Abstract
Parkinson’s disease (PD) research has largely focused on the disease as a single entity centred on the development of neuronal pathology within the central nervous system. However, there is growing recognition that PD is not a single entity but instead reflects multiple diseases, in which different combinations of environmental, genetic and potential comorbid factors interact to direct individual disease trajectories. Moreover, an increasing body of recent research implicates peripheral tissues and non-neuronal cell types in the development of PD. These observations are consistent with the hypothesis that the initial causative changes for PD development need not occur in the central nervous system. Here, we discuss how the use of neuronal pathology as a shared, qualitative phenotype minimises insights into the possibility of multiple origins and aetiologies of PD. Furthermore, we discuss how considering PD as a single entity potentially impairs our understanding of the causative molecular mechanisms, approaches for patient stratification, identification of biomarkers, and the development of therapeutic approaches to PD. The clear consequence of there being distinct diseases that collectively form PD, is that there is no single biomarker or treatment for PD development or progression. We propose that diagnosis should shift away from the clinical definitions, towards biologically defined diseases that collectively form PD, to enable informative patient stratification. N-of-one type, clinical designs offer an unbiased, and agnostic approach to re-defining PD in terms of a group of many individual diseases.
Subject terms: Genomics, Parkinson's disease, Translational research, Systems biology
Introduction
There is growing recognition that Parkinson’s disease is not a single entity1,2. Rather there are multiple different clinical, genetic and epidemiologically heterogeneous diseases that together are recognised within the one umbrella term of Parkinson’s disease3–5. Hereafter we refer to the multiple diseases as ‘PD’ for simplicity, and to prevent clouding the literature with a new term. Despite growing recognition of this concept, the majority of PD targeted research focuses on the ‘common’-pathological end-point of a linear PD storyline6: the physical manifestation of neuronal inclusions termed Lewy bodies, and the loss of dopaminergic neurons (DAn) within the central nervous system (CNS). This focus on the end-point pathology has proven its worth in the development of effective symptomatic therapies that include Levodopa7. However, the failure of nineteen phase 3 intervention trials8 targeting modification of disease progression illustrates a limitation of this focus. The restricted focus on endpoint pathology largely arises from issues including that PD diagnosis typically occurs many years after disease onset, predominantly on the basis of motor symptoms, and yet one can only study PD patients after this clinical diagnosis is made. The successful development of disease-modifying therapeutics has been further hindered by the absence of biomarkers, and more critically—the absence of informative, molecular mechanisms that define each of the individual diseases that collectively form PD. This is reflected in a lack of PD intervention trials that target specific mechanistic changes in groups of individual patients defined according to the mechanism(s) that contribute to disease development/progression. The SURE-PD3 trial is an exception that targeted only individuals with low serum urate concentrations9. However, beyond the SURE-PD3 trial, there is typically no specific measurable biological signal for the success of a disease-modifying intervention for each disease within the PD umbrella10. Instead, we remain reliant on relatively insensitive and variable clinical measures of PD progression8.
The advent of genome-wide association studies (GWAS) has enabled the identification of variants associated with risk of disease development11, different rates of cognitive decline12, and different rates of progression for PD13,14. However, the conglomeration of datasets needed to achieve the sample size and statistical power required for GWAS perpetuates the one-disease model of PD, and overlooks the presence of multiple different clinical, genetic and epidemiologically heterogeneous diseases. In these situations, the conglomeration of data across multiple different Parkinson diseases dilutes the frequency of specific disease-associated variants and thus reduces the ability to identify those variants that contribute to the trajectory of each individual disease (i.e., the aetiology). As such, the integration of genetic and standardised clinical data into a coherent coordinated approach to slow or prevent PD development, is yet to materialise. Achieving this requires that we move away from a dependence on the shared terminal pathology and clinical definitions and develop a means for patient stratification, using specific genetic information, that is based upon a sound understanding of the aetiology of each contributing molecular disease. But how can you achieve this, when to study the different diseases you must first define them? Here we will discuss how this circular argument can be broken using genetic, molecular and clinical information to identify the different trajectories within PD, from a prospective, disease risk-driven perspective, that stratifies patients and therapeutics without a priori assumptions.
Multiple disease trajectories beneath the Parkinson’s disease umbrella
In 2008, William Weiner wrote “there is no Parkinson disease”1 and suggested Parkinson diseases as a more fitting term for the observed multiple aetiologies. The term Parkinson diseases is consistent with the fact that there is no obvious, predictable disease trajectory following diagnosis, even in monogenic forms of the disease. Rather, each individual’s pathway is unique, or at most shared with a limited number of fellow patients15.
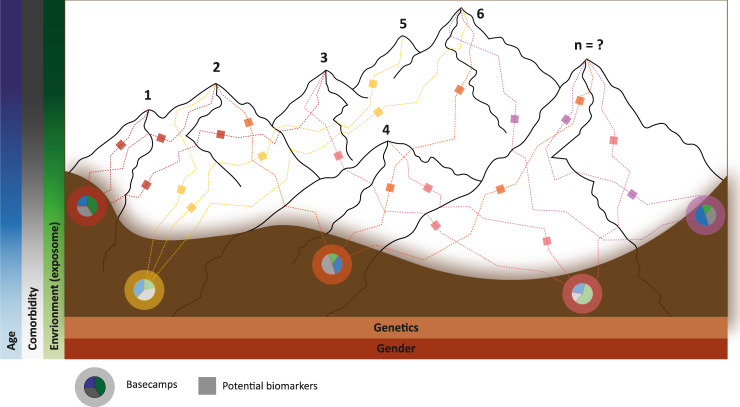
To illustrate the impact that treating PD as a group of diseases with different but overlapping aetiologies4,5,16 can have on our understanding of the disease, let us consider a conceptual model where each disease within PD is represented by a mountain within a range of mountains (Fig. 1). At present we are unable to accurately define the number of different diseases that collectively form PD, thus we have limited our model to seven mountains, for simplicity. In the PD mountain range model, an individual’s genetic risk is represented by the position in the valley (i.e., basecamp) where the individual starts climbing—this position naturally limits the mountain(s) that can be ascended and the route(s) that can be taken. PD patients cluster according to their basecamp, of which there are a limited number, defined by the potential and realised combinations of the risk variants within the genome. Environmental signals from the dynamic basecamp surroundings interact with the individual’s genetic factors to alter aspects of the disease, including onset age at which the patient begins climbing, or whether the individual even develops PD. These environmental signals include, among others: pesticides and pollutants17,18, diet19, viral infection20, head trauma21, inflammatory diseases22 (for an in-depth review on the role of environmental signals in relation to PD genetics see Johnson et al.23). Once an individual has begun ascending a mountain, the topology of the mountain, which represents the intrinsic (e.g. the gut microbiome24 or comorbid disease pathology25) and extrinsic (e.g. exercise26, diet19, and periodic fasting27) factors, influences how quickly each individual climbs the route (i.e., the rate of disease progression), and thus the presentation and severity of symptoms15.
Fig. 1. Parkinson Diseases Mountain Range model.
Conceptual model assimilating the different diseases within PD to mountains within a range. There are likely many more mountains (diseases) than presented in this conceptual model. The topology of the valley floor represents the total variation in interaction between age, environment, comorbidities, sex and genetics of the population. An individual’s genetic risk is represented by the position in the valley (i.e., basecamp) where the individual starts climbing. Different signals (environment, age, comorbidity) from the dynamic basecamp surroundings interact with the individual’s genetic factors to alter aspects of the disease including onset age at which the patient begins climbing, or whether the individual even develops PD (reflected in the pie charts at base camps). The topology of the mountain (e.g. intrinsic and extrinsic factors) affects how quickly each individual climbs the route (i.e., the rate of disease progression), and thus the range, presentation and severity of symptoms15. The small boxes (i.e., checkpoints) along the routes of ascent represent potential biomarkers that could be developed/used to provide an unbiased snap-shot that can be used to track disease development within individual patients. However, these ‘on route’ biomarkers will likely change over the course of the disease.
Individual diseases that together comprise PD are heterogeneous in and of themselves. This is represented in our model by the existence of multiple routes to each mountain summit. These routes are not independent, merging and diverging, meaning it is likely that individuals can switch between the routes dependent on their particular combination of intrinsic and extrinsic factors. Although heterogeneity likely exists within each disease, it would ideally be sufficiently homogeneous to provide a single therapeutic target for treatment development. Furthermore, each route has different markers, or checkpoints, at different stages—akin to the biomarkers that provide an unbiased snap-shot that can be used to track disease development within individual patients. It is important to note however that these ‘on route’ biomarkers will change over the course of the disease, and are likely to be influenced by the individual’s age, diet, and combination of predisposing comorbid diseases.
It can be argued that there are commonalities across individual diseases that contribute to PD (i.e., shared between the different mountains within the range). Treating these commonalities would provide treatment for a larger group of patients. This may be true. However, whilst potentially useful, treating these commonalities would have limited benefit, as the symptoms (e.g. resting tremor and bradykinesia) appear late in the disease course, and thus patients would be more disabled (closer to their respective summits) by the time the treatment is initiated. Notably, disease-modifying interventions that target PD based on this premise have yielded little success thus far.
Other models of PD have been presented before. Perhaps best known is William Langston’s elephant model28 which captures the idea of diverse symptomology but still presents PD as a single disease, or, elephant. In our model, the elephant would be represented as a single mountain within the PD mountain range. Thus Langston’s model does not capture the multiple diseases that collectively form PD, or the heterogeneity that is inherent to each disease.
Using ‘omics to inform origins and trajectories of Parkinson’s disease
It is the patient’s combination of genetic risk coding (e.g. LRRK2-G2019S or SNCA-A53T) and non-coding variants that initially “set the stage” and determine which basecamp and mountain an individual will start ascending in their journey towards PD. The application of GWAS to the study of PD enables unbiased population-level identification of the genetic basis of risk that exist long before the disease initiates. However, the genetic variants that have been associated with PD by GWAS (e.g. 90 genetic loci11) only explain between 16-36% of the heritability of PD. Additionally, apart from a few exceptions, the odds ratios of the individual variants are typically low (e.g. between 0.8 – 1.2)11. Indeed, the current predictive ability of the SNPs associated with PD is so low as to make meaningful risk score prognosis unfeasible29. The missing heritability can partly be explained by issues with merging the multiple different diseases that contribute to PD, into the single entity that is defined by late-stage pathological markers (i.e., performing GWAS from the perspective of PD being a single disease). Furthermore, the reliance on a clinical definition means that no two ‘omics studies yield similar results since they only represent those of the heterogeneous patient population from which they were applied (e.g.30). Averaging these different but related datasets results in the identification of only the most significant risk loci that are common across all the diseases reaching statistical significance. The issue of averaging signals across the heterogeneous diseases that contribute to PD, when undertaking a GWAS, can be addressed by stratifying PD patients according to their genetically defined start-point, in turn enabling selection of informative longitudinal biomarkers and effective therapeutic approaches (specific to each route). This stratification can be achieved through genomic approaches that explore the specificities of GWAS manifestation31,32, and inform the distinct routes of PD development. As such, GWAS-based patient stratification could indicate 1) which pathway(s) is dysregulated; 2) pathway biomarkers to be examined; and 3) which targets should be considered for therapeutic intervention. However, shifting from simply identifying GWAS signals to informative stratification requires in depth characterisation of the causative variant(s) function33.
Until recently33, our inability to functionally translate non-coding genetic variation and risk to biologically disease-relevant pathways has meant that the earlier stages of PD development have been primarily neglected as a means of stratification or therapeutic intervention. In contrast to the noncoding risk variants, coding mutations in GBA and LRRK2 genes have been explored and enabled patients with these specific mutations to be stratified for therapeutic intervention, targeting these genetic subgroups of patients34,35. Furthermore, Szwedo et al. demonstrated a role for APOE-ε4 and GBA mutations in the rate of cognitive decline in PD patients, but found no significant impact for common variants in SNCA and MAPT12. These findings raise the possibility for earlier identification and stratification of individuals at high risk of rapid cognitive decline, thus highlighting suitable candidates for future targeted trials. Despite progress, the known incomplete penetrance of these mutations is problematic36 and highlights a remaining knowledge gap surrounding the mechanistic role of some of these mutations, such as the role of LRRK2 mutations in disease progression14. This therefore raises the question as to whether such interventions will be effective against disease progression even in patients with these specific mutations. Nonetheless, with recent advancements, our understanding of how both coding and non-coding risk manifests is evolving33,37. Such understanding can be used to inform hypotheses which will aid in the identification and stricter classification of individual diseases within PD that could also lead to targeted therapeutics.
Functional characterisation requires that the associated molecular, cellular and physiological phenotyping is sufficiently deep to allow accurate assignment of the causal variants and their target genes38, and potentially what tissue(s), the disease risk is conveyed in. Panyard et al. applied an approach to functionally characterise and assign the action of causal genetic variants in Alzheimer’s disease (AD)39. Briefly, Panyard et al. integrated genomic and clinical data from two longitudinal AD cohorts with epigenetic annotations to develop cell-type-specific genomic functional annotations39. These annotations were used to identify which SNPs are likely to be functional in different tissues39. The authors demonstrated that effects of these SNPs in the liver were statistically associated with Alzheimer’s diagnosis39. In so doing, Panyard et al. highlighted a potential contribution from the liver towards AD, including associations with core AD cerebrospinal-fluid biomarkers, in what is widely considered a ‘brain-centric’ disease. Whilst a small study (n = 79 AD patients), the finding that changes in the liver were predictive for some, but not all, individuals is consistent with the hypothesis that the liver malfunction accounts for one of the heterogeneous diseases that collectively contribute to AD40.
Genomic approaches are also being applied in attempts to identify and understand the cell- and tissue- types where genetic risk manifests in PD41,42. For example, Coetzee et al.43 used histone modification data combined with enrichment analyses to demonstrate that many PD-associated genetic variants were enriched, and had expression quantitative trait loci (eQTL) associations, in non-neuronal cell-types, including lymphocytes, mesendoderm-, liver- and adipocyte- cells43. Similarly, we have used a discovery-based approach to identify putative regulatory impacts of non-coding PD-associated risk variants in both the CNS and peripheral tissues33,44. Notably, our analyses indicated that eQTL effects for a subset (28%) of the 90 PD-associated risk variants were only detected in peripheral tissues (e.g. thyroid and oesophagus)33 while only 2% of PD risk SNPs had identifiable eQTLs solely in CNS tissues. Given that tissues are complex mixtures of cell types, the oesophageal finding does not imply that the effect is due to the muscles at the exclusion of the nerves that innervate the oesophagus. However, the finding is consistent with peripheral symptomology (e.g. dysphagia), that is sometimes observed in the early stages of PD45.
In an attempt to determine which tissues, and subsequently cell-types, are responsible for PD heritability, Reynolds et al.41 used stratified Linkage Disequilibrium score regression46 (see box 1) to measure the contribution(s) that common genetic variation makes to the heritability of PD across 53 tissues (inc. 13 brain region tissues), using schizophrenia as a comparative measure. In contrast to schizophrenia in which all 13 brain tissues were significantly enriched for heritability, there was no enrichment for PD heritability across any of the 53 tissues (in the CNS or peripheral tissues). The lack of PD heritability enrichment across these bulk tissues led Reynolds et al. to question whether cellular heterogeneity within tissues may be masking signals, and thus sought to investigate cell-type-specific enrichment of heritability. However, across 6 human and 30 mouse CNS cell-types, Reynolds et al. identified no cell-type enrichment for PD heritability. The Lewy Body pathology in specific neuronal cell types, associated with PD, has encouraged researchers to focus efforts towards understanding risk in neuronal subtypes. However, the findings from Reynolds et al. provide reason to believe that risk loci are affecting non-CNS cell-types and/or cellular processes and pathways across multiple cell types, and to which different cell types have varying vulnerability41. Such varying vulnerability, consistent with the proposed threshold theory for PD47, could likely be a result of interactions with environmental factors and/or comorbid disease pathology.
In contrast to the lack of cell-type heritability enrichment identified by Reynolds et al., there have been multiple studies to date that implicate glial cell types, mostly microglia, in neuroinflammation and PD pathogenesis42,48,49. Given these implications, Bryois et al. combined cell-type-specific gene expression and GWAS data to explore the role of glial cells in PD pathogenesis49. Roles for microglia were indicated by the finding that cell-type-specific ATACseq identified functional PD risk loci that were enriched for autophagy and lysosomal processes50, both of which have been previously implicated in PD51. Furthermore, elevated LRRK2 expression, associated with the linked PD GWAS SNPs rs76904798 and rs7294619 (R2 = 0.842), has also been shown to occur specifically in microglia42. Collectively, these data are consistent with the hypothesis that PD genetic risk variants affect non-neuronal cell types of the CNS. However, while these studies highlight the importance of cell-type consideration, they are still driven by a priori assumptions that are CNS focused. As such, it is essential to extend these analyses to non-CNS cell-types, following a more discovery-based, hypothesis-free approach, to determine if such risk enrichment is truly specific to the microglia, or if other non-CNS cell-types may also be involved in disease initiation and propagation.
Together these studies highlight how multiple ‘omics approaches can be used to identify the tissue- and cell-type-specific manifestations for GWAS risk variants. The findings we have discussed support two potential, non-mutually exclusive, hypotheses: First, the individual diseases within the PD umbrella may arise through genetic variation-dependent mechanisms that dictate the tissue-of-origin(s) and thus the pathological pathways associated with the disease. This concept is reflected in the mountain range model, with each basecamp representing a different, genetically-informed, start-point. In the second hypothesis, variants impacting a specific peripheral tissue- or cell-type, cause dysregulation that adds to the disease complexity/symptoms without necessarily leading to the CNS pathology that is typically associated with PD. This second hypothesis aligns with the threshold theory for PD which was developed on the basis of parallel degeneration of both the central and peripheral nervous systems47. As such, there is a need to look beyond the tissue- and cell-types that are traditionally associated with PD pathology to gain a greater understanding of the mechanisms through which genetic risk may be manifested. Advances within the fields of single-cell transcriptomics52,53 and bulk-cell analyses54 will provide additional insights that begin to untangle the relative contributions of genetics and the environment to PD risk manifestation. But the question remains, how do we apply these approaches to a mechanistically-heterogeneous disease?
Box 1 Glossary of terms.
Expression quantitative trait loci (eQTL): A genetic locus that affects (or correlates with) the expression (mRNA) of one or more genes.
Genome wide association study (GWAS): An approach used to associate specific genetic variations with particular diseases or traits. The genomes of individuals with the disease or trait of interest are compared to the genomes of matched, control, individuals – to identify variants that are significantly associated with that particular disease or phenotypic trait.
Infinitesimal model: A model built on the premise that the inheritance of a quantitative trait is controlled by an infinite number of loci, and each locus has an infinitely small effect.
Linkage disequilibrium (LD): The non-random association of alleles at different loci.
Linkage disequilibrium score regression (LDSC): A statistical method for quantifying the separate contributions of polygenic effects and various confounding effects, such as population stratification, based on summary statistics from GWA studies.
N-of-1 approach: In this context, an n-of-1 analysis is a meta-analyses of deeply characterised single patient information, of individuals within a heterogeneous cohort, that explores genetic variation in the context of the measured phenotype(s). In effect, the characteristics of each participant are individually (and frequently where possible) noted and contrasted to each other individual. This approach accounts for the individual-level heterogeneity that is present in PD.
Omnigenic model of complex disease: The model is centred on the premise that human genome regulatory networks are hugely interconnected, and almost any gene with regulatory variants in at least one relevant tissue will contribute to the heritability of the phenotype.
Single nucleotide polymorphism (SNP): The most common type of variation among people. One SNP represents a variation at a single position (i.e. nucleotide) in the DNA sequence.
Using big data to identify individual trajectories in a heterogeneous disease
Conglomerating data from different cohorts provides a large sample size (n) which is otherwise unachievable from a single-centre cohort. As such, conglomerated data provides much-needed statistical power to address particular hypotheses. Despite providing statistical power, the conglomeration of different PD cohorts unfortunately also highlights the lack of strict diagnostic criteria for PD and related diseases, with different cohorts often using different diagnostic criteria30. A further confounding problem, that affects diagnosis even at the level of a single clinician, is misclassification55. Such misclassification raises the problem of inclusion of non-PD patients in cohorts, which may be skewing outcomes of observational studies and clinical trials. A third, and substantial, complicating factor is the likely multiple different mechanistic diseases that exist within the ‘homogenous’ clinical PD cohorts currently studied. This problem is particularly prevalent in cohorts that include patients with different genetic predispositions to diseases within PD, such as GBA-PD and LRRK2-PD patients, who typically present with different symptomatic trajectories56,57. Grouping these different individual diseases together is likely causing a loss of information. If data conglomeration is to achieve what is hoped, disease biomarkers, and more specifically biomarkers for the different diseases that collectively form PD are urgently needed. The need to define individual diseases as opposed to merging them into a single entity is in line with the prediction made by Espay and Lang that smaller, smarter clinical trials are needed to move away from this ‘homogeneous’ clinical Parkinson’s phenotype6.
As discussed earlier, genetic risk variants offer an option for such genetic stratification—with an individual’s risk profile determining their disease starting point (e.g. specific basecamp in the mountain range model). These genetic risk variants, or SNPs, do not however act independently33. Rather they act in a combinatorial manner within a much larger genetic background. In order to understand the full contribution that PD-associated SNPs make to PD, they need to be considered in the context of the omnigenic58 and infinitesimal59 models for disease (see box 1), and in terms of network medicine60. Network medicine approaches enable the disease to be contextualised as a sum of inter-connected perturbations, reflective of the underlying genetic and molecular risk drivers (i.e. studying PD risk variants in the context of an individual’s complete genotype). The utility of network medicine60 has being explored in other complex diseases, and has already aided in the identification of novel targets for therapeutic strategies and development61,62.
Exploring the impact and interconnectivity of genetic contributions to an individual’s disease risk profile, from a network medicine angle, has only become feasible following recent advancements. These include the reductions in costs for genome sequencing and computing63, and the development of machine learning approaches to detect complex patterns in genomes. Such advances have informed, and been enhanced by, the rapidly evolving post-GWAS genome-editing toolbox, including CRISPR screens64 and massively parallel reporter assays65 (to test observed patterns for functional significance). These tools will over time provide the data required to understand the complete genetic contributions to the development of the diseases that collectively form PD, amongst other complex diseases. Collaborative efforts, such as the Atlas of Variant Effects Alliance (https://www.varianteffect.org/)66, will be critical in enabling the curation and systematic collation of results from these functional post-GWAS studies. Another recent technological advance that will likely enhance genomic findings from a phenotypic perspective is the introduction of wearables67. Such devices have been shown to provide vital sign data (e.g. heart rate and electrodermal activity) at a level equivalent to that gained in a clinical setting68. The widespread uptake of these wearables enables individualised, longitudinal and continuous health monitoring. While identifying signal from noise in movement measurements is challenging, combining the in-depth phenotypic data that wearables provide with matched genetic data promises to aid in identifying clinical differences amongst the different genomic diseases within PD.
Information on genetic variation and drug responses can be used to help determine which drugs, are likely to be safe and efficacious in an individual. These approaches are leading to the emergence of ‘genetically-informed’ clinical trials (i.e., precision medicine approaches) in PD69–71. For example, Ambroxol has been repurposed to treat PD patients with a GBA coding mutation34. Despite having only been trialled in a small, open label, non-randomised group of individuals, Ambroxol shows promise for the treatment of this well-defined yet heterogeneous (i.e., it included multiple GBA coding mutations) subset of individuals34. The Ambroxol trial is an exemplar that paves the way for future precision-informed clinical trials in PD. Not only does it address the issue of treating patients according to genomic information, but also shows the potential of repurposing already licensed medication72, to accelerate the process of drug development. The Ambroxol trial also included some idiopathic PD patients—of whom also showed promising responses to the treatment. Identifying idiopathic PD patients who specifically have reduced GCase activity (i.e., those with GBA modifying genotypes30,44) may lead to better outcomes for patients.
Despite the obvious promise of a stratified approach to clinical testing and therapy, the lack of genotyping as a part of clinical assessment means that the identification of the relatively small numbers of individuals with genetic predispositions remains a major financial and temporal challenge. However, this is changing as initiatives, such as PD frontline (https://pdfrontline.com/en) and PD GENEration (https://www.parkinson.org/PDGENEration), are offering genetic testing for PD patients to ensure individuals carrying defined mutations are referred to the clinical trials best suited to them.
Concluding remarks & future perspectives
Recognising that many diseases contribute to PD highlights a challenge that is present in the search for a biomarker of PD progression and therapeutics. Specifically, if there are many diseases subsumed within the umbrella of PD, then we should be looking for biomarkers for each individual disease. That we continue selecting patients on the basis of clinical criteria rather than biological ones impairs our ability to do this. Even genetic risk for PD is currently viewed within the context of the shared pathology that connects the different Parkinson diseases. The utility of network medicine60 has been established in other complex diseases, aiding the identification of novel targets for therapeutic strategies and development61,62. While it is certainly true that further initiatives involving large-scale data conglomeration will aid in the molecular and clinical understanding of the disease, the lack of uniformity in PD diagnosis and disease trajectories will likely confound findings from genomic and biomarker studies30,73. Recent initiatives (e.g. PREDICT-PD74 and the Cincinnati Cohort Biomarker Program (CCBP)75) that incorporate discovery-based analyses of prospective cohorts are seeking to address this by defining PD developmental pathways and biomarkers. Furthermore, we contend that it is time to consider systematic n-of-176–78 approaches (see box 1) in PD research, to identify the combinations and relative contributions of the genetic, pathological and environmental factors in each unique circumstance3,79, for individuals within a heterogeneous population. The population’s use of wearables will contribute to the collection of relevant data for achieving such an approach80. Ultimately, the aggregated results of n-of-1 approaches will help elucidate the many diseases that contribute to the one complex Parkinson disease. Redefining the hypotheses driving PD research will enable movement away from the current focus on shared pathology and clinical definitions. This in turn will make way for the development of targeted diagnostic and therapeutic approaches that are based upon a molecular understanding of the aetiology of the individual diseases, and thus have the ability to slow, stop or reverse disease progression and ultimately achieve disease prevention.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
S.F., A.C., and J.M.O’S. were funded by the Michael J Fox Foundation for Parkinson’s research and the Silverstein Foundation for Parkinson’s with GBA—grant ID 16229 to J.M.O’S. S.F. and J.M.O’S. were funded by the Neurological Foundation—grant ID 3721588 (2008 SPG). S.F. was funded by the Dines Family Charitable Trust. A.C. received grant funding from the Australian Government.
Author contributions
S.F. conceived and wrote the first draft, and revised subsequent drafts. A.C. and J.M.O’S. contributed to conception, critical review, and revision of the paper. All authors approved the final draft for submission.
Data availability
There is no data to share that is specific to this perspective paper. All information is included in the references.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-022-00307-w.
References
- 1.Weiner WJ. There is no Parkinson disease. Arch. Neurol. 2008;65:705–708. doi: 10.1001/archneur.65.6.705. [DOI] [PubMed] [Google Scholar]
- 2.Espay AJ, et al. Disease modification and biomarker development in Parkinson disease. Neurology. 2020;94:481–494. doi: 10.1212/WNL.0000000000009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mestre TA, et al. Parkinson’s Disease Subtypes: Critical Appraisal and Recommendations. J. Parkinsons. Dis. 2021;11:395–404. doi: 10.3233/JPD-202472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espay AJ, Brundin P, Lang AE. Precision medicine for disease modification in Parkinson disease. Nat. Rev. Neurol. 2017;13:119–126. doi: 10.1038/nrneurol.2016.196. [DOI] [PubMed] [Google Scholar]
- 5.Bloem BR, Okun MS, Klein C. Parkinson’s disease. Lancet. 2021;397:2284–2303. doi: 10.1016/S0140-6736(21)00218-X. [DOI] [PubMed] [Google Scholar]
- 6.Espay AJ, Lang AE. Parkinson diseases in the 2020s and beyond: Replacing clinico-pathologic convergence with systems biology divergence. J. Parkinsons. Dis. 2018;8:S59–S64. doi: 10.3233/JPD-181465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paoletti FP, Tambasco N, Parnetti L. Levodopa treatment in Parkinson’s disease: earlier or later? Ann. Transl. Med. 2019;7:S189–S189. doi: 10.21037/atm.2019.07.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espay, A. J. & Stecher, B. Brain Fables: The hidden history of neurodegenerative diseases and a blueprint to conquer them. 10.1017/9781108888202 (Cambridge University Press, 2020).
- 9.Schwarzschild MA, et al. Effect of Urate-Elevating Inosine on Early Parkinson Disease Progression: The SURE-PD3 Randomized Clinical Trial. JAMA. 2021;326:926–939. doi: 10.1001/jama.2021.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espay AJ, et al. Biomarker‐driven phenotyping in Parkinson’s disease: A translational missing link in disease‐modifying clinical trials. Mov. Disord. 2017;32:319–324. doi: 10.1002/mds.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szwedo, A. A. et al. GBA and APOE Impact Cognitive Decline in Parkinson’s Disease: A 10-Year Population-Based Study. Mov. Disord. 10.1002/MDS.28932 (2022). [DOI] [PMC free article] [PubMed]
- 13.Tan MMX, et al. Genome‐Wide Association Studies of Cognitive and Motor Progression in Parkinson’s Disease. Mov. Disord. 2020;36:424–433. doi: 10.1002/mds.28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, et al. Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson’s disease. Nat. Genet. 2021;53:787–793. doi: 10.1038/s41588-021-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravan A, et al. Non-motor symptoms in an Indian cohort of Parkinson’s disease patients and correlation of progression of non-motor symptoms with motor worsening. Neurol. India. 2015;63:166. doi: 10.4103/0028-3886.156276. [DOI] [PubMed] [Google Scholar]
- 16.Berg D, et al. Prodromal Parkinson disease subtypes — key to understanding heterogeneity. Nat. Rev. Neurol. 2021;17:349–361. doi: 10.1038/s41582-021-00486-9. [DOI] [PubMed] [Google Scholar]
- 17.Pezzoli G, Cereda E. Exposure to pesticides or solvents and risk of Parkinson disease. Neurology. 2013;80:2035–2041. doi: 10.1212/WNL.0b013e318294b3c8. [DOI] [PubMed] [Google Scholar]
- 18.Kamel F. Paths from pesticides to Parkinson’s. Science. 2013;341:722–723. doi: 10.1126/science.1243619. [DOI] [PubMed] [Google Scholar]
- 19.Paknahad Z, Sheklabadi E, Derakhshan Y, Bagherniya M, Chitsaz A. The effect of the Mediterranean diet on cognitive function in patients with Parkinson’s disease: A randomized clinical controlled trial. Complement. Ther. Med. 2020;50:102366. doi: 10.1016/j.ctim.2020.102366. [DOI] [PubMed] [Google Scholar]
- 20.Olsen LK, Dowd E, McKernan DP. A role for viral infections in Parkinson’s etiology? Neuronal Signal. 2018;2:20170166. doi: 10.1042/NS20170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul KC, et al. The association between lifestyle factors and Parkinson’s disease progression and mortality. Mov. Disord. 2019;34:58–66. doi: 10.1002/mds.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villumsen M, Aznar S, Pakkenberg B, Jess T, Brudek T. Inflammatory bowel disease increases the risk of Parkinson’s disease: a Danish nationwide cohort study 1977–2014. Gut. 2019;68:18–24. doi: 10.1136/gutjnl-2017-315666. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ME, Stecher B, Labrie V, Brundin L, Brundin P. Triggers, Facilitators, and Aggravators: Redefining Parkinson’s Disease Pathogenesis. Trends Neurosci. 2019;42:4–13. doi: 10.1016/j.tins.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinzel S, et al. Gut Microbiome Signatures of Risk and Prodromal Markers of Parkinson Disease. Ann. Neurol. 2020;88:320–331. doi: 10.1002/ana.25788. [DOI] [PubMed] [Google Scholar]
- 25.Cheong JLY, de Pablo-Fernandez E, Foltynie T, Noyce AJ. The Association Between Type 2 Diabetes Mellitus and Parkinson’s Disease. J. Parkinsons. Dis. 2020;10:775–789. doi: 10.3233/JPD-191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crotty GF, Schwarzschild MA. Chasing Protection in Parkinson’s Disease: Does Exercise Reduce Risk and Progression? Front. Aging Neurosci. 2020;12:1–11. doi: 10.3389/fnagi.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neth BJ, Bauer BA, Benarroch EE, Savica R. The Role of Intermittent Fasting in Parkinson’s Disease. Front. Neurol. 2021;12:1–7. doi: 10.3389/fneur.2021.682184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langston JW. The Parkinson’s complex: Parkinsonism is just the tip of the Iceberg. Ann. Neurol. 2006;59:591–596. doi: 10.1002/ana.20834. [DOI] [PubMed] [Google Scholar]
- 29.Koch S, et al. Validity and Prognostic Value of a Polygenic Risk Score for Parkinson’s Disease. Genes (Basel) 2021;12:1859. doi: 10.3390/genes12121859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Sullivan JM, Heijer JM, Groeneveld GJ, Cooper AA. Intronic Haplotypes in GBA Modify Age at Diagnosis of Parkinson’s: Replication in a Subgroup. Mov. Disord. 2021;36:1468–1470. doi: 10.1002/mds.28620. [DOI] [PubMed] [Google Scholar]
- 31.Novikova G, et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 2021;12:1–14. doi: 10.1038/s41467-021-21823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, et al. The landscape of multiscale transcriptomic networks and key regulators in Parkinson’s disease. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrow SL, et al. Establishing gene regulatory networks from Parkinson’s disease risk loci. Brain. 2022;139:1–36. doi: 10.1093/brain/awac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullin S, et al. Ambroxol for the Treatment of Patients With Parkinson Disease With and Without Glucocerebrosidase Gene Mutations. JAMA Neurol. 2020;77:427–434. doi: 10.1001/jamaneurol.2019.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X, Ren F. Leucine-rich repeat kinase 2 inhibitors: a patent review (2014-present) Expert Opin. Ther. Pat. 2020;30:275–286. doi: 10.1080/13543776.2020.1729354. [DOI] [PubMed] [Google Scholar]
- 36.Anheim M, et al. Penetrance of Parkinson disease in glucocerebrosidase gene mutation carriers. Neurology. 2012;78:417–420. doi: 10.1212/WNL.0b013e318245f476. [DOI] [PubMed] [Google Scholar]
- 37.Ho D, et al. Machine Learning Identifies Six Genetic Variants and Alterations in the Heart Atrial Appendage as Key Contributors to PD Risk Predictivity. Front. Genet. 2022;12:2409. doi: 10.3389/fgene.2021.785436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karthik Jagadeesh, A. A. et al. Identifying disease-critical cell types and cellular processes across the human body by integration of single-cell profiles and human genetics. bioRxiv 2021.03.19.436212 10.1101/2021.03.19.436212 (2021).
- 39.Panyard, D. J. et al. Liver-specific polygenic risk score is more strongly associated than genome-wide score with Alzheimer’s disease diagnosis in a case-control analysis. medRxiv10.1101/2021.04.29.21256279 (2021).
- 40.Nho K, et al. Association of Altered Liver Enzymes With Alzheimer Disease Diagnosis, Cognition, Neuroimaging Measures, and Cerebrospinal Fluid Biomarkers. JAMA Netw. Open. 2019;2:e197978–e197978. doi: 10.1001/jamanetworkopen.2019.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds RH, et al. Moving beyond neurons: the role of cell type-specific gene regulation in Parkinson’s disease heritability. npj Park. Dis. 2019;5:1–14. doi: 10.1038/s41531-019-0074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langston, R. G. et al. Association of a Common Genetic Variant with Parkinson’s Disease is Propagated through Microglia. bioRxiv10.1101/2021.01.15.426824 (2021).
- 43.Coetzee SG, et al. Enrichment of risk SNPs in regulatory regions implicate diverse tissues in Parkinson’s disease etiology. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep30509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schierding W, et al. Common Variants Coregulate Expression of GBA and Modifier Genes to Delay Parkinson’s Disease Onset. Mov. Disord. 2020;35:1346–1356. doi: 10.1002/mds.28144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fasano A, Visanji NP, Liu LWC, Lang AE, Pfeiffer RF. Gastrointestinal dysfunction in Parkinson’s disease. Lancet Neurol. 2015;14:625–639. doi: 10.1016/S1474-4422(15)00007-1. [DOI] [PubMed] [Google Scholar]
- 46.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engelender S, Isacson O. The Threshold Theory for Parkinson’s Disease. Trends Neurosci. 2017;40:4–14. doi: 10.1016/j.tins.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Andersen MS, et al. Heritability Enrichment Implicates Microglia in Parkinson’s Disease Pathogenesis. Ann. Neurol. 2021;89:942–951. doi: 10.1002/ana.26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryois J, et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 2020;52:482–493. doi: 10.1038/s41588-020-0610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Booms, A., Pierce, S. E. & Coetzee, G. A. Parkinsons disease genetic risk evaluation in microglia highlights autophagy and lysosomal genes. bioRxiv 2020.08.17.254276 10.1101/2020.08.17.254276 (2020).
- 51.Senkevich K, Gan-Or Z. Autophagy lysosomal pathway dysfunction in Parkinson’s disease; evidence from human genetics. Park. Relat. Disord. 2019;73:60–71. doi: 10.1016/j.parkreldis.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Stuart T, Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 53.Aldridge S, Teichmann SA. Single cell transcriptomics comes of age. Nat. Commun. 2020;11:1–4. doi: 10.1038/s41467-020-18158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Przytycki PF, Pollard KS. CellWalker integrates single-cell and bulk data to resolve regulatory elements across cell types in complex tissues. Genome Biol. 2021;22:1–16. doi: 10.1186/s13059-021-02279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizzo G, et al. Accuracy of clinical diagnosis of Parkinson disease. Neurology. 2016;86:566–576. doi: 10.1212/WNL.0000000000002350. [DOI] [PubMed] [Google Scholar]
- 56.Malek N, et al. Features of GBA-associated Parkinson’s disease at presentation in the UK Tracking Parkinson’s study. J. Neurol. Neurosurg. Psychiatry. 2018;89:702–709. doi: 10.1136/jnnp-2017-317348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan-Or Z, et al. LRRK2 and GBA mutations differentially affect the initial presentation of Parkinson disease. Neurogenetics. 2010;11:121–125. doi: 10.1007/s10048-009-0198-9. [DOI] [PubMed] [Google Scholar]
- 58.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barton NH, Etheridge AM, Véber A. The infinitesimal model: Definition, derivation, and implications. Theor. Popul. Biol. 2017;118:50–73. doi: 10.1016/j.tpb.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee LY, Pandey AK, Maron BA, Loscalzo J. Network medicine in Cardiovascular Research. Cardiovasc. Res. 2020;117:2186–2202. doi: 10.1093/cvr/cvaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozturk K, Dow M, Carlin DE, Bejar R, Carter H. The Emerging Potential for Network Analysis to Inform Precision Cancer Medicine. J. Mol. Biol. 2018;430:2875–2899. doi: 10.1016/j.jmb.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Preston, J., VanZeeland, A. & Peiffer, D. A. Innovation at Illumina: The road to the $600 human genome. Nat. Portf. (2021).
- 64.Gasperini M, et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens HHS Public Access. Cell. 2019;176:377–390. doi: 10.1016/j.cell.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tewhey R, et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fowler, D. M. et al. The Atlas of Variant Effects (AVE) Alliance: understanding genetic variation at nucleotide resolution. Zenodo (2021).
- 67.DeGeurin, M. Wearables saw explosive—but conditional—growth in 2020 - Insider Intelligence Trends, Forecasts & Statistics. eMarketerhttps://www.emarketer.com/content/wearables-saw-explosive-conditional-growth-2020 (2021).
- 68.Dunn J, et al. Wearable sensors enable personalized predictions of clinical laboratory measurements. Nat. Med. 2021;27:1105–1112. doi: 10.1038/s41591-021-01339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheehan P, et al. PR001 gene therapy improved phenotypes in models of Parkinson’s disease with GBA1 mutation. Alzheimer’s Dement. 2020;16:e043614. [Google Scholar]
- 70.Skrahina V, et al. The Rostock International Parkinson’s Disease (ROPAD) Study: Protocol and Initial Findings. Mov. Disord. 2021;36:1005–1010. doi: 10.1002/mds.28416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider SA, Alcalay RN. Precision medicine in Parkinson’s disease: emerging treatments for genetic Parkinson’s disease. J. Neurol. 2020;267:860–869. doi: 10.1007/s00415-020-09705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pushpakom S, et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2018;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 73.Toffoli, M. et al. Intronic Haplotypes in the GBA Gene Do Not Predict Age at Diagnosis of Parkinson’s Disease. Mov. Disord. mds.28616 10.1002/mds.28616 (2021). [DOI] [PMC free article] [PubMed]
- 74.Noyce AJ, et al. PREDICT-PD: Identifying risk of Parkinson’s disease in the community: Methods and baseline results. J. Neurol. Neurosurg. Psychiatry. 2014;85:31–37. doi: 10.1136/jnnp-2013-305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sturchio A, et al. Phenotype-Agnostic Molecular Subtyping of Neurodegenerative Disorders: The Cincinnati Cohort Biomarker Program (CCBP) Front. Aging Neurosci. 2020;12:324. doi: 10.3389/fnagi.2020.553635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lillie EO, et al. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Per. Med. 2011;8:161–173. doi: 10.2217/pme.11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520:609–611. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 78.Margolis A, Giuliano C. Making the switch: From case studies to N-of-1 trials. Epilepsy Behav. Rep. 2019;12:100336. doi: 10.1016/j.ebr.2019.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patrick KL, Bell SL, Weindel CG, Watson RO. Exploring the ‘multiple-hit hypothesis’ of neurodegenerative disease: Bacterial infection comes up to bat. Front. Cell. Infect. Microbiol. 2019;9:1–18. doi: 10.3389/fcimb.2019.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riggare S, Hägglund M. Precision Medicine in Parkinson’s Disease – Exploring Patient-Initiated Self-Tracking. J. Parkinsons. Dis. 2018;8:441–446. doi: 10.3233/JPD-181314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There is no data to share that is specific to this perspective paper. All information is included in the references.