Summary
Background
Current microbiological methods lack the resolution to accurately identify multidrug-resistant organism (MDRO) transmission, however, whole genome sequencing can identify highly-related patient isolates providing opportunities for precision infection control interventions. We investigated the feasibility and potential impact of a prospective multi-centre genomics workflow for hospital infection control.
Methods
We conducted a prospective genomics implementation study across eight Australian hospitals over 15 months (2017,2018), collecting all clinical and screening isolates from inpatients with vanA VRE, MRSA, ESBL Escherichia coli (ESBL-Ec), or ESBL Klebsiella pneumoniae (ESBL-Kp). Genomic and epidemiologic data were integrated to assess MDRO transmission.
Findings
In total, 2275 isolates were included from 1970 patients, predominantly ESBL-Ec (40·8%) followed by MRSA (35·6%), vanA VRE (15·2%), and ESBL-Kp (8·3%).
Overall, hospital and genomic epidemiology showed 607 patients (30·8%) acquired their MDRO in hospital, including the majority of vanA VRE (266 patients, 86·4%), with lower proportions of ESBL-Ec (186 patients, 23·0%), ESBL-Kp (42 patients, 26·3%), and MRSA (113 patients, 16·3%). Complex patient movements meant the majority of MDRO transmissions would remain undetected without genomic data.
The genomics implementation had major impacts, identifying unexpected MDRO transmissions prompting new infection control interventions, and contributing to vanA VRE becoming a notifiable condition. We identified barriers to implementation and recommend strategies for mitigation.
Interpretation
Implementation of a multi-centre genomics-informed infection control workflow is feasible and identifies many unrecognised MDRO transmissions. This provides critical opportunities for interventions to improve patient safety in hospitals.
Funding
Melbourne Genomics Health Alliance (supported by State Government of Victoria, Australia), and National Health and Medical Research Council (Australia).
Keywords: Antimicrobial resistance, Whole genome sequencing, Infection prevention and control, Hospital epidemiology
Abbreviations: MRSA, Methicillin-resistant Staphylococcus aureus; VRE, Vancomycin-resistant Enterococcus; ESBL-Ec, Extended-spectrum beta-lactamase Escherichia coli; ESBL-Kp, Extended-spectrum beta-lactamase Klebsiella pneumoniae; MDRO, Multidrug-resistant organism; WGS, Whole genome sequencing
Research in context.
Evidence before the study
We searched PubMed for studies using the search terms “genomics”, “antimicrobial resistance”, “hospital”, infection control”, and “transmission” without date or language restrictions, and identified 262 results. We considered all studies from this search that used whole genome sequencing to assess likely transmission of multidrug-resistant organisms (MDROs) in the hospital environment, published before 5th September 2021. We excluded reviews, case reports and retrospective cohort analyses and outbreak investigations. We also excluded studies of fungi, mycobacteria, viruses, resistance to specific antimicrobial agents/classes not included in our study (e.g. carbapenems, colistin, linezolid), and studies not employing whole genome sequencing. Of the remaining nine studies with prospective sample collection, five focused on single species or resistance mechanisms (MRSA, E. coli, Enterococcus faecium/VRE, Klebsiella pneumoniae), one focused on VRE and multi-resistant Gram negatives, and two focussed on four different species. Study durations varied from three to 13 months; five studies included 1-2 wards, two studies included one hospital, and one study included five hospitals (single organism only). Only one study discussed results being reported to hospital infection control teams, or any changes made in response to genomic data; this was a pre- and post-intervention surveillance study to assess the effects of removing isolation requirements for patients colonised with MDR Gram negatives. No studies reported prospective genomic epidemiological investigations across multiple species across multiple hospital sites.
Added value of the study
This study is the first to implement and evaluate prospective genomic sequencing of more than one target MDRO across multiple institutions. We designed and implemented a study using prospective genomic surveillance for four MDROs, vanA VRE, MRSA, ESBL Escherichia coli (ESBL-Ec), and ESBL Klebsiella pneumoniae (ESBL-Kp), from all hospital inpatients across eight healthcare facilities over a period of 15 months. We combined genomic and epidemiologic data to identify the number of MDRO transmissions that occurred within and between hospitals, including up to 12 months prior to first MDRO identification. We demonstrated that vanA VRE was most likely to be acquired due to transmissions in hospitals. Hospital transmission rates for other MDROs (ESBL-Ec, ESBL-Kp and MRSA) were also significantly underestimated prior to the study, however colonisation with these pathogens is relatively common in our community. We identified key components of a prospective genomics workflow for successful implementation, and recommend mitigation strategies for challenges identified for implementation.
Implications of all the available evidence
This study addresses the lack of evidence for the prospective implementation of whole genome sequencing for uncovering and responding to MDRO transmissions across multiple healthcare sites. Taken together, studies of whole genome sequencing of MDROs in hospitals demonstrate the enormous potential for precision infection control interventions at the hospital level to be guided by this data. The next steps in transitioning from research to implementation require close engagement between hospital infection control teams, diagnostic microbiology laboratories and laboratories with expertise in genome sequencing and analysis, to create workflows that are efficient and sustainable in different settings, and accurately and rapidly communicate results to maximise the benefits to hospitals and patients. Our study also demonstrates that integrated data across multiple sites can inform higher level public health decision making regarding the notification and systematic surveillance of MDRO threats. Future implementation of whole genome sequencing for hospital infection control should consider and integrate a multi-layered approach (hospital and network/state level) to ensure that the data can inform precision infection control interventions at the hospital level as well as inform system-wide decision making.
Alt-text: Unlabelled box
Introduction
Antimicrobial resistance is a serious threat to human health, and disproportionately affects hospitalised patients, with many multidrug-resistant organisms (MDROs) acquired directly from other hospital patients or indirectly from the hospital environment.1, 2, 3 Whilst these MDROs can usually be adequately detected by routine diagnostic microbiology methods, these methods are insufficient to identify transmission chains, as multiple strains of each MDRO may be circulating in the hospital and the community.4,5
Whole genome sequencing (WGS, or 'genomics') of MDROs provides high-resolution typing data to identify transmission networks between patients and/or the environment.6, 7, 8 Genomics has increasingly been used in the research setting to identify hospital MDRO outbreaks, most often in a retrospective manner after a possible outbreak has already been identified by epidemiologic surveillance.9, 10, 11 However, the greatest potential for genomics for hospital infection control is to prospectively identify likely transmission of MDROs between a small number patients, allowing early infection control interventions before an outbreak has become established,12,13 therefore limiting patient morbidity, mortality, unnecessary antibiotic use, and waste of hospital resources.14, 15, 16 Although WGS costs have been steadily decreasing over the last decade, implementation should be directed to the most cost-effective targets with the greatest benefits to health and resources.17,18
Translation of genomics from the research setting to the real-world infection control setting requires consideration of the elements that are likely to be critical for successful implementation.5,17 Factors to consider include an optimised genomic workflow model to achieve clinically meaningful turnaround times, which patient populations and MDROs are likely to yield the greatest benefit from genomics, and how to effectively communicate complex genomic analyses to infection control teams at the coalface, to enable implementation of infection control interventions for maximal benefit to the patients and healthcare system.4,5
Here we present a study implementing a prospective genomics workflow to detect transmission of MDROs across multiple hospital networks over 15 months, with all MDRO samples from hospital inpatients being sequenced at a central laboratory, and results returned to the hospital infection control team for detailed epidemiological analysis and further management. We aim to establish the feasibility of such a workflow and potential impacts, and identify key components and potential barriers to implementation.
Methods
Study scoping and design
As a scoping exercise to design a genomics workflow for infection control, we first listed the components to be included, identified the key considerations for each component, then identified a strategy for implementation for each component, taking into account existing local infrastructure, MDRO patterns and resources (staffing, consumables, and financial resources), before settling on a study design for local implementation to establish the feasibility and potential impacts of infection control genomics.
We subsequently performed a prospective 'real-world' multi-centre genomics implementation study of MDRO transmission in eight hospitals from four hospital networks, including approximately 2800 acute and subacute inpatient beds, in Melbourne, Australia (population 4·9 million in 2018); hospital characteristics are described in Table 1. MDRO isolates from four defined species (see below) were included from patient samples (clinical or screening samples) collected routinely from hospital inpatients (>24 h) at any time during their admission. Duplicate screening isolates were excluded, and duplicate clinical isolates were only included if collected more than 14 days after the previous sample (see Supplementary Data for details).
Table 1.
Characteristics and infection control practices of hospital sites included in study.
| Hospital network | Hospital code | Hospital description | No. of inpatient bedsa | High-risk wards | MDRO screening practices during study period and changes during study | Management of patients colonised or infection with MDRO |
|---|---|---|---|---|---|---|
| A | A1 | Tertiary referral center, including ICU, solid-organ and bone marrow transplant | 560 | ICU, Haematology/BMT and Oncology , Renal Transplant & Liver Transplant (Spinal ward and respiratory ward added during study) |
vanA VRE and MRGN screening: ICU, on admission and twice weekly; haematology/oncology, renal and liver transplant wards screened on admission and weekly. Additional MRSA screening in ICU (on admission and twice weekly). Biannual point-prevalence survey for vanA VRE and MRGN. MRSA screening before critical surgeries (prosthetic joint, spinal and cardiac). Change during study: Added spinal ward and respiratory ward (ventilator support service) to high-risk wards for regular MDRO screening (October 2018) |
MRSA: patients only isolated in certain circumstances, e.g. highly exudative wound; single room, disposable apron, gloves. vanA VRE: single room with own bathroom, full gown, gloves; sometimes cohortedb with other vanA VRE-colonised patients. ESBL-Ec and ESBL-Kp (multidrug-resistant)c: single room, own bathroom, disposable apron |
| A2 | Subacute hospital, aged care and rehabilitation services | 150 | None | Biannual point-prevalence survey for vanA VRE and MRGN | ||
| A3 | Subacute hospital, rehabilitation services | 60 | None | Biannual point-prevalence survey for vanA VRE and MRGN | ||
| B | B1 | Tertiary referral center, including ICU and solid-organ transplant and specialist pediatric hospital (including neonatal ICU) | 640 | ICU, Renal Transplant | ICU and renal ward screened for vanA VRE and carbapenem-resistant Gram negatives (CRGN) on admission and weekly. MRSA screening before cardiac surgery. Changes during study: Stopped routine screening of renal ward for VRE (June 2018). Network-wide changes in cleaning practices to from microfibre/steam cleaning to bleach cleaning (September 2018) |
MRSA: single room, gloves and short-sleeved gown. vanA VRE: single room, own bathroom, occasional cohortingb with other vanA VRE-colonised patients; gloves and short-sleeved gown. Additional measures if diarrhoea (full gown). ESBL-Ec: no specific management measures. ESBL-Kp: single room, own bathroom, gloves and short-sleeve gown |
| B2 | Tertiary referral center, including ICU, trauma and some aged care & rehabilitation services | 573 | ICU | ICU patients screened for vanA VRE and carbapenem-resistant Gram negatives (CRGN) on admission and weekly. Changes during study: Cleaning protocol changes as above |
||
| C | C1 | Tertiary referral center, including ICU, solid-organ and bone marrow transplant | 571 | ICU, Haematology/BMT | ICU and haematology ward screened on admission and weekly for vanA VRE and MRGN | MRSA: single room only in certain situations, e.g. highly exudative wound. vanA VRE: single room, own bathroom, gloves and full gown. ESBL-Ec (multidrug-resistant)c and ESBL-Kp: single room, own bathroom, gloves and full gown |
| C2 | Subacute hospital, aged care and rehabilitation services | 150 | None | None | ||
| D | D1 | Specialized cancer care center. Located adjacent to Hospital C1 (ICU patients cared for at C1 before transfer back to hospital D1) |
96 | Haematology | Haematology ward patients screened on admission and weekly for vanA VRE and MRGN | MRSA: single room when possible; if open wounds, gown and gloves; if respiratory, mask and gloves. vanA VRE: single room, own bathroom, gloves and full gown. ESBL-Ec and ESBL-Kp: single room, own bathroom, no extra PPE but additional cleaning if incontinent or discharging wounds |
ICU, intensive care unit; MRGN, multi-resistant Gram negatives (includes ESBL and carbapenem-resistant phenotypes); CRGN, carbapenem-resistant Gram negatives; BMT, bone marrow transplant (allogeneic); PPE, personal protective equipment; VRE, vancomycin-resistant Enterococcus; ESBL-Ec, extended-spectrum beta-lactamase Escherichia coli; ESBL-Kp, extended-spectrum beta-lactamase Klebsiella pneumoniae; MRSA, methicillin-resistant Staphylococcus aureus.
Inpatient beds, excludes day cases, hospital-in-the-home and mental health.
Cohorting, practice of patients colonised with the same MDRO sharing a room.
Multidrug-resistant ESBL, ESBL isolates with additional resistance aminoglycosides, fluoroquinolones and trimethoprim-sulfamethoxazole (at least two classes).
The study was conducted in two phases: a pilot phase (24th April–18th June 2017), and an implementation phase (30th October 2017–30th November 2018), totalling 15 months. Results of transmission analyses were only reported to hospitals during the implementation phase. Pilot phase results have previously been reported,19 but are included here as isolates from these patients may be implicated in future transmission events detected in the implementation phase. Outbreak investigation support (including sequencing of environmental and relevant historical isolates) was also offered to participating sites during the study.
MDRO definitions
Four principal MDROs were included in the pilot phase, chosen as AMR pathogens of global significance (WHO global priority pathogen list20) with known transmission in hospitals; the selected MDROs were vanA vancomycin-resistant Enterococcus faecium (vanA VREfm), methicillin-resistant Staphylococcus aureus (MRSA), and extended-spectrum beta-lactamase phenotype Escherichia coli and Klebsiella pneumoniae (ESBL-Ec and ESBL-Kp) (see Supplementary data for details). Carbapenem-resistant Acinetobacter baumannii complex (CRAb) and carbapenem-resistant Pseudomonas aeruginosa (CRPa) were also included in the pilot phase,21 but excluded here due to low prevalence. Carbapenem-resistant Enterobacterales (CPE) were excluded, as these were already under genomic surveillance as part of the state-wide CPE surveillance program.22 As the pathogens included in the study were not notifiable by legislation, no specific public health interventions or reporting was in place that the time, and infection control units managed these independently.
Due to the large volume of ESBL-Ec included during the pilot phase, we modified inclusion criteria for the implementation phase to only include ESBL-Ec that were also resistant to fluoroquinolones, as this was more consistent with existing infection control practices at participating sites.
Infection control, clinical and epidemiologic data
Hospital infection control practices and MDROs screening protocols at each site were assessed regularly during the study, including any changes made in response to genomic data (Table 1 and Supplementary data). Basic demographic and isolate data were collected for all patients; epidemiologic data were collected for a subset of patients where genomics suggested potential MDRO transmission to assess the clinical likelihood of such cross-transmission having occurred. Transmission rates were estimated by calculating the number of putative transmissions per 100,000 occupied bed days (OBDs, inpatient beds occupied on daily bed count, excluding mental health).
Laboratory workflows
MDROs were isolated, worked up and reported by the hospital laboratories as per their usual protocols. For patients and isolates meeting inclusion criteria, a pure subculture was sent to the central laboratory for sequencing and isolate storage.
The sequencing laboratory is an ISO-accredited laboratory with high-throughput genomic sequencing capacity, located centrally between the participating hospitals. Once samples arrived, a single colony was subcultured onto horse blood agar, incubated overnight, then 1-2 colonies selected and placed into lysis buffer. DNA extraction, NexteraXT library preparation and quality control (QC) were performed as previous described,19 and isolates sequenced on Illumina NextSeq (San Diego, CA, USA).
Bioinformatics and transmission analysis
An overview of the combined genomic and epidemiologic analysis workflow to identify MDRO transmission can be found in Figure 1, with details included in Supplementary Data. Briefly, all sequences and assemblies underwent routine quality control checks, and underwent de novo genome assembly, species identification, multi-locus sequence typing (MLST), and identification of antimicrobial resistance (AMR) genes.
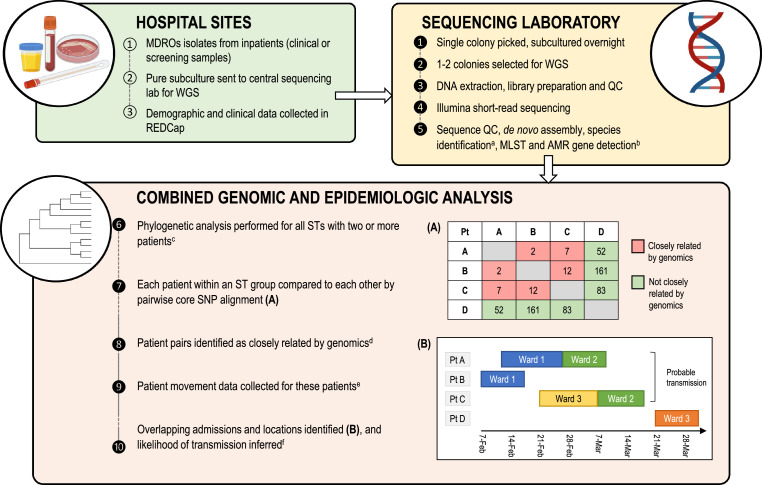
Figure 1.
Infection control genomics workflow implemented in this study.
(A). Example of pairwise SNP distance matrix for sequences from four patient isolates; patient pairs with pairwise SNP distances at or below the screening threshold (15 SNPs for MRSA, 25 SNPs for other MDROs) are classified as closely related by genomics (red), and pairs above these thresholds are designated as not closely related by genomics. (B) Example of Gantt chart demonstrating bed movements (ward admissions) of same four patients from (A) over time; patients A, B and C had closely related MDROs by genomics, and had overlapping admissions (same ward at the same time), therefore constituting probable MDRO transmission. Patient D's sequence was not related to the other patients’ sequences by genomics (above screening threshold), and not considered to be involved in MDRO transmission to or from these other patients, despite having an MDRO of the same sequence type (ST).
a Species identification by k-mer identification (kraken); Klebsiella further subspeciated using kleborate tool. b Sequences analysed for presence of complete vanA operon, ESBL/AmpC genes and mecA/mecC; if these were absent, isolates underwent further phenotypic testing to ensure they met inclusion criteria (phenotypic antimicrobial resistance). c Aligning isolates to reference genome of same ST, or de novo assembly of earliest isolate if this was not available; ST 131 E. coli analysed in two subclades due to large number of isolates. Recombinant sites were masked with gubbins for species other than S. aureus. d Each isolate sequence within an ST was compared to all others, and closely-related isolates were determined by core SNP differences; ≤15 SNPs for Staphylococcus aureus, ≤25 SNPs for other species. e Data collected for 12 months prior to first study sample until the end of study. f Likelihood of transmission inferred from combined genomic and epidemiologic data, categorised as ‘Probable’ (same ward at same time), ‘Possible’ (same ward at different time but within 60 days, or same hospital at the same time), ‘Unlikely’ (neither of the above), or ‘Above screening threshold’ (not closely related by genomics).
Abbreviations: MDRO, multidrug-resistant organism; WGS, whole genome sequencing; QC, quality control; MLST, multi-locus sequence typing; AMR, antimicrobial resistance; ST, sequence type; SNP, single-nucleotide polymorphism (single base difference between two or more isolates); Pt, patient.
Transmission analyses were performed within sequence type (ST) for all STs with two or more patients. Reads were aligned to a complete reference genome of the same ST, and recombinant sites were masked (except for S. aureus). Variant sites were detected from short reads for each ST, and pairwise single nucleotide polymorphism (SNP) differences were calculated for all isolate pairs.
To screen for potential MDRO transmission, we defined isolate pairs with ≤ 25 core SNP differences (E. coli, K. pneumoniae, and E. faecium) or ≤ 15 core SNP differences (MRSA) as ‘genomically related’. Isolate pairs with different key AMR genes were excluded from further analysis (i.e. mec type for MRSA, and ESBL or AmpC gene (allele) for ESBL-Ec and ESBL-Kp; vanA alleles were excluded as this was a requirement for study inclusion). Patients with isolates that were ‘genomically related’ then had further epidemiologic data collected, detailing their admission history (dates, hospitals and wards) for 12 months before the earliest isolate until the end of the study period.
Epidemiologic data were then compared for patients above the screening thresholds (‘genomically related’) within ST (or ST131 E. coli subclades). Patient pairs were classified according to a modification of previously published definitions23 as ‘probable transmission’ if on the same ward at the same time, ‘possible transmission’ if admitted to the same ward at a different time (within 60 days), or admitted to the same hospital at the same time; all other patients were classified as ‘unlikely transmission’. Statistical analyses were performed using Stata v16.
Reporting to infection control teams and report design
During the implementation phase of the study, genomic results for each hospital network were compiled and reported in person to infection control teams, with epidemiologic data where available; written reports were then supplied after each presentation. As this was the first time that most infection control teams had been exposed to genomic data, the reporting presentations included an informal education component to familiarise teams with genomic analyses and interpretation of hospital's genomic results. The study team also sought feedback from infection control teams during and after each reporting session, and made themselves available for any questions from infection control teams between reports or presentations.
Using the feedback from the reporting sessions, two focus group sessions were held at the end of the study with infectious diseases physicians, infection control practitioners, and medical microbiologists to collect feedback to design a genomics report for infection control use (see Supplementary Methods for details). A final report was designed based on feedback from the focus group sessions.
Ethics approval
This study was approved by the Melbourne Health Human Research Ethics Committee (HREC) and endorsed by the corresponding HREC at each participating site.
Role of funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or the decision to publish the work.
Results
Scoping and designing a workflow for implementation of genomics-informed infection control
To design a genomics workflow for infection control, we considered the critical elements for implementation: target population, target MDROs, MDRO screening strategy, sample types, clinical data collection, sequencing and analysis strategy, bioinformatics analysis, and communication of results (Table 2). The final workflow, as implemented in this study, is shown in Figure 1.
Table 2.
Considerations for implementation of genomics-informed infection control workflow.
| Component of genomics workflow | Key considerations | Implementation strategy selected for this study |
|---|---|---|
| Target population | Consider targeting high-risk populations (e.g. ICU, haematology/oncology, transplant) vs acute inpatients vs all inpatients (acute and subacute) | Included all inpatients due to frequent movement between wards, and unknown burden of disease and transmission in subacute care |
| Target MDRO/s | Consider AMR spectrum/high-risk organisms; MDROs not covered by existing surveillance programs; organisms where healthcare acquisition more likely than community acquisition; MDROs of high prevalence in local population | Selected van A VRE (high prevalence, recently emerged), ESBL-Ec and ESBL-Kp (unclear healthcare contribution) and MRSA (changing epidemiology, unclear healthcare contribution). Note CPE already covered by existing statewide surveillance system; CRPa and CRAb low prevalence |
| MDRO screening strategy | Consider using existing MDRO screening strategies vs harmonised screening approaches. Could target high-risk populations or all inpatients (e.g. ward-based or hospital-wide point-prevalence surveys) | Elected for pragmatic approach, with each hospital network continuing existing screening strategies, noting differences between networks in analysis |
| Sample types | All samples vs clinical samples only (latter would underestimate transmission) | All samples included to maximise likelihood of identifying transmission |
| Clinical data collection | Consider data required to identify likely transmission in hospital, including admissions at other sites; consider ability to accurately identify admissions at different networks by centralised electronic systems | Patient movement data (admissions, wards and beds) critical to identify likely transmissions in conjunction with genomic data. Minimal clinical data collected in implementation phase, as not required to infer transmission. |
| Sequencing and analysis strategy | (i) Sequencing location: centralised (sequencing laboratory) vs decentralised (hospital laboratories) | (i) Sequencing in central laboratory, as sequencing not established at study sites |
| (ii) Bioinformatics analysis location: centralised vs decentralised, based on resources and expertise | (ii) Centralised analysis at sequencing laboratory as not established at study sites | |
| (iii) Analysis of combined genomic and epidemiologic data, depending on IPC resources and training, and established systems for electronic data analysis | (iii) Genomic and epidemiologic data integrated centrally by study team, presented to IPC teams for further interpretation and action | |
| Bioinformatics analysis | Consider methods for transmission analysis. Examples: core genome SNP analysis, within species or within ST vs cgMLST approach, SNP thresholds, masking recombination Other considerations: frequency of analysis, inclusion of all data vs rolling window (e.g. last 6 or 12 months) |
Elected for core genome SNP analysis within ST based on existing accredited bioinformatics pipelines; recombination masked except for MRSA; SNP thresholds selected based on existing data at the time. All samples included in each analysis (approximately monthly), noting computational expense and personnel requirements |
| Communication of results | Consider audience (IPC/ID/microbiology teams) and understanding of genomics | Chose to engage with IPC/ID/microbiology teams using in-person presentations as part of initial implementation |
| Written reports (frequency) vs discussions in person vs interactive data portal with effective data visualisation | Written report format developed via focus group |
MDRO, multidrug-resistant organism; VRE, vancomycin-resistant Enterococcus; ESBL-Ec, extended-spectrum beta-lactamase-phenotype Escherichia coli; ESBL-Kp, extended-spectrum beta-lactamase-phenotype Klebsiella pneumoniae; MRSA, methicillin-resistance Staphylococcus aureus; CPE, carbapenemase-producing Enterobacterales; CRPa, carbapenem-resistant Pseudomonas aeruginosa; CRAb, carbapenem-resistant Acinetobacter baumannii; ST, sequence type; cgMLST, core genome multi-locus sequence type; SNP, single nucleotide polymorphisms; IPC, infection prevention and control; ID, infectious diseases.
Isolate and patient characteristics
Overall, 2641 bacterial isolates were submitted by hospital laboratories for WGS; 175 were excluded due to patients not meeting inclusion criteria, 116 isolates were excluded as duplicates, and a further 64 isolates did not meet microbiological inclusion criteria (see Supplementary Fig. S1 for details).
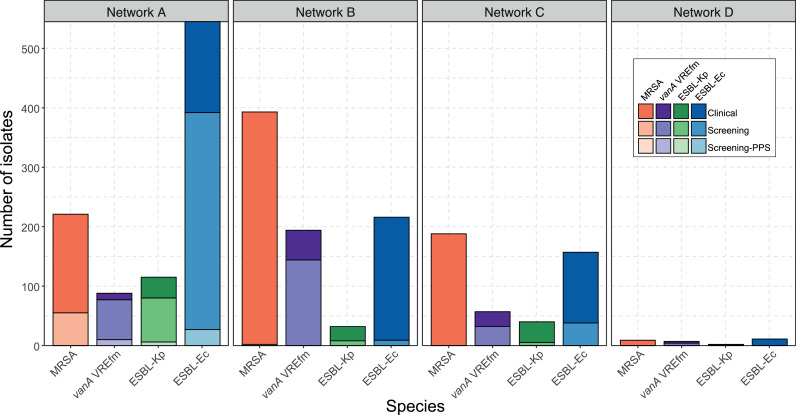
Ultimately, 2275 isolates from 1970 patients were included for analysis, predominantly ESBL-Ec (929 isolates, 47·2%), followed by MRSA (811 isolates, 41·2%), vanA VRE (346 isolates, 17·6%), and ESBL-Kp (189 isolates, 9·4%). Most isolates were collected for clinical reasons (62·8% overall), although this varied according to species and screening programs at each hospital network (Figure 2). After screening swabs (37·1%), urine (20·4%), and swabs from non-sterile sites (20·4%) were the most common sample types, with only 6·9% of isolates being from blood cultures (Supplementary Fig. S2). Median patient age was 65 (IQR 47-77), and 56·8% of patients were male. 297 patients (15·1%) had more than one isolate included (range 1-7 isolates)(Table 3). For patients with one screening and at least one clinical isolate with the same MDRO, median time between screening and first clinical isolate was 13 days (range 0–365 days, IQR 4–55 days)(Supplementary Table S2).
Figure 2.
MDRO isolates by network, species, and reason for sample collection. Bars are shaded by reason for sample collection with lightest shades representing clinical samples, middle shades representing screening samples, and darkest shades (only for Network A) representing screening samples performed as part of network-wide biannual point-prevalence surveys (PPS)(note, MRSA not included in PPS). MRSA, methicillin-resistant Staphylococcus aureus; VREfm, vancomycin-resistant Enterococcus faecium; ESBL-Kp, extended-spectrum beta-lactamase-phenotype Klebsiella pneumoniae; ESBL-Ec, extended-spectrum beta-lactamase-phenotype Escherichia coli.
Table 3.
Patient and isolate characteristics.
| Characteristic | N (%) or alternative parameter | ||
|---|---|---|---|
| Total no. of isolates included | 2275 | ||
| Total no. of patients included | 1970 | ||
| Age (median, IQR) | 65 years (47 – 77yrs) | ||
| Male sex (no., %) | 1063 (56·8%) | ||
| Sample collected for clinical purposes (suspected infection) | 1428 (62·8%) | ||
| Sample collected for MDRO screening purposes | 847 (37·2%) | ||
| Time from admission to isolate collection (median, IQR) | 3 days (1 – 12 days) | ||
| Ward where sample was collected | All samples (% total samples) | Clinical samples (% per ward) | Screening samples (% per ward) |
|---|---|---|---|
| Intensive care unit | 373 (16·4%) | 117 (31·4%) | 256 (68·6%) |
| Other high-risk warda | 434 (19·1%) | 121 (27·9%) | 313 (72·1%) |
| General acute wardb | 1090 (47·9%) | 865 (79·4%) | 225 (20·6%) |
| Emergency department | 209 (9·2%) | 204 (97·6%) | 5 (2·4%) |
| Subacute care wardc | 169 (7·4%) | 121 (71·6%) | 48 (28·4%) |
MDRO, multidrug-resistant organism; IQR, interquartile range.
High-risk wards, includes haematology, oncology, renal ward (including renal transplant), and liver transplant wards.
General acute wards, include all wards not designated as ICU, high-risk or subacute.
Subacute care, includes aged care, rehabilitation, palliative care and spinal wards.
vanA VREfm was transmitted more frequently in hospital than other MDROs in this study
Initial genomic analysis identified that 844 (42·8%) of isolates were genomically-related to at least one other isolate from the study (i.e. below screening pairwise SNP threshold, excluding isolates from same patient, and excluding key AMR gene mismatch) (Supplementary Table S3 and Supplementary Fig. S3).
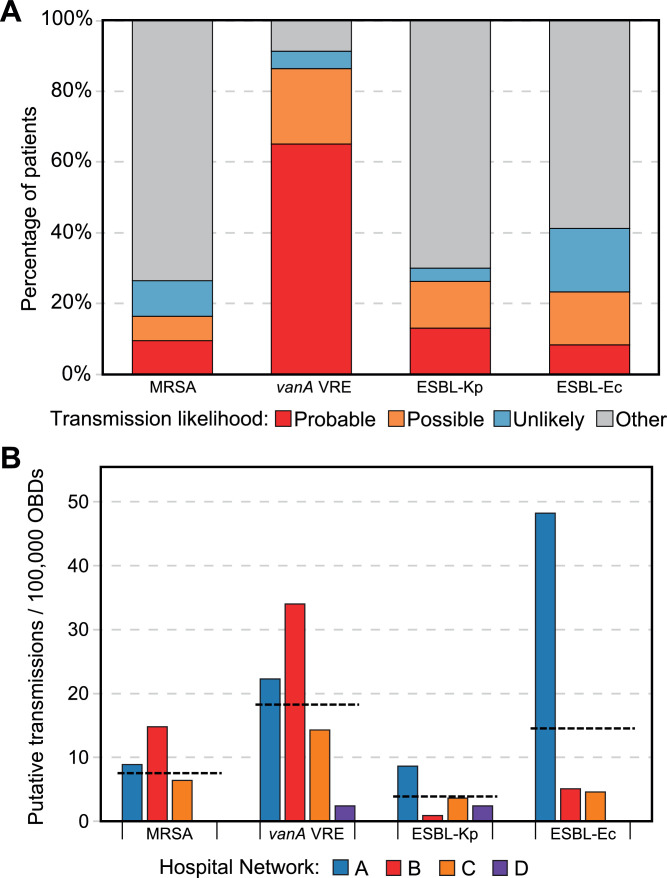
Aligning the patient admission history of each pair of patients with genomically-related isolates (see workflow Figure 1), MDRO isolates from 354 (18·0%) patients were identified as probable transmission in hospital, 253 patients (12·8%) as possible transmission in hospital, and 236 (12.0%) as unlikely acquired in hospital (Figure 3a, and pairwise SNP distributions shown Supplementary Fig. S4). This varied significantly by species: 266 vanA VREfm patients (86·4%) were identified as probable or possible transmission in hospital, whereas lower proportions of patients with ESBL-Ec, ESBL-Kp, and MRSA were identified as probable or possible in-hospital transmission (186 ESBL-Ec patients (23·0%), 113 MRSA patients (16·3%), 42 ESBL-Kp patients (26·3%); χ2 = 488·32 (VREfm vs other species), p < 0·001). Based on combined genomic and epidemiologic analysis, 44·8% of transmissions were thought to have occurred on a previous admission during the study period (up to 12 months prior to first sample collection), whilst another 17·5% of transmissions were likely to have occurred on a previous ward during the same admission as sample collection; only 34·7% of transmissions were likely to be from the same ward and admission when the MDRO sample was collected.
Figure 3.
MDRO transmission as assessed by combined genomics and epidemiologic assessment.
A/ Likelihood of transmission using combined genomic and epidemiologic data, by species. ‘Other’ (grey) includes patients with samples with no genomic links to other study samples (singleton STs, or above SNP screening thresholds). B. Putative transmission rates per 100,000 occupied bed days (OBDs) by species, coloured by hospital network. Horizontal dotted line represents the mean transmission rate for each species across all hospital networks. MRSA, methicillin-resistant Staphylococcus aureus; VREfm, vancomycin-resistant Enterococcus faecium; ESBL-Kp, extended-spectrum beta-lactamase-phenotype Klebsiella pneumoniae; ESBL-Ec, extended-spectrum beta-lactamase-phenotype Escherichia coli.
Hospital networks varied in the MDRO species with the highest transmission rates
Transmission rates per 100,000 OBDs varied widely between networks and species (Figure 3b). The greatest difference between networks was observed in ESBL-Ec, with network A having 47·5 transmissions per 100,000 OBDs, compared with the other two large networks (network B, 4·9 and network C, 5·4 transmissions per 100,000 OBDs) and zero transmissions for network D (incidence difference between network A and networks B and C, 43·14 transmissions per 100,000 OBDs (95% CI 37·54—48·75, p < 0·001)). This may partly reflect differences in screening practices (see Table 1), especially the network-wide point-prevalence surveys for Network A which contributed 27·2% of their transmissions (18·5% of ESBL-Ec, 62·9% of vanA VREfm and 11·1% of ESBL-Kp).
Conversely, network B registered a higher rate of vanA VRE transmission than other networks (transmissions per 100,000 OBDs: network B, 34·2; network A, 22·3; network C, 14·3; network D, 2·4; incidence difference between network B and networks A and C, 15·69 transmissions per 100,000 OBDs (95% CI 9·54–21·84, p < 0·001)). MRSA transmission was also higher for network B, but these data likely underestimate the true MRSA transmission rates for all networks due to low rates of routine MRSA screening.
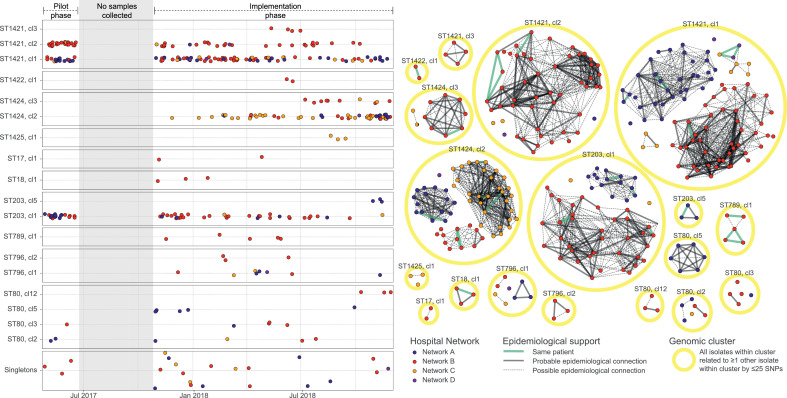
Successful vanA VREfm clones spread widely within and between hospital networks
Eight genomic clusters of vanA VREfm were detected in more than one hospital network (Figure 4); some were spread across networks from the commencement of the study (e.g. ST1421 cluster 1), whilst others emerged during the study, initially at one site, then other sites (e.g. ST1424 cluster 2). Whilst this study was not designed to capture transmissions between hospitals, and the state health information systems are decentralised (limiting tracking of patient movement between hospitals), this strongly suggests that patient movement between networks also contributes to the dissemination of vanA VRE in study sites, and likely more widely in Victoria.
Figure 4.
vanA VREfm cluster timeline and genomic transmission networks.
A. Timeline of vanA VREfm cases over the study periods, separated by ST and sub-cluster (cl) along the Y axis, and coloured by hospital network. Timeline is separated into pilot phase (left) and implementation phase (right); shaded timeframe shows the time between phases without sampling. Each point represents one case; only the first sample for each case in the study is included. Points have been separated vertically to allow for visualisation of closely-spaced cases. Sample sequences that did not cluster with any other study cases are shown in the ‘Singletons’ panel.
B. Network diagram of genomic and epidemiologic links between vanA VREfm cases. Each point represents one case; only the first sample for each case in the study is included, and is coloured by hospital network. The yellow circles outline genomic clusters, and lines between points represent epidemiologic links between cases (thick line, probable transmission; dotted line, possible transmission; green line, same patient. Absence of line indicates unlikely transmission by epidemiology, i.e. no known overlap between patients in space or time).
Abbreviations: VREfm, vancomycin-resistant Enterococcus faecium; ST, sequence type; cl, cluster; SNP, single-nucleotide polymorphism.
Impacts of genomics program
The implementation of genomic surveillance for hospital MDROs led to a wide range of impacts, falling into two main groups: identifying unexpected MDRO transmissions requiring targeted infection control interventions, or supporting the management of existing outbreaks through the provision of high-resolution genomic data to clearly define transmission networks (Table 4). The genomics program was particularly useful in identifying MDRO outbreaks occurring over a longer period of time, which had not been detected by conventional surveillance methods. Outbreak investigations were supported by including genomic analysis of historical and environmental isolates (including shared patient equipment), allowing infection control teams to trace the likely transmission pathways, and implement targeted measures to prevent further spread. This was especially useful where epidemiological links were complex, such as when a patient may have been exposed to vanA VREfm in more than one location, making attribution of MDRO acquisition impossible with epidemiologic data alone. Furthermore, genomics allowed the cessation of screening in one vanA VREfm outbreak, by demonstrating that new cases outside this ward were not related to the original outbreak, thus saving considerable resources and allowing the ward to re-open to new admissions.
Table 4.
Impacts of genomics program for hospital MDRO surveillance.
| Category | Impacts |
|---|---|
| Unexpected MDRO transmission identified, resulting in specific ward interventions | vanA VREfm cluster identified in haematology ward |
| ESBL-Kp ‘super-spreader’ patient identified in one ward.a | |
| Intensive ward cleaning instituted (‘super-clean’).b | |
| Review of patient placement/movements in affected wards. | |
| Increased surveillance in affected wards.c | |
| Outbreak investigation support | Supported patient-to-patient transmission and implication of shared patient equipment in outbreaks.d |
| Identified transmission where epidemiological links were complex. | |
| Able to show that new cases in other hospital wards were not linked to an outbreak, demonstrating that the outbreak management had been successful and allowing infection control interventions to be stepped down. | |
| Comparison of MDRO transmission rates between networks, resulting in network-wide changes | Identified higher rate of vanA VRE transmission in one network than others, resulting in network-wide change in cleaning practices.e |
| Increased awareness of MDRO transmission patterns | Increased awareness of MRSA and ESBL-Ec transmissions in hospital (previously thought to all be community-acquired). |
| Confirmed that MDRO identified through screening predicted the MDRO that subsequently caused clinical infection. Useful for advising empiric therapy e.g. in neutropaenic sepsis.38 | |
| Public health impacts | Large burden of hospital-acquired vanA VREfm demonstrated, contributing to new legislative requirement24 for all vanA VREfm cases to be notified to the state health department. |
| Low burden of carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii established as baseline before inclusion in state notifiable diseases list. |
VREfm, vancomycin-resistant Enterococcus faecium; ESBL-Ec, extended-spectrum beta-lactamase Escherichia coli; ESBL-Kp, extended-spectrum beta-lactamase Klebsiella pneumoniae; MRSA, methicillin-resistant Staphylococcus aureus; MDRO, multidrug-resistant organism.
‘Super-spreader’ indicating source patient with unusually high number of transmissions to other patients.
Ward ‘super-cleans’ included thorough terminal/discharge-type cleaning for all areas of the ward (including communal areas, corridors, offices, and storerooms) using a bleach-based detergent where possible.
Increased surveillance included additional environmental screening, additional patient screening (added to roster of high-risk wards with weekly patient screening for VREfm and MRGN, and additional focused hand hygiene surveillance.
Shared patient equipment includes any equipment that is not restricted to a single patient, e.g. blood glucometers, blood pressure cuffs, weigh chairs.
Cleaning practices changed from steam wands and microfibre cloth cleaning to bleach cleaning.
This study also had the unanticipated effect of encouraging data sharing between study sites, resulting in identification of one network with a higher rate of vanA VREfm transmissions than the others. This led to a review of cleaning practices, which were subsequently changed for the whole network towards the end of the study period. Additionally, data from this study also informed public health actions, with vanA VREfm newly added to the list of notifiable conditions by the state health department.24
Workflow implementation issues
A number of challenges for program implementation were identified throughout the study, primarily those causing delays in turnaround times or difficulty accessing and integrating electronic patient data (Table 5). Many of these issues relate specifically to this study, and would not be expected to limit the future implementation of a genomics workflow for infection control.
Table 5.
Areas for further development for implementation of infection control genomics workflow.
| Issue and domain | Description | Potential solutions |
|---|---|---|
| Delays in time to results | ||
| a. Hospital laboratory | ||
| Inadequate staffing | Dedicated staff (supervising microbiologist and technician) | |
| Batch processing of isolates (weekly or longer intervals) | More frequent processing of isolates for transit to sequencing lab | |
| Major disruptions from introduction of new LIS at two laboratories (inability to perform data extractions for study, affecting three sites) | Ensure robust data extraction capabilities available when implementing new LIS | |
| b. Sequencing laboratory | Minor delays, due to high volume of samples and lower relative priority | Increase in sequencing capacity, redundancy to account for surges in sample volume |
| c. Bioinformatic analysis | Delays in analysis due to inadequate staffing | Increase dedicated computing resources for complex bioinformatic analyses Work towards partial automation of transmission analysis |
| Increasing complexity of analysis and computational requirements towards end of study due to large isolate numbers (all study samples of same MLST included in each transmission analysis) | Develop alternative bioinformatics methods to deal with genomics ‘big data’ Explore effect of rolling windows (e.g. only including last 6 or 12 months of isolates) on detecting transmission | |
| d. Reporting process | In-person presentations to each network used in this study (to engage and educate staff at study sites) did not allow for results to be conveyed in a timely manner | Issue individual reports for each patient isolate, including MLST result (can be used by Infection Control teams to rapidly exclude or include patients in potential transmissions). New report designed for infection control genomics results (aggregated hospital or network-level data) from focus groups. Develop innovative approaches for reporting aggregated results on a hospital/network level e.g. interactive dashboards with advanced data visualisation capabilities |
| Impaired access to patient epidemiologic data (including admission, ward and bed data) | Each hospital network using different patient information management systems; no readily available way to extract patient data; data had to be collected manually (transcription errors, variable formatting requiring manual correction) | Develop informatics systems to reliably extract patient movement data, and integrate with results of transmission analyses to inform Infection Control interventions |
| Inability to track patient movements between hospital networks | No readily-available centralised system available for tracking admissions of patients across different hospital networks to identify inter-network MDRO transmission | Engage with health department to advocate for development of centralised admissions database, available in real-time |
| Perceived cost issues | Sequencing costs perceived to be prohibitive for widespread application currently | Optimise utility of prospective genomics by targeting to local MDROs or locations of concern; demonstrate cost-effectiveness of prospective genomics |
LIS, laboratory information system; MLST, multi-locus sequence type (a typing method to subset isolates of the same species into smaller, closely-related groups for targeted transmission analysis); MDRO, multidrug-resistant organism.
Whilst several factors contributed to delays in turnaround times (TAT), two factors dominated; (i) delays in hospital laboratory sending isolates to the sequencing laboratory (mostly due to unusual workflow disruptions in the hospital laboratories), and (ii) delays in reporting results to hospitals, as we elected to report results in person (to encourage engagement and education), rather than releasing results cumulatively (Supplementary Fig. S5). Established systems for rapid referral, sequencing and reporting of notifiable pathogens in our jurisdiction, such as carbapenemase-producing Enterobacterales (CPE) and Salmonella, are usually complete in 7 to 10 days. Once MDRO infection control sequencing is fully embedded using routine referral processes, it is anticipated that these delays experienced for some samples in this this study would be unlikely to occur.
Communication
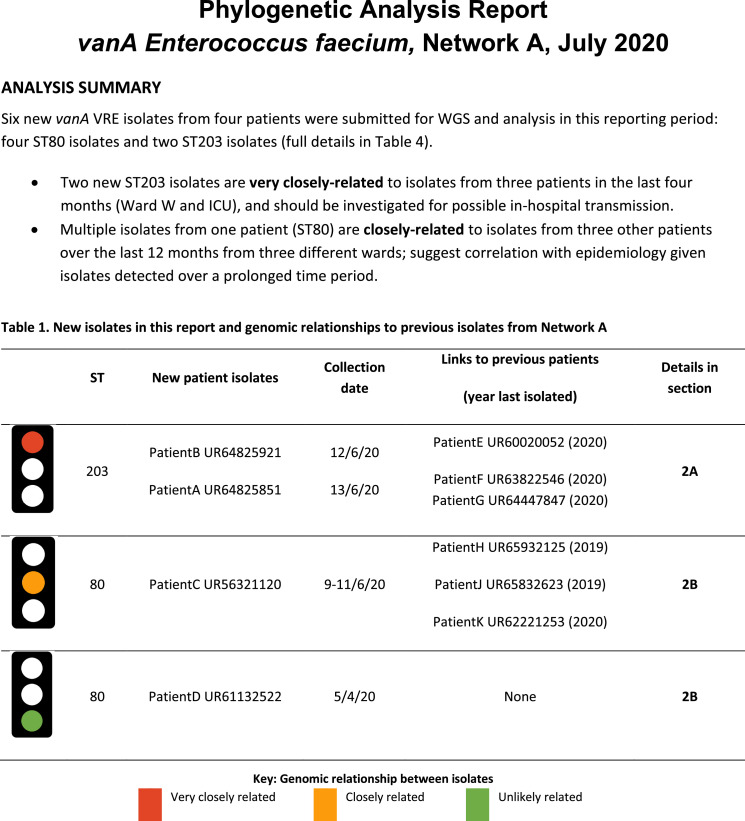
To design an effective written report format for infection control genomics, feedback from focus group participants at the conclusion of the study was used to design a template for reporting aggregated hospital/network-level genomics data (e.g. monthly report for each network)(Figure 5 and Supplementary Fig. S6). Recurrent themes included a desire for simplified results, strong preference for use of a ‘traffic light’ system to classify likelihood of transmission, general preference to avoid inclusion of phylogenetic trees (although not amongst more genomics-experienced participants), and specific comments directing infection control teams to which patients to investigate for possible transmission.
Figure 5.
Suggested format for reporting prospective genomics results to an infection control team
Excerpt of a suggested report format for intermittently reporting results from prospective genomic surveillance to an infection control team. This report was designed after feedback from focus groups of infection control, infectious diseases and microbiology staff, including some participants with experience in genomics, and some without. All data are fictitious. Full report included in Supplementary Data (Fig. S6).
Discussion
Here we have presented a large, multicentre study implementing a new workflow for prospective genomic surveillance of MDROs to inform infection control interventions in hospitals. We have demonstrated wide-ranging impacts of prospective genomic surveillance, identifying MDRO transmission where it was not suspected, providing support for outbreak investigations, and data to inform public health coordination of hospital-acquired infections. We believe that this is a critical step towards changing how we think about MDROs in hospitals, no longer relying on low-resolution traditional microbiological and epidemiologic surveillance, but proactively detecting MDRO transmission at a genomic level. This then enables early and tailored infection control interventions, potentially limiting the number of patients infected, morbidity and mortality, and saving limited hospital resources.4,18,25,26 This is most relevant with more common MDROs found in both hospitals and the community, as hospital transmission may be under-estimated without precision genomic data.4 Importantly, most of the MDRO transmission detected here would not have been identified by existing infection control surveillance, offering huge opportunities to detect silent MDRO transmission, and intervene.
Implementing a new workflow represents a major change for hospital laboratories and infection control teams, and identifies challenges to be overcome for successful implementation. One of the pressure points for successful implementation is minimising turnaround times, critical for timely and effective infection control responses. COVID-19 has demonstrated what TATs for sequencing may be achievable, with 5–7 days TATs routinely attained in our state (1–3 days for urgent samples).27 With adequate resourcing and buy-in from hospital laboratories, and capacity in sequencing laboratories, similar TATs could be achieved for MDRO surveillance. Alternatively, a decentralised genomics model (local WGS and analysis, or hybrid model with centralised analysis) may also optimise TATs; the choice of model ultimately depends on the available resources in each setting.28 Other key challenges include the optimisation of bioinformatics processes29 and integration of epidemiologic data, without which the genomic analyses are not interpretable.30,31 In our study, the linkage of genomic and epidemiologic data was impaired by poor integration with electronic medical records; this integration would be an important component for successful implementation in the future.32
Communication of complex genomic results to a new clinical setting is another challenge for implementation. Ideally, results should be easy to comprehend, and easily integrated with epidemiologic data to facilitate rapid interpretation, enabling rapid infection control intervention where required.33 As an early implementation study in a group of hospitals unfamiliar with genomics, we chose to engage infection control teams by presenting results in-person to educate and answer questions interactively, with the trade-off of delayed reporting times. Whilst this approach was very useful for early implementation and engagement, alternative reporting methods would be more suitable going forward, such as regular written reports designed for infection control (such as the example provided here). Further innovation to allow infection control teams rapid access to genomics results and interpretation would be welcome in future implementation studies.
Identifying strong epidemiologic evidence of transmission is difficult, particularly with common MDROs, but necessary to validate genomic inferences of MDRO transmission. In this study, we used genomic links (SNP thresholds) and epidemiologic data (overlapping admissions in space and time) to call probable or possible transmission, similar to other studies.25,34 SNP thresholds are a relatively blunt instrument to suggest potential genomic links, but core genome SNP analyses are accepted to be far higher resolution than existing typing methods (such as MLST), and at least equivalent to core genome MLST.35,36 Importantly, SNP thresholds are specific to the methods used to derive them, hence other studies using lower thresholds are not directly comparable to this study.32,34 In a pragmatic sense, if high-resolution core genome SNP methods are used for large-scale prospective genomics, then SNP thresholds are likely to be required; however, future studies in this area should be open to exploring alternative methods and thresholds.29,34,35,37
Our study has a number of limitations. Firstly, screening practices were not standardised between hospitals, and included one site performing a biannual hospital-wide point-prevalence survey. However, many transmissions were still identified at hospitals performing less screening; this suggests the potential advantages of additional screening to uncover unsuspected transmissions. Secondly, bed movement data would ideally have been acquired for all study patients; this was addressed in the pilot study and did not greatly change the interpretation of the transmission data.19 Third, these methods are not designed to detect transmission of AMR plasmids mechanisms, as short-read sequencing methods are limited in this ability.
Limited access to data (clinical data and centralised state admissions data) was also a limitation of the study, and largely prevented identification of inter-hospital transmission; with the increasing implementation of electronic medical records, we hope that these issues could be more comprehensively addressed in future studies.30 Turnaround times were also impacted by extraneous factors (laboratory delays); despite this, major impacts on infection control and public health were demonstrated, and future changes in resource management and system design have now been proposed to improve TATs. Lastly, the cost-effectiveness of this intervention has not been addressed in this study, but demonstrated elsewhere,18,25 and will be the subject of future work.
This study represents a framework considering the critical components and challenges for implementation of prospective genomics for infection control, and may be tailored to local needs and conditions, encompassing local MDROs of interest, different hospital and network structures, laboratory set-up, and screening practices. Further implementation studies should focus on reducing TATs for more real-time results, streamlined data integration and reporting systems, as well as furthering the education of hospital infection control, infectious diseases and laboratory teams to improve patient safety, and care through genomics-informed infection control in the near future.
Contributors
NLS, JCK, MLG & BPH conceived and designed the study. CG, RL, TS & CH designed and performed bioinformatic analyses and created some of the figures. SB & MLS coordinated sequencing laboratory workflows. TK, MG, ML & HC arranged for isolates to be sent to the sequencing laboratory and provided microbiological data. RS, CM, MAS, LW & JCK coordinated collection of clinical and epidemiologic data. NLS, CG, JCK, RS, CM, TK, MAS, RL, MG, ML, LW, HC, MLG & BPH reviewed the data during the study, had input into the ongoing performance of the study and provided feedback on the manuscript. NLS and CLG wrote the manuscript; BPH, MLG and JCK edited the manuscript.
Data sharing statement
Raw sequence data for all included study samples (Supplementary Table S1), are available in NCBI Sequence Read Archive (SRA), BioProject PRJNA565795. All software and code used is publicly available using links provided in the Methods section.
Declaration of interests
The authors declare no conflicts of interest.
Acknowledgements
Financial support. This work was supported by the Melbourne Genomics Health Alliance (funded by the State Government of Victoria, Department of Health and Human Services, and the 10 member organizations); and individual grants from National Health and Medical Research Council (Australia) to BPH (GNT1105905) and MAS (GNT1116576). NLS and JCK were supported by Australian Government Research Training Program (RTP) Scholarships.
Other members of the Controlling Superbugs Study Group include: Elizabeth Grabsch (Microbiology, Austin Health), Joanna Price and Carolyn Mason (Infection Control, Austin Health), Despina Kotsanas (Monash Infectious Diseases), Louise Wright (Infection Control, Monash Health), Dr Suraya Hanim Abdullah Hashim and Jennifer Mitchell (Infectious Diseases, Melbourne Health), Olivia Smibert and Victoria Madigan (Infectious Diseases, Peter MacCallum Cancer Centre) and Peter Pham (MDU Public Health Laboratory, University of Melbourne).
The authors would like to gratefully acknowledge the assistance of the following people: MDU PHL staff including Kerrie Stevens, Nicole Orlando, Samantha Tawil, Savitra Rambocus and Diane Daniel; John Greenough (infection control queries and patient movement data), Ben Rogers and Jean Lee (focus groups).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.lanwpc.2022.100446.
Appendix. Supplementary materials
References
- 1.World Health Organization . WHO Press; Geneva, Switzerland: 2014. Antimicrobial Resistance: Global Report on Surveillance. [Google Scholar]
- 2.Chia P., Sengupta S., Kukreja A., et al. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob Resist Infect Control. 2020;9:29. doi: 10.1186/s13756-020-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kritsotakis E., Kontopidou F., Astrinaki E., Roumbelaki M., Ioannidou E., Gikas A. Prevalence, incidence burden, and clinical impact of healthcare-associated infections and antimicrobial resistance: a national prevalent cohort study in acute care hospitals in Greece. Infect Drug Resist. 2017;10:317–328. doi: 10.2147/IDR.S147459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popovich K.J., Snitkin E.S. Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr Infect Dis Rep. 2017;19(4):15. doi: 10.1007/s11908-017-0570-0. [DOI] [PubMed] [Google Scholar]
- 5.Parcell B.J., Gillespie S.H., Pettigrew K.A., Holden M.T.G. Clinical perspectives in integrating whole-genome sequencing into the investigation of healthcare and public health outbreaks – hype or help? J Hosp Infect. 2021;109:1–9. doi: 10.1016/j.jhin.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruppe E., Olearo F., Pires D., et al. Clonal or not clonal? Investigating hospital outbreaks of KPC-producing Klebsiella pneumoniae with whole-genome sequencing. Clin Microbiol Infect. 2017;23(7):470–475. doi: 10.1016/j.cmi.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Willems S., Kampmeier S., Bletz S., et al. Whole-genome sequencing elucidates epidemiology of nosocomial clusters of Acinetobacter baumannii. J Clin Microbiol. 2016;54(9):2391–2394. doi: 10.1128/JCM.00721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang P., Croxen M.A., Hasan M.R., Hsiao W.W., Hoang L.M. Infection control in the new age of genomic epidemiology. Am J Infect Control. 2017;45(2):170–179. doi: 10.1016/j.ajic.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Conlan S., Thomas P.J., Deming C., et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbepenemase-producing Enterobacteriaceae. Sci Transl Med. 2014;6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris S.R., Cartwright E.J.P., Török M.E., et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13(2):130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Man T.J.B., Yaffee A.Q., Zhu W., et al. Multispecies outbreak of Verona integron-encoded metallo-ß-lactamase producing multidrug-resistant bacteria driven by a promiscuous incompatibility group A/C2 plasmid. Clin Infect Dis. 2021;72(3):414–420. doi: 10.1093/cid/ciaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peacock S.J., Parkhill J., Brown N.M. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology. 2018;164(10):1213–1219. doi: 10.1099/mic.0.000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuter S., Ellington M.J., Cartwright E.J., et al. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med. 2013;173(15):1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrasa-Villar J.I., Aibar-Remón C., Prieto-Andrés P., Mareca-Doñate R., Moliner-Lahoz J. Impact on morbidity, mortality, and length of stay of hospital-acquired infections by resistant microorganisms. Clin Infect Dis. 2017;65(4):644–652. doi: 10.1093/cid/cix411. [DOI] [PubMed] [Google Scholar]
- 15.Mollers M., Lutgens S.P., Schoffelen A.F., Schneeberger P.M., Suijkerbuijk A.W.M. Cost of nosocomial outbreak caused by NDM-1–containing Klebsiella pneumoniae in the Netherlands, October 2015–January 2016. Emerg Infect Dis. 2017;23(9):1574–1576. doi: 10.3201/eid2309.161710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otter J.A., Burgess P., Davies F., et al. Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: an economic evaluation from a hospital perspective. Clin Microbiol Infect. 2017;23(3):188–196. doi: 10.1016/j.cmi.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Balloux F., Bronstad Brynildsrud O., van Dorp L., et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26(12):1035–1048. doi: 10.1016/j.tim.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon L.G., Elliott T.M., Forde B., et al. Budget impact analysis of routinely using whole-genomic sequencing of six multidrug-resistant bacterial pathogens in Queensland, Australia. BMJ Open. 2021;11(2) doi: 10.1136/bmjopen-2020-041968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry N.L., Lee R.S., Gorrie C.L., et al. Pilot study of a combined genomic and epidemiologic surveillance program for hospital-acquired multidrug-resistant pathogens across multiple hospital networks in Australia. Infect Control Hosp Epidemiol. 2020;42(5):573–581. doi: 10.1017/ice.2020.1253. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization . WHO; Geneva: 2017. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. [Google Scholar]
- 21.Sherry N.L., Lane C.R., Kwong J.C., et al. Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing enterobacterales in Victoria, Australia, 2012 to 2016. J Clin Microbiol. 2019;57(9) doi: 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Department of Health and Human Services Victoria. Victorian guideline on carbapenemase-producing Enterobacteriaceae for health services (version 2.1). 2018. https://www2.health.vic.gov.au/public-health/infectious-diseases/infection-control-guidelines/carbapenemase-producing-enterobacteriaceae-management/cpe-for-health-services.
- 23.Voor In 't Holt A.F., Wattel A.A., Boers S.A., et al. Detection of healthcare-related extended-spectrum beta-lactamase-producing Escherichia coli transmission events using combined genetic and phenotypic epidemiology. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0160156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.State Government of Victoria. Public health and wellbeing regulations 2019, Statutory Rule no. 139/2019 v004. 2019.
- 25.Mellmann A., Bletz S., Boking T., et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol. 2016;54(12):2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clermont O., Christenson J.K., Denamur E., Gordon D.M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 27.Lane C.R., Sherry N.L., Porter A.F., et al. Genomics-informed responses in the elimination of COVID-19 in Victoria, Australia: an observational, genomic epidemiological study. Lancet Pub Health. 2021 doi: 10.1016/s2468-2667(21)00133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mintzer V., Moran-Gilad J., Simon-Tuval T. Operational models and criteria for incorporating microbial whole genome sequencing in hospital microbiology - a systematic literature review. Clin Microbiol Infect. 2019;25(9):1086–1095. doi: 10.1016/j.cmi.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Gorrie C.L., Da Silva A.G., Ingle D.J., et al. Key parameters for genomics-based real-time detection and tracking of multidrug-resistant bacteria: a systematic analysis. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00149-X. [DOI] [PubMed] [Google Scholar]
- 30.Pak T.R., Kasarskis A. How next-generation sequencing and multiscale data analysis will transform infectious disease management. Clin Infect Dis. 2015;61(11):1695–1702. doi: 10.1093/cid/civ670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong G.L., Maccannell D.R., Taylor J., et al. Pathogen genomics in public health. N Engl J Med. 2019;381(26):2569–2580. doi: 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward D.V., Hoss A.G., Kolde R., et al. Integration of genomic and clinical data augments surveillance of healthcare-acquired infections. Infect Control Hosp Epidemiol. 2019;40(6):649–655. doi: 10.1017/ice.2019.75. [DOI] [PubMed] [Google Scholar]
- 33.Crisan A., McKee G., Munzner T., Gardy J.L. Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory. PeerJ. 2018;6:e4218. doi: 10.7717/peerj.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouliouris T., Coll F., Ludden C., et al. Quantifying acquisition and transmission of Enterococcus faecium using genomic surveillance. Nat Microbiol. 2021;6(1):103–111. doi: 10.1038/s41564-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann B., Bender J.K., Maier B.F., et al. Comprehensive integrated NGS-based surveillance and contact-network modeling unravels transmission dynamics of vancomycin-resistant enterococci in a high-risk population within a tertiary care hospital. PLoS One. 2020;15(6) doi: 10.1371/journal.pone.0235160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinholt M., Larner-Svensson H., Littauer P., et al. Multiple hospital outbreaks of vanA Enterococcus faecium in Denmark, 2012-13, investigated by WGS, MLST and PFGE. J Antimicrob Chemother. 2015;70(9):2474–2482. doi: 10.1093/jac/dkv142. [DOI] [PubMed] [Google Scholar]
- 37.Schurch A.C., Arredondo-Alonso S., Willems R.J.L., Goering R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24(4):350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 38.Attwood L.O., Sherry N.L., Gorrie C., et al. Active surveillance to predict superbug infections (conference presentation). Proceedings of the Australian Society for Infectious Diseases (ASID) Conference; Melbourne, Victoria; 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.