralizing the TFMS acid. Using a Slide-A-Lyzer dialysis cassette with a 2,000 Da protein molecular weight cut-off (Thermo Fisher Scientific), the reaction solutions were removed from the sample with overnight dialysis at 4 °C into Tris/HCl (pH 8) buffer. After dialysis, small amounts of aggregates—presumably deglycosylated SMV1—were observed in the cassette. Centrifugation for 15 min at 4 °C and 20,000 x g was used to pellet the aggregates. The pellet was resuspended with 50 μl of Tris buffer (pH 8). SDS-PAGE was then performed at a constant voltage of 120 V using 16.5% precast polyacrylamide Mini-PROTEAN Tris-Tricine precast gels (Bio-Rad) and Tris/Tricine/SDS running buffer (Bio-Rad). For each sample, 15 μl of boiled sample in SDS was added to separate wells in the gel, and 5 μl of Precision Plus Protein Dual Xtra protein standard (Bio-Rad) was used as a marker. Silver staining was performed using a Pierce Silver Stain for Mass Spectrometry kit (Thermo Fisher Scientific). The detected gel bands were then excised, and mass spectrometry (Alphalyse) was performed to analyze the composition of each band. The protein samples were reduced and alkylated with iodoacetamide, and subsequently digested with chymotrypsin. The resulting peptides were concentrated by Speed Vac lyophilization. The peptides were dissolved in 0.1% formic acid and injected on a Dionex Ultimate 3000 nano-LC system (Thermo Scientific) coupled to a Bruker Maxis Impact QTOF mass spectrometer for MS/MS analysis. Each sample underwent a 30-minute-long gradient run on the instruments. As an external control of the instrument, tryptic BSA was analysed together with the samples. The results were run against the Sulfolobus monocaudavirus SMV1 proteins from the UniProt.
Analysis of SMV1 and host cell lipids
For lipid analyses, CsCl-purified virions were pelleted by ultracentifugation (Beckman Type 50.2 Ti rotor, 35000 rpm, 2h, 15 °C), resuspended in 20 mM Tris-acetate (pH6) and run on 5–20 % (wt/vol) sucrose gradient (Beckman SW32 Ti rotor, 24000 rpm, 30 min, 15 °C). The opalescent virus-containing band was collected, pelleted as above and resuspended in 20 mM Tris-acetate (pH6). The cellular and viral lipids were analyzed by UHPLC-MSD at NIOZ using an Agilent 1290 Infinity II ultra-high performance LC coupled to a 6230 Agilent MSD in selected ion mode as described (Besseling et al., 2020).
Supplementary Material
An alignment of one subunit (yellow) between two adjacent diameters: symmetry 7 (cyan) and symmetry 8 (magenta).
An alignment of one subunit (yellow) between two very different diameters: symmetry 2 (orange) and symmetry 12 (green).
The helical symmetry established for the tubes and extrapolated to the spindle-shaped bodies (Fig. 4C) has been used to construct a model for the full capsid. The seven strands of subunits are each shown in a different color. Although discrete subunits are not seen in this visualization, 98,000 copies of the capsid protein have been used to generate this model. The basis of the model is that the local packing throughout the structure is fixed with a spacing of 2.7 Å between subunits along a strand, and the helical pitch of each strand is also fixed at 203 Å. As the diameter increases due to internal pressure from the packaged genome, the strands slide past each other.
The role of hydrophobic (lipophilic) surfaces in the sliding of SMV1 strands past each other. This animation shows the continuous transformation of the same number of total subunits from the narrowest diameter tube to the widest diameter tube of the 12 that we have reconstructed. As the tube becomes wider, more subunits are added per turn to the left-handed 7-start helices. The hydrophobic surfaces (gold) at the interfaces between the strands allow for this sliding to take place, at the same time that solvent is excluded from passing through the capsid.
The top view of Supplemental movie 4.
A, SignalP (Almagro Armenteros et al., 2019) analysis did not reveal any signal peptides or signal peptidase cleavage sites in the cleaved N-terminus. B, The N-terminal regions which are processed in the mature proteins are not shown. The start site experimentally determined for SMV1 is indicated with an arrowhead. The secondary structure determined for the SMV1 protein is shown above the alignment, with the red ribbon representing α-helices. The N-glycosylation seqon is boxed, with the target Asn residue indicated with an asterisk. Each sequence is identified with the corresponding GenBank accession number followed by the virus name. C, The predictions of transmembrane helices within the MCPs of seven viruses were made using the TMHMM server v2.0.
A, Transmission electron micrographs of negatively stained SMV1 virions treated with 0.1% N-lauryl sarcosine. Bars, 200 nm. B, Distribution of the lipid species identified in SMV1 host, Saccharolobus islandicus, and SMV1 virions. GDGT, glycerol dibiphytanyl glycerol tetraether lipids; GTGT, glycerol tribiphytanyl glycerol tetraether lipids. Numbers following GDGT or GTGT represent the number of cyclopentane rings present in the lipid species.
A, SDS-PAGE with Coomassie blue staining of SMV1. The MW marker lane M is labeled on the left. A single band at ~ 13–14 kDa is observed for SMV1. B, SMV1 samples on silver-stained SDS-PAGE before and after deglycosylation using TFMS. M indicates the molecular weight markers lane. C, The band indicated by blue arrowhead in (B) was cut and used for tandem mass spectrometry analysis. Protein W0UU99 was detected. D, The band indicated by red arrowhead in (B) was cut and used for tandem mass spectrometry analysis. Both W0UU99 and the correct capsid protein, W0UUv5, were detected.
A, The symmetry ID, helical rise, helical twist and point group symmetry are labeled. B, The “gold-standard” half map FSCs were calculated in cryoSPARC using a 0.143 criterion.
A, Cryo-EM of SMV1 before and after the NaOH (pH=10) treatment. Scale bars are 50 nm. The arrow indicates a tube containing a viral genome, while all of the other tubes in this image are empty. B, SDS-PAGE and negative staining analysis of different fractions from the 5–20% sucrose gradient which were loaded with non-treated virus (left) and NaOH-treated virus (right). The non-treated virus migrated to the bottom of the gradient, where most of the protein and DNA were found (fractions 8–12), whereas following NaOH treatment, the viral DNA was found in fraction 1 in the form of amorphous aggregates, and the virion tubes were enriched in fraction 3.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Archaeal and virus strains | ||
| SMV1 | Uldahl et al., 2016 | N/A |
| ATV | Prangishvili et al., 2006 | N/A |
| Saccharolobus islandicus ΔC1C2 | Gudbergsdottir et al., 2011 | N/A |
| Acidianus covivator AA9 | Prangishvili et al., 2006 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Bacto Tryptone | Gibco | Cat #: 211705 |
| Bacto yeast extract | Gibco | Cat #: 212750 |
| Sucrose | Sigma-Aldrich | Cat #: S0389 |
| PEG6000 | Sigma-Aldrich | Cat #: 81260 |
| NaCl | Sigma-Aldrich | Cat #: S9888 |
| Trisma base | Sigma-Aldrich | Cat #: T1503 |
| Acetic acid | Sigma-Aldrich | Cat #: A6283 |
| CsCl | Euromedex | Cat #: EU0770 |
| Ethidium bromide | Eurobio Scientific | Cat #: GEPBET02-AF |
| InstantBlue | Abcam | Cat #: ab119211 |
| Deposited data | ||
| SMV1 Cryo-EM maps | This paper | EMDB: EMD-24585, EMD-24586, EMD-24587, EMD-24588, EMD-24589, EMD-24590, EMD-24591, EMD-24592, EMD-24593, EMD-24594, EMD-24595, EMD-24597 |
| SMV1 Cryo-EM atomic models | This paper | PDB: 7RO2, 7RO3, 7RO4, 7RO5, 7RO6, 7ROB, 7ROC, 7ROD, 7ROE, 7ROG, 7ROH, 7ROI |
| ATV Cryo-EM map | This paper | EMDB: EMD-24596 |
| APBV1 Cryo-EM model | Ptchelkine et al., 2017 | PDB: 5OXE |
| Software and algorithms | ||
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
| DeepTracer | Pfab et al., 2021 | https://deeptracer.uw.edu |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| Phenix | Afonine et al., 2018 | https://phenix-online.org |
| MolProbity | Williams et al., 2018 | http://molprobity.biochem.duke.edu |
| Psi-Blast | Altschul et al., 1997 | https://www.ebi.ac.uk/Tools/sss/psiblast/ |
| PROMALS3D | Pei and Grishin, 2014 | http://prodata.swmed.edu/promals3d/promals3d.php |
| AlphaFold2 | Jumper et al., 2021 | https://alphafold.ebi.ac.uk |
| FATCAT | Li et al., 2020 | http://fatcat.godziklab.org/ |
Highlights.
We present the first atomic structure of an archaeal spindle-shaped virus
Spindle-shaped viruses have evolved from archaeal rod-shaped viruses
Hydrophobic interactions underlie the pleomorphism of spindle-shaped capsids
Acknowledgments
We are grateful to Dr. Xu Peng (University of Copenhagen) for the kind gift of SMV1 and its host, Saccharolobus islandicus ΔC1C2. Part of the cryo-EM imaging of SMV1 was done at the Molecular Electron Microscopy Core Facility at the University of Virginia, which is supported by the School of Medicine. Part of the Cryo-EM and the Cryo-ET imaging were done at the Nano-Imaging Core Facility at Institut Pasteur, created with the help of a grant from the French Government’s Investissements d’Avenir program (EQUIPEX CACSICE - Centre d’analyse de systèmes complexes dans les environnements complexes, ANR-11-EQPX-0008). This work was supported by NIH Grant GM122510 (E.H.E.) and K99GM138756 (F.W.). The work in the M.K. laboratory was supported by grants from l’Agence Nationale de la Recherche (ANR-17-CE15-0005-01, ANR-20-CE20-009-02 and ANR-21-CE11-0001-01) and Ville de Paris (Emergence(s) project MEMREMA). We are grateful to Youssef Ghorbal for the help with data transfer, to Caglar Yildiz for support in the lipid analysis and to the Ultrastructural BioImaging unit of Institut Pasteur for access to electron microscopes.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, and Nielsen H. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37, 420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, et al. (2016). Membrane fission by dynamin: what we know and what we need to know. EMBO J 35, 2270–2284. 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C, and Dopson M. (2007). Life in acid: pH homeostasis in acidophiles. Trends in microbiology 15, 165–171. 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Baquero DP, Liu Y, Wang F, Egelman EH, Prangishvili D, and Krupovic M. (2020). Structure and assembly of archaeal viruses. Advances in virus research 108, 127–164. 10.1016/bs.aivir.2020.09.004. [DOI] [PubMed] [Google Scholar]
- Besseling MA, Hopmans EC, Bale NJ, Schouten S, Damste JSS, and Villanueva L. (2020). The absence of intact polar lipid-derived GDGTs in marine waters dominated by Marine Group II: Implications for lipid biosynthesis in Archaea. Sci Rep 10, 294. 10.1038/s41598-019-57035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, and Klug A. (1962). Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol 27, 1–24. [DOI] [PubMed] [Google Scholar]
- Costa TR, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, and Waksman G. (2016). Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell 166, 1436–1444 e1410. 10.1016/j.cell.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH, and Watson JD (1956). Structure of small viruses. Nature 177, 473–475. 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- Damodaran KV, Reddy VS, Johnson JE, and Brooks CL 3rd (2002). A general method to quantify quasi-equivalence in icosahedral viruses. J. Mol. Biol 324, 723–737. 10.1016/s00222836(02)01138-5. [DOI] [PubMed] [Google Scholar]
- Dellas N, Snyder JC, Bolduc B, and Young MJ (2014). Archaeal Viruses: Diversity, Replication, and Structure. Annu Rev Virol 1, 399–426. 10.1146/annurev-virology-031413-085357. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, and Egelman EH (2015). Virology. A virus that infects a hyperthermophile encapsidates A-form DNA. Science 348, 914–917. 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH (2000). A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr.D.Biol.Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, et al. (2014). A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18, 51–60. 10.1007/s00792-013-0591-z. [DOI] [PubMed] [Google Scholar]
- Forterre P, Krupovic M, and Prangishvili D. (2014). Cellular domains and viral lineages. Trends in microbiology 22, 554–558. 10.1016/j.tim.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De CP, and Unger VM (2008). Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet A, Vestergaard G, Felisberto-Rodrigues C, Campanacci V, Garrett RA, Cambillau C, and Ortiz-Lombardia M. (2010). Getting the best out of long-wavelength X-rays: de novo chlorine/sulfur SAD phasing of a structural protein from ATV. Acta crystallographica. Section D, Biological crystallography 66, 304–308. 10.1107/S0907444909051798. [DOI] [PubMed] [Google Scholar]
- Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, and Garrett RA (2011). Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79, 35–49. 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Vestergaard G, Rachel R, Chen L, Garrett RA, and Prangishvili D. (2005). Virology: independent virus development outside a host. Nature 436, 1101–1102. 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- Hay ID, and Lithgow T. (2019). Filamentous phages: masters of a microbial sharing economy. EMBO reports 20. 10.15252/embr.201847427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer MB, and Gibb BC (2016). Molecular Shape and the Hydrophobic Effect. Annu Rev Phys Chem 67, 307–329. 10.1146/annurev-physchem-040215-112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein R, Bollschweiler D, Dharmavaram S, Lintner NG, Plitzko JM, Bruinsma R, Engelhardt H, Young MJ, Klug WS, and Lawrence CM (2018). Structural studies of Acidianus tailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses. Proc. Natl. Acad. Sci. U.S.A 115, 2120–2125. 10.1073/pnas.1719180115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein RA, Amenabar MJ, Munson-McGee JH, Boyd ES, and Young MJ (2016). Acidianus Tailed Spindle Virus: a New Archaeal Large Tailed Spindle Virus Discovered by Culture-Independent Methods. J Virol 90, 3458–3468. 10.1128/JVI.03098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Pietila MK, Fu CJ, Schmid MF, Bamford DH, and Chiu W. (2015). Lemon-shaped halo archaeal virus His1 with uniform tail but variable capsid structure. Proc. Natl. Acad. Sci. U.S.A 112, 2449–2454. 10.1073/pnas.1425008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Koonin EV, Prangishvili D, and Krupovic M. (2016). Bipartite Network Analysis of the Archaeal Virosphere: Evolutionary Connections between Viruses and Capsidless Mobile Elements. J Virol 90, 11043–11055. 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson EA, Goodman DA, Gorchels ME, and Stedman KM (2017). Genetic Analysis of the Major Capsid Protein of the Archaeal Fusellovirus SSV1: Mutational Flexibility and Conformational Change. Genes (Basel) 8, 373. 10.3390/genes8120373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, and Olson AJ (2021). Icosahedral virus structures and the protein data bank. J Biol Chem 296, 100554. 10.1016/j.jbc.2021.100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, and Speir JA (1997). Quasi-equivalent viruses: a paradigm for protein assemblies. J. Mol. Biol 269, 665–675. 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Kim SJ, Cvirkaite-Krupovic V, Yu WJ, Gwak JH, Lopez-Perez M, Rodriguez-Valera F, Krupovic M, Cho JC, and Rhee SK (2019). Spindle-shaped viruses infect marine ammonia-oxidizing thaumarchaea. Proc. Natl. Acad. Sci. U.S.A 116, 15645–15650. 10.1073/pnas.1905682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knitsch R, Schneefeld M, Weitzel K, and Pfeifer F. (2017). Mutations in the major gas vesicle protein GvpA and impacts on gas vesicle formation in Haloferax volcanii. Mol Microbiol 106, 530–542. 10.1111/mmi.13833. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Dolja VV, and Koonin EV (2019). Origin of viruses: primordial replicators recruiting capsids from hosts. Nature reviews. Microbiology 17, 449–458. 10.1038/s41579-0190205-6. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Dolja VV, and Koonin EV (2020). The LUCA and its complex virome. Nature reviews. Microbiology 18, 661–670. 10.1038/s41579-020-0408-x. [DOI] [PubMed] [Google Scholar]
- Krupovic M, and Koonin EV (2017). Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U.S.A 114, E2401–E2410. 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Quemin ER, Bamford DH, Forterre P, and Prangishvili D. (2014). Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol 88, 2354–2358. 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, and Cannon MC (1998). Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. Journal of bacteriology 180, 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jaroszewski L, Iyer M, Sedova M, and Godzik A. (2020). FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res 48, W60–W64. 10.1093/nar/gkaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cvirkaite-Krupovic V, Baquero DP, Yang Y, Zhang Q, Shen Y, and Krupovic M. (2021). Virus-induced cell gigantism and asymmetric cell division in archaea. Proc. Natl. Acad. Sci. U.S.A 118, e2022578118. 10.1073/pnas.2022578118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, and Krupovic M. (2021). Viruses of Asgard archaea. bioRxiv, 2021.2007.2029.453957. 10.1101/2021.07.29.453957. [DOI]
- Mochizuki T, Yoshida T, Tanaka R, Forterre P, Sako Y, and Prangishvili D. (2010). Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 402, 347–354. 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- Newberry RW, and Raines RT (2019). Secondary Forces in Protein Folding. ACS Chem Biol 14, 1677–1686. 10.1021/acschembio.9b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, and Grishin NV (2014). PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol 1079, 263–271. 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti LE, Dharmavaram S, Klug WS, Marian J, Rudnick J, and Bruinsma RF (2016). Useful scars: Physics of the capsids of archaeal viruses. Phys Rev E 94, 012404. 10.1103/PhysRevE.94.012404. [DOI] [PubMed] [Google Scholar]
- Perotti LE, Zhang K, Rudnick J, and Bruinsma RF (2019). Kirigami and the Caspar-Klug construction for viral shells with negative Gauss curvature. Phys Rev E 99, 022413. 10.1103/PhysRevE.99.022413. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J.Comput.Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfab J, Phan NM, and Si D. (2021). DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl. Acad. Sci. U.S.A 118. 10.1073/pnas.2017525118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F, Gregor D, Hofacker A, Pl√∂sser P, and Zimmermann P. (2002). Regulation of gas vesicle formation in halophilic archaea. J. Mol. Microbiol. Biotechnol 4, 175–181. [PubMed] [Google Scholar]
- Pfitzner AK, Moser von Filseck J, and Roux A. (2021). Principles of membrane remodeling by dynamic ESCRT-III polymers. Trends in cell biology. 10.1016/j.tcb.2021.04.005. [DOI] [PubMed]
- Pietila MK, Atanasova NS, Oksanen HM, and Bamford DH (2013). Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environmental microbiology 15, 1674–1686. 10.1111/1462-2920.12030. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, and Krupovic M. (2017). The enigmatic archaeal virosphere. Nature reviews. Microbiology 15, 724–739. 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Vestergaard G, Haring M, Aramayo R, Basta T, Rachel R, and Garrett RA (2006). Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J. Mol. Biol 359, 1203–1216. 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Ptchelkine D, Gillum A, Mochizuki T, Lucas-Staat S, Liu Y, Krupovic M, Phillips SEV, Prangishvili D, and Huiskonen JT (2017). Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging. Nat. Commun 8, 1436. 10.1038/s41467-017-01668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- Quemin ER, Chlanda P, Sachse M, Forterre P, Prangishvili D, and Krupovic M. (2016). Eukaryotic-Like Virus Budding in Archaea. mBio 7, e01439–16. 10.1128/mBio.01439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ER, Pietila MK, Oksanen HM, Forterre P, Rijpstra WI, Schouten S, Bamford DH, Prangishvili D, and Krupovic M. (2015). Sulfolobus Spindle-Shaped Virus 1 Contains Glycosylated Capsid Proteins, a Cellular Chromatin Protein, and Host-Derived Lipids. J Virol 89, 11681–11691. 10.1128/JVI.02270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, and Forterre P. (2008). Redefining viruses: lessons from Mimivirus. Nature reviews. Microbiology 6, 315–319. 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- Rayment I, Baker TS, Caspar DL, and Murakami WT (1982). Polyoma virus capsid structure at 22.5 A resolution. Nature 295, 110–115. 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N. (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele U, Erdmann S, Ungewickell EJ, Felisberto-Rodrigues C, Ortiz-Lombardia M, and Garrett RA (2011). Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus. J Virol 85, 4812–4821. 10.1128/JVI.00072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevvana M, Klose T, and Rossmann MG (2021). Principles of Virus Structure. Encyclopedia of Virology, 257–277. 10.1016/B978-0-12-814515-9.00033-3. [DOI]
- Sheppard C, Blombach F, Belsom A, Schulz S, Daviter T, Smollett K, Mahieu E, Erdmann S, Tinnefeld P, Garrett R, et al. (2016). Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP. Nat. Commun 7, 13595. 10.1038/ncomms13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojar HT, and Bahl OP (1987). Chemical deglycosylation of glycoproteins. Meth. Enzymol 138, 341–350. [DOI] [PubMed] [Google Scholar]
- Stedman KM, DeYoung M, Saha M, Sherman MB, and Morais MC (2015). Structural insights into the architecture of the hyperthermophilic Fusellovirus SSV1. Virology 474, 105–109. 10.1016/j.virol.2014.10.014. [DOI] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, and Harrison SC (1994). Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369, 160–163. 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Strunk T, Hamacher K, Hoffgaard F, Engelhardt H, Zillig MD, Faist K, Wenzel W, and Pfeifer F. (2011). Structural model of the gas vesicle protein GvpA and analysis of GvpA mutants in vivo. Mol Microbiol 81, 56–68. 10.1111/j.1365-2958.2011.07669.x. [DOI] [PubMed] [Google Scholar]
- Uldahl KB, Jensen SB, Bhoobalan-Chitty Y, Martinez-Alvarez L, Papathanasiou P, and Peng X. (2016). Life Cycle Characterization of Sulfolobus Monocaudavirus 1, an Extremophilic Spindle-Shaped Virus with Extracellular Tail Development. J Virol 90, 5693–5699. 10.1128/JVI.00075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Baquero DP, Su Z, Beltran LC, Prangishvili D, Krupovic M, and Egelman EH (2020). The structures of two archaeal type IV pili illuminate evolutionary relationships. Nat. Commun 11, 3424. 10.1038/s41467-020-17268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Cvirkaite-Krupovic V, Kreutzberger MAB, Su Z, de Oliveira GAP, Osinski T, Sherman N, DiMaio F, Wall JS, Prangishvili D, et al. (2019). An extensively glycosylated archaeal pilus survives extreme conditions. Nat Microbiol 4, 1401–1410. 10.1038/s41564-019-0458-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MS, Eibauer M, Sivagurunathan S, Magin TM, Goldman RD, and Medalia O. (2021). Structural heterogeneity of cellular K5/K14 filaments as revealed by cryo-electron microscopy. eLife 10, e70307. 10.7554/eLife.70307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Science 27, 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H. (2007). 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol 157, 126–137. 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Xiang X, Chen L, Huang X, Luo Y, She Q, and Huang L. (2005). Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79, 8677–8686. 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Pena A, Low WW, Wong JLC, Frankel G, and Egelman EH (2020). Cryoelectron-Microscopic Structure of the pKpQIL Conjugative Pili from Carbapenem-Resistant Klebsiella pneumoniae. Structure 28, 1321–1328 e1322. 10.1016/j.str.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendörfer M, and Kristjansson JK (1993). Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Systematic and Applied Microbiology 16, 609–628. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An alignment of one subunit (yellow) between two adjacent diameters: symmetry 7 (cyan) and symmetry 8 (magenta).
An alignment of one subunit (yellow) between two very different diameters: symmetry 2 (orange) and symmetry 12 (green).
The helical symmetry established for the tubes and extrapolated to the spindle-shaped bodies (Fig. 4C) has been used to construct a model for the full capsid. The seven strands of subunits are each shown in a different color. Although discrete subunits are not seen in this visualization, 98,000 copies of the capsid protein have been used to generate this model. The basis of the model is that the local packing throughout the structure is fixed with a spacing of 2.7 Å between subunits along a strand, and the helical pitch of each strand is also fixed at 203 Å. As the diameter increases due to internal pressure from the packaged genome, the strands slide past each other.
The role of hydrophobic (lipophilic) surfaces in the sliding of SMV1 strands past each other. This animation shows the continuous transformation of the same number of total subunits from the narrowest diameter tube to the widest diameter tube of the 12 that we have reconstructed. As the tube becomes wider, more subunits are added per turn to the left-handed 7-start helices. The hydrophobic surfaces (gold) at the interfaces between the strands allow for this sliding to take place, at the same time that solvent is excluded from passing through the capsid.
The top view of Supplemental movie 4.
A, SignalP (Almagro Armenteros et al., 2019) analysis did not reveal any signal peptides or signal peptidase cleavage sites in the cleaved N-terminus. B, The N-terminal regions which are processed in the mature proteins are not shown. The start site experimentally determined for SMV1 is indicated with an arrowhead. The secondary structure determined for the SMV1 protein is shown above the alignment, with the red ribbon representing α-helices. The N-glycosylation seqon is boxed, with the target Asn residue indicated with an asterisk. Each sequence is identified with the corresponding GenBank accession number followed by the virus name. C, The predictions of transmembrane helices within the MCPs of seven viruses were made using the TMHMM server v2.0.
A, Transmission electron micrographs of negatively stained SMV1 virions treated with 0.1% N-lauryl sarcosine. Bars, 200 nm. B, Distribution of the lipid species identified in SMV1 host, Saccharolobus islandicus, and SMV1 virions. GDGT, glycerol dibiphytanyl glycerol tetraether lipids; GTGT, glycerol tribiphytanyl glycerol tetraether lipids. Numbers following GDGT or GTGT represent the number of cyclopentane rings present in the lipid species.
A, SDS-PAGE with Coomassie blue staining of SMV1. The MW marker lane M is labeled on the left. A single band at ~ 13–14 kDa is observed for SMV1. B, SMV1 samples on silver-stained SDS-PAGE before and after deglycosylation using TFMS. M indicates the molecular weight markers lane. C, The band indicated by blue arrowhead in (B) was cut and used for tandem mass spectrometry analysis. Protein W0UU99 was detected. D, The band indicated by red arrowhead in (B) was cut and used for tandem mass spectrometry analysis. Both W0UU99 and the correct capsid protein, W0UUv5, were detected.
A, The symmetry ID, helical rise, helical twist and point group symmetry are labeled. B, The “gold-standard” half map FSCs were calculated in cryoSPARC using a 0.143 criterion.
A, Cryo-EM of SMV1 before and after the NaOH (pH=10) treatment. Scale bars are 50 nm. The arrow indicates a tube containing a viral genome, while all of the other tubes in this image are empty. B, SDS-PAGE and negative staining analysis of different fractions from the 5–20% sucrose gradient which were loaded with non-treated virus (left) and NaOH-treated virus (right). The non-treated virus migrated to the bottom of the gradient, where most of the protein and DNA were found (fractions 8–12), whereas following NaOH treatment, the viral DNA was found in fraction 1 in the form of amorphous aggregates, and the virion tubes were enriched in fraction 3.
Data Availability Statement
The SMV1 maps were deposited in the Electron Microscopy Data Bank (EMDB) with entry codes EMD-24585, EMD-24586, EMD-24587, EMD-24588, EMD-24589, EMD-24590, EMD-24591, EMD-24592, EMD-24593, EMD-24594, EMD-24595, and EMD-24597 and the respective atomic models were deposited in the Protein Data Bank (PDB) with entry codes 7RO2, 7RO3, 7RO4, 7RO5, 7RO6, 7ROB, 7ROC, 7ROD, 7ROE, 7ROG, 7ROH and 7ROI. The ATV map was deposited in the EMDB with entry code EMD-24596.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request