Abstract
Developmental neural activity is a common feature of neural circuit assembly. Although glia have established roles in synapse development, the contribution of neuron-glia interactions to developmental activity remains largely unexplored. Here we show that astrocytes are necessary for developmental activity during synaptogenesis in Drosophila. Using wide-field epifluorescence and two-photon imaging, we show that the glia of the central nervous system participate in developmental activity with type-specific patterns of intracellular calcium dynamics. Genetic ablation of astrocytes, but not of cortex or ensheathing glia, leads to severe attenuation of neuronal activity. Likewise, inhibition of neuronal activity results in the loss of astrocyte calcium dynamics. By altering these dynamics, we show that astrocytic calcium cycles can influence neuronal activity but are not necessary per se. Taken together, our results indicate that, in addition to their recognized role in the structural maturation of synapses, astrocytes are also necessary for the function of synapses during development.
Keywords: synapse formation, neural activity, astrocytes, glia, Drosophila, neural circuit development
Introduction
Stimulus-independent activity during development contributes to synapse formation in both vertebrate and invertebrate systems. In vertebrates, such developmental activity has been observed in various regions of the brain, where they are generally initiated by local circuitry and, in some systems, have been shown to have broad roles in circuit maturation (Blankenship and Feller, 2010). In Drosophila, patterned, stimulus-independent neural activity (PSINA, ‘see-na’) accompanies synaptogenesis in the adult brain (Akin et al., 2019). PSINA engages the whole brain and manifests as cycles of coordinated active and silent phases which begin ~50 hours after pupal formation (hAPF) and continue until the last hour prior to eclosion. Perturbation of PSINA results in cell-type-specific alterations to synaptic structure in the visual system (Bajar B.T. et al., Nature In Revision).
Neuron-glia interactions are critical to neural activity in the mature nervous system, but the contribution of glia to developmental activity is less well understood. Glia play key roles in development, ranging from controlling neural proliferation and apoptosis (Corty and Freeman, 2013) to influencing axon guidance and synapse formation (Barres, 2008; Perez-Catalan et al., 2021). In Drosophila, astrocytes participate in PSINA with waves of activity complementary to neuronal activity (Akin et al., 2019). However, whether astrocytes or other glial cell types have specific roles in the regulation of developmental activity remains an open question.
There are five subtypes of glia in the Drosophila central nervous system (CNS): astrocytes, which penetrate the neuropil, ensheathing glia (EG) wrap neuropils and axonal tracts, cortex glia (CG) wrap the cortices, and the subperinuerial and perineurial glia cover the whole CNS (Kremer et al., 2017; Yildirim et al., 2019) (Figure 1a). In the visual system, these classes have been further stratified based on morphology and association with particular neuropils (Chotard & Salecker 2007). Astrocytes, in particular, are critical for synaptic development. These specialized glia elaborate peri-synaptic processes during synaptogenesis, and genetic ablation of astrocytes results in a 25––50% loss of synapses in different brain regions (Muthukumar et al., 2014). The co-dependence of synaptogenesis on glia and on PSINA (Bajar B.T. et al., Nature In Revision) suggests that neuron-glia interactions may regulate PSINA.
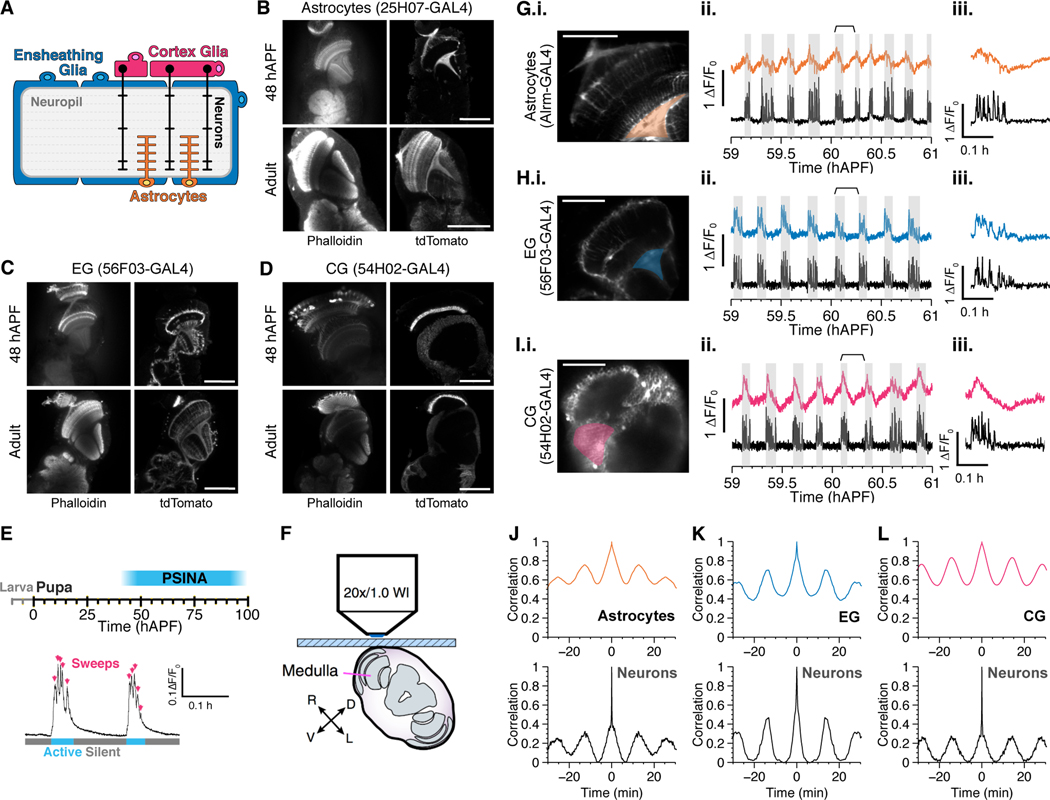
Figure 1.
Multiple glial cell types participate in PSINA.
A. Schematic showing location and morphology of glial cell types in the Drosophila central nervous system. Astrocytes infiltrate the neuropil and interact with neuronal processes (orange), ensheathing glia envelop the neuropil (blue), and cortex glia envelop the soma (magenta).
B-D. Developmental characterization of drivers for astrocytes (B), ensheathing glia (C), and cortex glia (D). Representative micrographs from 48 hours after pupal formation (hAPF, top row) or newly eclosed adult (bottom row). Left, neuropil counterstain. Right, cell membrane labeled with tdTomato and stained with anti-dsRed antibody. Note that R56F03-GAL4 shows sparse neuronal labeling. Scale bars, 100μm.
E. Above, timeline of PSINA during pupal development. Below, two representative cycles. Arrows indicate individual sweeps, while cyan and gray bars mark active and silent phases, respectively.
F. Schematic showing live 2P imaging preparation. Adapted from Akin et al. 2016.
G-H. Representative micrographs (i), traces (ii), and a single highlighted cycle (iii) showing calcium transients in astrocytes (G), cortex glia (H), and ensheathing glia (I) compared to neuronal activity. Highlighted region of interest on left used to generate glial traces on right. Shaded gray areas indicate neuronal active phases. Time-period indicated by the braces used to generate the single cycle in iii. Scale bars, 40μm.
J-L. Representative autocorrelograms from astrocytes (J), ensheathing glia (K), and cortex glia (L), with corresponding autocorrelograms from neurons measured in the same animal below.
For a complete list of genotypes used in this figure, see Table S1.
Here we show that astrocytes are necessary for developmental neuronal activity in Drosophila. In turn, calcium cycles in astrocytes are dependent on neuronal activity. These results indicate that neuron-glia interactions influence neural circuit formation through developmental activity.
Results
Glial cell types participate in PSINA
Drivers specific to glial cell types have been described for the adult fly (Kremer et al., 2017), and for larva (Li et al., 2014; Ackerman et al., 2021; Coutinho-Budd et al., 2017). We tested this set for developmental expression and identified drivers for astrocytes (25H07-GAL4), EG (56F03-GAL4), and CG (54H02-GAL4) (Figure 1b–d) that are largely specific from mid-pupa onwards. This expands the existing toolkit (Richier et al., 2017; Muthukumar et al., 2014) of type-specific glial drivers for studies of the developing adult nervous system.
PSINA comprises coordinated cycles of active and silent phases; between 55––70 hAPF these cycles arrive with a regular period of 12––15 minutes (Figure 1e). Each active phase consists of multiple bouts of activity termed ‘sweeps’. We have previously shown that astrocytes participate in PSINA with a pattern complementary to neuronal activity ((Akin et al., 2019) and see below): To ask whether other glial cell types also participate in PSINA, we expressed the green genetically-encoded calcium indicator (GECI) GCaMP6s (Chen et al., 2013) in glia and the red GECI jRCaMP1b (Dana et al., 2016) with orthogonal expression systems, and assessed activity in vivo using two-photon (2P) imaging of live pupae (Figure 1f) (Akin and Zipursky, 2016). We found that additional glial subtypes also participate in PSINA and do so in type-specific fashion (Figure 1g–i). As noted, astrocyte calcium cycles are complementary or anti-phase to neuronal activity: broadly, intracellular calcium levels rise during neuronal silent phases and fall during active phases. The reversal points, the peak before the fall and the trough before the rise, come after the beginning and end of neuronal active phases, respectively. CG follow a similar pattern but with a distinctive bi-phasic rise which transitions from gradual to abrupt with the onset of neuronal activity. By contrast to astrocytes and CG, EG have cycles that are in phase with neuronal activity and display calcium transients that appear to be delayed and broadened versions of the neuronal traces. Astrocytes, CG, and EG have cycle frequencies that match that of neuronal PSINA (Figure 1j–l). In sum, the three glial subtypes most closely associated with the neurons and neuronal processes of the CNS have type-specific calcium cycles during PSINA.
Astrocytes are necessary for PSINA
To ask whether any of these glial cell types are necessary for neuronal PSINA, we used the TARGET system (McGuire et al., 2003) to dis-inhibit expression of the proapoptotic gene hid during the second half of pupal development (Figure 2a). In the background of this type-specific ablation, we followed whole-brain neuronal activity from pan-neuronal jRCaMP1b using wide-field epifluorescence imaging. We confirmed ablation efficiency after imaging by quantifying surviving nuclei labeled with Stinger (Barolo et al., 2000).
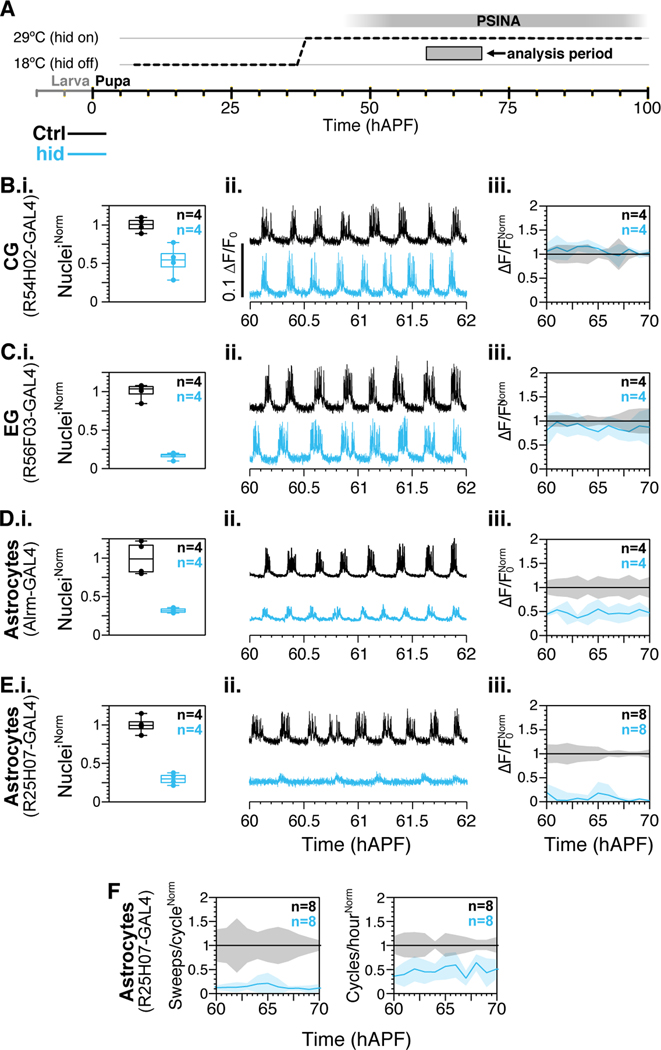
Figure 2.
Astrocytes are required for PSINA.
A. Expression control of UAS-hid with TARGET; animals shifted from 18°C to 29°C at 38 hAPF.
B-E. i. Quantification of glial subtype ablation in control (black) and hid-expressing (teal) animals. Sample counts indicated in top right corner. ii. Representative traces from neurons expressing jRCaMP1b in control (black) and in genetic background of glial subtype ablation (teal). iii. Active phase average amplitude binned by hour and normalized to wildtype activity between 60 and 70 hAPF. Shaded areas, standard deviation. Colors match genotypes in i. Sample sizes in top right corner.
F. Sweeps/cycle (left) and cycles/hour (right) when astrocytes are ablated with 25H07-GAL4 driven hid. Data binned by hour and normalized to control. Shaded areas, standard deviation. Genotypes color-matched to B-E. Sample sizes in top right corner.
For a complete list of genotypes used in this figure, see Table S1.
Ablation of CG or EG did not affect PSINA (Figure 2b–c). Although expression of hid results in a significant loss of glia (57±20% ablation for CG and 84±4% ablation for EG, assessed at 84 hAPF), there was no significant alteration to PSINA. By contrast, ablation of astrocytes resulted in strong attenuation of PSINA. With the alrm-GAL4 astrocyte driver, 68±3% of astrocytes were ablated by 84 hAPF, and ~50% of PSINA amplitude was lost (Figure 2d). We observed a similar but stronger effect with the 25H07-GAL4 astrocyte driver: 71±7% of astrocytes were ablated by 84 hAPF, and >80% of PSINA amplitude was lost (Figure 2e). Differences between the astrocyte drivers could be due to non-astrocyte expression in alrm-GAL4 (Kremer et al., 2017) or to driver-strength-dependent timing of ablation. Additionally, we found that ablation of astrocytes with 25H07-GAL4 led to a >80% decrease in sweeps per cycle and ~50% fewer cycles relative to controls (Figure 2f).
Taken together, these results indicate that astrocytes are required for wildtype PSINA.
Reciprocal influence of neuron-glia PSINA dynamics
Next, we asked whether PSINA in neurons and calcium cycles in astrocytes influence each other. To test for neuronal effects on glia, we expressed either the inward rectifying potassium channel, Kir2.1, or the inhibitor of synaptic release, tetanus toxin (TNT), in all neurons. As with the ablation experiments, we used the TARGET system to limit these severe perturbations to the second half of pupal development (Figure 3a). Pan-neuronal Kir2.1 or TNT expression results in near-complete loss of developmental activity (Figure 3b), consistent with previous observations (Bajar B.T. et al., Nature In Revision). Remarkably, targeted silencing of neuronal activity results in the loss of astrocytic calcium cycles (Figure 3c–d).
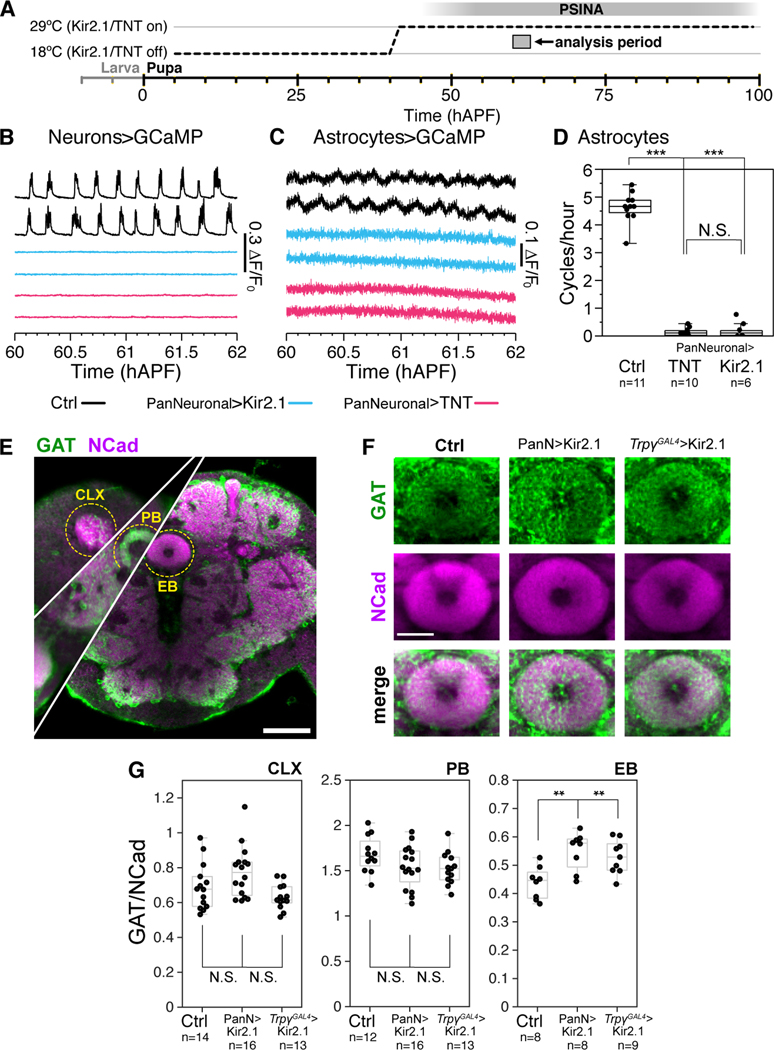
Figure 3.
Astrocyte calcium cycles depend on neuronal activity
A. Expression control of UAS-Kir2.1 or TNT with TARGET; animals shifted from 18°C to 29°C at 38 hAPF.
B-C. Representative traces of pupae expressing GCaMP6s in neurons (B) or astrocytes (C). Wildtype activity in the empty-GAL4>Kir2.1 condition (black, top), and reduced activity in the pan-neuronal-GAL4>Kir2.1 (blue, middle) and pan-neuronal-Gal4>TNT condition (magenta, bottom).
D. Astrocyte calcium cycles per hour from 60 hAPF to 70 hAPF. Box-and-whiskers mark 5th, 25th, 50th, 75th, and 95th percentiles. ***, p < 0.0001 by Tukey’s post-hoc test following ANOVA.
E. Representative micrograph of an empty-GAL4>Kir2.1 brain at 84 hAPF stained for GAT (green) and Ncad (magenta). Cuts from different depths through the central brain overlaid to show calyx (CLX), protocerebral bridge (PB), and ellipsoid body (EB). Scale bar, 50μm.
F. Representative micrographs of GAT expression in the EB in 84 hAPF pupae expressing Kir2.1 under the control of empty-GAL4, pan-neuronal -GAL4, or TrpγGAL4. For each channel, image display limits set to same values across genotypes. Scale bar, 20μm.
G. GAT to Ncad expression ratio in the CLX, PB, and EBs of 84 hAPF pupae expressing Kir2.1 under the control of empty-GAL4, pan-neuronal-GAL4, or TrpγGAL4. Data from 8––9 brains per condition. Box-and-whiskers mark non-outlier extrema, 25th, 50th, and 75th percentiles. **, p < 0.01 by Welch’s t-test.
For a complete list of genotypes used in this figure, see Table S1.
Astrocytes express the GABA transporter GAT in a manner dependent on neuronal GABA release (Muthukumar et al., 2014). To ask whether GAT expression is affected by PSINA, we silenced PSINA either with pan-neuronal Kir2.1 expression or with Kir2.1 expression in Trpγ expressing neurons. Trpγ is expressed in ~2,000 neurons during PSINA and is thought to be a marker for relay neurons that set up the brain-wide coordination of developmental activity: silencing this small population of Trpγ+ neurons leads to near-complete loss of PSINA (Bajar B.T. et al., Nature In Revision). We assessed GAT expression using immunofluorescence. The highly-specific anti-GAT antibody (Stork et al., 2014, Muthukumar et al., 2014) appears to have low mobility in whole-mount preparations and can produce highly variable labeling in superficial regions of the brain. As such, we focused our analysis on internal structures, where relative labeling of GAT and our reference marker, N-Cadherin, were proportional within each condition (Figure 3e). Of the three internal structures we assessed, in the mushroom body calyx and the protocerebral bridge GAT levels are unaffected in PSINA-silenced brains (Figure 3g). By contrast, in the ellipsoid body, both approaches to inhibiting neuronal PSINA result in a 20––25% increase in astrocytic GAT expression (Figure 3g–f). Previous work had shown that ablation of GABA neurons leads to a significant decrease in GAT labeling in the antennal lobes and subesophageal ganglion, suggesting a link between developmental activity in GABA neurons and astrocytic GAT expression (Muthukumar et al., 2014). Here we find that global loss of developmental activity, i.e. silencing both inhibitory and excitatory neurons, has region-specific effects on GAT expression; while we did not observe it in our sampling, it is possible that GAT would be reduced in other neuropils when PSINA is silenced.
Together, these data indicate that developmental neuronal activity is required for astrocytic calcium cycles and modulates wild-type GAT expression levels.
Finally, we asked if astrocyte calcium cycles affect neuronal activity. First, we expressed the thermosensitive calcium channel TrpA1 in astrocytes. While TrpA1 is vital for calcium homeostasis in mammalian astrocytes (Shigetomi et al., 2013), it is not natively expressed in Drosophila astrocytes (Kurmangaliyev et al., 2020). As such, heterologous TrpA1 expression can be used to induce calcium influx in these cells (Pulver et al., 2009; Zhang et al., 2017). We activated 25H07-driven TrpA1 at the onset of PSINA (Figure 4a) and observed two phases of altered astrocyte calcium dynamics (Figure 4b,c). In the first ~20 hours after the activating temperature shift, astrocytes exhibit a gradual increase in baseline intracellular calcium while maintaining the periodic cycles. During this first phase, neuronal PSINA amplitude increases alongside the astrocyte baseline, with modest increases to active phase length and sweeps per cycle (Figure 4b). After about 20 hours, calcium baseline returns to pre-activation levels, and, notably, the astrocytic calcium cycles are lost (Figure 4c). In this second phase, neuronal PSINA amplitude also returns to pre-activation values and activity persists despite the loss of astrocytic calcium cycles (Figure 4c). These changes to GECI-reported calcium dynamics do not affect GAT expression (Figure 4d), suggesting that this aspect of astrocyte physiology is not sensitive to the perturbation of PSINA-associated calcium cycles.
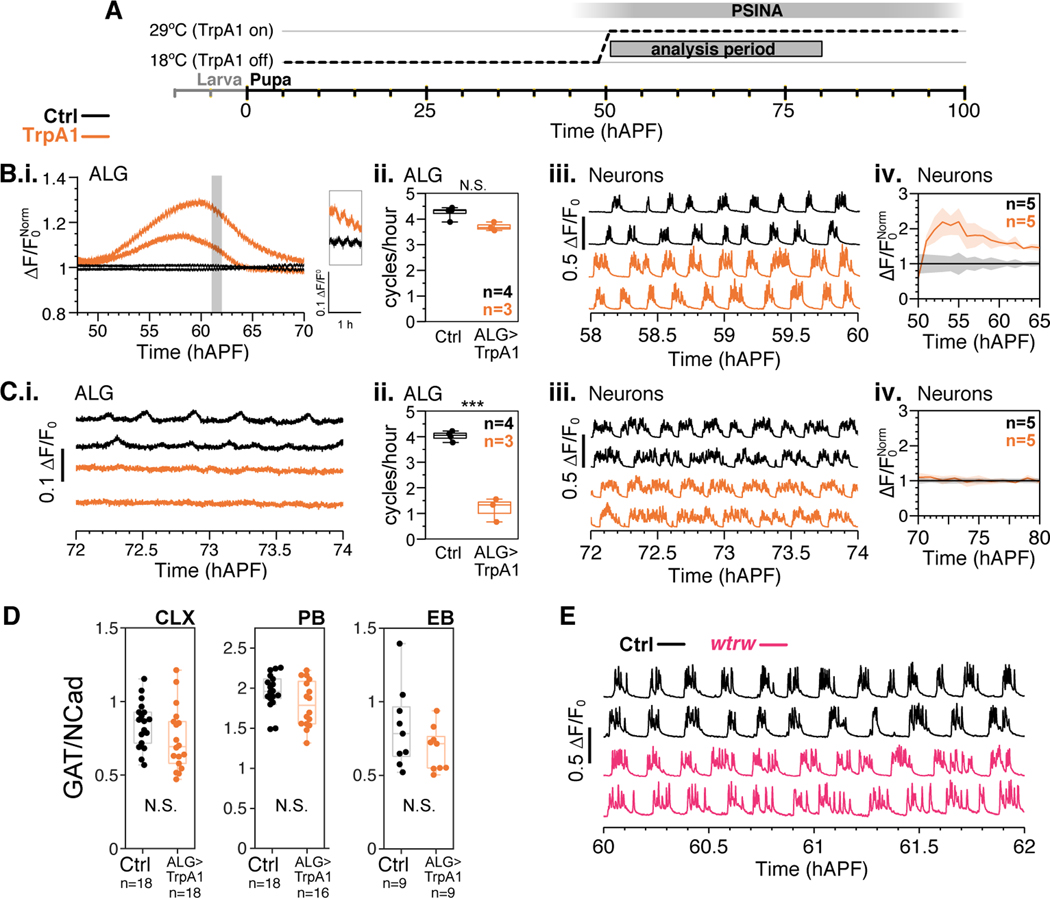
Figure 4.
Neuronal activity does not require astrocyte calcium cycles.
A. TrpA1 activation in astrocytes by temperature shift; animals shifted from 18°C to 29°C at 50 hAPF.
B-C.i. Representative traces of pupae expressing GCaMP6s in: (B) astrocytes in the 20 hours after temperature shift. Control (black) and TrpA1 (orange) traces normalized to average control trace. Inset shows time period highlighted in gray; or (C) astrocytes after 70 hAPF. Control (black) and TrpA1 (orange) traces plotted as ΔF/F0. ii. Astrocyte calcium cycles per hour from 60 to 70 hAPF. Box-and-whiskers mark 5th, 25th, 50th, 75th, and 95th percentiles. ***, p < 0.0001 by Welch’s unpaired t-test following Shapiro-Wilk test. Sample sizes indicated in top right corner. iii. Representative traces of pupae expressing GCaMP6s in neurons with control (black) or TrpA1 expression in astrocytes (orange). iv. Active phase average amplitude binned by hour and normalized to wildtype activity. Shaded error bar represents standard deviation. Colors match genotypes in i. Sample sizes indicated in top right corner.
D. GAT to Ncad expression ratio in the CLX, PB, and EBs of 84 hAPF pupae expressing TrpA1 under the control of empty-GAL4 or R25H07-GAL4 (astrocytes). Data from 8––9 brains per condition. Box-and-whiskers mark non-outlier extrema, 25th, 50th, and 75th percentiles. N.S., p > 0.05 by Welch’s t-test.
E. Representative traces of pupae expressing GCaMP6s in control (black) and wtrw (magenta) at 60 hAPF. Traces plotted as Δ F/F0.
For a complete list of genotypes used in this figure, see Table S1.
Next, we observed PSINA in mutants for water witch (wtrw), a calcium channel expressed in astrocytes known to mediate neuron-astrocyte interactions during larval stages (Ma et al., 2016). In wtrw mutants, we did not find a change in PSINA amplitude or cycle count, consistent with the notion that alterations to calcium signaling in astrocytes do not significantly affect the neuronal PSINA (Figure 4e).
We conclude that astrocyte calcium cycles can modulate PSINA but are dispensable for ongoing neuronal activity.
Discussion
Here we show that astrocytes specifically are necessary for PSINA. Ablation of other glial cell types did not have a significant effect on developmental activity, indicating either that they are dispensable for PSINA, or that the survivors are able to compensate for the lost cells. Both cortex glia and astrocytes have been shown to be able to elaborate processes to compensate for ablated cells (Stork et al., 2014; Coutinho-Budd et al., 2017). As such, it remains possible that cortex glia and ensheathing glia play a role in PSINA that is masked by subtotal ablation in the present study. For astrocytes, the extent of ablation achieved with the current protocol does not result in membrane coverage compensation (Muthukumar et al., 2014), suggesting that loss of astrocyte neuropil coverage contributes to attenuated PSINA.
Glial cell types show type-specific calcium dynamics. In astrocytes, these dynamics depend on neuronal PSINA, but a reciprocal dependence does not hold: while manipulating astrocytic calcium dynamics does modestly alter PSINA, robust neuronal activity persists even when astrocyte calcium cycles are nearly eliminated. These data suggest that astrocyte calcium dynamics are unlikely to be driving PSINA; instead, the glial calcium cycles are likely a response to neuronal activity.
Astrocytes have established roles in synapse formation, pruning, and maintenance (Perez-Catalan et al., 2021). During synaptogenesis in Drosophila, astrocytes extend numerous branches into the neuropil that closely interact with neuronal processes (Muthukumar et al., 2014; Richier et al., 2017). The developmental time-course of astrocytic morphological maturation closely tracks the accumulation of synapses in the fly brain (Muthukumar et al., 2014). Indeed, ablation of astrocytes during the second half of pupal development results in a ~25–50% reduction of synapses in different regions of the brain (Muthukumar et al., 2014). Here we show that PSINA, which takes place over the same time period, requires astrocytes, indicating that astrocytes are also necessary for synapse function during development. These data are consistent with in vitro results showing that spontaneous activity in retinal ganglion cells depend on the presence of glia (Pfrieger and Barres, 1997). And, as in the fly, loss of astrocytes in mammalian models results in fewer and functionally immature synapses (Ullian et al., 2001). In adult neuronal activity, astrocytes have multiple roles in influencing synaptic transmission and synaptic strength, ranging from modulation of the peri-synaptic extracellular matrix to neuron-glia communication via gap junctions (Perez-Catalan et al., 2021; Allen and Eroglu, 2017). How or whether these mechanisms are responsible for astrocytic control of PSINA remain open questions.
Recent work in Drosophila has established clear interplay between astrocytes and neural activity. In larvae, astrocytes modulate dopaminergic neurons in response to tyramine and octopamine via calcium-dependent signaling (Ma et al., 2016). Additionally, astrocytes are required to close a critical period for a motor circuit plasticity occurring during the first hours of larval life (Ackerman et al., 2021). In the adult antennal lobe, astrocytes display calcium transients, and activation of astrocytes modulate the synaptic strength between olfactory receptor neurons and projection neurons (Liu et al., 2014).
Despite the established relation between neural activity and glia, little is known whether glia plays a role in stimulus-independent, developmental activity. A transient population of astrocyte-like supporting cells initiates developmental activity in the sensory neurons of the cochlea via release of ATP (Tritsch et al., 2007). In the retina, Müller glia display calcium transients alongside retinal waves that depend on neurotransmitter spillover (Zhang et al., 2019; Rosa et al., 2015). These observations suggest that astrocytes may play varied roles in initiating or maintaining developmental activity. Our results support two possible mechanisms by which developmental activity depends on astrocytes in Drosophila. First, given that astrocytes promote synaptic function in the adult (Perez-Catalan et al., 2021), developmental synaptic function may also rely on astrocytes. In the absence of astrocytes, synapses appear to experience either arrested or delayed development (Muthukumar et al., 2014). The attenuation of PSINA in this background could thus be due to a global loss or reduction in synaptic function. Secondly, astrocytes may be directly involved in the rhythmogenesis of PSINA, perhaps through release of neuromodulators that stimulate a central pattern generator that initiates activity. Our data suggest that such a mechanism should be independent of glial calcium cycles (Figure 4). Identifying the molecular mechanisms that link astrocyte function to developmental activity will be key to understanding the relationship between glia, neuronal activity, and synaptic development.
Materials and Methods
Experimental Model and Subject Details
Flies were reared at 18°, 25°, or 29°C on standard cornmeal/molasses medium; developmental time was matched to the 25°C standard (1x) using relative rates of 0.5x and 1.25x for 18°C and 29°C, respectively. Pupal development was staged with respect to white pre-pupa formation (0 hAPF) or head eversion (12 hAPF). GAL4/UAS (Brand and Perrimon, 1993) and LexA/LexAop (Lai and Lee, 2006) expression systems were used to drive cell-type-specific transgene expression.
2P imaging
2P imaging was performed as described previously (Akin et al., 2019).
Wide-field imaging
Pupae were staged for white pre-pupa (0 hAPF) formation or head eversion (12 hAPF) and reared at 25°C unless otherwise noted. The cuticle around the heads were removed with fine forceps and the animals were affixed to a metal plate (McMaster-Carr, CA, USA) with double-stick adhesive tape (3M, MN, USA). The metal plate was placed in a custom environmental chamber to maintain temperature and humidity. This chamber comprised: a PTC1 temperature-controlled breadboard (Thorlabs, NJ, USA) to maintain sample temperature at 18°C, 25°C, or 29°C; a set of four 35mm dishes filled with deionized water to maintain humidity; and a 150mm petri-dish lid to provide enclosure. To improve imaging quality, a large format coverslip was fitted into a rectangular opening made in the lid. This coverslip sat in the optical path, between the pupae and the objective lens, and was treated with Barbasol shaving cream (Perio, OH, USA) to prevent condensation during temperature shifts.
Images were captured using an Axio Zoom.V16 epifluorescence microscope (Zeiss, Germany) equipped with a Retiga R1 CCD Camera (QImaging, Canada) and X-Cite TURBO LED 6-Channel Light Source (Excelitas Technologies, MA, USA). Images were acquired at 0.4 Hz with a PlanNeoFluar Z 1X objective (Zeiss, Germany) with 100ms exposure time for green fluorophores and 500ms exposure time for red fluorophores. Acquisition was controlled by Slidebook 6 software (Intelligent Imaging, CO, USA). Time series were processed with Fiji 42 and analyzed using MATLAB (Mathworks, MA, USA).
Immunohistochemistry
For data in Figures 1––2, brains were dissected in cold Schneider Medium (SM, Gibco #21720–024) and fixed with 3% v/v glyoxal solution (Electron Microscopy Sciences #16525) for 30 minutes at room temperature (RT) or with 4% v/v PFA (Electron Microscopy Sciences #15710) in SM for 20 minutes at RT. Brains were then washed out of fixative into PBS (Quality Biological), solubilized in PBST (0.5% Tween-20 (Sigma #P9416) in PBS) for 1 h, and blocked in PBTN (5% Normal Donkey Serum (Jackson ImmunoResearch #017–000-121) in PBST) for 1––2 h, all at RT. Brains were sequentially incubated in primary and secondary antibodies diluted in PBTN for 24––48h at 4°C, with at least 3 washes through PBST over 2 h at RT in between and afterwards. Brains were post-fixed with 3% v/v glyoxal for 30 minutes at RT or with 4% v/v PFA in SM for 20 minutes at RT, followed by multiple washes into PBST over 10 minutes. Brains were finally transferred to Everbrite mounting medium (Biotium #23001) and mounted on to slides for imaging. For anti-GAT data in Figures 3––4, the following modifications were made to improve the penetration of the anti-GAT antibody: Brains were fixed in 4% v/v PFA, solubilized in PBSX (0.5% Triton X-100 in PBS) with 300 mM NaCl, blocked in and incubated with primary antibodies in PBXN (5% Normal Donkey Serum in PBSX) with 300 mM NaCl. Post-primary washes and secondary antibody incubations were carried out in PBSX and PBXN without additional salt. Antibody incubations were carried out in PCR tubes nutating at 4°C; brains were kept in primary for 4 days and secondary for 2 days.
Primary antibodies and dilutions used in this study were as follows: Mouse monoclonal anti-V5 (Novus Biologicals #NBP2–52703-0.2mg, 1:150), rat monoclonal anti-FLAG (DYKDDDDK) (Novus Biologicals #NBP1–06712, 1:100), mouse monoclonal anti-c-MYC (Developmental Studies Hybridoma Bank (DSHB) #9E10-concentrate, 1:100), rabbit polyclonal anti-dsRed (Clontech #632496, 1:125), chicken anti-GFP (Abcam #ab13970, 1:1000), rabbit monoclonal anti-HA (Cell Signaling Technology #3724, 1:300), rat monoclonal anti-Ncad (DSHB #DN-Ex #8-c, 1:100), rabbit anti-GAT ((Muthukumar et al., 2014), 1:5000).
Secondary antibodies and dilutions used in this study were as follows: Alexa 488 donkey polyclonal anti-chicken (Jackson ImmunoResearch # 703-545-155, 1:400), Alexa 488 donkey polyclonal anti-mouse (Jackson ImmunoResearch #715-545-151, 1:400), Alexa 568 donkey polyclonal anti-rabbit (Invitrogen #A10042, 1:400), Alexa 647 donkey polyclonal anti-rat (Jackson ImmunoResearch #712-605-153, 1:400), Alexa 647 goat polyclonal anti-rabbit (ThermoFisher # A-21244, 1:200).
Immunofluorescence images were acquired using Zeiss LSM 780 confocal microscope with 20x/0.8 air or 40x/1.2 glycerol immersion objectives (Zeiss). To ensure consistent imaging conditions for the GAT expression studies (Figures 3e–g and 4d), one control brain in each experimental set was used to establish laser power depth corrections for each channel; all brains were imaged using these settings.
Quantification and Statistical Analysis.
Analysis of 2P imaging data was carried out as previously described (Akin et al., 2019).
Analysis of pupal wide-field imaging data:
Preparation of time lapse imaging data was performed in Fiji (ImageJ). Per frame pixel averages of user-defined ROIs were used to define raw signal (F) traces from the image time series. The raw signal was baseline subtracted and the resulting net signal (ΔF) was used in subsequent analysis, performed in MATLAB. ΔF/F0 is defined as the baseline-subtracted net signal divided by the raw baseline signal.
For each time series, sweeps were defined by growing the ‘domain’ of each signal local maximum (i.e. peak) through preceding and succeeding time-points until another peak at least 75% as large as the original was reached in both directions. Ordered signal peaks were processed iteratively in this fashion, starting from the largest on down, and lesser peaks which were subsumed in the sweep domain of a larger one were removed from separate evaluation. Clusters of sweeps were used to define active phases; silent phases were defined by the span between active phases; cycles were defined as the sum of an active phase and subsequent silent phase.
Processing and quantification of confocal images.
Counting nuclei: Three-dimensional binary masks of the half-brain were manually generated for each confocal stack using Fiji (ImageJ). Labeled nuclei within these masks were segmented from confocal stacks using custom scripts written in MATLAB (Mathworks); a function critical to this task was sourced from the MathWorks File Exchange repository (Tim Jerman (2021). Jerman Enhancement Filter (https://github.com/timjerman/JermanEnhancementFilter), GitHub.).
Quantifying GAT immunofluorescence:
Fiji (ImageJ) was used to trace out masks for the CLX, PB, and EB out of the confocal stacks; the slice range containing each feature was also recorded during this process. The resulting 3D sub-volumes were extracted from the anti-GAT channel of each stack and used to create a more refined segmentation of the features using a custom-written MATLAB script. The ratio of the average pixel intensities in the anti-GAT and anti-Ncad channels was calculated for each slice of each segmented feature. This ratio was found to be comparable through the depth of each feature and between brains for each experimental condition, indicating consistent staining and imaging conditions. The single value ratios for each feature reported in the figures are averaged from the slices in the center half-thickness of each feature (i.e. 25––75% of full z-range). Features near areas damaged during dissection were likely surface exposed during primary antibody incubation and were stained much more strongly by anti-GAT, similar to typically superficial regions of the brain. Such features were not included in the analysis. The two calyces and two sides of the PB from each brain were analyzed independently.
Statistical analysis.
Statistical analysis was performed on RStudio or MATLAB. We used either Welch’s t-test following the Shapiro-Wilk test for normality for comparisons between two groups, or Tukey’s post-hoc test following ANOVA to assess statistical significance of differences between groups. Population averages are given as mean ± standard deviation (SD).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
V. Acknowledgments:
We thank S. Lawrence Zipursky and members of the Zipursky lab for valuable feedback and advice. For generously sharing reagents, we thank Marc R. Freeman and Samuel A. LoCascio. This work was supported by NIH grants T32GM008042 and F30EY029952 to B.T.B, and R01NS123376 to OA.
Footnotes
Conflict of Interest statement: The authors declare no conflicts of interest.
Viii. References
- Ackerman SD, Perez-Catalan NA, Freeman MR, & Doe CQ (2021). Astrocytes close a motor circuit critical period. Nature, 592(7854), 414––420. 10.1038/s41586-021-03441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin O, Bajar BT, Keles MF, Frye MA, & Zipursky SL (2019). Cell-type-Specific Patterned Stimulus-Independent Neuronal Activity in the Drosophila Visual System during Synapse Formation. Neuron, 101(5), 894––904.e5. 10.1016/j.neuron.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin O, & Zipursky SL (2016). Frazzled promotes growth cone attachment at the source of a Netrin gradient in the. Elife, 5, e20762. 10.7554/eLife.20762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, & Eroglu C. (2017). Cell Biology of Astrocyte-Synapse Interactions. Neuron, 96(3), 697––708. 10.1016/j.neuron.2017.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Carver LA, & Posakony JW (2000). GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques, 29(4), 726, 728, 730, 732. 10.2144/00294bm10 [DOI] [PubMed] [Google Scholar]
- Barres BA (2008). The mystery and magic of glia: a perspective on their roles in health and disease. Neuron, 60(3), 430––440. 10.1016/j.neuron.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Blankenship AG, & Feller MB (2010). Mechanisms underlying spontaneous patterned activity in developing neural circuits. Nat Rev Neurosci, 11(1), 18––29. 10.1038/nrn2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, & Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118(2), 401––415. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, … Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499(7458), 295––300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, & Salecker I. (2007). Glial cell development and function in the Drosophila visual system. Neuron Glia Biol, 3(1), 17––25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, & Freeman MR (2013). Cell biology in neuroscience: Architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol, 203(3), 395––405. 10.1083/jcb.201306099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Budd JC, Sheehan AE, & Freeman MR (2017). The secreted neurotrophin Spätzle 3 promotes glial morphogenesis and supports neuronal survival and function. Genes Dev, 31(20), 2023––2038. 10.1101/gad.305888.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, … Kim DS (2016). Sensitive red protein calcium indicators for imaging neural activity. Elife, 5, e12727. 10.7554/eLife.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer MC, Jung C, Batelli S, Rubin GM, & Gaul U. (2017). The glia of the adult Drosophila nervous system. Glia, 65(4), 606––638. 10.1002/glia.23115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmangaliyev YZ, Yoo J, Valdes-Aleman J, Sanfilippo P, & Zipursky SL (2020). Transcriptional Programs of Circuit Assembly in the Drosophila Visual System. Neuron, 108(6), 1045––1057.e6. 10.1016/j.neuron.2020.10.006 [DOI] [PubMed] [Google Scholar]
- Lai SL, & Lee T. (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci, 9(5), 703––709. 10.1038/nn1681 [DOI] [PubMed] [Google Scholar]
- Li HH, Kroll JR, Lennox SM, Ogundeyi O, Jeter J, Depasquale G, & Truman JW (2014). A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep, 8(3), 897––908. 10.1016/j.celrep.2014.06.065 [DOI] [PubMed] [Google Scholar]
- Liu H, Zhou B, Yan W, Lei Z, Zhao X, Zhang K, & Guo A. (2014). Astrocyte-like glial cells physiologically regulate olfactory processing through the modification of ORN-PN synaptic strength in Drosophila. Eur J Neurosci, 40(5), 2744––2754. 10.1111/ejn.12646 [DOI] [PubMed] [Google Scholar]
- Ma Z, Stork T, Bergles DE, & Freeman MR (2016). Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature, 539(7629), 428––432. 10.1038/nature20145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, & Davis RL (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science, 302(5651), 1765––1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Muthukumar AK, Stork T, & Freeman MR (2014). Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat Neurosci, 17(10), 1340––1350. 10.1038/nn.3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Catalan NA, Doe CQ, & Ackerman SD (2021). The role of astrocyte-mediated plasticity in neural circuit development and function. Neural Dev, 16(1), 1. 10.1186/s13064-020-00151-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger FW, & Barres BA (1997). Synaptic efficacy enhanced by glial cells in vitro. Science, 277(5332), 1684––1687. 10.1126/science.277.5332.1684 [DOI] [PubMed] [Google Scholar]
- Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, & Griffith LC (2009). Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol, 101(6), 3075––3088. 10.1152/jn.00071.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier B, Vijandi CM, Mackensen S, & Salecker I. (2017). Lapsyn controls branch extension and positioning of astrocyte-like glia in the Drosophila optic lobe. Nat Commun, 8(1), 317. 10.1038/s41467-017-00384-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa JM, Bos R, Sack GS, Fortuny C, Agarwal A, Bergles DE, … Feller MB (2015). Neuron-glia signaling in developing retina mediated by neurotransmitter spillover. Elife, 4, e09590. 10.7554/eLife.09590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Jackson-Weaver O, Huckstepp RT, O’Dell TJ, & Khakh BS (2013). TRPA1 channels are regulators of astrocyte basal calcium levels and long-term potentiation via constitutive D-serine release. J Neurosci, 33(24), 10143––10153. 10.1523/JNEUROSCI.5779-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, & Freeman MR (2014). Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron, 83(2), 388––403. 10.1016/j.neuron.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Yi E, Gale JE, Glowatzki E, & Bergles DE (2007). The origin of spontaneous activity in the developing auditory system. Nature, 450(7166), 50––55. 10.1038/nature06233 [DOI] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, & Barres BA (2001). Control of synapse number by glia. Science, 291(5504), 657––661. 10.1126/science.291.5504.657 [DOI] [PubMed] [Google Scholar]
- Yildirim K, Petri J, Kottmeier R, & Klämbt C. (2019). Drosophila glia: Few cell types and many conserved functions. Glia, 67(1), 5––26. 10.1002/glia.23459 [DOI] [PubMed] [Google Scholar]
- Zhang RW, Du WJ, Prober DA, & Du JL (2019). Müller Glial Cells Participate in Retinal Waves via Glutamate Transporters and AMPA Receptors. Cell Rep, 27(10), 2871––2880.e2. 10.1016/j.celrep.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Ormerod KG, & Littleton JT (2017). Astrocyte Ca2+ influx negatively regulates neuronal activity. eNeuro, 4(2), ENEURO.0340–16.2017. 10.1523/ENEURO.0340-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.