Abstract
We investigated hearing impairment (HI) in 51 families from Ghana with at least two affected members that were negative for GJB2 pathogenic variants. DNA samples from 184 family members underwent whole-exome sequencing (WES). Variants were found in 14 known non-syndromic HI (NSHI) genes [26/51 (51.0%) families], five genes that can underlie either syndromic HI or NSHI [13/51 (25.5%)], and one syndromic HI gene [1/51 (2.0%)]. Variants in CDH23 and MYO15A contributed the most to HI [31.4% (16/51 families)]. For DSPP, an autosomal recessive mode of inheritance was detected. Post-lingual expression was observed for a family segregating a MARVELD2 variant. To our knowledge, seven novel candidate HI genes were identified (13.7%), with six associated with NSHI (INPP4B, CCDC141, MYO19, DNAH11, POTEI, and SOX9); and one (PAX8) with Waardenburg syndrome. MYO19 and DNAH11 were replicated in unrelated Ghanaian probands. Six of the novel genes were expressed in mouse inner ear. It is known that Pax8-/- mice do not respond to sound, and depletion of Sox9 resulted in defective vestibular structures and abnormal utricle development. Most variants (48/60; 80.0%) have not previously been associated with HI. Identifying seven candidate genes in this study emphasizes the potential of novel HI genes discovery in Africa.
Subject terms: Medical genetics, Molecular medicine
Whole-exome sequencing for 51 families in Ghana reveals 7 candidate genes associated with hearing impairment, 6 of which have previously been demonstrated to be expressed in the mouse inner ear.
Introduction
Hearing impairment (HI) is a sensory disorder that can be mild to profound and has an incidence of 2–3 out of 1000 live births in developed countries and twice as high in developing countries1. HI is often caused by acquired factors such as infectious diseases and antibiotic exposure2–4. Genetic factors are known to cause ~50% of congenital HI2,5, and to date, >120 non-syndromic (NS) HI genes have been identified6. GJB2 is the most common gene associated with autosomal recessive (AR) NSHI among European populations7–9.
Although connexin genes are the most widely reported HI genes, their contribution to HI in sub-Saharan African populations is negligible10. However, Ghana is an exception with a high frequency of the founder GJB2-p.R143W variant3,11,12, which accounts for at least 27% of HI in Ghanaian families segregating ARNSHI13, 8% of isolated NSHI cases, and a population carrier frequency of 0.7%3.
Considering the highly heterogeneous status of HI6, next-generation sequencing platforms and particularly whole-exome sequencing (WES) have recently enhanced the discovery of novel HI gene variants14,15. In the current study of families from Ghana, we used WES to investigate rare variants in GJB2-negative families segregating HI.
Results
Patients’ demographics and phenotypic descriptions
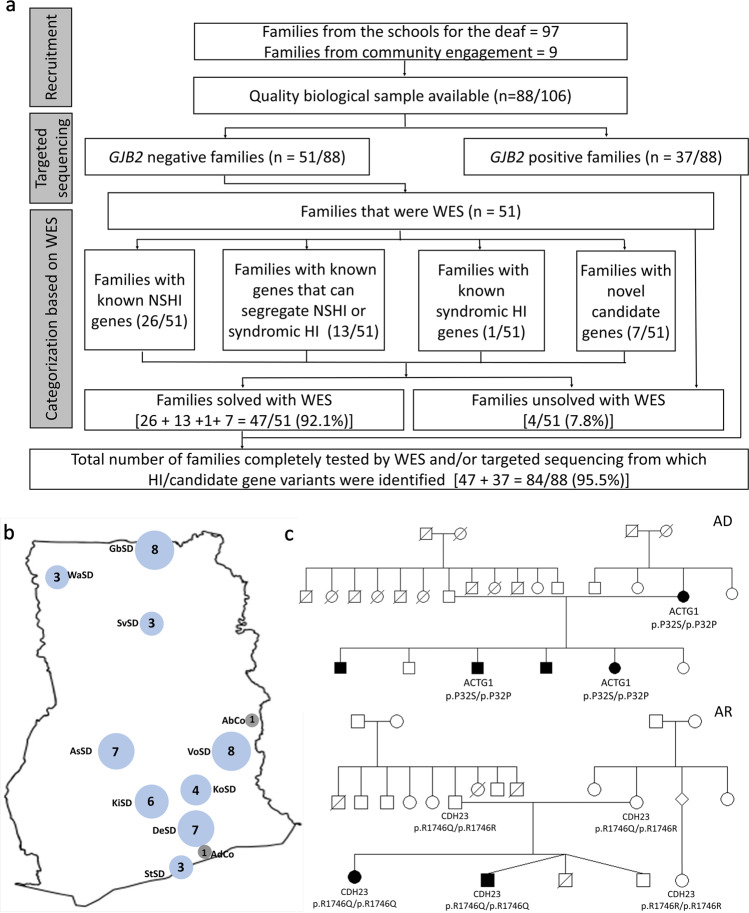
The study participants were recruited across the country with the majority from the Eastern region of Ghana (Fig. 1a, b), which has the highest number of schools for the deaf. These are boarding schools with students mostly from the surrounding towns and villages. A total of 88 families were ascertained, of which 37 could be explained by GJB2 mutations leaving 51 families for further study (Fig. 1a). The number of affected females and males included in the study (from whom WES was performed) who are negative for GJB2 variants was 48 (46.2%) and 56 (53.8%) (Supplementary Fig. 1), respectively.
Fig. 1. Summary of patient enrollment in Ghana.
a Flow diagram of the study. b National recruitment sites for families with HI (n = 51) considered for WES. Blue circles denote schools for the deaf and the number of recruited families written in the circles. State School for the Deaf (StSD) in Greater Accra Region, Gbeogo School for the Deaf (GbSD) in Upper East Region, Volta School for the Deaf (VoSD) in Volta Region, Ashanti School for the Deaf (AsSD) in Ashanti Region, Koforidua School for the Deaf (KoSD), Kyebi School for the Deaf (KiSD), and Demonstration School for the Deaf (DeSD) in Eastern Region, Savelugu School for the Deaf (SvSD) in Northern Region, and Wa School for the Deaf (WaSD) in Upper West Region. Gray circles represent communities, Abotoase (AbCo), and Adamorobe (AdCo). c Representative families with autosomal dominant (AD) and autosomal recessive (AR) mode of inheritance.
The analysis of the student records retrieved from the schools for the deaf showed that all the students had bilateral HI. For the 51 pedigrees for which exome sequence data were generated, 50 (98.0%) families have members with prelingual HI while one family (Fam18, Supplementary Fig. 1g) has affected members with post-lingual HI, that developed HI at >8 years of age. Except for the post-lingual family, none of the hearing-impaired participants have verbal communication and are only able to communicate through sign language.
Although the student records confirmed that all the students had sensorineural HI, we were only able to retrieve audiograms for 39 study participants. The audiogram for most of the participants could not be obtained from the schools’ archives, and several of them had written audiological reports without an attached audiogram from the hearing assessment center. For the retrieved audiograms, the hearing threshold across all frequencies tested was 101.8 dB (standard deviation = 10.3) in the better ear. There was no significant difference in the pure tone average of the left and right ears for the hearing-impaired participants. Two unrelated families 2/51 (3.9%) included in the study clinically expressed phenotypes of Waardenburg syndrome (Supplementary Fig. 1f).
Pedigree analysis
The analysis of the families enrolled in the study showed that the majority [37/51, (72.5%)] families) exhibited a likely AR mode of inheritance (Fig. 1c, Tables 1–3 and Supplementary Figs. 2–6). Six families showed a pattern of inheritance compatible with an autosomal dominant (AD) mode of inheritance (11.8%) (Fig. 1d, Tables 1, 3 and Supplementary Figs. 2–6). One family (2.0%) had a possible X-linked recessive mode of inheritance. The mode of inheritance in seven families was inconclusive since the pattern of inheritance is compatible with AR and X-linked recessive. None of the families segregated HI in a fashion that is compatible with mitochondrial inheritance. The average number of family members for whom a DNA sample was obtained is four. The maximum number of affected individuals per family was five (n = 2 families), with most families having two affected members (n = 38 families) (Supplementary Fig. 1). WES data were generated on DNA samples obtained from one affected member for two families, two affected members for 45 families, and three affected members for four families (n = 104; Supplementary Fig. 1). From all families, a total of 9 affected and 31 unaffected participants had available DNA but were not exome sequenced.
Table 1.
Ghanaian families with variants in known genes underlying non-syndromic hearing impairment.
| Gene | F-ID | Nucleotide change | Protein change | Rs-number | Inh | GT | Other HI gene variants detected that do not segregate with HI |
|---|---|---|---|---|---|---|---|
| MYO15A | Fam9 | c.4778A>G | p.(E1593G) | – | AR | Het | – |
| c.6011C>T | p.(P2004L) | rs756172590 | AR | Het | – | ||
| MYO15A | Fam11 | c.8767C>T | p.(R2923X) | rs373462792 | AR | Het | – |
| c.6551G>C | p.(C2184S) | – | AR | Het | – | ||
| MYO15A | Fam13 | c.8340G>A | p.(T2780T) (splicing) | rs878853228 | AR | Het | C1QBP: c.241G>A: p.(A81T) (rs145848155) (Hom) |
| c.4216G>A | p.(E1406K) | rs759810756 | AR | Het | |||
| MYO15A | Fam21 | c.6716A>C | p.(H2239P) | rs760577812 | AR | Het | MYO15A: c.1196A>G, p.(Y399C) (rs368682932); GREB1L: c.4840A>G: p.(I1614V) (rs1032115325) (Het) |
| c.8065delT | p.(W2689Gfs*49) | rs1567654885 | AR | Het | |||
| MYO15A | Fam22 | c.6518C>T | p.(S2173F) | – | AR | Het | SRCAP: c.2821A>T: p.(I941L) (rs1176169022) (Het) |
| c.6863C>T | p.(S2288L) | rs886052676 | AR | Het | – | ||
| MYO15A | Fam23 | c.4519C>T | p.(R1507X) | rs549138385 | AR | Het | – |
| c.6011C>T | p.(P2004L) | rs756172590 | AR | Het | – | ||
| MYO15A | Fam34 | c.6302T>C | p.(L2101P) | rs201908493 | AR | Het | – |
| c.6634G>A | p.(E2212K) | rs371352836 | AR | Het | – | ||
| MYO15A | Fam50 | c.8340G>A | p.(T2780T) (splicing) | rs878853228 | AR | Het | – |
| c.9947A>G | p.(Q3316R) | AR | Het | – | |||
| GIPC3 | Fam10 | c.241C>T | p.(L81F) | rs1487341857 | AR | Hom | – |
| GIPC3 | Fam30 | c.216_ 225del | p.(P73Ffs*21) | – | AR | Hom | – |
| GIPC3 | Fam31 | c.241C>T | p.(L81F) | rs1487341857 | AR | Hom | – |
| OTOF | Fam32 | c.1051C>T | p.(Q351X) | rs1558492758 | AR | Hom | – |
| OTOF | Fam14 | c.5813+1G>A | AR | Het | – | ||
| c.1747C>G | p.(P583A) | rs756894987 | AR | Het | – | ||
| c.1479_ 1481del | p.(K493_ R494delinsN) | – | AR | Het | – | ||
| OTOF | Fam36 | c.1479_ 1481del | p.(K493_ R494delinsN) | – | AR | Hom | PCDH7: c.3659A>G: p.(N1220S) (Hom) |
| ESPN | Fam1 | c.2254_2275del+c.2279G>Ta | p.(E731Cfs*6) + p.(R760L) | rs371306949 | AR | Hom | – |
| ESPN | Fam3 | c.2254_2275del+c.2279G>Ta | p.(E731Cfs*6) + p.(R760L) | rs371306949 | AR | Hom | – |
| GRXCR1 | Fam5 | c.784C>T | p.(R262X) | rs761349153 | AR | Hom | – |
| MARVELD2 | Fam18b | c.1058dupT | p.(V354Sfs*5) | AR | Hom | – | |
| KARS | Fam19 | c.1685G>C | p.(C562S) | rs1156833108 | AR | Hom | – |
| TRRAP | Fam27 | c.1156G>A | p.(V386I) | – | AD | Het | [TMEM132E: c.470G>C: p.(G157A) (rs151214976) (het)], [SLC17A8: c.904-6C>A (Het)] |
| MYH14 | Fam28 | c.1775G>A | p.(R592Q) | rs1445498283 | AD | Het | MYO7A: c.2095-1G>A (Hom) |
| OTOG | Fam33 | c.6982T>A | p.(S2328T) | rs1274985459 | AR | Het | – |
| c.8513G>A | p.(R2838H) | rs544815967 | AR | Het | TPRN: c.107delG: p.(G36Afs*414) (rs1011757302) (Hom) | ||
| DSPP | Fam37 | c.2306A>G | p.(D769G) | rs370931212 | ARc | Het | AHCY: c.1075G>T: p.(V359L) (Het) |
| c.2502_2518del | p.(D834Efs*2) | – | ARc | Het | – | ||
| LOXHD1 | Fam38 | c.3734G>A | p.(G1245D) | – | AR | Hom | – |
| c.5050G>A | p.(A1684T) | rs376122149 | AR | Hom | – | ||
| TMC1 | Fam47 | c.1622T>A | p.(I541N) | – | AR | Hom | – |
| SLC12A2 | Fam35 | c.2935G>A | p.(E979K) | rs1581138934 | AD | Het | – |
Rs-number reference number, Inh mode of inheritance, hom homozygote, het heterozygote, GT patient genotype.
aVariants are both on the same haplotype, the missense variant (R760L) is predicted to be at position 753 as part of the frameshift sequence.
bThese families feature a HI with post-lingual expression, an expansion of the phenotype of MARVELD2.
cThese variants are inherited via a different inheritance model than those known for this gene.
Table 3.
Novel non-syndromic and syndromic hearing impairment candidate genes.
| Gene | F-ID | Phenotype | Nucleotide change | Protein change | Rs-number | Inh | GT | Expression in the mouse inner ear | Previous/animal studies |
|---|---|---|---|---|---|---|---|---|---|
| INPP4B | Fam2 | NSHI | c.1848G>C | p.(Q616H) | rs147919355 | AR | Het | Yes |
Depletion of INPP4B suppresses callosal axon formation in the developing mice and may cause mild to severe cognitive impairment INPP4B regulates nerve conduction velocity Mouse models and rat models express the gene in the inner ear. |
| c.1271T>C | p.(I424T) | rs747224392 | AR | Het | |||||
| CCDC141 | Fam26 | NSHI | c.704A>G | p.(D235G) | rs1029313097 | AR | Het | Yes | CCDC141 variants have been implicated in Kallmann Syndrome, and though rare, HI is reported in some Kallmann Syndrome patients. Mouse models express the gene in the inner ear. Ccdc141-null mice showed impaired recognition memory and spatial reference memory. |
| c.202G>A | p.(E68K) | rs540836199 | AR | Het | |||||
| MYO19 | Fam40 | NSHI | c.949G>T | p.(A317S) | rs199866785 | AR | Hom | Yes | Mouse models express the gene in the inner ear. The ortholog of the gene was associated with autistic disorder in different animals such as Canis lupus familiaris, Pan paniscus, Sus scrofa, and Chinchilla lanigera. In humans, Myosin 19 functions as an actin-based motor for mitochondrial movement in vertebrate cells. |
| Case1 | NSHI | c.1300G>A | p.(A434T) | rs375962449 | AR | Het | |||
| c.2552C>T | p.(P851L) | rs749344013 | AR | Het | |||||
| DNAH11 | Fam44 | NSHI | c.11232C>G | p.(I3744M) | rs201120788 | AR | Het | Yes | DNAH11 was implicated in primary ciliary dyskinesia and Kartagener syndrome which has HI as one of its symptoms. |
| c.12969G>C | p.(Q4323H) | rs191802172 | AR | Het | |||||
| Case2 | c.6118C>T | p.(R2040C) | rs199772877 | AR | Het | ||||
| c.846G>A: | p.(M282I) | rs375023124 | AR | Het | |||||
| Case3 | c.7295G>A | p.(R2432Q) | rs769003090 | AR | Het | ||||
| c.6131G>A | p.(R2044Q) | rs372051486 | AR | Het | |||||
| SOX9 | Fam45 | NSHI | c.432-3C>A | – | rs1033320617 | AD | Het | Yes | SOX9 is essential for cochlear development in mice. Mutant mice had a HI phenotype. Depletion of Sox9 resulted in defective vestibular structures, semi-circular canals, and utricle developments. |
| PAX8 | Fam39 | Waardenburg syndrome | c.968C>G | p.(P323R) | rs1573435665 | AD | Het | Yes | (Pax8−/− mice did not respond to sound when examined by the auditory brain stem response (ABR) test. |
| POTEI | Fam6 | NSHI | c.1676G>C | p.(G559A) | rs1254207451 | AR | Het | – | – |
| c.409C>T | p.(R137X) | rs536831847 | AR | Het |
Cases 1–3 are isolated cases (families with only one affected individual) that were examined as added participants from Ghana.
Rs-number reference number, Inh mode of inheritance, hom homozygote, het heterozygote, GT patient genotype.
Bioinformatic and molecular analysis
Principal component analysis
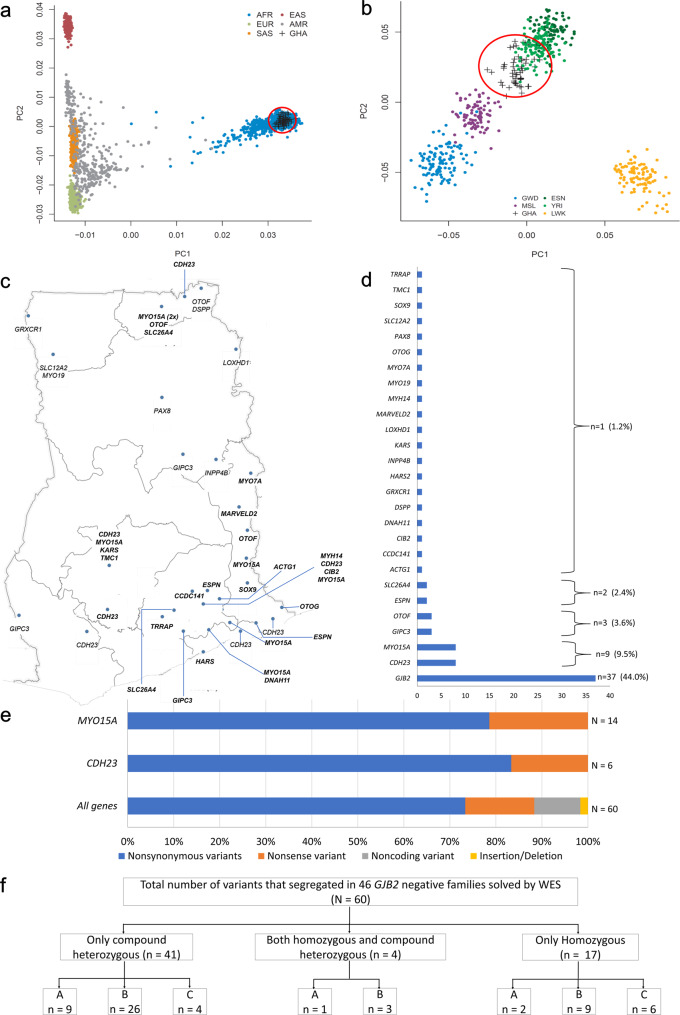
Principal components (PCs) constructed using genotype data obtained from families in this study were projected against data extracted from participants of continental ancestries in the 1000 Genomes reference panel (phase 3 version 5) showed that our samples clustered with other African populations, as expected (Fig. 2a). When projected only against African populations, our samples clustered between samples from Nigeria [YRI (Yoruba) and ESN (Esan)], and Sierra Leone [MSL (Mende)] as geographically expected, with closer proximity to the YRI population of Nigeria (Fig. 2b). Projecting all other family members against the coordinates of one member per family showed that members of the same family clustered close to each other (Supplementary Fig. 1h).
Fig. 2. Principal component analysis and overview of gene variants found in Ghana.
PCA plot of samples from Ghana projected against the a global populations and b African populations. GHA samples from Ghana (highlighted with red circles), AFR Africa, EUR European, SAS South Asian, EAS East Asian, AMR Admixed Americans, GWD Gambian in Western Division, MSL Mende in Sierra Leone, ESN Esan in Nigeria, YRI Yoruba in Ibadan, Nigeria, and LWK Luhya in Webuye, Kenya. c Geographical distribution and d frequency of the known HI and candidate genes in Ghanaian families. e Proportions of variant types in the two most prevalent (MYO15A and CDH23) and other known and novel candidate HI genes were identified. f A flow diagram showing variants absent from the databases “A”, present in the databases with new HI association “B” and known HI variants “C” identified in the families studied.
Known and candidate HI genes identified within the Ghanaian population
DNA samples from a total of 180 individuals from 51 GJB2-negative families underwent WES, including 104 hearing-impaired individuals and 76 unaffected family members. In these 51 families, variants in known human HI genes were found in 40/51 (78.4%) families, and highly likely candidate genes in 7/51 (13.7%). A total of 60 variants were found in 26 genes identified in the families studied (Fig. 2c, d and Tables 1, 2). The identified genes were randomly distributed across the territories of Ghana (Fig. 2c). Forty-four of the variants were missense variants and the remaining variants were insertion/deletion (1), nonsense (9), or non-coding variants (6) (Fig. 2e and Tables 1, 2). Of all the variants, 12 (20.0%) were not observed in dbSNP16, gnomAD17, Ensembl18, and TOPMed19 databases. Thirty-eight of the 60 variants (63.3%) were present in databases but were not previously reported to be associated with HI, including 24 of 48 (50.0%) variants in known HI genes. Only 10 (16.6%) of the observed variants were previously reported to be associated with HI (Fig. 2f).
Table 2.
Known genes associated with both non-syndromic and syndromic HI found in Ghanaian families.
| Gene | F-ID | Nucleotide change | Protein change | Rs-number | Inh | GT | Other HI gene variants found in the listed families, but not segregating with HI | Syndrome |
|---|---|---|---|---|---|---|---|---|
| Genes associated with both non-syndromic and syndromic HI | ||||||||
| CDH23 | Fam15 | c.2206C>T | p.(R736X) | rs1230303971 | AR | Hom | Usher syndrome, type 1D | |
| CDH23 | Fam7 | c.5237G>A | p.(R1746Q) | rs111033270 | AR | Hom | Usher syndrome, type 1D | |
| CDH23 | Fam12 | c.2746G>A | p.(D916N) | rs1318444606 | AR | Het | Usher syndrome, type 1D | |
| c.4562A>G | p.(N1521S) | rs780987516 | AR | Het | Usher syndrome, type 1D | |||
| CDH23 | Fam24 | c.3181G>A | p.(E1061K) | rs1060499793 | AR | Het | Usher syndrome, type 1D | |
| c.6514C>T | p.(P2172S) | – | AR | Het | Usher syndrome, type 1D | |||
| CDH23 | Fam41 | c.4562A>G | p.(N1521S) | rs780987516 | AR | Hom | Usher syndrome, type 1D | |
| CDH23 | Fam46 | c.2746G>A | p.(D916N) | rs1318444606 | AR | Hom | Usher syndrome, type 1D | |
| CDH23 | Fam43 | c.4562A>G | p.(N1521S) | rs780987516 | AR | Hom | Usher syndrome, type 1D | |
| CDH23 | Fam29 | c.2206C>T | p.(R736X) | rs1230303971 | AR | Het | Usher syndrome, type 1D | |
| c.4562A>G | p.(N1521S) | rs780987516 | AR | Het | Usher syndrome, type 1D | |||
| MYO7A | Fam8 | c.1996C>T | p.(R666X) | rs121965085 | AR | Het | Usher syndrome, type 1B | |
| c.5101C>T | p.(R1701X) | rs111033182 | AR | Het | ||||
| CIB2 | Fam42 | c.556C>T | p.(R186W) | rs370359511 | AR | Hom | [CIB2:c.409C>T: p.(R137W) (Hom)], [CIB2: c.427C>T: p.(R143W) (Hom)], [CIB2: c.571C>T: p.(R191W) (Hom)] | Usher syndrome, type IJ |
| ACTG1 | Fam20 | c.94C>T | p.(P32S) | rs1598551290 | AD | Hom | Baraitser–Winter syndrome 2 | |
| SLC26A4 | Fam17 | c.2089+1G>A | – | rs727503430 | AR | Hom | Pendred syndrome | |
| SLC26A4 | Fam25 | c.1225C>T | p.(R409C) | rs147952620 | AR | Hom | Pendred syndrome | |
| Associated with syndromic HI only | ||||||||
| HARS2 | Fam16 | c.346G>A | p.(A116T) | rs1312606802 | AR | Het | Perrault syndrome 2 | |
| c.659A>G | p.(Y220C) | rs746757469 | AR | Het | ||||
Most families investigated (30/51) had unique variants (Tables 1–3), confirming the high level of allelic heterogeneity for HI genes within the Ghanaian population. Variants in CDH23 (n = 8 families) and MYO15A (n = 8 families) accounted for the majority (31.4%) in this GJB2-negative group of families and represent 18.2% (16/88) of all families in the entire Ghanaian cohort initially recruited (Tables 1 and 2). GIPC3 and OTOF variants were found in three families each, while variants in ESPN and SLC26A4 were each found in two families, and variants in 20 other genes were only observed in one family (Tables 1, 2 and Fig. 2d). Thirty-one of the variants in known HI genes were classified as pathogenic (P) or likely pathogenic (LP) and 32/51 families were found to segregate at least one pathogenic and likely pathogenic (PLP) variant (Supplementary Table 1).
Non-syndromic hearing impairment gene variants
Fourteen known NSHI genes were identified in 25 of the 51 families (49.0%). These genes included MYO15A that was one of the most common genes observed (n = 8 families: Table 1). For the NSHI genes, 13 compound heterozygote variants were identified, and each variant was unique to a family except for MYO15A c.6011C>T: p.(P2004L) and c.8340G>A: p.(T2780T) that were found in two families (Table 1). The other 12 NSHI genes were GIPC3, OTOF, ESPN, DSPP, GRXCR1, KARS, LOXHD1, MARVELD2, MYH14, OTOG, TMC1, and TRRAP (Table 1).
Expansion of phenotypes for NSHI: detection of recessive inheritance for DSPP, and post-lingual expression of HI for MARVELD2
For DSPP, an ADNSHI gene, an AR mode of inheritance was detected in Fam37 (Table 1 and Supplementary Fig. 6).
For MARVELD2, we found a post-lingual expression of ARNSHI in Fam18, with variable age of onset with two family members with onset at 12 years of age and a third family member with onset at 8 years of age. All the affected family members could efficiently read lips, and since the condition developed post-language acquisition, they were able to engage in verbal communications (Table 1 and Supplementary Fig. 1).
Genes that cause both non-syndromic, and syndromic hearing impairment
Eight families segregated variants in CDH23 (OMIM: 601067) (Table 2), which has been implicated in Usher syndrome type 1D20, ARNSHI21, and age-related HI22. All the variants (n = 6) identified in these families were classified as PLP based on the ACMG-AMP classification guidelines for HI (Supplementary Table 2). CDH23: c.4562A>G: p.(N1521S) and c.2746G>A: p.(D916N) were found in four and two families, respectively. The four families, which segregated the CDH23: c.4562A>G: p.(N1521S) variant, were ascertained in different geographical regions in Ghana. Three families were found to have compound heterozygote variants. In addition, Fam42 was found to have a CIB2 (OMIM: 605564): c.556C>T: p.(R186W) variant segregating with the HI phenotype (Table 2). The above variant was classified as likely pathogenic with a combined annotation dependent depletion (CADD) score of 32, suggesting it is deleterious. The CIB2 [c.556C>T: p.(R186W)] variant was previously found in an Usher syndrome type 1J family23, although it is currently questioned whether CIB2 is involved in Usher syndrome etiology. Moreover, MYO7A (OMIM: 276903) is associated with Usher Syndrome type 1B (OMIN: 276900), ARNSHI (OMIM: 600060), and ADNSHI (OMIM: 601317), and PLP variants in this gene were found in Fam8 (Table 2). Two families (Fam17 and Fam25) segregate SLC26A4 (OMIM: 605646) variants [c.2089+1G>A and c.1225C>T p.(R409C)]. SLC26A4 has been associated with ARNSHI and Pendred syndrome [(OMIM: 274600) (Table 2)]. Fam20 segregates ACTG1 (OMIM: 102560) variant c.94C>T: p.(P32S), ACTG1 has been associated with ADNSHI, and AD Baraitser–Winter syndrome 2 BWS2 [(OMIM: 614583); (Table 2)]. BWS2 is mostly likely always due to de novo variants, contrary to the Fam20 pedigrees that feature ADNSHI with an affected mother, four affected and two unaffected children of both sexes (Supplementary Fig. 4). For the families that segregate variants in genes associated with both NSHI and syndromic HI, there were no clinical signs observed during sample collection that would indicate that affected family members have syndromic HI.
Hearing impairment gene associated with Perrault syndrome
HARS2 compound heterozygote variants were identified as the likely cause of HI in Fam16 (Table 2). HARS2 (OMIM: 600783) has been implicated in AR Perrault syndrome 2 (OMIM: 614926) that is characterized by HI in both males and females. Affected females also display primary amenorrhea, streak gonads, and infertility, while affected males show normal pubertal development and are fertile. Since the two HI family members are male, it is not possible to clinically diagnose this syndrome.
Seven candidate HI genes
Six candidate genes identified in Ghanaian families with NSHI
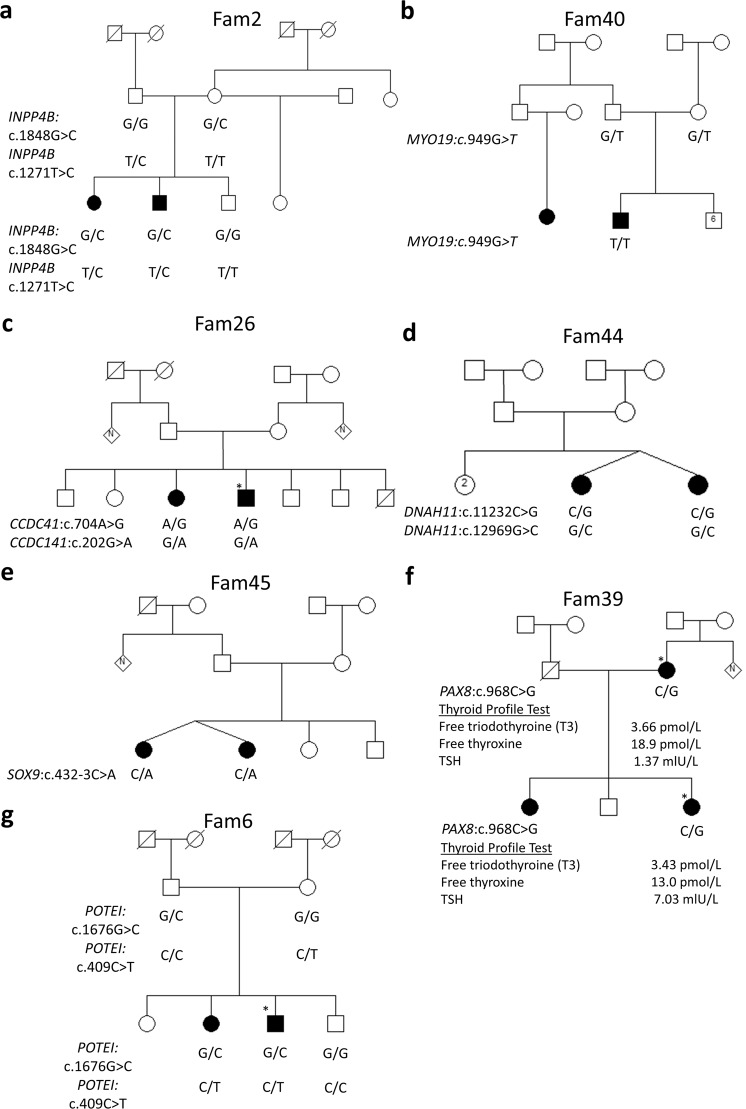
Six genes (INPP4B, CCDC141, MYO19, DNAH11, POTEI, and SOX9) whose variants segregate with NSHI were found in six families (11.8%). Five of the six families had compound heterozygote variants which segregated with ARNSHI (Table 3, Fig. 3, and Supplementary Table 3).
Fig. 3. Pedigrees of families which segregate candidate genes.
The segregation of candidate gene variants was shown in the respective families, a Fam2, b Fam40, c Fam26, d Fam44, e Fam45, f Fam39, and g Fam6. *Individuals whose audiogram was obtained.
Two variants were intronic: In family 40, a splicing variant in MYO19 (c.2464-8T>C) segregated with HI but was found to be likely benign (Supplementary Table 4), but another biallelic missense variant in the same haplotype was classified as likely pathogenic (MYO19: c.949G>T). While a likely pathogenic splicing variant (SOX9: c.432-3C>A) was found in Fam45 segregated with an AD mode of inheritance (Supplementary Table 4). The gnomAD and TOPMed allele frequencies for these variants identified in candidate genes were either all zero or close to zero (Supplementary Table 3). CADD scores are also provided in Supplementary Table 3.
PAX8: a candidate gene for Waardenburg syndrome
A variant in a candidate gene [PAX8: c.968C>G: p.(P323R)] was found in one of the two families clinically presenting with Waardenburg syndrome (Table 3 and Fig. 3). The patients presented with the typical features of Waardenburg syndrome: HI, striking blue eyes, and premature gray hair phenotype (Supplementary Fig. 1f). Because, in addition to HI Pax8 knockout mice model was reported to be athyroid24, we investigated thyroid hormones profile in the family and found it to be normal in the affected mother and daughter, except for thyroid-stimulating hormone that was high in the affected daughter, suggesting a trend to hypothyroidism (Fig. 3f). Although the variant was not in gnomAD it was present with a low frequency in TOPMed (minor allele frequency [MAF] = 1.13 × 10−5) and had a CADD score of 14.55. In addition, it has been reported that Pax8−/− mice did not respond to sound when tested using auditory brain stem response (ABR). Moreover, abnormalities in the outer and middle ear structures were found in a high percentage of Pax8−/− mice. Maturation of the inner ear also appeared delayed by about 1 week with respect to euthyroid controls24.
Other variants of unknown significance
Additional variants were found in some of the families, but these variants either did not segregate with the phenotype, or a second heterozygote variant was not found for families with ARHI. One of such families was Fam26 (Fig. 3c) in which the LOXHD1 (OMIM:613072): c.4202C>T variant was found in the heterozygous state without a second variant. Fam39 (Fig. 3f) had only one affected family member positive for HGF (OMIM: 142409): c.776A>G; c.278G>C compound heterozygote variants. The same family member also had FOXI1 (OMIM:601093): c.776G>A heterozygote variant. In addition, a heterozygote COQ8A (OMIM: 606980): c.692A>C variant was segregated in Fam39 but a second variant was not identified. In Fam40 (Fig. 3b), a heterozygote MED12 (OMIM:300188): c.5348A>G variant was found. Compound heterozygote variants in GRXCR1 (OMIM:613283): c.74G>A; c.645G>T were found in only one affected Fam45 member. Other heterozygote variants observed in Fam45 were TECTA (OMIM:602574): c.545A>G and GREB1L (OMIM: 617782): c.2498C>A.
Validation of candidate gene variants among isolated African HI cases
We investigated candidate genes in Ghanaian exomes data of non-familial/isolated HI cases (n = 153) and found two probands with compound heterozygous variants in DNAH11 and in one proband with MYO19 variants (Table 3 and Supplementary Fig. 7). Using Sanger sequencing, we validated the variants found in DNAH11 and MYO19 in the three probands (Supplementary Fig. 7).
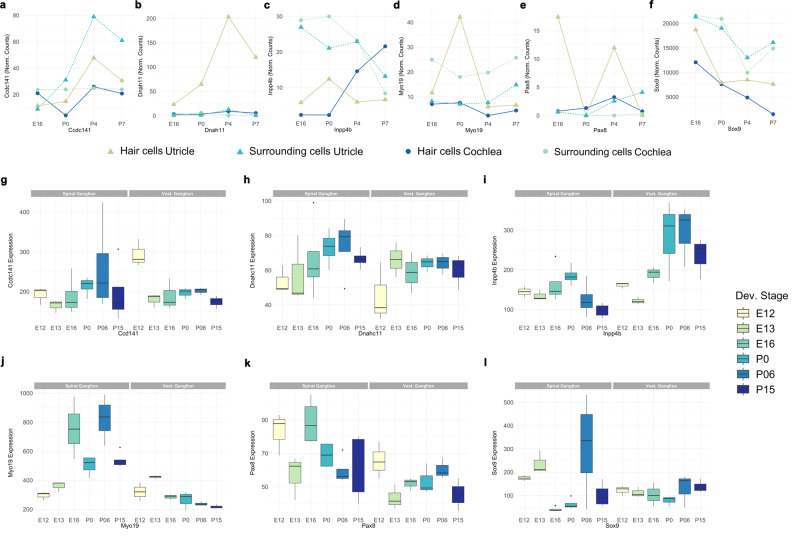
Expression of HI candidate genes in the developing and adult mouse inner ear
We investigated the expression of the six HI candidate genes in the inner ear via the interrogation of various expression atlases and found evidence of their expression. These genes all show expression in the inner ear hair cells, and in the spiral and vestibular ganglion during development [Fig. 4, Ccdc141 (panels a and g), Dnah11 (panels b and h), Inpp4b (panels c and i), Myo19 (panels d and j), Pax8 (panels e and k), Sox9 (panels f and l)]. In addition, there was evidence of expression in various other cell types of the cochlear floor epithelium (Supplementary Figs. 8–16). Myo19a shows a wide-spread expression in the cochlear epithelium throughout E14, E16, and P1, and is predominantly expressed in the inner hair cells at P7 (Supplementary Figs. 8 and 13–15). Sox9 shows a high and ubiquitous expression pattern across the entire cochlear epithelium, particularly during E14, E16, and P1. It displays a more restricted pattern during P7 with a predominant expression in the outer and inner pillar cells (Supplementary Figs. 9 and 13–15). Ccdc141 displays a wide-spread but low expression in the cochlear tissues throughout development, with high expression at E14 in the lesser epithelial ridge cells and the basilar membrane cells at P7 (Supplementary Figs. 10 and 13–15), Inpp4b and Dnah11 show a more restricted expression in the cochlear tissues throughout development, with Inpp4b expressed in the developing lateral great epithelial ridge cells at P1, the basilar membrane cells at P7; and Dnah11 expressed in developing inner hair cells and mature inner hair cells at P1 and P7 respectively (Supplementary Figs. 11 and 12).
Fig. 4. Expression of novel candidate genes in the cochlea and utricle during mouse development.
a–f RNA sequencing data of hair cells and surrounding cells from the cochleae and utricles of mice at four developmental stages: E16, P0, P4, and P7 are presented for the mouse orthologs of six novel candidate genes104: Ccdc141 (a), Dnah11 (b), Inpp4b (c), Myo19 (d), Pax8 (e), Sox9 (f). All genes show expression in the inner ear hair cells. Data are presented as normalized RNA-seq counts. g–l Expression of novel candidate genes in spiral and vestibular ganglion neurons of the inner ear during mouse development at six developmental stages: E12, E13, E16, P0, P06, and P15105. Expression is presented for Ccdc141 (g), Dnah11/ Dnahc11 (h), Inpp4b (i), Myo19 (j), Pax8 (k), and Sox9 (l) with all genes showing expression in the spiral and vestibular ganglion of the inner ear during development. Expression microarray data is presented by the perfect match and mismatch probe differences (PM/MM). Data for (a–l) were obtained from SHIELD (Shared Harvard Inner-Ear Laboratory Database).
Moreover, five candidate genes i.e. Ccdc141, Inpp4b, Myo19, Sox9, and Pax8 also showed expression in various craniofacial tissues during early mouse development. Specifically, there was the greatest expression of Ccdc141, Inpp4b, Myo19, and Sox9 at E10.5, within the maxillary arch epidermal ectoderm for Ccdc141and Inpp4b; the lateral prominence neural epithelium for Myo19, and the central neural epithelium for Sox9 (Supplementary Fig. 15). The greatest expression of Pax8 occurs at E8.5 (Supplementary Figs. 15 and 16), within the caudal brain neural epithelium (Supplementary Fig. 15). Dnah11 was not assessed in this latter data set and could not be interrogated.
In Fam6, compound heterozygous variants in POTEI: (c.1676G>C and c.409C>T) were found to segregate with ARNSHI. This gene does not have a mouse ortholog and expression data could not be investigated in the available data sets.
Protein structure analysis
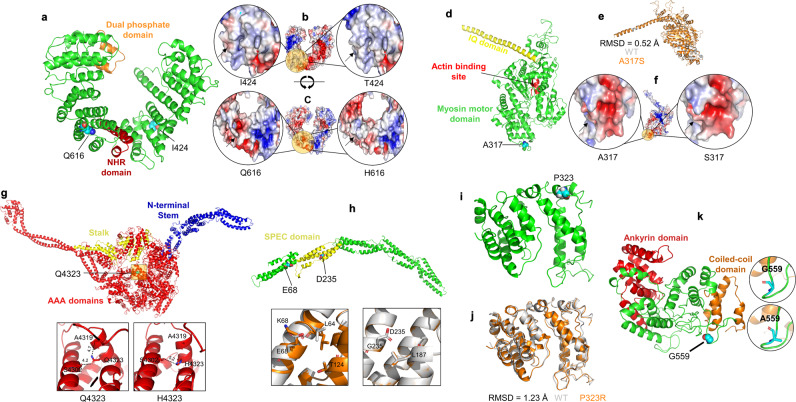
Homology modeling techniques were utilized to model the wild-type (WT) and the mutant missense variant(s) of three-dimensional structures of INPP4B, MYO19, DNAH11, CCDC141, and PAX8 proteins via the MODELLER program25. In the case of INPP4B, the two mutated residues, I424 and Q616, reside at the helix and loop region, respectively (Fig. 5a). Both residues are located on the surface of the protein and do not interact directly with nearby residues. The substitution of nonpolar hydrophobic amino acid isoleucine into polar residue threonine at position 424 displays different surface patches of electrostatic potential (Fig. 5b). In the case of Q616H, the replacement of neutral polar charged glutamine into basic polar charged histidine causes a shift in electrostatic potential and surface area, as indicated by the arrow (Fig. 5c).
Fig. 5. Structural modeling missense variants in novel hearing impairment candidate genes.
a Homology model of INPP4B. The mutated residues (I424T and Q616H) are represented with a sphere model. The NHR and dual phosphate domains are indicated with red and orange colors, respectively. b, c The electrostatic potential surface of wild-type and mutant protein is highlighted in the circle zoom-up view showing different patches (indicated by the arrow) around the mutated residue. d Three-dimensional structure of MYO19 showing the mutated residue, A317, in a sphere model. The actin-binding pocket, myosin motor, and IQ domain are represented by green, red, and yellow colors, respectively. e The superposed structure of the WT (gray) and mutant (orange) structure of MYO19 shows the root-mean-square deviation. f The electrostatic potential surface of wild-type and mutant protein is highlighted in a circle zoom-up view showing different patches (indicated by the arrow) around the mutated residue. g Three-dimensional structure of DNAH11 showing the N-terminal stem (blue), stalk (yellow), and the mutated residue Q4323 (sphere model) of the AAA domain (red). The difference in the interaction network due to the mutation is highlighted in the bottom panel. The residue is represented with a stick model and the distance between the residues is labeled in angstrom. h Predicted structure of CCDC141 showing the mutated residues in a sphere model. The bottom row represents the zoom-up view of WT (gray) and mutant type structures displaying the different orientations of residues (stick model). i Three-dimensional structure of PAX8. j The superposed structure of WT (gray) and mutant (orange) structure showing the difference in overall conformation due to the P323R mutation. k Predicted structure of POTEI showing the wild-type and mutated residues in a stick model in the right panel.
For MYO19, residue A317 is located at the surface of the protein in the myosin motor domain (Fig. 5d). The RMSD of WT (A317) and mutant (S317) MYO19 show a value of 0.52 Å, suggesting a slight change in overall conformation (Fig. 5e). The electrostatic potential of mutant protein compared with WT displays a change in surface charge distribution as shown in Fig. 5f.
The mutated residue Q4323 in DNAH11 is located in the helix region of the AAA domain (Fig. 5g). The Q4323 residue is involved in an interaction with nearby residues, A4319, and S4302. The substitution of WT glutamine with a histidine residue changes the interaction network of nearby residues. We were unable to reliably model the I3744M variant for DNAH11.
For CCDC141, E68K and D235G both alter the nature of the side chain (Fig. 5h). The close views of WT and mutant CCDC141 protein display the change in the secondary structure and side chain of the nearby residues.
The superimposed structure of the WT and mutant structure of PAX8 structure displays a 0.52 Å RMSD value suggesting the effect of the mutation on the overall conformation (Fig. 5i, j).
The POTEI p.(G559A) missense variant was modeled (Fig. 5k) and the predicted structure shows a minimal difference between the WT and mutated residues. The p.(R137X) variant in POTEI was not modeled as that one is predicted to lead to nonsense-mediated decay and there is likely no protein expression.
Unsolved families
In four families (7.8%), a possible gene for HI was not identified. Although rare variants in HI genes were identified in the exome sequence data for these families, they were unlikely to be the cause of HI (Supplementary Table 5 and Supplementary Fig. 17). An example is one family presenting with Waardenburg syndrome (Fam51) that had variants in three HI genes, but these variants did not segregate with the phenotype (Supplementary Table 5). Assuming an AD inheritance model (with a possible affected father), we did not find any variants in a known HI or Waardenburg gene. There are multiple possible candidate variants that will need confirmation in the future (Supplementary Table 5).
Fam4 segregates a previously reported BCORL1 (OMIM:300688) variant c.4079_ 4081del: p.(D1360_ L1361delinsV) with HI. BCORL1 causes Shukla–Vernon syndrome (OMIM: 301029), an X-linked recessive neurodevelopmental disorder characterized by global developmental delay, variably impaired intellectual development, and behavioral abnormalities, including autism spectrum disorder and ADHD26. This variant is unlikely to be responsible for HI because it has not been reported as part of this syndrome before. Moreover, Fam4 features NSHI. In addition, there is no inner ear expression data available for this gene. Fam4 segregates a heterozygous MYO15A variant that could have explained the HI, but a second variant could not be identified (Supplementary Fig. 17).
Fam48 had a heterozygous GJB2 p.(R143W) founder variant in one of the two affected family members, but a second GJB2 variant was not identified (Supplementary Table 5).
In Fam52, we identified compound heterozygous variants segregating with HI in DACT2 (c.796-12C>T and c.1021T>G). DACT2 is an important mediator involved in the regulation of intracellular signaling pathways during development27. DACT2 is expressed in the outer pillar cells of the inner ear28,29, but its role in HI remains to be fully characterized. Moreover, the CADD score was low for one of the variants found in DACT2, and the other variant is located in the splicing region with no predicted impact on splicing. Also, for Fam52, compound heterozygous variants were found to segregate with HI in NCL [c.2065G>A: p.(G689S), and c.188A>G: p.(K63R)]. Ncl is widely expressed in the mouse and zebrafish ear. However, there is not enough evidence to support it as a good candidate and one of the variants was also found in the heterozygous state in three members of another family in this study, suggesting its frequency may be high in this population.
Last, no relevant copy number variations (CNVs) encompassing HI genes were identified in any of these families.
Discussion
In this study, we investigated the largest exome sequence data set for sub-Saharan African families segregating HI. This study will inform and enhance genetic medicine practice locally in Ghana, and African populations in the diaspora, with putative West African/Ghanaian ancestry. The principal component analysis (PCA) plot shows the similarity between the data obtained from the studied families and those obtained for the Nigerian population (Fig. 2a, b). Our previous investigations identified GJB2: p.R143W variant as the most frequent cause of familial NSHI in Ghana3. Putting together, the GJB2 positive rate from our previous studies3,13 and some additional families, we re-evaluated the contribution of GJB2 to 42.0% (37/88) among all families investigated (Fig. 1a). However, the majority of GJB2 positive cases originated from the same region (Eastern Region) where the village with a high prevalence of deafness (Adamorobe) is located11,13. A frequency rate of 34.1% was estimated without the inclusion of cases from Adamorobe village.
The present data identified variants in 20 known HI genes of which CDH23 (8/88; 9.1%) and MYO15A (8/88; 9.1%) were observed to be the cause of NSHI in multiple families, supporting future prioritization of these two genes in clinical practice. Moreover, the overall combined frequency of 18.2 % (16/88) for CDH23 and MYO15A amongst hearing-impaired families in Ghana is likely underestimated due to an oversampling of GJB2 positive families, with the frequency amongst GJB2-negative families at 31.4% (16/51). Indeed, we probably oversampled families from the Eastern region of Ghana where the GJB2-p.(R143W) founder variant was first described and is more prevalent3,11,12, and consequently where most schools for the deaf are established (Fig. 1b).
Similar to a previous study in Cameroon using a targeted panel sequencing30, within the Ghana population there is a high degree of locus and allelic heterogeneity. Five out of the seven families (71.4%) solved in the Cameroonian study had compound heterozygote variants in the identified gene, likely due to the absence of consanguinity. In the current report, 22/47 solved (46.8%) GJB2-negative families display compound heterozygosity. This will pose a great challenge in the development of a single affordable diagnostic tool that can be widely used in clinical practice, specifically for compound heterozygotes, which are harder to detect than homozygous variants and also parents need to be investigated to ensure when two variants within a gene are on different haplotypes. Targeted sequencing panels are useful in diagnosing genetic HI, however, only known genes can be investigated. In addition, there is a need for constant upgrade and revalidation of the targeted sequencing panels whenever new HI genes are discovered31,32. The above limitations, and the evidence illustrated by the present study advocate for the use of clinical WES and whole-genome sequencing31,33–36. It is also worth noting that additional variants in known HI genes as found in this study (Tables 1 and 2) can complicate interpretation. In addition, the handling of incidental findings, in so-called actionable genes, and policies toward them is still to be implemented in African settings37,38.
CDH23 has been implicated in ARNSHI and Usher syndrome type 1D (Table 2)20, and was reported as an important contributor to HI in several global populations39,40 with high frequencies in Eastern Asian countries such as Japan41 and Korea42. Two of the identified CDH23 variants were previously associated with HI, c.5237G>A: p.(R1746Q) in Spain43 and Ireland44 and c.2206C>T: p.(R736X) in French Canadians from Quebec45.
MYO15A has been ranked as the third to the fourth most common cause of severe-to-profound ARNSHI, worldwide46,47. The global frequency of HI-associated MYO15A variants was calculated at 6.2% by Farjami et al. underscoring the importance of this gene in the development of HI48. The frequency observed in Ghana is higher than what was previously reported for some African countries such as Nigeria (2.0%), South Africa (2.0%), Tunisia (2.3%)49, and Morocco (3.0%)50 using a custom capture panel (MiamiOtoGenes) and WES. On the other hand, one study of 61 Egyptian families reported a frequency of 15.3%51. Recent WES data from a Cameroonian study identified variants in MYO15A in 22.2% of hearing-impaired cases of putative genetic origin; with three variants identified in multiplex families and one variant detected in a simplex family. These data suggest that MYO15A may be the most common gene underlying HI in Cameroon52. A review on MYO15A has shown that the majority of studies reported novel variants48, which is consistent with our study. To the best of our knowledge, we identified four novel MYO15A variants [c.4778A>G:p.(E1593G), c.6551G>C:p.(C2184S), c.9947A>G:p.(Q3316R), and c.6518C>T:p.(S2173F)]. The large number of exons in MYO15A coupled with the fact that more than 2000 missense variants have been identified in this gene46, although not all involved in HI, makes routine screening of MYO15A variants in patients difficult.
An important novelty of this study is the identification of seven new HI candidate genes, six of which are associated with NSHI, and the AD variant, PAX8: c. 968C>G: p.(P323R), found in a family with Waardenburg syndrome. The involvement of these candidate genes in HI is also supported by clinical and segregation analyses (Table 3 and Fig. 3). Interestingly, variants in two of the candidate genes were also observed in unrelated HI patients from Ghana (i.e., DNAH11, MYO19). In the latest genome-wide association data browser from the FinnGen research project (Release 5; 218,792 individuals; https://r5.finngen.fi/), which aims to study genetic variation associations with various traits in the isolated population of Finland, we found variants in or near these seven novel candidate genes that are possibly associated to HI as well (+/− 100 kb). This includes variants in or near CCDC141 (rs144697379; p = 2.6 × 10−5; intronic); DNAH11 (rs2965393; p = 4.5 × 10−4; intronic); PAX8 (rs115708270; p = 3.6 × 10−4; upstream); and SOX9 (rs16977126; p = 5.7 × 10−5; downstream) for sensorineural HI, and in MYO19 (rs143245472; p = 6.6 × 10−6; upstream); INPP4B [rs184880581; p = 2.6 × 10−5; intronic) and POTEI (rs1337496945; p = 5.6 × 10−4; downstream) for sudden idiopathic HI. Although these results support our findings that these genes are involved in HI, without access to the data it is impossible to correct for multiple testing which may render the finding insignificant. Moreover, unlike congenital HI described in Ghanaian patients, most of these participants are adults suggesting these genes could be also associated with late-onset HI.
For six candidate genes, publicly available RNA sequence and microarray data sets were used for in silico investigation of gene expression in the developing and adult mouse inner ear (Fig. 4 and Supplementary Figs. 8–13, Ccdc141, Dnah11, Inpp4b, Myo19, Pax8, and Sox9) and also for protein modeling studies (Fig. 5).
Paired box gene 8, PAX8 (OMIM:167415), is located on chromosome 2q14.1 and encodes a transcription factor53. Cloning and expression experiments have shown that PAX8 is mainly expressed in the thyroid gland and can cause hypothyroidism54. In addition, in Pax8 mutant mice, major abnormalities of the outer and middle ear structures were found and an overall delay in the maturation of the auditory system of Pax8 knockout mice compared to the WT mice24.
Previous studies have shown that SOX9 is part of a family of transcription factors that are involved in the regulation of inner ear development. In mice, Sox9 is required for invagination of the otic placode55 and the loss of Sox9 severely compromises expansion, differentiation, and remodeling of the otic fibrocyte compartment56. Sox9‐depletion also results in major defects in the development of vestibular structures, semi-circular canals, and utricle in Xenopus and in failure of otic placode invagination in the mouse55,57. In humans, previous molecular studies showed that hair cell differentiation in humans was consistently present from 12 weeks, coinciding with downregulation of SOX958. Heterozygous variants in SOX9 cause campomelic dysplasia (OMIM: 114290) that can include HI as one of its clinical expressions. However, campomelic dysplasia is often lethal in the first year of age, due to respiratory insufficiency related to small chest size life and tracheobronchial hypoplasia. Moreover, the Ghanaian family with monoallelic variant in SOX9 only presented with HI and there was no evidence of shortness and bowing of long tubular bones, or sex reversal. Nevertheless, a careful follow-up with an evaluation of evolving discrete phenotypes of campomelic dysplasia could be considered in this family.
There is a relatively large number of HI genes that encode myosin superfamily members (MYO3A, MYO6, MYO7A, MYO15A, MYH14, and MYH9)59; therefore, it might not be surprising that MYO19 is a HI gene. Human myosin 19 functions as an actin-based motor for mitochondrial movement in vertebrate cells and promotes their localization to stress-induced filopodia60–62. In mice, Myo19 is a high-duty ratio molecular motor moving to the plus-end of the actin filament63.
A previous study in mice has indicated that Ccdc141 is required for radial migration and myosin II-mediated centrosome positioning during neuronal development64. Loss-of-function analysis in primitive neural stem cells derived from mouse embryonic stem cells demonstrated that Ccdc141 plays an important function in the regulation of early neural commitment65. Ccdc141 mouse mutants have normal ABR thresholds, but there are other possible explanations for this, such as incomplete knockout of gene function. Ccdc141-deficient mice exhibited impaired recognition memory and spatial reference memory66. In human, biallelic variants in CCDC141 was associated with pituitary stalk interruption syndrome (a rare disorder characterized by an absent or ectopic posterior pituitary, interrupted pituitary stalk, and anterior pituitary hypoplasia)67; and in normonosmic or anosmic hypogonadotropic hypogonadism, due to neuronal migration disorders the results in a defect in the development of the GnRH and the olfactory system68–70.
Dynein heavy chain 11 (DNAH11), an essential component of ciliary structure and function and left-right asymmetry in mice71–73. Biallelic variants in the DNAH11 gene are associated with primary ciliary dyskinesia in multiple populations74–77, and in heterotaxy syndrome, and laterality defect78–80. The present study adds NSHI to the spectrum of pathologies associated with variants in DNAH11, likely due to the alteration of primary cilia in hearing function found in numerous ciliopathies81.
INPP4B overexpression is associated with human achalasia, which is a rare motility disorder characterized by myenteric neuron and interstitial cells of Cajal abnormalities82. Depleting INPP4B by in utero electroporation in mice suppressed medially directed callosal axon formation and significantly attenuated the formation of pyramidal neurons and axon polarization in cortical neurons during cortical development83. However, the mechanisms of INPP4B dysregulation in hearing function will require further investigation.
In the future, it is desirable to perform in vitro functional analysis, particularly for the 17 missense and the one splice variants found in the novel candidate genes (Table 3), and if possible, to develop and investigate animal models when applicable, to complement the data provided in this study: families’ variants segregations (Fig. 3 and Supplementary Fig. 7), in silico analysis (Fig. 5 and Supplementary Table 4), and gene expressions profiles in the inner ear (Fig. 4 and Supplementary Figs. 8–16). It is also worthwhile to refine the phenotyping for the existing mutant hearing-impaired mice, i.e., Pax8, Sox9, and Ccdc141, with regard to the new clinical evidence described in this study that strongly supports their relevance to the genetics of human hearing.
In addition to the seven new candidate genes identified, a phenotypic expansion was seen for MARVELD2, and a family was found with a different inheritance model of DSPP. A high proportion of HI-associated variants that were not previously described was seen as well. Other strengths of this study are the number and the size of families and the ability to recruit a substantial portion of the family members. Exome sequence data generated for multiple family members facilitated segregation analysis and variant identification. There was no significant difference in the size of the solved versus unsolved families. The data set contributes to refining HI gene curation since most of the identified variants have not previously been associated with HI. The data will also contribute to available exomes from African populations, facilitate future exploration of pathogenic variants in actionable genes and refine their relevance and importance in African populations.
Using WES, this study of families from Ghana obtained a high-solve rate of genetic causes among GJB2-negative families segregating HI. WES should be considered for routine investigations in clinical settings. Six new genes were associated with NSHI: INPP4B, CCDC141, MYO19, DNAH11, SOX9, and POTEI; and one new gene, PAX8, was associated with Waardenburg syndrome. The study reveals and emphasizes the high level of allelic heterogeneity for HI genes. Most identified variants, 48/60 (80.0), had not previously been reported to be associated with HI, and to the best of our knowledge, 12 (20%) of the variants are also novel. Most of the GJB2-negative families that had variants in a known HI gene (29/48) had a unique variant segregating with HI, suggesting that future clinical diagnostic approaches should use next-generation sequencing, and ideally clinical WES for GJB2-negative families. This study will contribute to the global knowledge on the genetics of HI in understudied African populations and has provided the opportunity for novel HI genes discovery.
Methods
Ethical approval
We observed and adhered strictly to the guiding principles of the Declaration of Helsinki. Ethical approvals were obtained from Noguchi Memorial Institute for Medical Research Institutional Review Board (NMIMR-IRB CPN 006/16-17) and College of Basic and Applied Sciences, Ethics Committee for Basic and Applied Sciences (ECBAS 053/19-20) the University of Cape Town, Faculty of Health Sciences’ Human Research Ethics Committee (HREC 104/2018), and the Institutional Review Boards of Columbia University (IRB-AAAS2343). The study was clearly explained to each participant in the language in which they are fluent and informed consent was signed prior to participation. Parents/guardians provided signed consent for their children who were ≤18 years of age. In addition, assent was obtained for children older than 7 years of age. Where applicable, informed consent was obtained from the study participants for publishing their images.
Participants’ recruitment
Patient recruitment procedures were previously described3. The probands of Ghanaian families segregating HI were identified through nine schools for the deaf across the country (Fig. 1), and additional family members were recruited thereafter. Families with HI were also identified and recruited through our community engagement activities13 (Fig. 1). Medical records of all our participants were reviewed by a medical geneticist, and an ear, nose, and throat specialist. Detailed personal and family histories were obtained through a rigorous clinical interview. A structured questionnaire was used to interview each participant to rule out potential environmental causes of HI. The studied families had at least two members affected with HI that followed a Mendelian mode of inheritance. For those participants that were ascertained from schools for the deaf, before being admitted to the school they had undergone a systemic general and otological examination, that included pure tone audiometry.
A total of 5 ml of peripheral blood was obtained from each participant and genomic DNA (gDNA) was extracted from the samples using the QIAamp DNA Blood Maxi Kit® (Qiagen, USA).
Exclusion of GJB2 and the del(GJB6-D13S1830) deletion
All hearing-impaired participants were first screened using targeted sequencing as described previously3, and shown to be negative for PLP GJB2 variants including the del(GJB6-D13S1830) deletion. Allele-specific primers were used to amplify the coding region of GJB2 and GJB6. By the use of BigDye™ Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific), the amplicons were Sanger sequenced and resolved using ABI 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). A total of 88 families were ascertained altogether and 37/88 were found with GJB2 variants and excluded from the set of 51 families to be studied by WES (Fig. 1a).
Whole-exome sequencing and data analyses
gDNA samples from 51 families with at least two affected hearing-impaired family members (Fig. 1) underwent WES. The DNA concentration and quality checks were conducted using the QuantiFluor dsDNA System on a Quantus Fluorometer (Promega, Madison, WI). The exome library for 129 samples (batch one) was prepared using the Nextera Rapid Capture Exome kit (Illumina, San Diego, CA), 50 ng of gDNA was fragmented using the Nextera transposomes and the resultant libraries were hybridized with a 37 Mb probe pool to enrich exome sequences. Libraries were sequenced on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA) with the use of the pair-end 100 bp run format. The exome library preparation of the rest of the samples (n = 51, batch two) was performed using SureSelect V4 + UTR 71 Mb All Exon Capture Kit (Agilent Technologies, Inc., Santa Clara, CA, USA), ~3–5 µg of the DNA was fragmented with ultrasound using a Covaris® instrument (Covaris, Inc., Woburn, MA, USA). The libraries were sequenced on the Illumina HiSeq 2000 (Illumina, San Diego, CA) and produced paired-end reads of 100 bp. The exome sequence data mapping and variant calling were performed using the Illumina BaseSpace app suite. The Illumina DRAGEN Germline Pipeline v3.2.8 was used to align the sequence reads to the human reference genome (hg19) and variants were jointly called using the Genome Analysis Toolkit (GATKv4.1.7) software package84. Variant quality was assessed using variant quality score recalibration (VQSR) using the ApplyVQSR function of GATK.
Annotation and filtering strategy
An in-house pipeline that uses ANNOVAR85, dbNSFP86, and dbscSNV was used to annotate and filter single-nucleotide and insertion/deletion (indels) variants. Filtering was performed using Genome Aggregation Database (gnomAD)17 population-specific MAF of <0.005 [for AR and X-linked (XL)] and <0.0005 for AD with variants meeting these criteria being further ranked based on the bioinformatics prediction scores from SIFT87; polymorphism phenotyping v2 (PolyPhen-2)88 MutationTaster89; CADD90; deleterious annotation of genetic variants using neural networks (DANN)91; and Genomic Evolutionary Rate Profiling (GERP++)92. Information from the Hereditary Hearing Loss Homepage (HHL)6, Online Mendelian Inheritance in Man (OMIM)93, Human Phenotype Ontology (HPO)94, and ClinVar95 databases and deafness animal models were also used to prioritize identified variants. The MAFs of variants were further evaluated using the TOPMed Bravo database96. In families for which candidate variant(s) were not identified exome sequence data were also analyzed for CNV using CoNIFER97. These variants were assessed for their clinical significance based on the ACMG-AMP classification98, considering CADD scores90, and their allele frequencies in gnomAD and TOPMed databases (Supplementary Tables 1 and 2).
Principal component analysis (PCA)
Joint calling was performed on samples in each batch separately and two VCF files were generated that included only variants with read depth >8, genotype quality >20%, and that passed VQSR filters. Biallelic single-nucleotide variants (SNVs) for both batches (176,820 for batch one and 326,250 for batch two) were then extracted into separate VCF files. Further quality control to remove variants that were present in only one batch was performed by applying a genotyping call rate of 95%. The phased batch one and two data sets were then merged using bcftools and contained 1,141,838 biallelic SNVs of the phased 1KGP3 data, that were polymorphic in the Mende population from Sierra Leone (MSL). The MSL population has been previously reported to harbor a large proportion of basal West African ancestry99. Further quality control includes the removal of variant sites missing more than 5% of their genotypes and samples missing more than 10% of their data, as well as the removal of variants that failed the Hardy–Weinberg equilibrium test at a threshold of 1 × 10−6, and SNVs with MAF < 0.05 was applied to the merged data set using PLINK2100 leaving a total of 18,880 SNPs. Linkage disequilibrium-pruning including only SNPs with r2 < 0.2 within a 50 bp region (window size = 10 bp) left 14,987 high-quality SNVs for inclusion in the PCA, for which five PCs were computed using PLINK2.
Expression of candidate genes in the developing and adult mouse inner ear
Various publicly available RNA sequencing and microarray data sets were used for an in silico investigation of the expression of CCDC141, DNAH11, INPP4B, MYO19, PAX8, and SOX9 in the developing and adult mouse inner ear. To study the expression during mouse craniofacial development, series GSE55966 from the Gene Expression Omnibus database was evaluated101,102. This data set includes RNA sequence data of CD1 mouse embryos at three stages: E8.5, E9.5, and E10.5102. Craniofacial gene expression data were presented as a set of FPKM (fragments per kilobase million) values for 13 different tissue/stage pairs102 that were converted to TPM (transcripts per kilobase million) values for our analysis.
We also studied expression levels during mouse inner ear development in previously generated data sets present in the Shared Harvard Inner-Ear Laboratory Database (SHIELD)103. The first data set detailed expression over four developmental stages: E16, P0, P4, and P7104. Data were obtained from the cochlea and utricles of mice that expressed EGFP under the Pou4f3 promoter104. Fluorescence-activated cell sorting was used to separate cells into hair cells (GFP+) and surrounding cells (GFP–) prior to RNA extraction104. To supplement these data, a second data set was downloaded, which contained expression data for six developmental stages: E12, E13, E16, P0, P6, and P15105. The expression data were produced by Affymetrix Mouse 420 v.2 GeneChips and subset into expression in spiral ganglion neurons and vestibular ganglion neurons105.
Lastly, the gene Expression Analysis Resource (gEAR) suite was used to visualize expression in cells of the cochlear epithelium during mouse development. gEAR includes single-cell RNA sequence data obtained from CD1 mouse embryos at four developmental stages: E14, E16, P1, and P7106. CCDC141, DNAH11, INPP4B, MYO19, and SOX9 expression was also grouped based on cell groups in four overarching classes: developing supporting cells, developing prosensory cells, developing sensory cells, and developing greater epithelial ridge cells of which each were further divided into subclasses106.
Sanger sequencing validation of MYO19 and DNAH11 variants
Allele-specific primers (Supplementary Table 6) were designed using NCBI primer BLAST® and optimized for polymerase chain reaction (PCR) and sequencing of the regions of MYO19 and DNAH11. The initial denaturation temperature was 95 °C for 3 min and 30 s for subsequent denaturation. An annealing temperature was 60 °C for 30 s and an extension temperature of 72 °C for 1 min for 35 cycles was employed. The PCR amplicons were sanger sequencing using BigDye™ Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific). The Sanger sequences were resolved using ABI 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Homology modeling
The human INPP4B (ID: O15327), MYO19 (Q96H55), DNAH11 (ID: Q96DT5), CCDC141 (ID: Q6ZP82), and PAX8 (ID: Q06710) protein sequences were obtained from the UniProt database for homology modeling107. To find appropriate structural templates for these proteins, PSI-BLAST was run against the Protein Data Bank (PDB). The crystal structure of PDB ID: 2Q5D108, PDB ID: 5IOI109, PDB ID: 3VKG110, PDB ID: 3EDV111, and PDB ID 4K6J112 were used as a template for the construction of INPP4B (residue 215-877), MYO19 (residue 30-804), DNAH11 (residue 1372-4515), CCDC141 (residue 36-562), and PAX8 (residue 120-398) protein models. Homology modeling was carried out for WT and mutant proteins using MODELLER113 based on the sequence alignment generated between the template and target sequences. The Adaptive Poisson-Boltzmann Solver Electrostatics plugin of PyMOL was used for protein surface electrostatic potentials. PyMOL114 version 2.0.3 was used for visualization and the processing of figures.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are grateful to the leadership of the Special Education Unit of the Ghana Education Service, all staff of Schools for the Deaf, patients, and their family members for their participation in this research project. We are grateful to Prof. Karen Steel (Wolfson Centre for Age-Related Diseases, King’s College London, UK) to have critically revised this manuscript. This study was possible thanks to funding from the Wellcome Trust, grant number 107755Z/15/Z to G.A.A. and A.W.; NIH, USA, grant number U01-HG-009716 to A.W., the African Academy of Science/Wellcome Trust, grant number H3A/18/001 to A.W., and the National Institute of Deafness and other Communication Disorders grants R01 DC01165, R01 DC003594, R01 DC016593, and R01 DC017712 to S.M.L. The funders were not involved in the study design, data collection, and analysis, the decision to publish, or the preparation of the manuscript.
Author contributions
Conception of the project: A.W., S.M.L., and G.A.A.; Ghanaian family recruitment and molecular experiments: S.M.A. and E.T.A.; exclusion of GJB2 and GJB6 variants: E.W.-T., N.M., and S.M.A.; bioinformatics analysis: I.S., T.B., S.M.L., L.M.N.-S., A.A., and K.E.; in silico analysis of the pathogenicity of variants: S.M.A., I.S., and K.E.; protein modeling and mouse inner ear expression data analysis102: I.S., D.M.C.S., T.B., A.N., J.L.E., M.K.K., L.M.N.-S., K.E., and S.M.A.; writing of the first draft of the manuscript: A.W. and S.M.A.; review and editing: all authors; project management and oversight: K.P., C.deK., and V.N.; supervision of the whole project: G.A.A., G.K.A., O.Q., S.M.L., and A.W. All authors have agreed to the final version of the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: George Inglis. Peer reviewer reports are available.
Data availability
The Sanger sequences generated were submitted to GenBank with the following accession codes: OM965691, OM965692, OM965693, OM965694, OM965695, and OM965696. SNV data have been submitted to dbSNP (https://www.ncbi.nlm.nih.gov/snp/). Source data for the graphs can be found in Supplementary Data 1 and Supplementary Data 2. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. Due to lack of ethical approval, individual-level whole-exome sequence data cannot be made publicly available; however, they can be obtained from the corresponding authors [A.W. and S.M.L.] upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ambroise Wonkam, Samuel Mawuli Adadey.
Contributor Information
Ambroise Wonkam, Email: awonkam1@jhmi.edu.
Suzanne M. Leal, Email: sml3@cumc.columbia.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03326-8.
References
- 1.Tucci D, Merson MH, Wilson BS. A summary of the literature on global hearing impairment: current status and priorities for action. Otol. Neurotol. 2010;31:31–41. doi: 10.1097/MAO.0b013e3181c0eaec. [DOI] [PubMed] [Google Scholar]
- 2.Wonkam A, et al. Aetiology of childhood hearing loss in Cameroon (sub-Saharan Africa) Eur. J. Med. Genet. 2013;56:20–25. doi: 10.1016/j.ejmg.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Adadey SM, et al. GJB2 and GJB6 mutations in non-syndromic childhood hearing impairment in Ghana. Front Genet. 2019;10:1–10. doi: 10.3389/fgene.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Prevention of blindness and deafness In: Prevention of blindness and deafness (PBD). World health Organization. https://www.who.int/pbd/deafness/hearing_impairment_grades/en/ (2020).
- 5.Morton NE. Genetic epidemiology of hearing impairment. Ann. N. Y. Acad. Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Camp G. & Smith R. Hereditary Hearing Loss Homepage. https://hereditaryhearingloss.org/ (2020).
- 7.Koohiyan M. Genetics of hereditary hearing loss in the Middle East: a systematic review of the carrier frequency of the GJB2 mutation (35delG) Audiol. Neuro-Otol. 2019;24:161–165. doi: 10.1159/000502201. [DOI] [PubMed] [Google Scholar]
- 8.Azadegan-Dehkordi F, et al. Mutations in GJB2 as major causes of autosomal recessive non-syndromic hearing loss: first report of c.299-300delAT mutation in Kurdish Population of Iran. J. Audio. Otol. 2019;23:20–26. doi: 10.7874/jao.2018.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koohiyan M, et al. An update of spectrum and frequency of GJB2 mutations causing hearing loss in the south of Iran: a literature review. Int J. Pediatr. Otorhinolaryngol. 2019;119:136–140. doi: 10.1016/j.ijporl.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Wonkam A. Letter to the editor regarding “GJB2, GJB6 or GJA1 genes should not be investigated in routine in non syndromic deafness in people of sub-Saharan African descent”. Int J. Pediatr. Otorhinolaryngol. 2015;79:632–633. doi: 10.1016/j.ijporl.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N. Engl. J. Med. 1998;338:548–550. doi: 10.1056/NEJM199802193380813. [DOI] [PubMed] [Google Scholar]
- 12.Hamelmann C, et al. Pattern of connexin 26 (GJB2) mutations causing sensorineural hearing impairment in Ghana. Hum. Mutat. 2001;18:84–85. doi: 10.1002/humu.1156. [DOI] [PubMed] [Google Scholar]
- 13.Adadey SM, et al. Enhancing genetic medicine: rapid and cost-effective molecular diagnosis for a GJB2 founder mutation for hearing impairment in Ghana. Genes. 2020;11:132. doi: 10.3390/genes11020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan D, Tekin M, Blanton SH, Liu XZ. Next-generation sequencing in genetic hearing loss. Genet Test. Mol. Biomark. 2013;17:581–587. doi: 10.1089/gtmb.2012.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 16.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. doi: 10.1101/gr.9.8.677. [DOI] [PubMed] [Google Scholar]
- 17.Karczewski K, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe KL, et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.TOPMed. The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program. BRAVO variant browser. University of Michigan and NHLBI. https://bravo.sph.umich.edu/freeze5/hg38/ (2018).
- 20.Siemens J, et al. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl Acad. Sci. USA. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 22.Burghard AL, Morel NP, Oliver DL. Mice heterozygous for the Cdh23/Ahl1 mutation show age-related deficits in auditory temporal processing. Neurobiol. Aging. 2019;81:47–57. doi: 10.1016/j.neurobiolaging.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riazuddin S, et al. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat. Genet. 2012;44:1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christ S, et al. Hearing loss in athyroid pax8 knockout mice and effects of thyroxine substitution. Audiol. neuro-Otol. 2004;9:88–106. doi: 10.1159/000076000. [DOI] [PubMed] [Google Scholar]
- 25.Fiser A., Šali A. Modeller: generation and refinement of homology-based protein structure models. In: Methods in Enzymology. (Elsevier, 2003). [DOI] [PubMed]
- 26.Muthusamy B, Bellad A, Girimaji SC, Pandey A. Shukla-Vernon syndrome: a second family with a novel variant in the BCORL1 gene. Genes (Basel) 2021;12:452. doi: 10.3390/genes12030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DH, Kim EJ, Kim DH, Park SW. Dact2 is involved in the regulation of epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2020;524:190–197. doi: 10.1016/j.bbrc.2019.12.090. [DOI] [PubMed] [Google Scholar]
- 28.Lewis MA, Di Domenico F, Ingham NJ, Prosser HM, Steel KP. Hearing impairment due to Mir183/96/182 mutations suggests both loss-of-function and gain-of-function effects. Dis. Models Mechanisms. 2021;14:dmm047225. doi: 10.1242/dmm.047225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith CM, et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019;47:D774–D779. doi: 10.1093/nar/gky922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebeko K, et al. Targeted genomic enrichment and massively parallel sequencing identifies novel nonsyndromic hearing impairment pathogenic variants in Cameroonian families. Clin. Genet. 2016;90:288–290. doi: 10.1111/cge.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sang S, et al. Proband based whole exome sequencing identified genes responsible for autosomal recessive non-syndromic hearing loss in thirty-three Chinese nuclear families. Front Genet. 2019;10:639. doi: 10.3389/fgene.2019.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, et al. Increased diagnostic yield by reanalysis of data from a hearing loss gene panel. BMC Med. Genomics. 2019;12:76. doi: 10.1186/s12920-019-0531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis MA, et al. Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Med. Genomics. 2018;11:77. doi: 10.1186/s12920-018-0395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung J, et al. Whole-exome sequencing identifies two novel mutations in KCNQ4 in individuals with nonsyndromic hearing loss. Sci. Rep. 2018;8:16659. doi: 10.1038/s41598-018-34876-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atik T, Bademci G, Diaz-Horta O, Blanton SH, Tekin M. Whole-exome sequencing and its impact in hereditary hearing loss. Genet Res. (Camb.) 2015;97:e4. doi: 10.1017/S001667231500004X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downie L, et al. Exome sequencing in infants with congenital hearing impairment: a population-based cohort study. Eur. J. Hum. Genet. 2020;28:587–596. doi: 10.1038/s41431-019-0553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wonkam A, de Vries J. Returning incidental findings in African genomics research. Nat. Genet. 2020;52:17–20. doi: 10.1038/s41588-019-0542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bope CD, et al. Dissecting in silico mutation prediction of variants in African genomes: challenges and perspectives. Front Genet. 2019;10:601. doi: 10.3389/fgene.2019.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Astuto LM, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu T, Zhu W, Wang P. The p.P240L variant of CDH23 and the risk of nonsyndromic hearing loss: a meta-analysis. Eur. Arch. Oto-Rhino-Laryngology. 2019;276:11–16. doi: 10.1007/s00405-018-5160-8. [DOI] [PubMed] [Google Scholar]
- 41.Wagatsuma M, et al. Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin. Genet. 2007;72:339–344. doi: 10.1111/j.1399-0004.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim BJ, et al. Discovery of CDH23 as a significant contributor to progressive postlingual sensorineural hearing loss in Koreans. PLoS One. 2016;11:e0165680. doi: 10.1371/journal.pone.0165680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bujakowska KM, et al. Targeted exon sequencing in Usher syndrome type I. Invest Ophthalmol. Vis. Sci. 2014;55:8488–8496. doi: 10.1167/iovs.14-15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao L, et al. Next-generation sequencing-based molecular diagnosis of 82 retinitis pigmentosa probands from Northern Ireland. Hum. Genet. 2015;134:217–230. doi: 10.1007/s00439-014-1512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebermann I, et al. Deafblindness in French Canadians from Quebec: a predominant founder mutation in the USH1C gene provides the first genetic link with the Acadian population. Genome Biol. 2007;8:R47. doi: 10.1186/gb-2007-8-4-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehman AU, et al. Mutational spectrum of MYO15A and the molecular mechanisms of DFNB3 human deafness. Hum. Mutat. 2016;37:991–1003. doi: 10.1002/humu.23042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, et al. Genotype-phenotype correlation analysis of MYO15A variants in autosomal recessive non-syndromic hearing loss. BMC Med. Genet. 2019;20:60. doi: 10.1186/s12881-019-0790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farjami M, et al. The worldwide frequency of MYO15A gene mutations in patients with autosomal recessive non-syndromic hearing loss: a meta‐analysis. Iran J. Basic Med. Sci. 2020;23:841–848. doi: 10.22038/IJBMS.2020.35977.8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan D, et al. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum. Genet. 2016;135:953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salime S, et al. Homozygous mutations in PJVK and MYO15A genes associated with non-syndromic hearing loss in Moroccan families. Int. J. Pediatr. Otorhinolaryngol. 2017;101:25–29. doi: 10.1016/j.ijporl.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Budde BS, et al. Comprehensive molecular analysis of 61 Egyptian families with hereditary nonsyndromic hearing loss. Clin. Genet. 2020;98:32–42. doi: 10.1111/cge.13754. [DOI] [PubMed] [Google Scholar]
- 52.Wonkam A, Manyisa N, Bope CD, Dandara C, Chimusa ER. Whole exome sequencing reveals pathogenic variants in MYO3A, MYO15A and COL9A3 and differential frequencies in ancestral alleles in hearing impairment genes among individuals from Cameroon. Hum. Mol. Genet. 2020;29:3729–3743. doi: 10.1093/hmg/ddaa225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corona RI, et al. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat. Commun. 2020;11:2020. doi: 10.1038/s41467-020-15951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwahashi-Odano M, et al. Congenital hypothyroidism due to truncating PAX8 mutations: a case series and molecular function studies. J. Clin. Endocrinol. Metab. 2020;105:dgaa584. doi: 10.1210/clinem/dgaa584. [DOI] [PubMed] [Google Scholar]
- 55.Barrionuevo F, et al. Sox9 is required for invagination of the otic placode in mice. Developmental Biol. 2008;317:213–224. doi: 10.1016/j.ydbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 56.Trowe MO, et al. Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Dev. Biol. 2010;342:51–62. doi: 10.1016/j.ydbio.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 57.Park BY, Saint-Jeannet JP. Long-term consequences of Sox9 depletion on inner ear development. Dev. Dyn. 2010;239:1102–1112. doi: 10.1002/dvdy.22259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Locher H, et al. Neurosensory development and cell fate determination in the human cochlea. Neural Dev. 2013;8:20. doi: 10.1186/1749-8104-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman T. B., Belyantseva I. A. & Frolenkov G. I. Myosins and hearing. In: Myosins. (Springer, 2020). [DOI] [PubMed]
- 60.Quintero OA, et al. Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 2009;19:2008–2013. doi: 10.1016/j.cub.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rohn JL, et al. Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr. Biol. 2014;24:2598–2605. doi: 10.1016/j.cub.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usaj M, Henn A. Kinetic adaptation of human Myo19 for active mitochondrial transport to growing filopodia tips. Sci. Rep. 2017;7:11596. doi: 10.1038/s41598-017-11984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu ZK, et al. Mouse myosin-19 is a plus-end-directed, high-duty ratio molecular motor. J. Biol. Chem. 2014;289:18535–18548. doi: 10.1074/jbc.M114.569087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukuda T, Sugita S, Inatome R, Yanagi S. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J. Biol. Chem. 2010;285:40554–40561. doi: 10.1074/jbc.M110.179481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsang WH, et al. LIF-dependent primitive neural stem cells derived from mouse ES cells represent a reversible stage of neural commitment. Stem Cell Res. 2013;11:1091–1102. doi: 10.1016/j.scr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Fukuda T, Nagashima S, Inatome R, Yanagi S. CAMDI interacts with the human memory-associated protein KIBRA and regulates AMPAR cell surface expression and cognition. PLoS One. 2019;14:e0224967. doi: 10.1371/journal.pone.0224967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brauner R, Bignon-Topalovic J, Bashamboo A, McElreavey K. Pituitary stalk interruption syndrome is characterized by genetic heterogeneity. PLoS One. 2020;15:e0242358. doi: 10.1371/journal.pone.0242358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turan I, et al. CCDC141 mutations in idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2017;102:1816–1825. doi: 10.1210/jc.2016-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hutchins BI, et al. CCDC141 mutation identified in anosmic hypogonadotropic hypogonadism (Kallmann Syndrome) alters GnRH neuronal migration. Endocrinology. 2016;157:1956–1966. doi: 10.1210/en.2015-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou Q, et al. Genotypic and phenotypic spectrum of CCDC141 variants in a Chinese cohort with congenital hypogonadotropic hypogonadism. Eur. J. Endocrinol. 2020;183:245–254. doi: 10.1530/EJE-19-1018. [DOI] [PubMed] [Google Scholar]
- 71.Essner JJ, et al. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- 72.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/S0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 73.Lai M, et al. Gene editing of DNAH11 restores normal cilia motility in primary ciliary dyskinesia. J. Med. Genet. 2016;53:242–249. doi: 10.1136/jmedgenet-2015-103539. [DOI] [PubMed] [Google Scholar]
- 74.Kurokawa A, et al. Multifaceted analysis of Japanese cases of primary ciliary dyskinesia: Value of immunofluorescence for ciliary protein detection in patients with DNAH5 and DNAH11 mutations. Respir. Investig. 2021;59:550–554. doi: 10.1016/j.resinv.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 75.Guan Y, et al. Clinical and genetic spectrum of children with primary ciliary dyskinesia in China. Chest. 2021;159:1768–1781. doi: 10.1016/j.chest.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shoemark A. et al. Topological data analysis reveals genotype-phenotype relationships in primary ciliary dyskinesia. Eur. Respir. J.58, 2002359. 10.1183/13993003.02359-2020 (2021). [DOI] [PubMed]
- 77.Schultz R, Elenius V, Lukkarinen H, Saarela T. Two novel mutations in the DNAH11 gene in primary ciliary dyskinesia (CILD7) with considerable variety in the clinical and beating cilia phenotype. BMC Med. Genet. 2020;21:237. doi: 10.1186/s12881-020-01171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu S, et al. DNAH11 variants and its association with congenital heart disease and heterotaxy syndrome. Sci. Rep. 2019;9:6683. doi: 10.1038/s41598-019-43109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Namavarian A, Eid A, Goh ES, Thakur V. A novel DNAH11 variant segregating in a sibship with heterotaxy and implications for genetic counseling. Mol. Genet Genom. Med. 2020;8:e1358. doi: 10.1002/mgg3.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loges NT, et al. Recessive DNAH9 loss-of-function mutations cause laterality defects and subtle respiratory ciliary-beating defects. Am. J. Hum. Genet. 2018;103:995–1008. doi: 10.1016/j.ajhg.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moon K. H., et al. Dysregulation of sonic hedgehog signaling causes hearing loss in ciliopathy mouse models. Elife9, e56551. 10.7554/eLife.56551 (2020). [DOI] [PMC free article] [PubMed]
- 82.Bonora E, et al. INPP4B overexpression and c-KIT downregulation in human achalasia. Neurogastroenterol. Motil. 2018;30:e13346. doi: 10.1111/nmo.13346. [DOI] [PubMed] [Google Scholar]
- 83.Ji L, Kim NH, Huh SO, Rhee HJ. Depletion of inositol polyphosphate 4-phosphatase II suppresses callosal axon formation in the developing mice. Mol. Cells. 2016;39:501–507. doi: 10.14348/molcells.2016.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diaz-Horta O, et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7:e50628. doi: 10.1371/journal.pone.0050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variations from next-generation sequencing data. Nucl. Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu X, Li C, Mou C, Dong Y, Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103. doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen‐2. Curr. Prot. Hum. Genet. 2013;76:1–52. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 90.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP plus. PLoS Comput. Biol. 2010;6:1–13. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robinson PN, Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 95.Landrum MJ, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krumm N, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oza AM, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018;39:1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skoglund P, et al. Reconstructing prehistoric African population structure. Cell. 2017;171:59–71.e21. doi: 10.1016/j.cell.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Purcell S., Chang C. PLINK2 (v1. 90b6. 9). Accessed 1 May 2019. www.cog-genomics.org/plink/20/.
- 101.Clough E, Barrett T. The gene expression omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brunskill EW, et al. A gene expression atlas of early craniofacial development. Dev. Biol. 2014;391:133–146. doi: 10.1016/j.ydbio.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen J, Scheffer DI, Kwan KY, Corey DP. SHIELD: an integrative gene expression database for inner ear research. Database. 2015;2015:bav071. doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scheffer DI, Shen J, Corey DP, Chen ZY. Gene expression by mouse inner ear hair cells during development. J. Neurosci. 2015;35:6366–6380. doi: 10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu CC, Appler JM, Houseman EA, Goodrich LV. Developmental profiling of spiral ganglion neurons reveals insights into auditory circuit assembly. J. Neurosci. 2011;31:10903–10918. doi: 10.1523/JNEUROSCI.2358-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kolla L, et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bateman A, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 109.Burger D, et al. Crystal structures of the human doublecortin C- and N-terminal domains in complex with specific antibodies. J. Biol. Chem. 2016;291:16292–16306. doi: 10.1074/jbc.M116.726547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kon T, et al. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 111.Davis L, et al. Localization and structure of the ankyrin-binding site on beta2-spectrin. J. Biol. Chem. 2009;284:6982–6987. doi: 10.1074/jbc.M809245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ouyang Z, et al. Structure of the human cohesin inhibitor Wapl. Proc. Natl Acad. Sci. USA. 2013;110:11355–11360. doi: 10.1073/pnas.1304594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Webb B., Sali A. Protein structure modeling with MODELLER. Functional Genomics 39–54 (2017). [DOI] [PubMed]
- 114.DeLano W. L. Pymol: an open-source molecular graphics tool. In: CCP4 Newsletter on Protein Crystallography. http://148.79.162.84/newsletters/newsletter40/11_pymol.pdf (2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The Sanger sequences generated were submitted to GenBank with the following accession codes: OM965691, OM965692, OM965693, OM965694, OM965695, and OM965696. SNV data have been submitted to dbSNP (https://www.ncbi.nlm.nih.gov/snp/). Source data for the graphs can be found in Supplementary Data 1 and Supplementary Data 2. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. Due to lack of ethical approval, individual-level whole-exome sequence data cannot be made publicly available; however, they can be obtained from the corresponding authors [A.W. and S.M.L.] upon reasonable request.