Abstract
The vomeronasal type 2 receptor (V2R, also called OlfC) multigene family is found in a broad range of jawed vertebrates from cartilaginous fish to tetrapods. V2Rs encode receptors for food-related amino acids in teleost fish, whereas for peptide pheromones in mammals. In addition, V2Rs of teleost fish are phylogenetically distinct from those of tetrapods, implying a drastic change in the V2R repertoire during terrestrial adaptation. To understand the process of diversification of V2Rs in vertebrates from “fish-type” to “tetrapod-type”, we conducted an exhaustive search for V2Rs in cartilaginous fish (chimeras, sharks, and skates) and basal ray-finned fish (reedfish, sterlet, and spotted gar), and compared them with those of teleost, coelacanth, and tetrapods. Phylogenetic and synteny analyses on 1897 V2Rs revealed that basal ray-finned fish possess unexpectedly higher number of V2Rs compared with cartilaginous fish, implying that V2R gene repertoires expanded in the common ancestor of Osteichthyes. Furthermore, reedfish and sterlet possessed various V2Rs that belonged to both “fish-type” and “tetrapod-type”, suggesting that the common ancestor of Osteichthyes possess “tetrapod-type” V2Rs although they inhabited underwater environments. Thus, the unexpected diversity of V2Rs in basal ray-finned fish may provide insight into how the olfaction of osteichthyan ancestors adapt from water to land.
Subject terms: Molecular evolution, Evolutionary biology, Comparative genomics
Introduction
Olfaction is a critical chemosensory system for eliciting social behaviors in vertebrates, including reproduction, kin recognition, aggression, and feeding. The vertebrate olfaction system has experienced drastic changes in anatomy and neurophysiology during adaptation from water to land, one of the most important events is vertebrate evolution. Specifically, aquatic vertebrates detect water-soluble chemicals using their olfactory epithelium (OE), whereas terrestrial vertebrates detect both volatile and nonvolatile chemicals by differentiating their OE into main olfactory epithelium (MOE) and vomeronasal organ (VNO). Accompanied by the terrestrial adaptation of olfaction, the chemosensory receptors are proposed to have undergone major innovative diversification, although the detailed evolutionary process of diversification remains to be poorly understood at the DNA level.
Olfaction of vertebrates is mainly composed of four types of G protein-coupled receptors (GPCRs), namely, the olfactory receptor (OR), vomeronasal receptor type I (V1R), vomeronasal receptor type II (V2R), and trace amine-associated receptor (TAAR), all forming multigene families1. In teleost fish, V2Rs, also referred to as OlfCs (olfactory receptors classified as type C GPCRs)2,3, are expressed in OE of the nasal cavity. Several independent studies have shown that teleost V2Rs detect amino acids that elicit certain feeding behaviors. For example, V2Rs are expressed in microvillous sensory neurons of zebrafish and respond to amino acids, but not bile acids or sex pheromones4. In addition, the genetic blockage of neural transmission in the V2R-expressing neurons abolishes the attractive response to a mixture of amino acids5. However, given that Yang et al.6 have proposed a possible contribution of some V2Rs to elicit fright reactions, it is premature to rule out the possibility that V2Rs detect some chemicals other than amino acids. In tetrapods, VNO is a specific organ that mainly detect pheromones1. V2Rs are specifically expressed in the VNO of mice7, frogs8, and reptiles9. Hence, they are believed to encode pheromone receptors. Indeed, it has been shown that V2Rs detect peptide pheromones10,11 and peptides for the major histocompatibility complex12.
Until now, phylogenetic analyses have revealed that teleost fish possess 20–60 V2Rs, which are divided into 16 subfamilies2,13,14. On the other hand, mammalians possess variable number of V2Rs (0–121), which are genetically closely related to each other1,13,15,16. In the phylogenetic tree, V2Rs of teleost fish are distinct from those of tetrapods, which were described as “fish-like” and “tetrapod-like”17. Similar distinctive evolution was also observed between V1Rs of teleost fish and tetrapods18, which were named “fish-type” (f-V1Rs) and “tetrapod-type” (t-V1Rs), respectively19–21. In accordance with previous studies, we hereafter use the name of "fish-type" V2Rs (f-V2Rs) for the putative amino acid receptor in teleost fish and "tetrapod-type" V2Rs (t-V2Rs) for the putative peptide receptor in tetrapods. The f-V2Rs and t-V2Rs are also found to be distinct in their synteny relationships in that the f-V2Rs form single large cluster in particular chromosomes13,14,16 and t-V2Rs are scattered on several chromosomes22. It is notable that the coelacanth, which is the close relatives of tetrapods, have both f-V2Rs and t-V2Rs17, showing that the coelacanth is an important organism as it serves as a missing link to fill an evolutionary gap between vertebrates under water and land19,21. Recent studies on several shark genomes have shown that olfaction in cartilaginous fish is dominated by f-V2Rs rather than conventional ORs23–25, which is consistent with the ultrastructural observation that the presence of only microvillous sensory neurons in OE26 and with the immunohistochemical observations that most neurons in OE are positive for Go antibodies27. In addition, a previous study has shown that V2Rs were absent in lamprey genomes28. Based on the above findings, V2Rs are considered to have originated in the jawed vertebrate ancestor before the split between the two extant descendant lineages, that is, cartilaginous fish and Osteichthyes (ray-finned and lobed-finned fish including tetrapods).
In this study, we have performed a comprehensive exploration and phylogenetic analyses for V2Rs in nine cartilaginous fish species (one elephant shark, one rabbit fish, four sharks, two skates, and sawfish), three basal ray-finned fish (reedfish, sterlet, and spotted gar), two teleost fish (eel, zebrafish), a coelacanth, two amphibians (western clawed frog and caecilian), an anole lizard, and a mouse to elucidate the process of diversification of f-V2Rs and t-V2Rs in vertebrates. Especially, focusing on basal ray-finned fish, which retain many ancestral characters of Osteichthyes, is of primary importance in understanding the evolution and diversification of V2Rs from fish to tetrapods29. As a result, we have characterized 9–42 V2Rs in cartilaginous fish, and a large amount (47–189) of V2Rs in basal ray-finned fish. Phylogenetic analyses of V2Rs of 19 various vertebrate species have revealed the existence of t-V2Rs in the genomes of reedfish and sterlet, implying the presence of an evolutionary seed for mammalian peptide pheromone receptors in the basal ray-finned fish. The results of this study including; (1) genomic survey and identification of V2Rs, (2) fine-scale phylogenetic and synteny analyses, and (3) mRNA expression profiles on the OE, would provide important insights into understanding the process of diversification of V2Rs during vertebrate evolution.
Results
Characterization of V2Rs from the genomes of a broad range of vertebrates
We explored V2R gene repertoires from the genomes of nine cartilaginous fish (one elephant shark, one rabbit fish, four sharks, and three rays), three basal ray-finned fish (reedfish, sterlet, and spotted gar), two teleost fish (zebrafish, Japanese eel), a coelacanth, two amphibians (western clawed frog, caecilian), an anole lizard, and a mouse using a software “fate”30. The copy numbers of V2Rs in individual species are summarized and provided in Table 1. The number of V2Rs in teleost fish was mostly comparable to previous studies with some update. For example, zebrafish that possessed 72 V2Rs, which was larger than in previous studies2,13. The numbers of V2Rs in cartilaginous fish ranged from 8 (rays) to 41 (chimera), which were found to be smaller than those of the teleost fish. In contrast, basal ray-finned fish possessed an unexpectedly large number of V2Rs. In reedfish, we identified 188 intact V2Rs, which was the largest in all ray-finned fish studied until now. The numbers of V2Rs in sterlet and spotted gars were also high (46 and 49, respectively). The copy number of V2Rs in the western clawed frog (691), which far exceeded that previous studies15, was the largest among vertebrates studied so far; it is much higher than those of other tetrapods, such as caecilian (275), anole lizard (64), and mouse (154). It is noteworthy that we found no V2Rs in the genomes of agnathans and amphioxus.
Table 1.
Number of intact V2R genes identified in the genomes of broad range of vertebrates.
| Scientific name | Common name | Assembly name | V2R2 | ancV2R | t-V2R | f-V2R | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum | Sum | Sum | Sum | Sub 1 | Sub 2 | Sub 3 | Sub 4–9 | Sub 10 | Sub 11 | Sub 12 | Sub 13 | Sub 14 | Sub 15 | Sub 16 | Sub a1 | Sub a2 | Sub a3 | Unplaced | |||
| Callorhinchus milii | Elephant shark | Callorhinchus_milii-6.1.3 | 1 | 1 | 0 | 26 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 19 | 2 | 2 |
| Hydrolagus affinis | Small-eyed rabbitfish | UP_Haf | 1 | 1 | 0 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 36 | 0 | 2 |
| Scyliorhinus torazame | Cloudy catshark | Storazame_v1.0 | 1 | 1 | 0 | 21 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 13 | 1 | 0 |
| Carcharodon carcharias | Great white shark | ASM360424v1 | 1 | 1 | 0 | 12 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 1 |
| Rhincodon typus | Whale shark | ASM164234v2 | 1 | 0 | 0 | 21 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 16 | 0 | 1 |
| Chiloscyllium punctatum | Brownbanded bambooshark | Cpunctatum_v2.1 | 1 | 1 | 0 | 24 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 18 | 1 | 0 |
| Leucoraja erinacea | Little skate | LER_WGS_1 | 1 | 1 | 0 | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| Amblyraja radiata | LER_WGS_1 | sAmbRad1.pri | 1 | 1 | 0 | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| Pristis pectinata | Smalltooth sawfish | sPriPec2.pri | 1 | 1 | 0 | 22 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 1 | 0 |
| Erpetoichthys calabaricus | Reedfish | Reedfish | 1 | 1 | 3 | 183 | 1 | 2 | 1 | 0 | 4 | 1 | 1 | 2 | 1 | 1 | 67 | 101 | 0 | 0 | 1 |
| Acipenser ruthenus | Sterlet | ASM1064508v1 | 1 | 1 | 3 | 41 | 1 | 11 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 1 | 21 | 0 | 0 | 1 |
| Lepisosteus oculatus | Spotted gar | LepOcu1 | 1 | 1 | 0 | 47 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 9 | 29 | 0 | 0 | 1 |
| Anguilla japonica | Japanese eel | Ajp_01 | 1 | 0 | 0 | 63 | 1 | 4 | 2 | 39 | 1 | 4 | 1 | 2 | 1 | 3 | 5 | 0 | 0 | 0 | 0 |
| Danio rerio | Zebrafish | GRCz11 | 1 | 0 | 0 | 71 | 2 | 6 | 1 | 24 | 1 | 3 | 1 | 6 | 2 | 2 | 23 | 0 | 0 | 0 | 0 |
| Latimeria chalumnae | Coelacanth | LatCha1 | 1 | 0 | 75 | 13 | 1 | 1 | 1 | 0 | 0 | 1 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 3 |
| Xenopus tropicalis | Western clawed frog | UCB_Xtro_10.0 | 1 | 1 | 689 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Geotrypetes seraphini | Caecilian | aGeoSer1.1 | 1 | 0 | 273 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anolis carolinensis | Anole lizard | AnoCar2.0 | 1 | 0 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mus musculus | Mouse | GRCm38.p6 | 7 | 0 | 147 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
The bold highlighted the cells in which no genes were found in the corresponding subfamilies.
Evolution of f-V2Rs of vertebrates
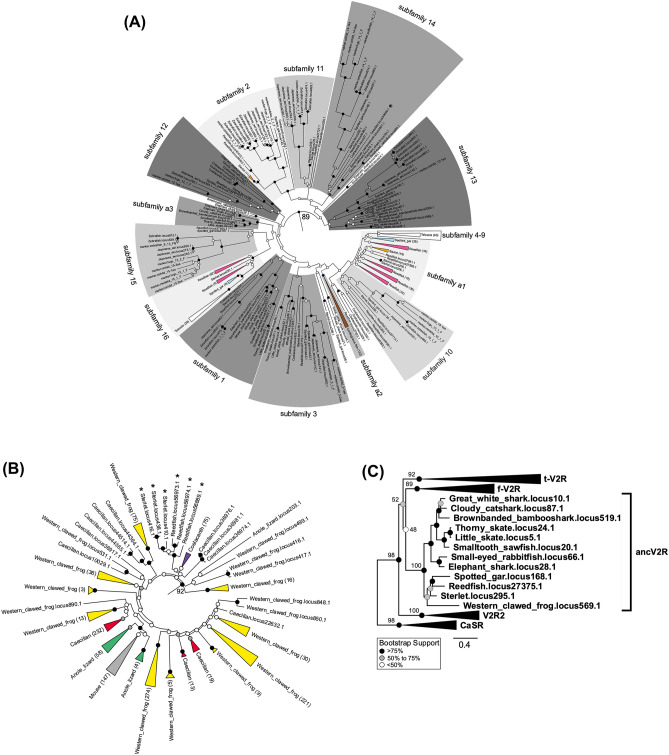
To classify and infer the evolutionary history of V2Rs in vertebrates, we constructed a phylogenetic tree of all V2Rs identified in the study (1897 from 19 species) using maximum likelihood algorithm in RAxML31. The phylogenetic tree with all sequences shows a clear separation of the two V2R clades, each of which consists of the sequences of f-V2Rs of teleost fish and t-V2Rs of tetrapods, respectively (Supplementary File 2). Therefore, the phylogenetic trees of the two clades consisting of f-V2Rs (Fig. 1A) and t-V2Rs (Fig. 1B) were shown separately for better visibility of the details. A simplified phylogenetic tree of f-V2Rs, t-V2Rs, and other two additional clades are shown in Fig. 1C.
Figure 1.
Phylogenetic relationships of V2Rs of a broad range of vertebrates from cartilaginous fish to tetrapods constructed using RAxML-NG v.1.0.1 (https://github.com/amkozlov/raxml-ng). (A) Phylogenetic tree of f-V2Rs. Note that f-V2Rs were subdivided into 16 known and 3 novel subfamilies, as indicated by gray thick bars. Triangles in red, orange, blue, white, and brown indicate the expanded V2R clusters specific to reedfish, sterlet, spotted gar, cartilaginous fish, and teleost fish, respectively. Only one f-V2Rs found in the caecilian species was marked using an asterisk. (B) Phylogenetic tree of t-V2Rs. Triangles in violet, pink, yellow, green, and gray indicate expanded V2R clusters specific to coelacanth, caecilian, western clawed frog, anole lizard, and mouse, respectively. Asterisks were used to mark the t-V2Rs identified in reedfish and sterlet. Note that the t-V2Rs, in contrast to the f-V2Rs, are composed of many clusters that are expanded in a species-specific manner. (C) Overview of the phylogenetic tree of all V2Rs showing novel orthologous clade ancV2R. The calcium-sensing receptor (CaSR) gene was used for outgrouping all V2Rs. The OTU names consist of the common name and locus as summarized in Supplementary Table S1. The f-V2Rs, t-V2Rs, and V2R2 clades were compressed into black triangles. The number on the branches indicates the bootstrap support values for particular nodes. Note that the grouping of the orthologous ancV2Rs of cartilaginous fish, basal ray-finned fish, and western clawed frog was suggested by maximum bootstrap support (100%). The numbers next to triangles indicate the copy number of V2Rs included in the clusters. The filled circles on each node indicate bootstrap supports (black > 75, 75 ≧ gray ≧ 50, 50 > white). Scale bar indicates the number of amino acid substitutions per site.
In the phylogenetic tree of f-V2R clade, we identified all known 16 subfamilies13,14 in addition to the newly identified subfamilies “a1”, “a2” and “a3” (Fig. 1A). Notably, over half of the V2Rs of the basal ray-finned fish were included “a1”. Also, we have found a cartilaginous fish-specific subfamily “a2” and “a3” (Table 1, Figs. 1A, S1). In contrast to ray-finned fish in which V2R subfamilies were highly diverse, cartilaginous fishes were relatively less diverse (Table 1). One exceptional finding was that only one sequence of caecilian V2R was classified as being a member of the f-V2R subfamily 14 (Table 1, Fig. 1A, marked with an asterisk).
Evolution of t-V2Rs of vertebrates
The t-V2Rs clade were shown to be mainly dominated by closely related V2Rs of tetrapods that were genetically closely related each other9,13,15. Notably, V2Rs of amphibians are highly diverse in that they are subdivided into many species-specific clusters, while all V2Rs of mouse belonged to a single cluster (Fig. 1B, Table 1). The existence of 75 V2Rs of coelacanth in t-V2R clade was consistent with a previous study (Fig. 1A, Table 1)17. One of the most striking results of this study was that six V2R sequences of ray-finned fishes (reedfish and sterlet) were classified into t-V2Rs. Indeed, in the phylogenetic tree, these V2Rs were sister group of the coelacanth V2R cluster (Fig. 1B and Supplementary File S2). Although the existence of V2Rs of basal ray-finned fish in t-V2R clade leads to a slight confusion in definitions, we will continue to adopt this nomenclature, taking into account that; 1. classification in previous studies used these names for each clade17,19,21, and 2. dual character (fish-like as well as tetrapod-like) of the basal ray-finned fish is meaningful in discussing vertebrate evolution (see discussion).
Newly identified V2R orthologs conserved from cartilaginous fish to amphibians
Previous studies have shown that V2Rs were divided into three well-supported clades, namely, the V2R2s, f-V2Rs, and t-V2Rs17,22,28. The orthologous V2R2s were shared in all vertebrates with a few species-specific duplications in mice28,32. In this study, V2R2 ortholog of all vertebrates was located at the basal position of the V2R tree (Table 1, Fig. 1C, Supplementary Fig. 1). In addition to the three clades, we identified a novel clade, in which V2Rs of cartilaginous fish, basal ray-finned fish, and amphibians were included (Fig. 1C). A closer inspection of the genome assembly did not reveal any apparent errors that could lead to the generation of the artifact sequences. Since this clade contained just one V2R from each species, and the tree topology was identical to the species tree, these V2R sequences were considered to be orthologous. This orthologous V2Rs were evolutionarily distinct from the f-V2Rs and t-V2Rs, which were amplified in a species-specific manner. Considering that only one ortholog has been retained since quite a long time ago, we named them anc (ancient) V2R. In this study, highly conserved ancV2R sequences were found in the genomes of cartilaginous fish to amphibians, but not in teleost fish, coelacanth, and mammals (Fig. 1C).
Conserved gene clusters of f-V2Rs
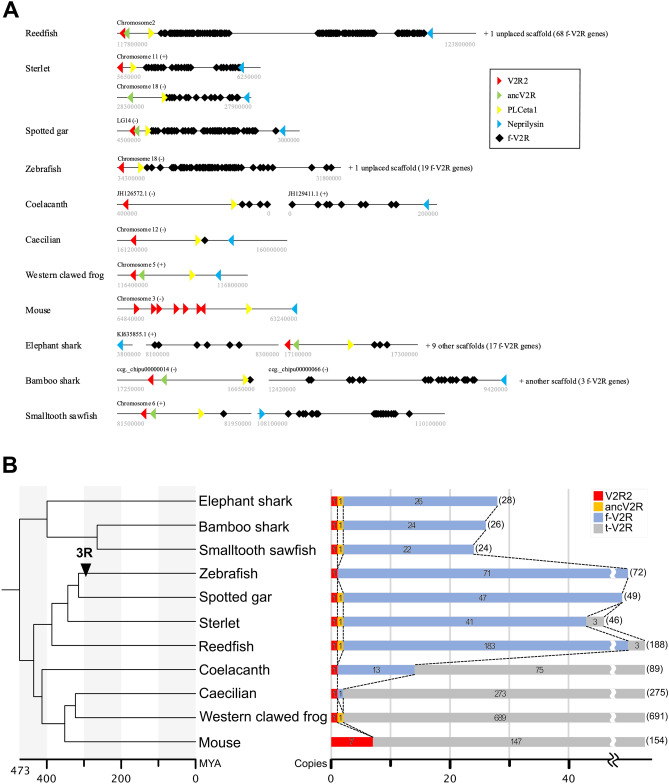
In addition to phylogenetic analysis, the synteny relationships have provided important insight into the classification of V2Rs. Previous studies have shown that the V2Rs in teleost were clustered in one particular chromosomal region, which was flanked by two landmark genes, phospholipase C (PLC) eta1 and neprilysin13,14,16. In contrast, no V2Rs were found between these two genes in tetrapods, and t-V2Rs were scattered into several chromosomes22. Therefore, to ascertain if the V2Rs are f-V2Rs or t-V2Rs, we examined the synteny relationships, in which the gene arrangements in the genomic region of two landmark genes in various vertebrates from cartilaginous fish to mammals were summarized (Fig. 2A). Notably, V2R2 and ancV2R was found in tandem of the neighboring regions of the PLC eta1. No V2R was found in tetrapods in this region, except for only one caecilian V2R, which was classified as f-V2Rs in the phylogenetic tree (Fig. 1A). Importantly, we revealed that f-V2Rs were located in this cluster region. In addition, three t-V2Rs identified in reedfish and sterlet were located on different chromosomes (chr.3 in reedfish, chr.52, 53 and VTUV01000346.1 in sterlet, Supplementary Table S1). In sterlet, we identified two distinct chromosomal regions of the f-V2R clusters, which were due to polyploidization specific to this group33. The synteny of coelacanth also showed a conservation of the f-V2R cluster. Overall, the phylogenetic and synteny analyses both supported the conservation of the cluster for f-V2Rs as well as the existence of the t-V2Rs in basal ray-finned fish.
Figure 2.
(A) Synteny relationships for the f-V2R clusters among basal ray-finned fish (reedfish, sterlet, spotted gar), teleost fish (zebrafish), lobe-finned fish (coelacanth), tetrapods (caecilian, western clawed frog, mouse), and cartilaginous fish (elephant shark, bamboo shark, and smalltooth sawfish), respectively. Triangles in yellow, blue, red, and green indicate two landmark genes (PLC eta1, neprilysin), V2R2, and ancV2R, respectively. Black squares indicate f-V2Rs. Indicated at the upper-left of each line were several chromosomes or scaffolds and its directions. Indicated below the ends of the line are the start and end of the cluster regions. Unplaced scaffolds are not shown in this figure. Note that f-V2Rs were flanked by two landmark genes and that V2R2s and ancV2Rs were located in tandem close to the clusters. No t-V2Rs were observed in these cluster regions. In the elephant shark, some f-V2Rs are located outside the cluster because the cluster regions were not properly assembled. (B) Changes in the number of V2Rs during vertebrate evolution. The phylogenetic tree with timescale for 11 representative vertebrates (left) and the number of V2Rs in these species (right) are shown. The timing of teleost-specific third round whole genome duplication (3R) is indicated by arrow. The color and number on the bar graph indicate the clade and copy number, respectively. The total copy number of V2Rs is shown in parentheses on the right of the graph. MYA: million years ago.
The copy number of V2Rs in vertebrate evolution
We investigate the evolutionary trends of V2Rs in terms of copy number changes by mainly focusing on f-V2Rs and t-V2Rs (Fig. 2B). The copy numbers of V2R2 and ancV2R were constant, which were mostly one in any species. The cartilaginous fish possess relatively small number of V2Rs, which are dominated by f-V2Rs. The V2Rs suddenly increased in ray-finned fish, which were also dominated by f-V2Rs. Notably, three copies of t-V2Rs emerged in basal ray-finned fish, represented by reedfish and sterlet, but were lost in teleost fish. Then, f-V2Rs decreased and t-V2Rs increased in coelacanth and tetrapods. Thus, the changes in the proportion of f-V2Rs and t-V2Rs in the genomes appears to coincide with evolutionary transition of vertebrates from water to land. Overall, it is obvious that sudden increase of V2Rs occurred three times, namely, in the common ancestor of Osteichthyes, in the common ancestor of tetrapods, and in Western clawed frog. By contrast, sudden increase of V2Rs was not observed in the lineage of teleost fish.
Expression of V2Rs in the olfactory epithelium of basal ray-finned fish
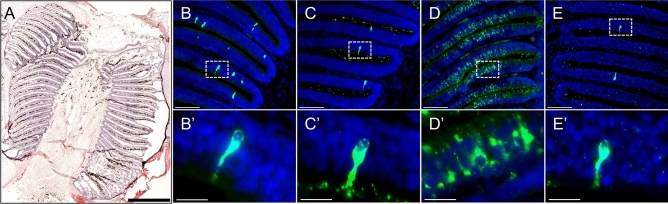
To evaluate the functional role of V2Rs found in the genomes of basal ray-finned fish, we examined the cellular expression patterns for these receptors in the OE. The locations of transcripts for four different V2Rs were detected by in situ hybridization on frozen sections of the OE of Polypterus senegalus (bichir), which is a basal ray-finned fish closely related to reedfish (Fig. 3). The probes of the four V2Rs f-V2R, t-V2R, V2R2, and ancV2R each of which has been identified as a distinct clade in the phylogenetic tree (Fig. 1C), were used in the experiments. The expression of a member of the f-V2Rs showed a sparse pattern in the sensory cells of the OE, typical of canonical V2Rs (Fig. 3B). The expression of a member of t-V2Rs, which was newly identified in basal ray-finned fish, also showed similar sparse pattern in the OE of P. senegalus (Fig. 3C). The expression of V2R2 has showed widespread patterns in their OE (Fig. 3D). The expression pattern of V2R2 in P. senegalus was consistent with the ubiquitous expression in zebrafish34 and mouse32. The expression of ancV2R showed a sparse pattern in their OE (Fig. 3E). Overall, the V2Rs belonging to four clades were all expressed in the OE, suggesting their functions as olfactory receptors. However, the patterns of expressions were ubiquitous in V2R2, while they were sparse in V2Rs of other clades (Table 2).
Figure 3.
The expression patterns of V2Rs in the olfactory epithelium of the basal ray-finned fish, Polypterus senegalus. Overall view of a HE-stained horizontal section of the olfactory organ, showing that lamellae of olfactory organ are stained blue-violet, and the nerve bundles are stained red-violet (A). Expression of four V2R genes were confirmed by FISH on horizontal sections of the olfactory organ of the bichir using DIG-labeled RNA antisense probes for f-V2R (B) and t-V2R (C), V2R2 (D), and ancV2R (E). Green indicated the expression signals. The blue area indicates the cell nucleus stained with DAPI. V2R2 was globally expressed in the deep layers of olfactory lamellae (D). In contrast, f-V2R, t-V2R, and ancV2R were sparsely expressed in a small number of neurons in the deeper layers of the olfactory lamellae (B, C, and E). (B’-E') High magnification view of the dotted squares in B–E. Scale bars show 1 mm (A), 100 µm (B–E) and 20 µm (B'–E').
Table 2.
Summary of current knowledge on four V2R clades.
| V2R2 | ancV2R | f-V2R | t-V2R | |
|---|---|---|---|---|
| Specific characters | ||||
| Expression | Broada,32 | Sparseb* | Sparse5,35 | Sparse7,10 |
| Gene number | One or a few32 | One* | Expanded2,13,14 | Expanded7,9,15 |
| Ligands | Amino aicds34 | Unknown | Amino acids5 | Peptides10–12 |
| Habitat (water/land) | Both | Both | Mainly water | Mainly land |
| Presence (P)/absence (A) | ||||
| Cartilaginous fish | P | P* | P | A |
| Basal ray-finned fish | P | P* | P* | P* |
| Teleost fish | P | A | P | A |
| Coelacanth | P | A | P | P |
| Amphibians | P | P* | P* | P |
| Reptiles | P | A | A | P |
| Mammals | P | A | A | P |
*Newly found in this study.
abroadly expressed in all vomeronasal neurons and co-expressed with the other V2Rs.
bsparsely expressed according to one neuron one receptor rule.
Discussion
In this study, we have conducted a comprehensive exploration of V2R sequences from the genomes of 19 vertebrate species. Phylogenetic analyses of a large number of V2Rs allowed us to gain a panoramic view of the diversity in terms of copy number and repertoire of V2Rs across vertebrates. According to a recent study by Bi et al.29, more than 50 V2R-like sequences in hagfish and a few in amphioxus were observed. However, our phylogenetic analyses revealed that they did not form a cluster with known V2Rs. Therefore, we need to be still cautious to designate these sequences as V2Rs, reaching a conclusion that no typical V2Rs exist in agnathans and amphioxus. In addition, we need to remain careful about the detail copy numbers, as it depends on the quality of the genome assemblies, which were relatively low in some cartilaginous fish. Based on the current data set obtained in this study, we here discuss the evolution of V2Rs and how these factors drive the adaptive evolution of the olfactory system in vertebrates.
It is obvious that V2Rs were abundant in ray-finned fish compared to cartilaginous fish, which is achieved by a species-specific expansion of f-V2Rs. Notably, species-specific expansions of V2Rs did not occur uniformly in all subfamilies, but were rather concentrated in certain ones. For example, the expansion of V2Rs was mainly observed in subfamilies 4–9, 16, ‘a1’, and ‘a2’, while the copy numbers of other subfamilies remained one or two. At present, V2Rs are expected to detect amino acids and their derivatives, eliciting feeding behaviors in teleost fish4,5,35. It is reasonable to assume that a limited number of amino acids in diets were received by evolutionarily conserved V2R subfamilies. However, it is plausible to assume that the V2R subfamily with frequent lineage-specific gene duplications is responsible for detecting some species-specific variable chemicals for social communication. For example, Yambe et al.36 showed that an amino acid derivative, L-kynurenine, secreted in the female urine, acts as the male-attracting pheromone in masu salmon. In addition, a previous study showed a possible correlation between expansions of V2Rs in subfamily 9 and the evolution of fright reactions in teleost fish6. Thus, to elucidate the function of V2Rs in ray-finned fish in addition to amino acid detection, it is necessary to further examine the V2Rs from a multidisciplinary framework, including the ligand binding, and behavioral experiments using candidate chemicals.
We examined the possible link between V2R expansion and whole genome duplication (WGD). First, we precisely examined the presence/absence of 2:1 relationship for Western clawed frog, in which V2Rs were expanded (Fig. 1B, Supplementary File 1). As a result, we failed to find such relationship between V2Rs of Western clawed frog and the other species. The results suggested that the expansion of V2Rs in Western clawed frog was not by WGD but by species-specific gene expansion. Indeed, it has been shown that Western clawed frog did not experience WGD specific to this species39. Previous studies have revealed that the third round of WGD has occurred in the common ancestor of Teleostei, namely, after the split of teleost fish and spotted gar about 300 million years ago (called 3R, Fig. 2B)37,38. However, expansion of V2Rs was not observed in the lineage of teleost fish. We precisely examined the 2:1 relationship for each V2R of teleost fish and spotted gar by checking the phylogenetic tree, again failed to find such relationship (Fig. 1A, Supplementary File 1). In addition, V2Rs of teleost fish were located at only one genomic region (Supplementary Table S1, Fig. 2A). Therefore, WGD is unlikely to have made a significant impact on the expansion of V2Rs during vertebrate evolution. Recent study on the keratin multigene family also revealed no apparent link between the gene expansion and WGD40. In multigene families such as chemoreceptor genes and keratin genes, tandem duplication, not WGD, may be the main driving force to increase copy number.
We showed that orthologous sequences of V2R2 and newly identified ancV2R have long been conserved during the evolution of vertebrates. Although the order of divergence among ancV2R, t-V2Rs and f-V2Rs are uncertain due to the lack of sufficient bootstrap support, ancV2R is expected to have first emerged by tandem duplication from V2R2. The existence of V2R2 and ancV2R in close genomic proximity may also imply this scenario (Fig. 2A). Studies by Silvotti et al32. in the mouse and DeMaria et al34. in zebrafish have revealed that highly conserved V2R2 was expressed in a broad area of the OE and was co-expressed with one of the many canonical V2Rs. Consistent with previous studies, V2R2 of Polypterus senegalus showed widespread expression patterns in the OE (Fig. 3D), while the f-V2Rs and t-V2Rs showed sparse patterns (Fig. 3B, C). Although ancV2R was similar to V2R2 in terms of evolutionary conservation and genomic proximity, the pattern of expression in the OE was sparse rather than widespread (Table 2). It is implicative to note that ancV2R has characteristics between V2R2 and canonical f-V2Rs and t-V2Rs. Thus, taking the evolutionary conservation as well as the sparse pattern of expression into account, ancV2R may have retained ancestral nature inherited from a protogene before the split between f-V2Rs and t-V2Rs, which are now highly diversified in jawed vertebrate genomes. It was of interest that the expression patterns of V2R2, ancV2R, and f-V2Rs were distinct despite their location on the same genomic cluster. A detailed investigation into this genomic region would lead to the elucidation of a cis-regulatory mechanism that controls the expression of canonical V2Rs, as to say “one neuron one receptor” rule41.
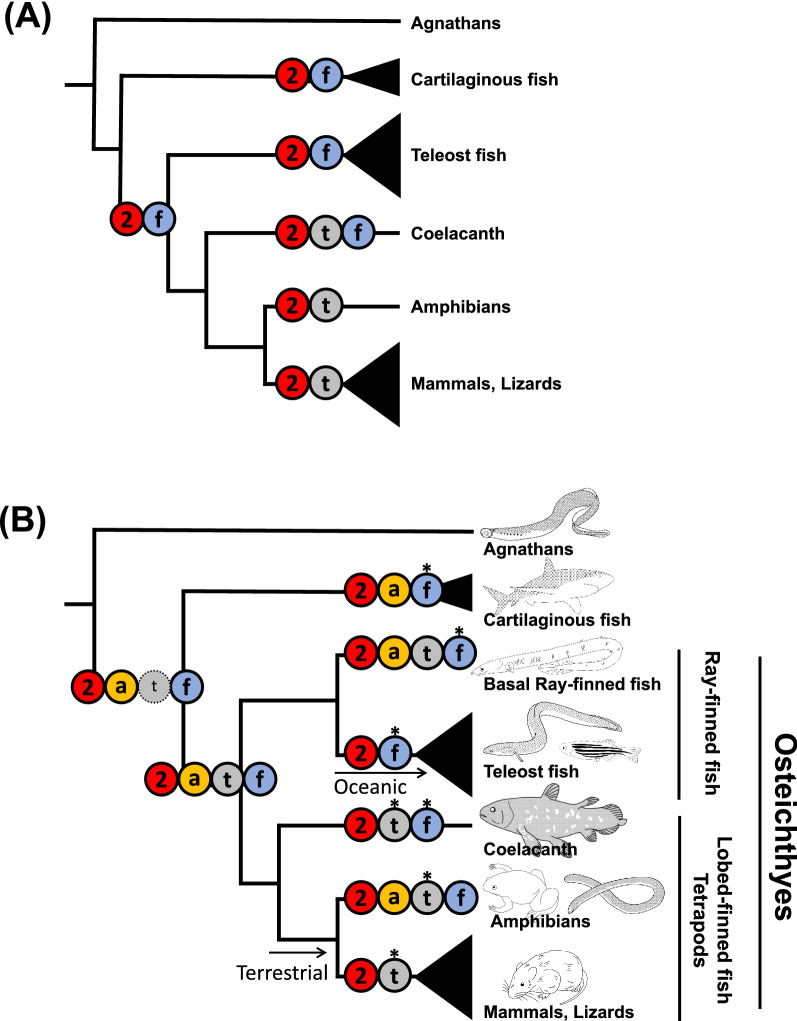
The previous (A) and present (B) hypotheses for evolutionary scenarios of the four clades of V2R in vertebrates from agnathans (lamprey and hagfish) to mammals were shown in Fig. 4, by marking the distribution of these four V2R clades in extant species and two ancestral nodes. In addition, the character of the four clades of V2Rs were summarized in Table 2. The newly characterized V2Rs in cartilaginous fish, basal ray-finned fish and caecilian provided new insights into V2R evolution in vertebrates. No V2R was found in the agnathans, implying the acquisition of a V2R-mediated olfaction as a common ancestor of jawed vertebrates (cartilaginous fish and Osteichthyes). Given that agnathans have three other types of chemoreceptor genes, namely, ORs, TAARs, and V1Rs, it is likely that V2Rs were originated later than these three receptors28,42. V2R2 was conserved in all extant jawed vertebrates with no exceptions, which suggests the highly important and fundamental role of V2R2 in the detection and subsequent signaling pathway for olfactory substances in both underwater and terrestrial environments. Importantly, except for V2R2, mammal and teleost fish possessed only specific clades of V2Rs, namely, t-V2Rs and f-V2Rs, respectively. In contrast, basal ray-finned fish and one amphibian (caecilian) possessed both t-V2Rs and f-V2Rs. This finding suggests that all four clades of V2Rs were present at least in the common ancestor of Osteichthyes, or even earlier in the common ancestor of jawed vertebrates. However, subsequently, the repertoires of V2Rs were lost in teleost fish and mammals during adaptation to their specific environments. It is interesting to note that one copy of f-V2R is exceptionally retained in caecilian. This may reflect its ancestral position in Amphibia, its water-dependent habitat, and/or subterranean lifestyle, but further studies are needed to examine these possibilities. Because the previous studies of V2Rs have been limited to teleost fish and mammalians, the diversity of V2Rs was underestimated. Thus, the genome sequences of non-model animals, which become available due to the recent advancement of sequence technique, allowed us to uncover the hidden and remarkable diversity in the genomes of vertebrates.
Figure 4.
Evolutionary scenarios of V2Rs during vertebrate evolution proposed by previous (A) and this (B) studies. The presence/absence of the four major V2R clades was plotted on the phylogenetic tree of vertebrates from agnathans to mammals. The red circle with “2,” yellow with “a,” gray with “t,” and blue with “f” indicate V2R2, ancV2R, t-V2Rs, and f-V2Rs, respectively. (A) Except for V2R2, teleost fish and tetrapods possess only f-V2Rs or t-V2Rs, respectively, while the coelacanth possesses both V2Rs. Common ancestor of Osteichthyes was expected to possess V2R2 and f-V2Rs. (B) The large-scale analyses revealed the existence of t-V2Rs in basal ray-finned fish and identified a novel clade of ancV2R. In contrast to basal ray-finned fish with all four clades of V2Rs, teleost fish, mammals, and lizards possess only two of them. The reduction of specific V2R clades in these lineages would be due to adaptation to specific oceanic and terrestrial environments. Note that the origin of t-V2Rs dates back to the common ancestor of extant Osteichthyes, but its antiquity in the jawed vertebrate ancestor remains to be examined with complete genome sequences of more cartilaginous fish (dotted circle with “t” inside). Asterisks above the circles indicate clades with substantial copy numbers due to gene expansions in each extant species. The illustration of the animals was drawn by using free software Vectr (https://vectr.com).
One of the important discussions is the timing of V2Rs diversification. The repertoires of the subfamily of the f-V2Rs are abundant in ray-finned fish and coelacanth compared with those in cartilaginous fish (Table 1). In addition, all four clades of V2Rs were shown to be present in basal ray-finned fish and amphibians. The abundance of copy numbers and repertoires in these groups suggests that V2Rs were highly diversified in the common ancestor of Osteichthyes. The diversification of V2Rs is not caused by WGD, but is most likely a signature of adaptive evolution. Given that the olfaction-related genes also emerge in the common ancestor of the Osteichthyes (e.g., ancV1R43; OMP44), it might be possible that an innovative evolution of the olfactory system occurred in this timing.
It is also worth mentioning here that the polypterids (bichir and reedfish) possesses large paired openings (spiracles) on top of their head, in which they use for breathing air45. Similar spiracle-like structures were observed in the fossil records of stem tetrapods46. Thus, breathing air using spiracles may have been an important respiratory strategy in the stem Osteichthyes, which inhabit shallow freshwater environments and use lungs in addition to gills for respiration47. Specifically, the evolution of air-breathing by spiracles may increase the opportunity to raise their head above water, which led to the acquisition of the primitive capabilities of detecting airborne chemicals before terrestrial adaptation. Thus, such dual functional roles of the olfactory system in stem Osteichthyes were related to the diversification of V2Rs, including f-V2Rs and t-V2Rs. However, at this moment, both types of V2Rs are expected to detect non-volatile ligands such as amino acids and peptides (Table 2), thus the adaptive significance of the emergence of t-V2Rs in stem Osteichthyes remains unclear. Deorphanization of V2Rs and/or the exploration of V2R expressions in various organs including non-OE in basal ray-finned fish in the near future is necessary to evaluate the above possibility.
Materials and methods
Sequence retrieval
To estimate the evolutionary history of V2Rs, we conducted a comprehensive exploration of V2R sequences in the genome assemblies of a broad range of vertebrates, including nine cartilaginous fishes and three basal ray-finned fishes. In addition to two teleost fishes (Japanese eel and zebrafish), coelacanth, two amphibians (caecilian and western clawed frog), anole lizard, and mouse were explored (Table 1). To identify the V2R sequences from the genome assemblies, we performed tBLASTn searches using the transmembrane (TM) domain of V2Rs as queries against all of the genomic sequences. The query sequences were generated by aligning V2R sequences of mouse, anole lizard, tropical clawed frog, coelacanth, spotted gar, and zebrafish deposited in the Ensembl database and of elephant shark and cloudy catshark24 using the MA–T –dash option48. Then, the TM domains, from 50-aa sequences 7n1upstream of the 1st TM region to 50-aa downstream of the 7th TM region, were extracted using a protein structure of the glutamate receptor (PDB: 4OR2)49 as reference.
Next, the V2R-coding sequences were predicted for tBLASTn hit regions using GeneWise v.2.4.150. The above procedures were conducted by using a software “fate”, which is developed specifically for the identification of multiple gene families30. Sequences shorter than 600 nucleotides were discarded. The homology of intact outputs was then determined by the next phylogenetic analysis. The queries used in the BLAST search were critical to the estimated number of V2Rs. Indeed, the number of V2Rs identified in this study was consistent with previous studies, whereas a few differences were observed in some cases. For example, the numbers of V2Rs in elephant shark and catshark identified in this study (29 and 24 copies) were slightly smaller than those reported by Sharma et al.24. The difference in query sequences used for the BLAST search reflects results. Specifically, we used the seven-transmembrane regions of the V2R, whereas Sharma et al.24 used the entire region of V2R, including the extracellular Venus flytrap module region (VFTM)51 The VFTM region of the V2R protein has been determined to be highly diverse and difficult in predicting the exon–intron structure of the gene. This result affects the judgment of intact or pseudogenes. Therefore, we compared the number of V2Rs estimated under a unified condition using the seven-transmembrane region as a query.
Phylogenetic and synteny analyses
Sequences were translated into amino acid sequences and aligned using MAFFT with the -ginsi option52 and default parameters. The sites with < 50% coverage among all sequences were removed. Maximum likelihood trees for V2R genes were then inferred using RAxML-NG v.1.0.131 with the JTT + G + F amino acid substitution model. This result was the best fitting model selected by the ModelTest-NG53,54 based on AIC scores. Rapid bootstrap analyses were performed using 100 replicates to assess the reliability of nodes. The V2R sequences for coelacanth, western clawed frog, and mouse were then clustered based on the criteria of 80% similarity at the amino acid level to save computational costs. To avoid false positives in identification of the V2R genes and prevent long branch attractions, we obtained other GPCR family C sequences from GenBank or Ensembl for use as outgroups, including CaSR (NM_013803.3), Tas1r1 to Tas1r3 (ENSMUSG00000028950, ENSMUSG00000028738, ENSMUSG00000029072), GPCR6 (NM_153071.1), GRM1a to GRM8 (NM_016976.1, NM_001160353.1, NM_181850.2, NM_001081414.2, NM_001081414.2, NM_173372.2, NM_177328.3, NM_001361125.1), and GABA B1 to GABA B2 (NM_019439.3 and NM_001081141.2), for constructing an initial gene tree. All genes in the sister clade to CaSR were named homologs of V2R (including V2R2). Using these genes, we constructed the V2R gene tree again (CaSR was used as an outgroup). We also included sequences of all 16 teleost fish subfamilies classified in previous studies13,14, as markers to indicate V2R subfamilies.
The synteny relationships of f-V2Rs of vertebrates were then illustrated based on the genomic location of the identified V2Rs (Supplementary Table S1). The numeric data for the genomic position of V2Rs, which were identified as f-V2Rs, V2R2, and ancV2R in the phylogenetic tree, were then compiled, with those of PLC eta1 and neprilysin.
Histology and in situ hybridization
Bichir (Polypterus senegalus) individuals of 11–25 cm long, which were used for the preparation of frozen sections and the extraction of total RNA from the olfactory organs, were purchased from a commercial supplier; they were kept under standard conditions suitable for tropical fish breeding until experimental manipulations. All experimental studies using the animals were approved by the Institutional Animal Experiment Committee of the Tokyo Institute of Technology were performed in accordance with the institutional, governmental ARRIVE guidelines. In addition, all methods were performed in accordance with the relevant guidelines and regulations. TRIzol (Invitrogen) was then used for total RNA extraction from the olfactory organs of the bichir. Using the total RNA extracted from the olfactory organs of the bichir, cDNA was synthesized by reverse transcription reaction using SuperScript III RTase (Invitrogen). Coding regions of V2R were amplified by PCR using the primer sets, which were designed on the basis of V2R sequences of reedfish, as has been summarized in Supplementary Table S2. The PCR products were cloned using the pGEM-T vector (Promega) and the DH5α strain of E. coli. Digoxigenin-labeled RNA probes were synthesized using the plasmid vector as a template using T7 or SP6 RNA Polymerase (Roche) and DIG RNA labeling mix (Roche) as well. The olfactory organs of the bichir were then fixed with 4% PFA, replaced with sucrose, and embedded in an O.C.T compound (Sakura Finetek). In situ hybridization was performed according to the method as previously described43,44. Briefly, hybridization was performed using DIG-labeled RNA probes. The antibody reaction was conducted using anti-digoxigenin-POD, Fab fragments (Sigma-Aldrich), or anti-fluorescein-POD, Fab fragments (PerkinElmer). The signal was then amplified with Tyramide Signal Amplification Plus Biotin kit (Kiko Tech), and detected by streptavidin, Alexa Fluor 488 conjugate (Thermo Fisher). Finally, the sections were sealed using a VECTASHIELD Mounting Medium with DAPI (VECTOR). The sealed sections were observed using a fluorescence microscope Axioplan (Carl Zeiss). All fluorescence photographs were taken using an Axiocam 503 color (Carl Zeiss) and optimized for brightness and contrast in Adobe Photoshop.
For histological observation, the frozen sections of the bichir olfactory organs were stained with hematoxylin for 4 min and washed with tap water. The sections were then stained with eosin for 10 min and treated with 70% ethanol for 1 min, 80% ethanol for 1 min, 90% ethanol for 1 min, and 100% ethanol for 5 min three times. The sections were then treated with xylene for 5 min and three times and sealed in ENTELLAN NEW (MERCK).
Supplementary Information
Acknowledgements
This work was funded by JSPS KAKENHI (20H03307) and Asahi Glass Foundation to M.N. We thank the researchers, who determined and assembled the genome sequences of cartilaginous fish and reedfish as part of the Vertebrate Genome Project (VGP), for releasing their unpublished data. We also thank Yujiro Kawabe of the Tokyo Institute of Technology for the animal illustration.
Author contributions
Project design, coordination: M.N.; bioinformatics and evolutionary analyses: Z.Z.; molecular experiments: A.S.; manuscript writing: Z.Z., S. K., M.N.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-10428-0.
References
- 1.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 2.Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: Candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnstone KA, et al. Genomic organization and evolution of the vomeronasal type 2 receptor-like (OlfC) gene clusters in Atlantic salmon Salmo salar. Mol. Biol. Evol. 2009;26:1117–1125. doi: 10.1093/molbev/msp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speca DJ, et al. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/S0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- 5.Koide T, et al. Olfactory neural circuitry for attraction to amino acids revealed by transposon-mediated gene trap approach in zebrafish. Proc. Natl. Acad. Sci. USA. 2009;106:9884–9889. doi: 10.1073/pnas.0900470106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, et al. Expansion of vomeronasal receptor genes (OlfC) in the evolution of fright reaction in Ostariophysan fishes. Commun. Biol. 2019;2:235. doi: 10.1038/s42003-019-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/S0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 8.Hagino-Yamagishi K, et al. Expression of vomeronasal receptor genes in Xenopus laevis. J. Comp. Neurol. 2004;472:246–256. doi: 10.1002/cne.20073. [DOI] [PubMed] [Google Scholar]
- 9.Brykczynska U, Tzika AC, Rodriguez I, Milinkovitch MC. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol. Evol. 2013;5:389–401. doi: 10.1093/gbe/evt013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- 11.Haga S, et al. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature. 2010;466:118–122. doi: 10.1038/nature09142. [DOI] [PubMed] [Google Scholar]
- 12.Leinders-Zufall T, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- 13.Hashiguchi Y, Nishida M. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol. Biol. 2006;6:76. doi: 10.1186/1471-2148-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido M, et al. Lineage-specific expansion of vomeronasal type 2 receptor-like (OlfC) genes in cichlids may contribute to diversification of amino acid detection systems. Genome Biol. Evol. 2013;5:711–722. doi: 10.1093/gbe/evt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva L, Antunes A. Vomeronasal receptors in vertebrates and the evolution of pheromone detection. Annu. Rev. Anim. Biosci. 2017;5:353–370. doi: 10.1146/annurev-animal-022516-022801. [DOI] [PubMed] [Google Scholar]
- 17.Picone B, et al. Taste and odorant receptors of the coelacanth: A gene repertoire in transition. J. Exp. Zool. B Mol. Dev. Evol. 2014;322:403–414. doi: 10.1002/jez.b.22531. [DOI] [PubMed] [Google Scholar]
- 18.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido M, et al. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013;23:1740–1748. doi: 10.1101/gr.158105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zapilko V, Korsching SI. Tetrapod V1R-like ora genes in an early-diverging ray-finned fish species: The canonical six ora gene repertoire of teleost fish resulted from gene loss in a larger ancestral repertoire. BMC Genomics. 2016;17:83. doi: 10.1186/s12864-016-2399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido M. Evolution of V1R pheromone receptor genes in vertebrates: diversity and commonality. Genes Genet. Syst. 2019;94:141–149. doi: 10.1266/ggs.19-00009. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Hara Y, et al. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2018;2:1761–1771. doi: 10.1038/s41559-018-0673-5. [DOI] [PubMed] [Google Scholar]
- 24.Sharma K, Syed AS, Ferrando S, Mazan S, Korsching SI. The chemosensory receptor repertoire of a true shark is dominated by a single olfactory receptor family. Genome Biol. Evol. 2019;11:398–405. doi: 10.1093/gbe/evz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marra NJ, et al. White shark genome reveals ancient elasmobranch adaptations associated with wound healing and the maintenance of genome stability. Proc. Natl. Acad. Sci. USA. 2019;116:4446–4455. doi: 10.1073/pnas.1819778116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theisen B, Zeiske E, Breucker H. Functional morphology of the olfactory organs in the spiny dogfish (Squalus acanthias L.) and the small-spotted catshark (Scyliorhinus canicula (L.)) Acta Zoologica. 1986;67:73–86. doi: 10.1111/j.1463-6395.1986.tb00851.x. [DOI] [Google Scholar]
- 27.Ferrando S, et al. Immunolocalization of G-protein alpha subunits in the olfactory system of the cartilaginous fish Scyliorhinus canicula. Anat. Rec. 2009;292:1771–1779. doi: 10.1002/ar.21003. [DOI] [PubMed] [Google Scholar]
- 28.Grus WE, Zhang J. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol. Biol. Evol. 2009;26:407–419. doi: 10.1093/molbev/msn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi X, et al. Tracing the genetic footprints of vertebrate landing in non-teleost ray-finned fishes. Cell. 2021;184:1377–1391. doi: 10.1016/j.cell.2021.01.046. [DOI] [PubMed] [Google Scholar]
- 30.Moriya-Ito K, Hayakawa T, Suzuki H, Hagino-Yamagishi K, Nikaido M. Evolution of vomeronasal receptor 1 (V1R) genes in the common marmoset (Callithrix jacchus) Gene. 2018;642:343–353. doi: 10.1016/j.gene.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 31.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silvotti L, Moiani A, Gatti R, Tirindelli R. Combinatorial co-expression of pheromone receptors. J. Neurochem. 2007;103:1753–1763. doi: 10.1111/j.1471-4159.2007.04877.x. [DOI] [PubMed] [Google Scholar]
- 33.Du K, et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020;4:841–852. doi: 10.1038/s41559-020-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeMaria S, et al. Role of a ubiquitously expressed receptor in the vertebrate olfactory system. J. Neurosci. 2013;33:15235–15247. doi: 10.1523/JNEUROSCI.2339-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J. Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yambe H, et al. L-Kynurenine, an amino acid identified as a sex pheromone in the urine of ovulated female masu salmon. Proc. Natl. Acad. Sci. USA. 2006;103:15370–15374. doi: 10.1073/pnas.0604340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 38.Braasch I, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura Y, Nikaido M. Conserved keratin gene clusters in ancient fish: An evolutionary seed for terrestrial adaptation. Genomics. 2021;113:1120–1128. doi: 10.1016/j.ygeno.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/S0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 42.Poncelet G, Shimeld SM. The evolutionary origins of the vertebrate olfactory system. Open Biol. 2020;10:200330. doi: 10.1098/rsob.200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H, et al. A single pheromone receptor gene conserved across 400 million years of vertebrate evolution. Mol. Biol. Evol. 2018;35:2928–2939. doi: 10.1093/molbev/msy186. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki H, Nikaido M, Hagino-Yamagishi K, Okada N. Distinct functions of two olfactory marker protein genes derived from teleost-specific whole genome duplication. BMC Evol. Biol. 2015;15:245. doi: 10.1186/s12862-015-0530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham JB, et al. Spiracular air breathing in polypterid fishes and its implications for aerial respiration in stem tetrapods. Nat. Commun. 2014;5:3022. doi: 10.1038/ncomms4022. [DOI] [PubMed] [Google Scholar]
- 46.Clack JA. Devonian climate change, breathing, and the origin of the tetrapod stem group. Integr. Comp. Biol. 2007;47:510–523. doi: 10.1093/icb/icm055. [DOI] [PubMed] [Google Scholar]
- 47.Graham JB. Air-Breathing Fishes: Evolution, Diversity, and Adaptation. Academic Press; 1997. [Google Scholar]
- 48.Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019;47:W5–W10. doi: 10.1093/nar/gky874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Hara PJ, et al. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-W. [DOI] [PubMed] [Google Scholar]
- 52.Katoh K, Kuma KI, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flouri T, et al. The phylogenetic likelihood library. Syst. Biol. 2014;64:356–362. doi: 10.1093/sysbio/syu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darriba D, et al. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.