Aminoglycosides are among the most commonly used broad-spectrum antibiotics in the anti-infective armamentarium. The vast majority of aminoglycosides are bactericidal, they have predictable pharmacokinetics, and they often act in synergy with other antibiotics, properties that make them valuable as anti-infectives. Furthermore, despite the potential for renal toxicity, ototoxicity, and bacterial resistance, several members of this family of antibiotics have enjoyed clinical use for several decades.
Aminoglycosides are multifunctional hydrophilic sugars that possess several amino and hydroxy functionalities. The amine moieties are mostly protonated in biological media; hence, these antibiotics can be considered polycationic species for the purpose of understanding their biological interactions. Since they are polycationic, they show a binding affinity for nucleic acids. Specifically, aminoglycosides possess high affinities for certain portions of RNAs, especially the prokaryotic rRNA (12–14, 38, 39). In addition, aminoglycosides bind to the hammerhead ribozyme (45, 49), tRNAPhe (26), the Rev response element (RRE) transcriptional activation region in human immunodeficiency virus (HIV) (8, 18), the ribozyme from hepatitis delta virus (41), and group I self-splicing introns (6, 51, 52).
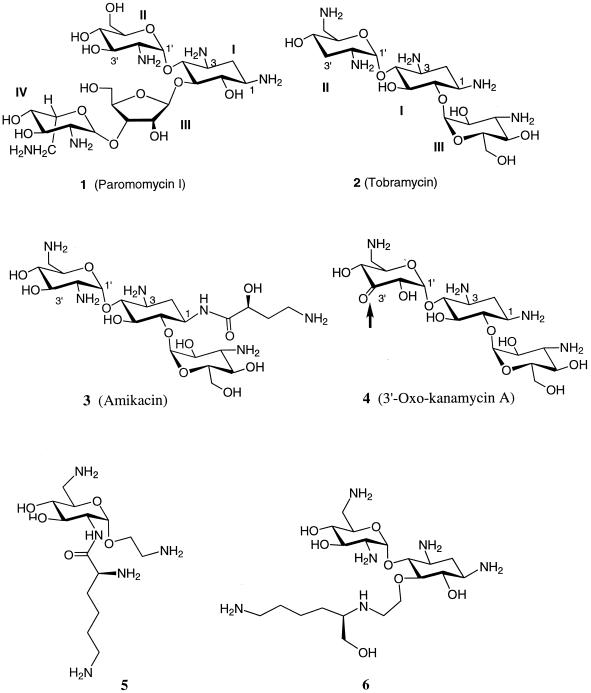
There are only a handful of structural studies on the interactions of aminoglycoside antibiotics with specific RNA sequences. One such study aimed at understanding the molecular interactions of paromomycin (Fig. 1) with the aminoacyl site (A-site)-decoding region of rRNA (12–14, 39). Another dealt with the interactions of tobramycin (Fig. 1) with an RNA aptamer (23). In 1998, Yoshizawa et al. (57) elucidated the structure of gentamicin Cla bound to an A-site RNA complex. These studies used nuclear magnetic resonance techniques to elucidate the three-dimensional structures of the complexes of RNA and aminoglycosides. In addition to these structural investigations, several footprinting studies and efforts with semisynthetic aminoglycosides have been reported for RNA interactions (1, 7, 8, 30, 48). In this minireview, structural determinants that are responsible for the specific recognition of certain RNA folds by aminoglycosides, which have implications for the mechanisms of action of these antibiotics, will be examined. Recent determination of the low-resolution X-ray structure of the 70S ribosome from Thermus thermophilus has shed light on the global features of various domains in rRNAs and their interactions with mRNA and tRNA (5). In light of the structural information presented above, mechanistic features of aminoglycoside binding to rRNA will be discussed as well.
FIG. 1.
Structures of aminoglycosides.
The topics of the biological activities, resistance, and toxicities of aminoglycoside antibiotics have been reviewed (4, 10, 31, 32, 44). The present report addresses the complementary subjects of the molecular interactions responsible for the activities of this class of antibiotics, the structural factors responsible for the specificity, and the efforts made to design novel derivatives of aminoglycoside antibiotics.
STRUCTURAL BASES FOR MECHANISM OF ACTION
The 16S rRNA from Escherichia coli is well studied among the rRNA subunits, and in particular, the interactions of various aminoglycoside antibiotics with the 16S rRNA and their effects on the process of translation of mRNA into polypeptide have been scrutinized (35). Similar rRNA structures exist in other organisms, such as yeast and Tetrahymena (33). Treatment of rRNA with an aminoglycoside protects several nucleic bases in rRNA from chemical modification, implying that these molecules possess high affinities for certain sites in rRNA. This mode of binding was likened by Noller (35) to that of enzyme inhibitors, which usually bind to the active sites of enzymes and interfere with their activities. Different classes of aminoglycoside antibiotics bind to different sites on the rRNA, depending on the structural complementarity between the two. For example, neomycin, paromomycin (Fig. 1), gentamicin, and kanamycin are believed to bind to the A-site on the 16S rRNA in E. coli in a similar fashion and were shown to protect bases A1408 and G1494 in chemical footprinting experiments (Fig. 2) (33). Four bases, A1408, A1492, A1493, and G1494, in the rRNA A-site interact with tRNA, although with different affinities. The binding of the aforementioned aminoglycosides to the A-site in the decoding region (i.e., the site of codon and anticodon recognition) interferes with the accurate recognition of cognate tRNA by rRNA during translation (35). These interactions are also thought to interfere with the translocation of tRNA from the A-site to the peptidyl-tRNA site (P-site).
FIG. 2.
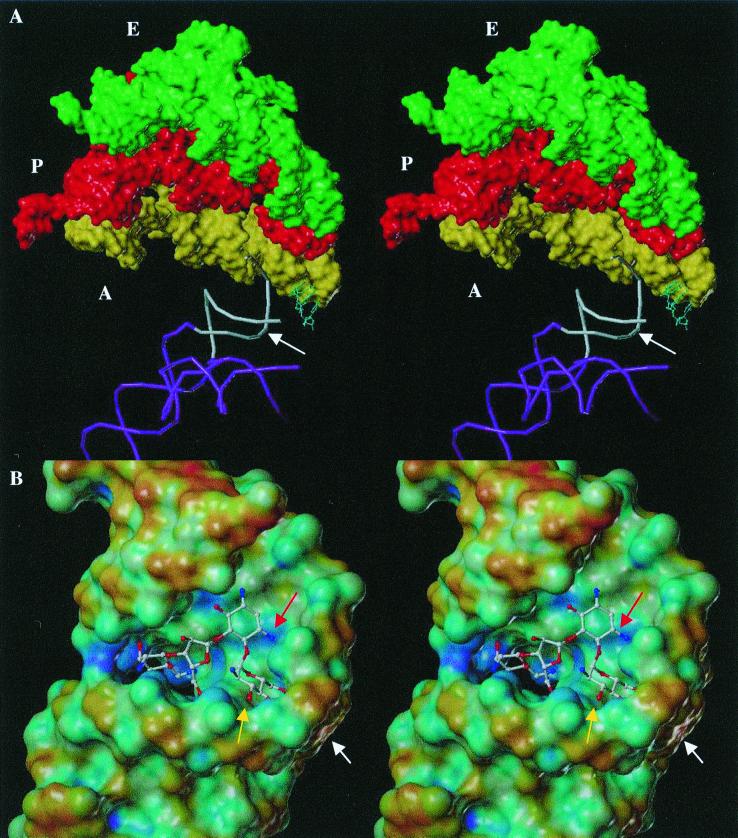
(A) Stereo view of the partial structure of the 70S rRNA complexed with three tRNA molecules (Protein Databank code, 486D [available at http://www.rcsb.rutgers.edu]). The A-site region on the 16S rRNA is shown in white, where aminoacyl tRNA ‘A‘ (in yellow) is bound near the A-site of rRNA. Two other tRNAs, peptidyl and exit, “P” (in red) and “E” (in green), respectively, are shown as well. The backbone of the penultimate stem of the 16S rRNA molecule and the 900-loop are shown in violet. The binding site of paromomycin at the A-site is indicated by the white arrow. (B) Stereo view of the solution structure of RNA A-site template bound by paromomycin, which approximately corresponds to the A-site region on the 16S rRNA in white in panel A. The Connolly surface of the A-site RNA is rendered according to the electrostatic potential by using the MOLCAD program (Tripos, Inc., St. Louis, Mo.), and the aminoglycoside is shown in a ball-and-stick representation. The most electronegative potential is rendered in blue, and the most electropositive potential is rendered in red on the surface; all other colors show the potentials between blue and red. The arrow in white shows the kink generated by A1492, which does not have a base-pairing partner. The arrow in yellow shows the pocket generated by the A1408 · A1493 base pair and A1492. The arrow in red shows the location of the 3-amine on ring II, the site for acetylation by AAC(3).
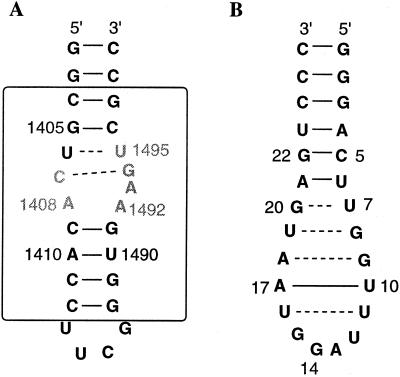
Puglisi and coworkers (12–14, 39) recently provided structural evidence on the mode of interactions of paromomycin, a representative aminoglycoside of the neomycin class, with a 27-nucleotide RNA template that was designed to mimic the A-site region of the 16S rRNA in E. coli (Fig. 3A). The design of the RNA template was based on previous knowledge that paromomycin interacts with the C1407 · G1494 base pair, A1408, A1493, and U1495 and that these bases are absolutely necessary for high-affinity binding (39) (shown in gray in Fig. 3A). Additional structural features, such as the pocket created by the asymmetry in the internal loop region due to the presence of A1492 and the base pairing of C1409 · G1491 at the lower stem region, are also important. These structural characteristics collectively create a pocket that is optimal for the binding of paromomycin (see below).
FIG. 3.
(A) Model of the A-site RNA template used to study the interactions of paromomycin. The box represents the portion of the rRNA that is homologous to the A-site. (B) RNA aptamer template used to study the interactions of tobramycin.
In the native A-site RNA template, the stem has base-pairing interactions at U1406 · U1495 (noncanonical) and C1407 · G1494 (Fig 3A). Upon binding of paromomycin, the distinct structure formed by A1408, A1492, and A1493 is stabilized (Fig. 2B) and bases A1408 and A1493 form a noncanonical base pair (12, 39). Nucleotide A1492, which does not have any base-pairing interactions, creates a kink in the RNA structure, and the combined effects of A1492 and the A1408 · A1493 base pair creates a bulge in the A-site where paromomycin binds and further extends the angle of the kink (Fig. 2B). The functional groups on paromomycin, such as the hydroxyl and amino groups, participate in specific interactions with the RNA molecule (see below).
The pocket created by A1492 and the A1408 · A1493 base pair is occupied by ring II of paromomycin, and this ring stacks above base G1491 (shown by a yellow arrow in Fig. 2B) (12). Ring I of paromomycin makes specific contacts with the “universally” conserved base pairs U1406 · U1495 and C1407 · G1494 in the rRNA. It is noteworthy that ring I is absolutely necessary for specific binding of aminoglycoside antibiotics to rRNA. Rings III and IV of paromomycin extend these interactions further into the major groove of rRNA. The amino and hydroxyl moieties mostly contribute to the nonspecific interactions of paromomycin with rRNA; thus, they are not sequence-dependent interactions. Another important point is that the base pair C1409 · G1491 provides the seat for binding of aminoglycoside in the pocket, and a mismatch base pair in this position results in the loss of binding (12). In general, aminoglycosides that share structural features with paromomycin bind to rRNA similarly (13). However, different aminoglycoside antibiotics appear to bind to the same binding site in more than one conformation (28). In essence, the conformation of the aminoglycoside that does bind to RNA must satisfy the electronic and steric constraints of the binding site. In a different study on the complex of gentamicin Cla and the A-site RNA template, rings I and II of gentamicin Cla (which are similar to those of paromomycin) exhibited binding interactions similar to those in the complex of paromomycin and the A-site RNA template (57). However, ring III in gentamicin Cla interacts with base pairs U1406 · U1495 and G1405 · C1496 in the upper stem region (Fig. 3A). From these observations, Puglisi and coworkers (57) proposed that all the aminoglycosides that target the A-site of the 16S rRNA bind in a common manner, similar to rings I and II in paromomycin and gentamicin.
Although the overall structure of rRNA is conserved among all species in an evolutionary sense, there are differences that make binding of aminoglycosides more specific—by at least a 10-fold higher affinity—to the rRNA of prokaryotes than to that of eukaryotes (19, 35, 38). This is not a large difference in binding affinity and may in part explain the toxic effects of these antibiotics in mammalian systems. Eukaryotic rRNA contains a guanine in place of A1408, resulting in a G1408 · A1493 base pair. In addition, the matched base pair at C1409 · G1491 does not exist in eukaryotes. These differences collectively result in lowered affinities of aminoglycosides for the eukaryotic rRNA (12, 19, 35, 38). Having outlined these differences, binding of aminoglycosides to the A-site of rRNA in prokaryotes alters the conformation of the A-site and affects the specific interactions of mRNA and tRNA at this site, resulting in erratic codon-anticodon interactions. There is a paucity of structural information on the specifics of these interactions at the ribosomal level to date (see below), but the clear and ultimate consequence is disruption of the translation process.
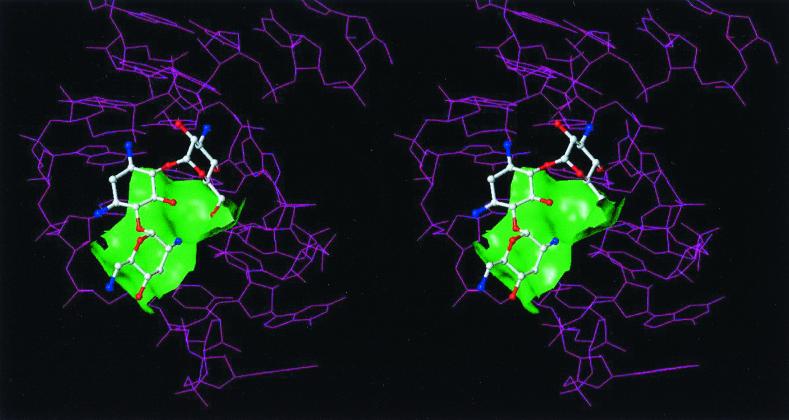
Another structural study was performed on binding of tobramycin (Fig. 1) to an RNA aptamer (23). The RNA aptamer that was used in this study was a 26-nucleotide stem-loop RNA (Fig. 3B). There are four mismatch pairs, U7 · G20, G8 · U19, G9 · A18, and U11 · U16, in this RNA aptamer that are part of the zippered hairpin loop. Tobramycin binds in this groove partially encapsulated by the surface of the deep groove and the guanine base of the residue G15 (Fig. 4). In this complex, ring I of tobramycin sits on the floor of the deep groove. One of the amino groups on ring II of tobramycin interacts with the phosphate backbone in the deep groove, and the other amino group is exposed to the solvent. Ring III is positioned in the center of the deep groove, with hydroxyl groups directed toward the floor of the groove. The conformation of the RNA aptamer described above was suggested to be similar to those of the hairpin loops in tRNA and rRNA (23).
FIG. 4.
Stereo view of the complex of tobramycin bound to the RNA aptamer. The green Connolly surface represents a portion of the aminoglycoside binding site.
The 7.5-Å-resolution X-ray structure of the functional complex of T. thermophilus 70S rRNA containing tRNA and mRNA is helpful to put the above discussion in perspective (5). From this structure, one could envision how well the model studies with smaller RNA templates such as the tobramycin-RNA aptamer and the A-site–RNA with paromomycin (see above) would fit into the complete structure of rRNA. The A-site in the 16S rRNA subunit of the 70S rRNA is seen near the interface of tRNA and the 50S rRNA subunit, in the proximity of the codon-anticodon pair (Fig. 2A). Upon comparison of the solution structure of the A-site RNA template to the A-site in the X-ray structure of 70S rRNA, the X-ray structure appeared to be closely related to the paromomycin-bound RNA template but not to the native solution structure of the A-site RNA template (5). This is intriguing and suggests that perhaps in the functional form the bulge or kink near bases A1492 and A1493 always exists in the 70S rRNA. If this were true, it implies that the binding pocket for paromomycin is already in existence when the 70S rRNA becomes functional; hence, it is predisposed for inhibition by paromomycin. This contradicts the assertion that binding of paromomycin increases the kink angle at the binding site. Evidence in support of this idea comes from a recent study that suggested that the affinities of different aminoglycosides for the A-site RNA template are different, and the ability of these antibiotics to inhibit protein synthesis in vitro varies as well (15). Gentamicin and several other related antibiotics interact with the A-site RNA with dissociation constants (Kd) in the micromolar range, but they inhibited an in vitro translation process, with 50% inhibitory concentrations in the nanomolar range (15). The latter finding was inferred as the result of aminoglycoside binding to the intact rRNA in the decoding region (A-site), and the difference in binding to intact rRNA versus that to the A-site template RNA may be due to the differences in the conformations of these two RNAs, as discussed above.
The A-site makes weak contacts with the mRNA and tRNA, implying that this region plays a role in recognition of appropriate tRNA via subtle changes in the free energy (5). The binding of aminoglycoside near this site may affect the delicate process of interactions between codon and anticodon. It was also proposed that the presence of an aminoglycoside stabilizes the complex of mRNA and tRNA at the A-site, which in turn affects the process of translation (5). It is difficult to surmise all the effects of aminoglycosides on the rRNA structure, and further structural studies with the aminoglycosides bound to the complexes, such as the 70S rRNA, will be helpful in elucidating and understanding the subtle changes that lead to the antibiotic actions of aminoglycosides.
A number of investigations have used synthetic probes to understand the interactions between RNA templates and aminoglycosides. It has been suggested that aminoglycosides bind to more than one target site in the ribozyme (6, 30). Recently, several aminoglycoside antibiotics such as neomycin B, tobramycin, and kanamycin A have been dimerized either symmetrically or asymmetrically by using a “tether,” and their binding affinities were compared to those of the monomeric parent aminoglycosides (30). It was suggested that if there were multiple binding sites on the RNA, the dimerized aminoglycosides should bind with a higher affinity than the parent antibiotic, provided that multiple binding sites are accessible. It was indeed observed that the dimerized aminoglycosides bind to the Tetrahymena ribozyme 20- to 1,200-fold better than the parent aminoglycosides. One explanation for the higher binding affinity could be the increased number of positively charged amino groups on the dimerized aminoglycoside, but this effect seems to be synergistic with the entropic advantage gained by dimerization (30). It also indicated the presence of multiple high-affinity binding sites for aminoglycoside antibiotics in an RNA molecule. Another study attempted to exploit the RNA binding properties of paromomycin and the intercalating behaviors of certain compounds such as pyrene and thiazole orange (48). This strategy envisioned aminoglycosides as a means for the delivery of intercalating agents to the RNA. The conjugate of paromomycin with thiazole orange or pyrene showed better binding properties to the 27-nucleotide A-site RNA template. In fact, the dissociation constant for the paromomycin-thiazole orange conjugate was measured at 46 nM, which was reported as the highest affinity that the rRNA A-site has shown for any ligand.
The structural requirements for RNA binding by aminoglycosides indicated that a bulge in the RNA sequence is necessary to allow binding of aminoglycosides (7). By using a specific stem-loop derivative of the RNA aptamer, a series of chemical interference, chemical modification, and mutation studies was performed to understand the structural requirements for binding of tobramycin to the RNA aptamer. This aminoglycoside appeared to interact mainly with the nucleic bases in the RNA aptamer but not with the phosphate backbone. The presence of a bulge, however, was proposed to be important for the high-affinity binding of tobramycin in a stoichiometric ratio, and it was concluded that a bulge creates a cavity for interactions of the aminoglycoside and the nucleic base (7). This analogy can be applied to other RNA sites such as the hammerhead region and the A-site, where a cavity is present due to the noncanonical base-pairing or loops or bulges that create a suitable site for the aminoglycosides to interact with the anionic phosphate groups and the nucleic bases. Along these lines, Westhoff and colleagues (49) put forward a proposal that the interaction of aminoglycosides with RNA is likely to be shape specific rather than sequence specific.
Consistent with this concept, the electrostatic fields in the RNA folds were deemed as the guiding force for binding (see below). Hermann and Westhoff (20) were able to identify the docking confirmations of several aminoglycoside antibiotics in various RNA templates, such as tobramycin-RNA aptamers and the A-site region in the 16S RNA, for which structural information was available. On the basis of those observations, the binding mode of aminoglycosides to the trans-activating response element region in HIV was predicted (20). In another study on RRE in HIV, the binding region for the Rev protein, Cho and Rando (8) investigated the role of a single-base bulge and a cavity for binding of aminoglycosides. Consistent with previous hypotheses, it was deduced that grooves in the nonduplex regions of RNA are important for high-affinity binding of aminoglycosides to RNA. In this case, the single-base bulge did not affect binding, but the cavity, a G-rich region consisting of two noncanonical base pairs and one single bulged U, has a high affinity for aminoglycosides. Tampering with the cavity decreased the affinity of the RRE RNA for aminoglycosides, thus indicating that the noncanonical base pair-containing bulges are the primary aminoglycoside-binding sites in this RNA template (8).
RESISTANCE TO AMINOGLYCOSIDES AND STRATEGIES TO COUNTER IT
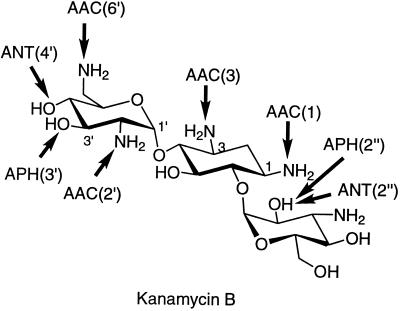
Even though the binding site for aminoglycosides is in rRNA, resistance to aminoglycosides is not manifested by alteration of this target in general. This is in part due to the fact that the function of the rRNA is central in the protein biosynthetic process and that this function is so well preserved across genera that it cannot be impaired by the possibility of such structural alteration. In addition, all organisms have multiple copies of the genes that encode rRNA. In order to generate an rRNA resistant to a certain antibiotic that binds to it, all these genes will have to be mutated, and the probability of the occurrence of such an event is virtually nonexistent. Moreover, it is easier for an organism either to modify the rRNA target posttranslationally or to produce resistance enzymes (see below). On the other hand, there are some reports of mutations in the ribosomal proteins that confer resistance to certain antibiotics. For example, mutations in the ribosomal protein L10e lead to resistance to sordarin, a tetracyclic diterpene glycoside antifungal agent that selectively inhibits fungal protein biosynthesis (24). Mutations in another ribosomal protein, L22, confer resistance to erythromycin, and recently, the X-ray crystal structure of this protein from T. thermophilus was determined (50). Mutations that confer resistance to erythromycin are on a beta-hairpin loop on L22, and it was postulated that this region may be in proximity to the erythromycin-binding site near the peptidyl transfer center (50). However, these types of mechanisms for resistance are not seen for aminoglycosides. It is also observed that certain mutations in the highly conserved 530 stem-loop region in rRNA result in “streptomycin dependence,” perturbing (actually reducing) the translational error frequency (22, 37). However, there is no three-dimensional structural information on this observation for us to be able to comment on it further. Resistance to aminoglycosides is widely reported, but the preponderance of the cases of resistance to these agents has not been as overwhelming as that of the cases of resistance to β-lactam antibiotics. This may be due in part to the more frequent use of β-lactams in the clinic, but it may also be due to the differences in the mechanisms of dissemination of the resistance determinants (34, 55). Whereas methylation of 16S rRNA in aminoglycoside-producing organisms gives high-level resistance to the actions of these antibiotics (3, 46), the most common mechanism for clinical resistance to aminoglycosides is their structural modification by specific enzymes expressed in resistant organisms. The sites of modifications in kanamycin B by various enzymes are shown schematically in Fig. 5. The binding of the modified aminoglycoside antibiotics to their target sites is compromised. There are three classes of these enzymes: aminoglycoside phosphotransferases (APHs), aminoglycoside nucleotidyltransferases (ANTs), and aminoglycoside acetyltransferases (AACs). Within each class, there are enzymes with different regiospecificities for aminoglycoside modifications: there are four nucleotidyltransferases [ANT(6), ANT(4′), ANT(3"), and ANT(2")], seven phosphotransferases [APH(3′), APH(2"), APH(3"), APH(6), APH(9), APH(4), and APH(7")], and four acetyltransferases [AAC(2′), AAC(6′), AAC(1), and AAC(3)]. There also exists a bifunctional enzyme, AAC(6′)–APH(2"), that can acetylate and phosphorylate its substrates sequentially (2, 9). The issue of the origins of these resistance enzymes has been discussed in recent reviews and will not be repeated here in the interest of brevity (32, 55).
FIG. 5.
Sites of modification on kanamycin B by various aminoglycoside-modifying enzymes. The arrows point to the sites of modification by the specific enzymes, namely, acetyltransferases, phosphotransferases, and nucleotidyltransferases.
Crystal structures for four aminoglycoside-modifying enzymes have been reported (21, 36, 42, 53, 56). These are for ANT(4′), AAC(3), AAC(6′), and APH(3′) type IIIa [APH(3′)-IIIa]. It is interesting that the same forces that drove the evolution of aminoglycoside biosynthesis for binding to the ribosomal sites also would appear to have driven evolution of the resistance enzymes. Namely, electrostatic interactions appear to be quite significant. It was noted that the positively charged aminoglycoside is attracted to APH(3′)-IIIa, since the active site of the enzyme provides favorable interactions due to the excessive negative charge distribution in this region of the surface (21, 55). Once the complex forms between the two (i.e., the substrate and the enzyme), this electrostatic attraction on the substrate is complemented by specific electrostatic interactions between the active site and the aminoglycoside in APH(3′)-Ia, -IIa, and -IIIa, as documented by studies of specific deaminated aminoglycosides (29, 40). In the same sense that the amines in the structures of aminoglycosides are important for binding to rRNA, they would appear to be significant—perhaps indispensable—for recognition of the drugs by the resistance enzymes.
The structural modifications of aminoglycosides result in a severe reduction of the ability of the modified antibiotic to bind to the target RNA due to unfavorable steric and/or electrostatic interactions. For example, acetylation of the amine group at position 3 on paromomycin (Fig. 1) results in unfavorable steric clashes in the A-site RNA, as can be surmised from the structure of the complex between paromomycin and A-site RNA (shown by a red arrow in Fig. 2B). In addition, acetylation in this case would interfere with the necessary electrostatic attraction between the RNA and the aminoglycoside as well. Another example is phosphorylation of aminoglycoside at the 3′ position (corresponding to the 3′ site in ring II of paromomycin [Fig. 1]) due to the action of APH(3′). Considering that kanamycin A [a substrate for APH(3′) enzymes] binds in a fashion similar to that for paromomycin at the A-site of rRNA, ring II should fit into the pocket created by A1492. However, the phosphate group at the 3′ position would interfere with this binding due to the repulsive electrostatic and steric interactions between the 3′ phosphate and the phosphate backbone of the rRNA, resulting in poor binding of the 3′-phosphorylated aminoglycosides at the A-site of rRNA.
Gentamicin C is susceptible to at least five or six modifying enzymes, and the same pattern can be seen for the semisynthetic antibiotic tobramycin. A major breakthrough was the preparation of amikacin (Fig. 1), a kanamycin A derivative with the amino group at position 1 acylated by 4-amino-2-hydroxybutyrate. The presence of the aminohydroxybutyryl group in general prevents the enzymatic modification of amikacin at multiple positions. This antibiotic undergoes acetylation by different types of AAC(6′), as well as by a few other aminoglycoside-modifying enzymes such as AAC(2′), APH(2"), and ANT(2"). The activity of the parent antibiotic is not compromised by this synthetic alteration because the aminohydroxybutyryl group does not interfere with binding to the A-site of rRNA. The aminohydroxybutyryl functionality at position 1 of amikacin appears to fit well into the extended cleft, as seen in the model generated from the nuclear magnetic resonance structure of the A-site RNA; the amino group in this moiety may also enhance binding of amikacin to RNA through electrostatic interactions (unpublished results from our group). Consequently, greater than 80% of the gentamicin-resistant members of Enterobacteriaceae and 25 to 85% of Pseudomonas aeruginosa-resistant strains are sensitive to amikacin (25, 27). A recent surveillance study conducted in North America indicated that amikacin is still an active antimicrobial agent against P. aeruginosa (11). Similar studies in Europe showed that amikacin exhibited activity against gram-negative bacilli superior to those of gentamicin and tobramycin (43).
To date, all attempts to make semisynthetic aminoglycosides have focused on the synthesis of derivatives that circumvent resistance enzymes. Examples of such antibiotics are dibekacin and tobramycin, which lack the 3′-hydroxyl group and are therefore not substrates for APH(3′) compounds. Other examples are isepamycin and amikacin, both of which have an acylated N-1 group, which makes them poorer substrates for a number of the modifying enzymes. Recently, 3′-oxo-kanamycin A (Fig. 1) was designed and prepared (16). This prototypic aminoglycoside serves as a good substrate for the common APH(3′). However, the product of phosphorylation is inherently unstable, and it releases the phosphoryl group to the solution as inorganic phosphate, regenerating the original antibiotic. Such an antibiotic renders these bacterial resistance enzymes obsolete for the resistant bacterium (16). Further applications such as this await future experimentation.
As described earlier, recent investigations have revealed that rings I and II of the neomycin class of aminoglycosides are essential for RNA binding and that they are sufficient to direct the binding of aminoglycosides to the unique binding pocket in the RNA A-site (1, 12, 13, 17, 41). Wong and coworkers (1, 17) suggested that the 1,3-hydroxylamine moiety present in almost all aminoglycoside antibiotics may be an important recognition motif for RNA binding. They investigated small molecules that recognize RNA with the 1,3-hydroxylamine motif as a core (1, 17). To test this idea, a series of aminoglycoside derivatives bearing either one or two of these recognition motifs for RNA interaction was synthesized (1, 15). Some of these new compounds showed antibiotic activities, and a few were found to be effective in binding to the rRNA A-site sequence (1, 15, 54). Although these derivatives (such as structures 5 and 6 in Fig. 1) bind to the truncated model of the rRNA A-site in the submicromolar range, there is only a weak correlation between RNA binding affinity and antibacterial activity. For instance, despite stronger binding affinities than neamine, some of these compounds were weaker antibiotics. On the contrary, compounds such as gentamicin and ribostamycin, even though they contain a neamine-like core moiety, exhibit good antibiotic activity but are poor A-site binders (54). A possible explanation is that in vitro binding to this RNA construct does not precisely mimic the in vivo binding event in the ribosome in every respect.
In parallel to these investigations, Tok and Rando (47) described simple 1,3(2)-aminoalcohol-containing molecules with potencies similar to or greater than that of paromomycin. The structural simplicity of these 1,2- and 1,3-aminoalcohols and their potential as effective substituents for structurally complicated aminoglycoside antibiotics could stimulate further interest in the design and rapid syntheses of a series of new and potent antibacterial agents.
CONCLUSIONS
The discovery of the first aminoglycoside in 1944 stimulated considerable interest in these antibiotics. A number of subclasses of these antibiotics have been identified, and the structures of the parent drugs have been further elaborated by chemists to generate potent derivatives with expanded spectra of antibacterial activity. Several of these semisynthetic aminoglycosides have also been less prone to structural alterations by the resistance enzymes. The current knowledge of the activities of these antibiotics, their pharmacokinetics, their structural diversity, and chemistry for their preparation is considerable indeed. There has been a perception that investigations of aminoglycosides have reached maturity and that the prospects for novel insights are perhaps remote. However, structural and mechanistic information on the target(s) of these antibiotics and their respective resistance enzymes has only begun to emerge in the past few years. This information should stimulate novel developments in the de novo design of molecules that bind to the ribosomal target site or molecules that would serve as inhibitors for the resistance enzymes in the near future. The structural and mechanistic investigations reviewed here represent the starting point for future developments in this important area.
ACKNOWLEDGMENT
We are indebted to Philip Cunningham for insightful critique of the manuscript.
REFERENCES
- 1.Alper P B, Hendrix M, Sears P, Wong C-H. Probing the specificity of aminoglycoside-ribosomal RNA interactions with designed synthetic analogs. J Am Chem Soc. 1998;120:1965–1978. [Google Scholar]
- 2.Azucena E, Grapsas I, Mobashery S. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2"-phosphorylation and acetyl-CoA-dependent 6′-acetylation of aminoglycosides. J Am Chem Soc. 1997;119:2317–2318. [Google Scholar]
- 3.Beauclerk A A, Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987;193:661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- 4.Brummett R, Fox K. Aminoglycoside-induced hearing loss in humans. Antimicrob Agents Chemother. 1989;33:797–800. doi: 10.1128/aac.33.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cate J H, Yusupov M M, Yusupova G Z, Earnest T E, Noller H F. X-ray crystal structure of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 6.Cech T R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 7.Cho J, Hamasaki K, Rando R R. The binding site of a specific aminoglycoside binding RNA molecule. Biochemistry. 1998;37:4985–4992. doi: 10.1021/bi972757h. [DOI] [PubMed] [Google Scholar]
- 8.Cho J, Rando R R. Specificity in the binding of aminoglycosides to HIV-RRE RNA. Biochemistry. 1999;38:8548–8554. doi: 10.1021/bi990273a. [DOI] [PubMed] [Google Scholar]
- 9.Daigle D M, Hughes D W, Wright G D. Prodigious substrate specificity of AAC(6′)-APH(2"), an aminoglycoside antibiotic resistance determinant in enterococci and staphylococci. Chem Biol. 1999;6:99–110. doi: 10.1016/S1074-5521(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 10.Davis B D. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doern G V, Jones R N, Pfaller M A, Kugler K C, Beach M L. Bacterial pathogens isolated from patients with skin and soft tissue infections: frequency of occurrence and antimicrobial susceptibility patterns from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 1997). SENTRY Study Group (North America) Diagn Microbiol Infect Dis. 1999;34:65–72. doi: 10.1016/s0732-8893(98)00162-x. [DOI] [PubMed] [Google Scholar]
- 12.Fourmy D, Recht M I, Blanchard S C, Puglisi J D. Structure of the A-site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 13.Fourmy D, Recht M I, Puglisi J D. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA. J Mol Biol. 1998;277:347–362. doi: 10.1006/jmbi.1997.1552. [DOI] [PubMed] [Google Scholar]
- 14.Fourmy D, Yoshizawa S, Puglisi J D. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J Mol Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg W A, Priestley E S, Sears P S, Alper P B, Rosenbohm C, Hendrix M, Hung S-C, Wong C-H. Design and synthesis of new aminoglycoside antibiotics containing neamine as an optimal core structure: correlation of antibiotic activity with in vitro inhibition of translation. J Am Chem Soc. 1999;121:6527–6541. [Google Scholar]
- 16.Haddad J, Vakulenko S, Mobashery S. An antibiotic clocked by its own resistance enzyme. J Am Chem Soc. 1999;121:11922–11923. [Google Scholar]
- 17.Hendrix M, Alper B P, Priestly E S, Wong C-H. Hydroxyamines as a new motif for the molecular recognition of phosphodiesters: implications for aminoglycoside-RNA interactions. Angew Chem Int Ed Engl. 1997;36:95–98. [Google Scholar]
- 18.Hendrix M, Priestley E S, Joyce G F, Wong C-H. Direct observation of aminoglycoside-RNA interactions by surface plasmon resonance. J Am Chem Soc. 1997b;119:3641–3648. doi: 10.1021/ja964290o. [DOI] [PubMed] [Google Scholar]
- 19.Hermann T, Westhoff E. Saccharide-RNA recognition. Biopolymers. 1998;48:155–165. doi: 10.1002/(SICI)1097-0282(1998)48:2<155::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 20.Hermann T, Westhoff E. Docking of cationic antibiotics to negatively charged pockets in RNA folds. J Med Chem. 1999;42:1250–1261. doi: 10.1021/jm981108g. [DOI] [PubMed] [Google Scholar]
- 21.Hon W C, McKay G A, Thompson P R, Sweet R M, Yang D S, Wright G D, Berghuis A M. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 22.Honore N, Marchal G, Cole S T. Novel mutation in 16S rRNA associated with streptomycin dependence in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1995;39:769–770. doi: 10.1128/AAC.39.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Patel D J. Solution structure of the tobramycin-RNA aptamer complex. Nat Struct Biol. 1998;5:769–774. doi: 10.1038/1804. [DOI] [PubMed] [Google Scholar]
- 24.Justice M C, Ku T, Hsu M J, Carniol K, Schmatz D, Nielsen J. Mutations in ribosomal protein L10e confer resistance to the fungal-specific eukaryotic elongation factor 2 inhibitor sordarin. J Biol Chem. 1999;274:4869–4875. doi: 10.1074/jbc.274.8.4869. [DOI] [PubMed] [Google Scholar]
- 25.King J W, White M C, Todd J R, Conrad S A. Alterations in the microbial flora and in the incidence of bacteremia at a university hospital after adoption of amikacin as the sole formulary aminoglycoside. Clin Infect Dis. 1992;14:908–915. doi: 10.1093/clinids/14.4.908. [DOI] [PubMed] [Google Scholar]
- 26.Kirk S R, Tor Y. tRNAPhe binds aminoglycoside antibiotics. Bioorg Med Chem. 1999;7:1979–1991. doi: 10.1016/s0968-0896(99)00170-4. [DOI] [PubMed] [Google Scholar]
- 27.Kucers A, Bennett N M. Gentamicin, tobramycin, amikacin, sisomicin and netilmicin. In: Kucers A, Bennett N M, editors. The use of antibiotics. London, United Kingdom: William Heinemann Medical Books; 1987. p. 619. [Google Scholar]
- 28.Llano-Sotelo B, Chow C S. RNA-aminoglycoside antibiotic interactions: fluorescence detection of binding and conformational change. Bioorg Med Chem Lett. 1999;9:213–216. doi: 10.1016/s0960-894x(98)00718-5. [DOI] [PubMed] [Google Scholar]
- 29.McKay G A, Roestamadji J, Mobashery S, Wright G D. Recognition of aminoglycoside antibiotics by the enterococcal/staphylococcal aminoglycoside 3'-phosphotransferase type IIIa: role of substrate amino groups. Antimicrob Agents Chemother. 1996;40:2648–2650. doi: 10.1128/aac.40.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael K, Wang H, Tor Y. Enhanced RNA binding of dimerized aminoglycosides. Bioorg Med Chem. 1999;7:1361–1371. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 31.Mingeot-Leclercq M-P, Tulkens P M. Aminoglycosides: nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003–1012. doi: 10.1128/aac.43.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mingeot-Leclercq M-P, Glupczynski Y, Tulkens P M. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moazed D, Noller H F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 34.Mobashery, S., and E. F. Azucena, Jr. Mechanisms of bacterial antibiotic resistance. In Encyclopedia of life sciences, in press. Macmillan Reference Ltd., London, United Kingdom.
- 35.Noller H F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen L C, Benning M M, Holden H M. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyl transferases. Biochemistry. 1995;34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- 37.Powers T, Noller H F. Selective perturbation of G530 of 16S rRNA by translational miscoding agents and a streptomycin-dependence mutation in protein S12. J Mol Biol. 1994;235:156–172. doi: 10.1016/s0022-2836(05)80023-3. [DOI] [PubMed] [Google Scholar]
- 38.Recht M I, Douthwaite S, Puglisi J D. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Recht M I, Fourmy D, Blanchard S C, Dahlquist K D, Puglisi J D. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J Mol Biol. 1996;262:421–436. doi: 10.1006/jmbi.1996.0526. [DOI] [PubMed] [Google Scholar]
- 40.Roestamadji J, Grapsas I, Mobashery S. Mechanism-based inactivation of bacterial aminoglycoside 3′-phosphotransferases. J Am Chem Soc. 1995;117:80–84. [Google Scholar]
- 41.Rogers J, Chang A H, von Ashen U, Schroeder R, Davies J. Inhibition of the self-cleavage reaction of the human hepatitis delta virus ribozyme by antibiotics. J Mol Biol. 1996;259:916–925. doi: 10.1006/jmbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- 42.Sakon J, Liao H H, Kanikula A M, Benning M M, Rayment I, Holden H M. Molecular structure of kanamycin nucleotidyl transferase determined to 3 Å resolution. Biochemistry. 1993;32:11977–11984. doi: 10.1021/bi00096a006. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz F J, Verhoef J, Fluit A C. Prevalence of aminoglycoside resistance in 20 European university hospitals participating in the European SENTRY Antimicrobial Surveillance Programme. Eur J Clin Microbiol Infect Dis. 1999;18:414–421. doi: 10.1007/s100960050310. [DOI] [PubMed] [Google Scholar]
- 44.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistant genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stage T K, Hertel K J, Uhlenbeck O C. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J, Skeggs P A, Cundliffe E. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol Gen Genet. 1985;201:168–173. doi: 10.1007/BF00425655. [DOI] [PubMed] [Google Scholar]
- 47.Tok J B-H, Rando R R. Simple aminols as aminoglycoside surrogates. J Am Chem Soc. 1998;120:8279–8280. [Google Scholar]
- 48.Tok J B-H, Cho J, Rando R R. Aminoglycoside hybrids as potent RNA antagonists. Tetrahedron. 1999;55:5741–5758. [Google Scholar]
- 49.Tor Y, Hermann T, Westhoff E. Deciphering RNA recognition: aminoglycoside binding to the hammerhead ribozyme. Chem Biol. 1998;5:R277–R283. doi: 10.1016/s1074-5521(98)90286-1. [DOI] [PubMed] [Google Scholar]
- 50.Unge J, Berg A, Al-Kharadaghi S, Nikulin A, Nikonov S, Davydova N, Nevskaya N, Garber M, Liljas A. The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance. Structure. 1998;6:1577–1586. doi: 10.1016/s0969-2126(98)00155-5. [DOI] [PubMed] [Google Scholar]
- 51.von Ashen U, Noller H F. Footprinting the sites of interaction of antibiotics with catalytic group I intron RNA. Science. 1991;260:1500–1503. doi: 10.1126/science.8502993. [DOI] [PubMed] [Google Scholar]
- 52.von Ashen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 53.Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley S K. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 54.Wong C-H, Hendrix M, Manning D D, Rosenbohm C, Greenberg W A. A library approach to the discovery of small molecules that recognize RNA: use of a 1,3-hydroxyamine motif as core. J Am Chem Soc. 1998;120:8319–8327. [Google Scholar]
- 55.Wright G D, Berghuis A M, Mobashery S. Aminoglycoside antibiotics: structures, functions and resistance. In: Rosen B P, Mobashery S, editors. Resolving the antibiotic paradox. New York, N.Y: Kluwer Academic and Plenum Publishers; 1998. pp. 27–69. [PubMed] [Google Scholar]
- 56.Wybenga-Groot L E, Draker K, Wright G D, Berghuis A M. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure Fold Des. 1999;7:497–507. doi: 10.1016/s0969-2126(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 57.Yoshizawa S, Fourmy D, Puglisi J D. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]