Abstract
Eukaryotic mRNAs possess a poly(A) tail at their 3′-end, to which poly(A)-binding protein C1 (PABPC1) binds and recruits other proteins that regulate translation. Enhanced poly(A)-dependent translation, which is also PABPC1 dependent, promotes cellular and viral proliferation. PABP-interacting protein 2A (Paip2A) effectively represses poly(A)-dependent translation by causing the dissociation of PABPC1 from the poly(A) tail; however, the underlying mechanism remains unknown. This study was conducted to investigate the functional mechanisms of Paip2A action by characterizing the PABPC1–poly(A) and PABPC1–Paip2A interactions. Isothermal titration calorimetry and NMR analyses indicated that both interactions predominantly occurred at the RNA recognition motif (RRM)2–RRM3 regions of PABPC1, which have comparable affinities for poly(A) and Paip2A (dissociation constant, Kd = 1 nM). However, the Kd values of isolated RRM2 were 200 and 4 μM in their interactions with poly(A) and Paip2A, respectively; Kd values of 5 and 1 μM were observed for the interactions of isolated RRM3 with poly(A) and Paip2A, respectively. NMR analyses also revealed that Paip2A can bind to the poly(A)-binding interfaces of the RRM2 and RRM3 regions of PABPC1. Based on these results, we propose the following functional mechanism for Paip2A: Paip2A initially binds to the RRM2 region of poly(A)-bound PABPC1, and RRM2-anchored Paip2A effectively displaces the RRM3 region from poly(A), resulting in dissociation of the whole PABPC1 molecule. Together, our findings provide insight into the translation repression effect of Paip2A and may aid in the development of novel anticancer and/or antiviral drugs.
Keywords: poly(A)-binding protein C1, PABP-interacting protein 2A, RNA recognition motif, NMR, surface plasmon resonance, isothermal titration calorimetry
Abbreviations: CITED2, CBP/p300 interacting transactivator with ED-rich tail 2; CSP, chemical shift perturbation; FL, full length; HIF-1α, hypoxia-inducible factor 1α; HSQC, heteronuclear single quantum coherence; ITC, isothermal titration calorimetry; PABC, PABP C-terminal domain; PABPC1, poly(A)-binding protein C1; Paip2A, PABP-interacting protein 2A; PAM, PABPC1-interacting motif; poly(A), polyadenylate; RRM, RNA recognition motif; SPR, surface plasmon resonance; TROSY, transverse relaxation optimized spectroscopy
mRNAs synthesized in the nuclei of eukaryotic cells are transported to the cytoplasm after addition of a 5′ cap and polyadenylate (poly(A)) tail to their 5′- and 3′-ends, respectively (1). Addition of the poly(A) tail to mRNA, known as poly(A)-dependent translation, enhances translation (2). The poly(A) tail contains multiple molecules of poly(A)-binding protein C1 (PABPC1), which form a one-dimensional array on the poly(A) tail, where each PABPC1 molecule recognizes 24 to 27 bases (3, 4). Poly(A)-bound PABPC1 molecules play important roles in poly(A)-dependent translation by recruiting various factors such as the eukaryotic translation initiation factor 4G (5) and translation termination eukaryotic peptide release factor 3 (6).
The expression of PABPC1 appears to be positively correlated with the proliferation of cells and viruses. For instance, human and mouse heart cells proliferate immediately after birth, during which the PABPC1 expression is enhanced. After their growth, cell proliferation is repressed, and PABPC1 expression is reduced (7). In addition, the mRNA levels of PABPC1 are increased in colon cancer cells, where cell proliferation is active (8). Some viruses, such as dengue virus, utilize PABPC1 in their host cells to translate viral mRNA, thereby resulting in virus proliferation (9). PABPC1 also accumulates in cytomegalovirus-infected cells (10).
Among the PABPC1-binding proteins that regulate translation, Paip2A exerts a unique function, in that it downregulates poly(A)-dependent translation by dissociating PABPC1 from the poly(A) tail (11). Reportedly, the expression of Paip2A represses the proliferation of oncogene-transformed NIH3T3 cells (12), which increases the intracellular concentrations of Paip2A and represses the replication of dengue virus and cytomegalovirus (9, 13). This represents a mechanism by which Paip2A dissociates PABPC1 from the poly(A) tail and may provide a molecular/structural basis for the development of anticancer or antiviral drugs.
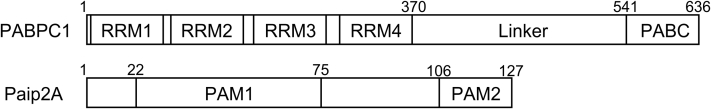
PABPC1 is a 636-residue protein with a molecular weight of 71 kDa (Fig. 1). It possesses four tandemly repeated RNA recognition motifs (RRM1–4) at the N terminus, each of which consists of approximately 90 residues, and a PABP C-terminal domain (PABC) (also known as MLLE [Mademoiselle]) domain with 75 residues at the C terminus, connected by a 170-residue unstructured linker region (14). The RRM region is responsible for the poly(A)-binding ability of PABPC1, where the dissociation constant, Kd value, for the interaction of the truncated PABPC1 containing four RRMs (residues 1–370, hereafter referred to as RRM1/2/3/4) with poly(A) is approximately 0.15 nM (15), which is comparable to that of full-length (FL) PABPC1 (0.69 nM) (15). The crystal structure of another truncated PABPC1 containing RRM1 and RRM2 (residues 1–190, hereafter referred to as RRM1/2) in complex with a poly(A) RNA with nine bases (A9) has been reported (16), where each RRM consists of two α-helices and a four-stranded β-sheet. The two α-helices are positioned on one side of the β-sheet, whereas the poly(A) binds to the β-sheet surface on the other side of the α-helices (16).
Figure 1.
Domain composition of PABPC1(14) and Paip2A(17). PABPC1 is a 636-residue protein with four RNA recognition motifs (RRMs) in the N terminus. Residues 11 to 89, 99 to 175, 191 to 268, and 294 to 370 are RRM1, RRM2, RRM3, and RRM4, respectively. The C terminus contains an unstructured region named as the linker and a PABC (PABP C-terminal domain). Paip2A is a 127-residue protein with PABP-interacting motif 1 (PAM1) in the N terminus and PABP-interacting motif 2 (PAM2) in the C terminus. PABPC1, poly(A)-binding protein C1; Paip2A, PABP-interacting protein 2A.
Paip2A, a 127-residue protein with a molecular weight of 15 kDa (Fig. 1), possesses two PABPC1-interacting motifs, PAM1 and PAM2, at its N and C termini, respectively (17). The Kd value for the Paip2A–PABPC1 interaction was reported to be approximately 0.66 nM. PAM1 binds primarily to the RRM2–RRM3 region of PABPC1 (17); in contrast, PAM2 binds to the C-terminal PABC domain of PABPC1 with a Kd of 74 to 400 nM (17, 18). Although the PAM2–PABC interaction plays a role in Paip2A function (18), the more than 100-fold difference in the Kd values between PAM1 and PAM2 for PABPC1 suggests that the interaction between the RRM region and PAM1 plays a major role in PABPC1–Paip2A binding. Particularly, the function of Paip2A to dissociate PABPC1 from poly(A) is likely ascribed to binding of PAM1 to the RRM region (17), as the RRM region is responsible for poly(A) binding of PABPC1.
The binding affinity of PABPC1 for Paip2A is comparable to that for poly(A), with a Kd value of approximately 0.7 nM (15, 17). However, the mechanism by which Paip2A dissociates poly(A) from PABPC1 remains unclear. Here, we examined the functional mechanisms of Paip2A by characterizing the PABPC1–poly(A) and PABPC1–Paip2A interactions using surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), and NMR spectroscopy. We propose a functional mechanism to explain how Paip2A effectively dissociates PABPC1 from poly(A), despite its comparable affinities of PABPC1 for Paip2A and poly(A).
Results
Paip2A dissociates PABPC1 from poly(A)
We previously reported the poly(A)-binding affinity of human PABPC1 and RRM1/2/3/4, wherein the Kds for poly(A) with a length of 24 bases (A24) were 0.69 and 0.15 nM, respectively, using SPR (15). The comparable Kd values indicate that the region from RRM1 to RRM4 is primarily responsible for the poly(A)-binding affinity of PABPC1.
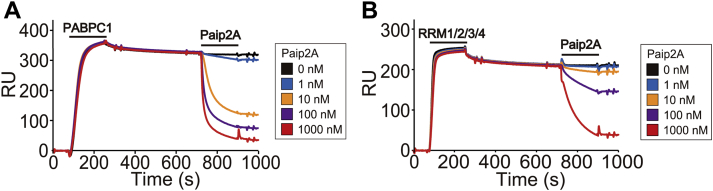
Here, we expressed and purified human Paip2A(full-length) (hereafter, Paip2A[FL]) and examined its ability to dissociate PABPC1 or RRM1/2/3/4 from A24 using SPR. The purity of the prepared samples was determined by SDS-PAGE (Fig. S1, A, B and H). When PABPC1 or RRM1/2/3/4 were applied to the A24-attached sensor chip, the resonance unit, that is, response, was increased and became saturated, approaching the maximum value of the sensorgram (Fig. 2). In our previous study, subsequent dissociation was observed using running buffer. Together with the association, the Kd values for poly(A) were 0.69 and 0.15 nM, respectively (15). Here, rather than using running buffer, Paip2A(FL) was applied to PABPC1 and RRM1/2/3/4 bound to poly(A). Upon application of Paip2A(FL), the resonance unit decreased in a Paip2A(FL) concentration–dependent manner, indicating that the purified Paip2A(FL) could dissociate PABPC1 and RRM1/2/3/4 from A24. Notably, Paip2A dissociated PABPC1 more efficiently than RRM1/2/3/4, which does not possess the linker and PABC (Fig. 2). This may be because of the interactions between PAM2 and PABC.
Figure 2.
Surface plasmon resonance results for the activity of prepared Paip2A(FL) that dissociates PABPC1 and RRM1/2/3/4 from poly(A). 5′-Biotinylated A24 was immobilized on a streptavidin-coated sensor chip. Application periods for 128 nM PABPC1 (A) and RRM1/2/3/4 (B) (81–260 s) and Paip2A(FL) (721–900 s) are indicated as bars above the sensorgrams. PABPC1, poly(A)-binding protein C1; Paip2A(FL), PABP-interacting protein 2A (full length); RRM, RNA recognition motif.
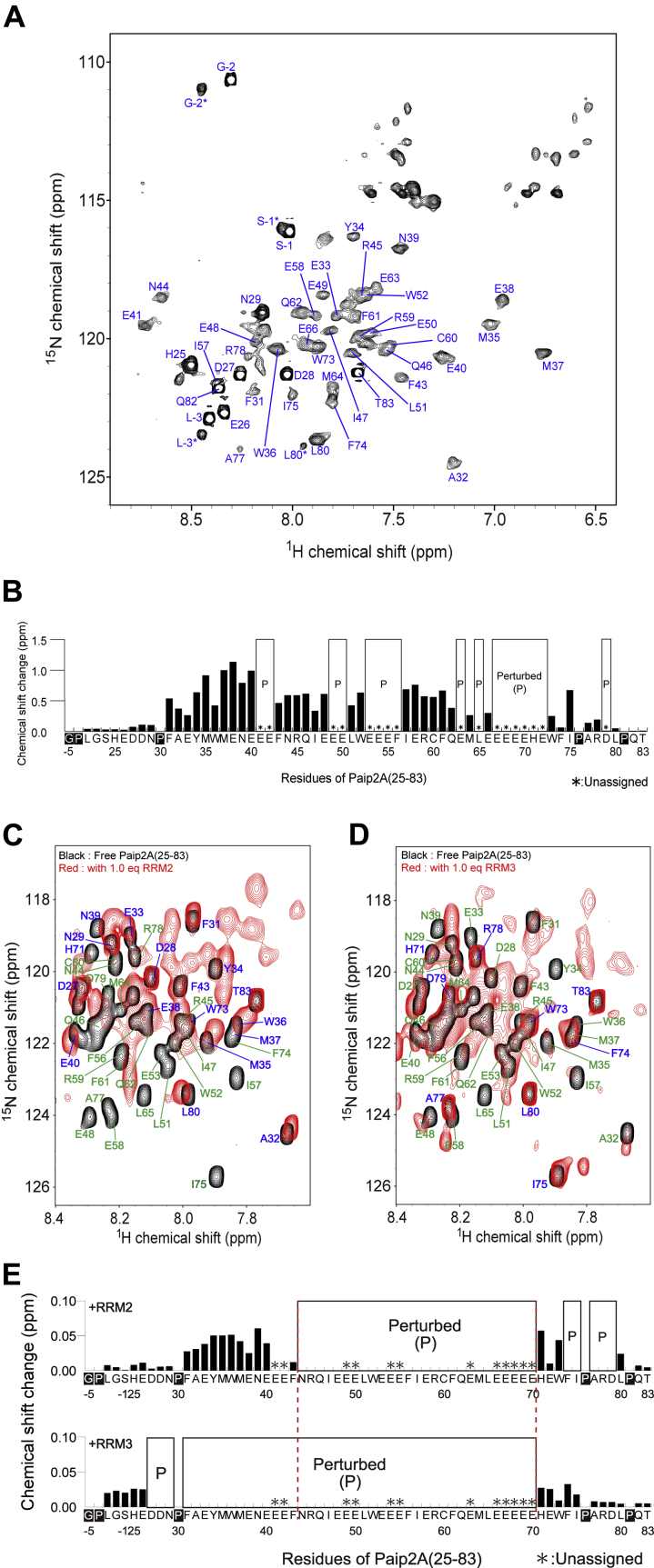
Paip2A residues 26 to 83 are involved in interactions with the RRM region of PABPC1
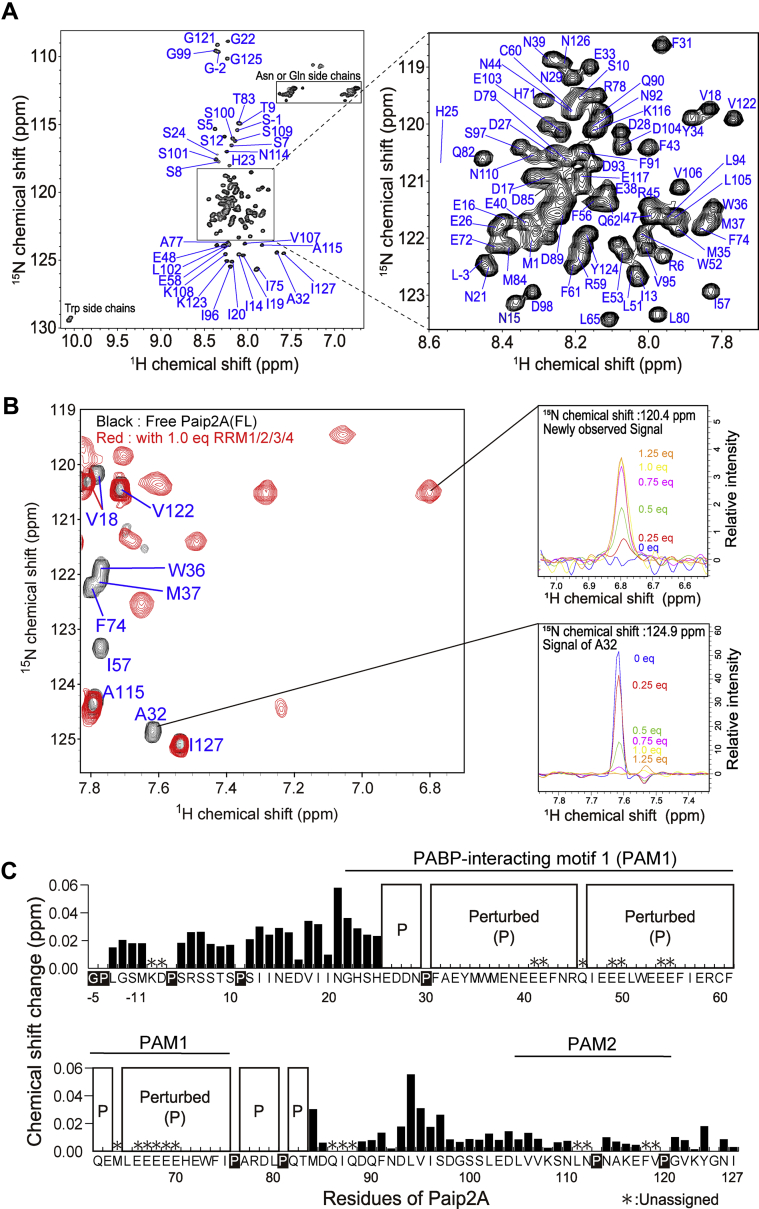
As RRM1/2/3/4 is responsible for the poly(A)-binding affinity of PABPC1, we investigated the interactions of Paip2A(FL) with RRM1/2/3/4 using solution NMR. First, we observed the 1H–15N heteronuclear single quantum coherence (HSQC) spectrum of uniformly 15N-labeled Paip2A(FL) (Fig. 3A). We assigned 98 backbone NMR resonances (80% of 123 theoretically observed resonances) by analyzing triple resonance experiments, whereas 16 signals were present but remained unassigned. The narrow dispersion of the 1H chemical shifts of the backbone amide groups ranging from 7.8 to 8.6 ppm (Fig. 3A), as well as the chemical shift index of Paip2A(FL) (Fig. S2) (19), strongly suggest that Paip2A is a natively disordered protein.
Figure 3.
NMR spectral changes in Paip2A(FL) after adding RRM1/2/3/4. A, backbone NMR assignments of Paip2A(FL) identified on the 1H–15N HSQC spectrum. B, overlay of 1H–15N TROSY spectra for 2H, 15N-labeled Paip2A(FL) in the presence (red) and absence (black) of nonlabeled RRM1/2/3/4. The top left panel shows a cross-section of the A32 signals after adding RRM1/2/3/4. The bottom left panel shows unassigned signals that appeared after adding RRM1/2/3/4. The equivalent numbers for RRM1/2/3/4 are shown in each figure, colored as blue (0 equivalent), red (0.25 equivalent), green (0.5 equivalent), purple (0.75 equivalent), yellow (1.0 equivalent), or orange (1.25 equivalent). C, chemical shift changes in each residue of Paip2A(FL). “Perturbed (P)” indicates the residues whose signals disappeared after adding RRM1/2/3/4 and appeared at different positions. Chemical shift changes were calculated using the following equation: , where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively. HSQC, heteronuclear single quantum coherence; Paip2A(FL), PABP-interacting protein 2A (full-length); RRM, RNA recognition motif; TROSY, transverse relaxation optimized spectroscopy.
We then observed a series of 1H–15N transverse relaxation optimized spectroscopy (TROSY) spectra of uniformly 2H, 15N-labeled Paip2A(FL) with sequential addition of RRM1/2/3/4, which exhibited large spectral changes (Figs. 3B and S3). When 1.0 to 1.25 equivalents of RRM1/2/3/4 were added, 53 Paip2A(FL) signals decreased in intensity and finally disappeared: 41 signals were assigned, and 12 signals were unassigned but identified as Glu based on the side-chain 1H and 13C chemical shifts. The 51 new signals appeared at different chemical shifts, which were smaller than 7.4 ppm or larger than 8.6 ppm in the 1H dimension (Figs. 3B and S3). In addition, the spectral changes were saturated, suggesting that Paip2A forms conformations by binding directly to RRM1/2/3/4 with a binding stoichiometry of 1:1. Although the assignments of the signals could not be transferred from those for the free state of Paip2A(FL) through the titrations, these changes reflect the chemical shift changes in a slow exchange regime, where the chemical shift differences between the free-bound and RRM1/2/3/4-bound states were much larger than their exchange rates. Consequently, we regarded these signals in a slow exchange regime as the “perturbed” signals. In contrast, the chemical shift changes for residues 1, 5 to 10, 12 to 25, 84, 85, 89 to 110, 114 to 117, and 121 to 127 were very small (≤0.06 ppm, Fig. 3C), which were regarded as unperturbed signals.
The spectral changes in Paip2A(FL) after adding RRM1/2/3/4 are summarized in Figure 3C. The signals altered in a slow exchange regime (shown as “perturbed [P]”) were within residues 26 to 83 of Paip2A, indicating that these residues are involved in the interactions with RRM1/2/3/4.
Poly(A) binds mainly to the region from RRM2 to RRM3 of PABPC1
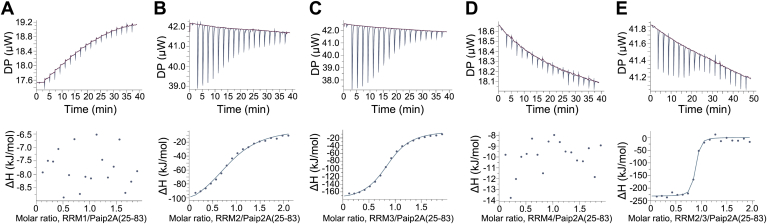
To compare the contributions of each RRM to the interactions with poly(A) and Paip2A, ITC analyses were performed using each of the isolated motifs: RRM1 including the RRM1–2 linker (residues 1–99), RRM2 including the RRM2–3 linker (residues 100–190), RRM3 including the RRM3–4 linker (residues 191–289), RRM4 (residues 290–371), and Paip2A(25–83) (residues 25–83). These proteins were expressed and purified to homogeneity (Fig. S1, D–G, I). The chemical shift index of Paip2A(25–83) (Fig. S4) (19) strongly suggests that Paip2A(25–83) is a natively disordered protein.
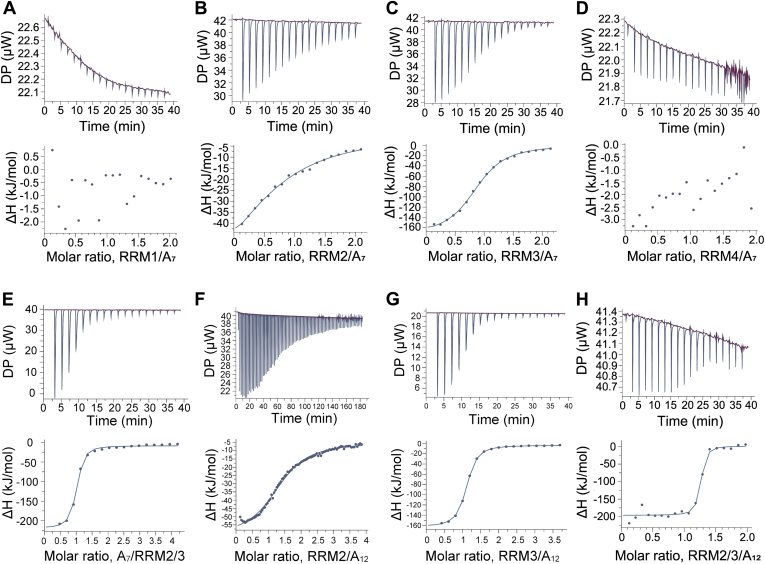
First, we investigated the interactions of poly(A) (A7) with these RRMs using ITC. As shown in Figure 4, significant heat exchange was observed during titration for RRM2 and RRM3, whereas essentially no heat exchange was observed for RRM1 and RRM4 (Fig. 4, A–D). Fitting the integrated isotherm to a one-site binding model resulted in Kd values of 200 and 4.7 μM for RRM2 and RRM3, respectively, with a binding stoichiometry of 1:1 (Fig. 4, B and C, Table 1). We further investigated the binding affinity of the region from RRM2 to RRM3 for A12. We prepared a protein corresponding to the region of PABPC1 (residues 100–289, hereafter referred to as RRM2/3; Fig. S1C). ITC analysis of the RRM2/3 binding to A12 revealed a Kd value of 1.3 nM with a 1:1 stoichiometry (Fig. 4H and Table 1), which is similar to the reported Kd value of 0.69 nM for the PABPC1–A24 interaction (15). These results indicate that the region from RRM2 to RRM3 in PABPC1 mainly contributes to poly(A) interactions.
Figure 4.
ITC analysis of the interactions of RRM of PABPC1 with poly(A). ITC analysis of the interactions of each RRM ((A) RRM1, (B) RRM2, (C) RRM3, and (D) RRM4) with A7, (E) RRM2/3 with A7, (F) RRM2 with A12, (G) RRM3 with A12, (H) RRM2/3 with A12. Upper panel, A–E, traces of the 19 titrations of 2 μl aliquots of A7 into cells containing RRM, (F) trace of the 91 titrations of 0.4 μl aliquots of RRM2 into cells containing A12, (G) traces of the 19 titrations of 2 μl aliquots of RRM3 into cells containing A12, and (H) traces of the 19 titrations of 2 μl aliquots of RRM2/3 into cells containing A12. Lower panel, the integrated binding isotherms obtained from the experiments were fitted using a “One Set of Sites” model. The parameters obtained from the best fit (solid line) with the error values calculated from the fitting are summarized in Table 1. ITC, isothermal titration calorimetry; PABPC1, poly(A)-binding protein C1; RRM, RNA recognition motif.
Table 1.
Thermodynamic parameters obtained via ITC analysis of each RRM–poly(A) and RRM2/3–poly(A) interaction
| Poly(A) | RRM | Na | Kd (M) | ΔH (kJ/mol) | ΔG (kJ/mol) | ΔS (kJ mol−1 K−1) |
|---|---|---|---|---|---|---|
| A7 | RRM1 | ND | ND | ND | ND | ND |
| RRM2 | 1.4 ± 0.1 | (2.0 ± 0.4) × 10−4 | −60 ± 2 | −21 | −0.13 | |
| RRM3 | 1.2 | (4.7 ± 0.3) × 10−6 | −1.5 × 102 | −30 | −0.41 | |
| RRM4 | ND | ND | ND | ND | ND | |
| RRM2/3 | 0.92 ± 0.01 | (0.71 ± 0.1) × 10−6 | −2.1 × 102 | −35 | −0.59 | |
| A12 | RRM2 | 1.4 | (70 ± 3) × 10−6 | −60 ± 1 | −24 | −0.11 |
| RRM3 | 1.0 | (0.52 ± 0.3) × 10−6 | −1.6 × 102 | −36 | −0.41 | |
| RRM2/3 | 1.1 | (1.3 ± 0.6) × 10−9 | −2.0 × 102 | −51 | −0.49 |
Abbreviation: ND, not detected.
A7: Poly(A) containing seven bases of adenine; A12: Poly(A) containing 12 bases of adenine.
For N, Kd, and ΔH, if the value of error is less than significant, the error is not stated.
N is the molar ratio of RRM to poly(A) or A7 to RRM2/3.
Paip2A binds to the RRM2 and RRM3 of PABPC1
Next, the contributions of each RRM to Paip2A binding were evaluated using ITC. For Paip2A(25–83), significant heat exchange was observed during titration for RRM2 and RRM3, whereas no heat exchange was observed for RRM1 and RRM4 (Fig. 5, A–D). Fitting of the integrated isotherm to a one-site binding model resulted in Kd values of 4.0 and 1.3 μM for RRM2 and RRM3, respectively, with binding stoichiometries of 1:1 in both cases (Table 2). We then investigated the interactions of RRM2/3 with Paip2A(25–83). ITC analysis revealed a Kd value of 1.9 nM and 1:1 stoichiometry (Fig. 5E and Table 2), which is consistent with the reported Kd value of 0.85 nM (17). We compared the NMR spectra of 2H, 15N-Paip2A(25–83) obtained following addition of 1.1 equivalents of RRM2/3 with those containing added RRM1/2/3/4 (Fig. S5). The backbone assignments of Paip2A(25–83) bound to RRM2/3 indicated that the signals from the N-terminal region of Paip2A (residues 25–29, 31, 32, 34–41, and 43–48) overlapped well, whereas there were slight differences in signals from the C-terminal region of Paip2A (residues 49–52, 57, 58, 60–65, 73–75, 77, 78, 80, 82, and 83). The slight differences were likely caused by differences in the sequence of RRM2, with a cloning artifact (GPLGS) in the RRM2/3 versus a linker between RRM1 and RRM2 in RRM1/2/3/4; however, the overall good spectral overlap suggests that the RRM2–RRM3 region of PABPC1 is mainly responsible for Paip2A binding.
Figure 5.
ITC analysis of interactions of the RRM of PABPC1 with Paip2A(25–83). ITC results are shown for the interactions of Paip2A(25–83) with (A) RRM1, (B) RRM2, (C) RRM3, (D) RRM4, and (E) RRM2/3. Upper panel, traces of the 19 titrations of 2 μl aliquots of RRM into cells containing Paip2A(25–83). Lower panel, integrated binding isotherms obtained from the experiments were fitted using a “One Set of Sites” model. The parameters obtained from the best fit (solid line), with error values calculated from the fitting, are summarized in Table 1. ITC, isothermal titration calorimetry; PABPC1, poly(A)-binding protein C1; Paip2A, PABP-interacting protein 2A; RRM, RNA recognition motif.
Table 2.
Thermodynamic parameters obtained via ITC analysis of the interaction of RRM or RRM2/3 with Paip2A(25–83)
| RRM | Na | Kd (M) | ΔH (kJ/mol) | ΔG (kJ/mol) | ΔS (kJ mol−1 K−1) |
|---|---|---|---|---|---|
| RRM1 | ND | ND | ND | ND | ND |
| RRM2 | 0.87 ± 0.03 | (4.0 ± 0.5) × 10−6 | −1.2 × 102 | −31 | −0.31 |
| RRM3 | 0.87 | (1.3 ± 0.1) × 10−6 | −1.8 × 102 | −34 | −0.50 |
| RRM4 | ND | ND | ND | ND | ND |
| RRM2/3 | 0.83 | (1.9 ± 0.6) × 10−9 | −2.3 × 102 | −50 | −0.61 |
Abbreviation: ND, not detected.
Paip2A(25–83): The peptide with residues 25 to 83 of PABP-interacting protein 2A.
For N, Kd, and ΔH, if the value of error is less than significant, the error is not stated.
N is the molar ratio of RRM to Paip2A(25–83).
Notably, the triple resonance spectra of 2H,13C,15N-labeled Paip2A(25–83) in complex with RRM2/3 showed line broadening, and thus 64 scans on the NMR of 800 MHz with a cryogenic probe were required to observe broadened 13Cα resonances in the HNCA and HN(CO)CA spectra even at the high sample concentration of 830 μM. This feature likely reflects a local conformational exchange in the complex. Using the backbone assignments of the complex (Fig. 6A), the chemical shift perturbation (CSP) of the interaction between Paip2A(25–83) and RRM2/3 was analyzed. As shown in Figure 6B, residues 31 to 79 of Paip2A(25–83) underwent chemical shift changes or disappearance upon RRM2/3 binding.
Figure 6.
Chemical shift perturbation (CSP) analysis of Paip2A(25–83) upon RRM2/3, RRM2, and RRM3 titration.A, backbone NMR assignments of [2H, 13C, 15N] Paip2A(25–83) bound to RRM2/3 identified on the 1H–15N TROSY spectrum. The L-3∗, G-2∗, S-1∗, and L80∗ are derived from the cis isomer of proline. B, chemical shift changes in each residue of Paip2A(25–83) bound to RRM2/3. Chemical shift changes were calculated using the following equation: , where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively. Asterisks indicate unassigned residues. However, the amino-acid types are identified for all unassigned peaks. Empty boxes indicate that the signal for the residue disappeared after RRM2/3 addition. Overlays of the 1H–15N HSQC spectra of 15N-labeled Paip2A(25–83) in the absence (black) or the presence (red) of (C) RRM2 and (D) RRM3. The perturbed and not perturbed signals are indicated in green and blue, respectively. E, chemical shift changes are plotted versus the residue numbers for Paip2A(25–83) after adding RRM2 (upper) or RRM3 (lower). Signals from residues 44 to 70 and 74 to 79 were perturbed after adding RRM2 and those from residues 27 to 70 were perturbed after adding RRM3. Residues 44 to 70 were perturbed after adding either RRM2 or RRM3. Chemical shift changes were calculated using the following equation: , where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively. HSQC, heteronuclear single quantum coherence; Paip2A, PABP-interacting protein 2A; RRM, RNA recognition motif; TROSY, transverse relaxation optimized spectroscopy.
To determine which RRM caused perturbations in Paip2A(25–83) residues 31 to 79, we observed the 1H–15N HSQC spectra of 15N-labeled Paip2A(25–83) in the RRM2-bound or RRM3-bound state. Figures 6, C and D and S6 show the spectral changes after addition of the isolated RRM2 and RRM3, respectively, which indicated that the different Paip2A regions were perturbed by binding of the isolated RRM2 and RRM3 but that the perturbed residues partially overlapped. The Paip2A(25–83) residues perturbed by RRM2 binding were residues 44 to 70 and 74 to 79, whereas those perturbed by RRM3 binding were residues 27 to 70, and residues 44 to 70 overlapped (Fig. 6E). Notably, the 1H–15N HSQC spectra of Paip2A(25–83) in the RRM2-bound or RRM3-bound state did not overlap with the corresponding signals in the NMR spectrum of Paip2A(25–83) in the RRM2/3-bound state (Fig. S7, A and B). These findings suggest that the binding mode of isolated RRM2 and RRM3 differs from that of RRM2/3. This is consistent with the NMR observation of RRMs, in which overlay of the HSQC spectra of Paip2A(25–83)-bound RRM2 and RRM3 did not reproduce the HSQC spectrum of Paip2A(25–83)-bound RRM2/3 (Fig. S7C). Thus, the isolated RRM2 and RRM3 preferentially bind to the C-terminal and N-terminal regions of Paip2A, with an overlapping region at the central part of Paip2A (residues 44–70); however, the binding mode of RRMs slightly differs from that in the RRM2/3-bound state.
Poly(A)-binding interface of RRM2 and RRM3
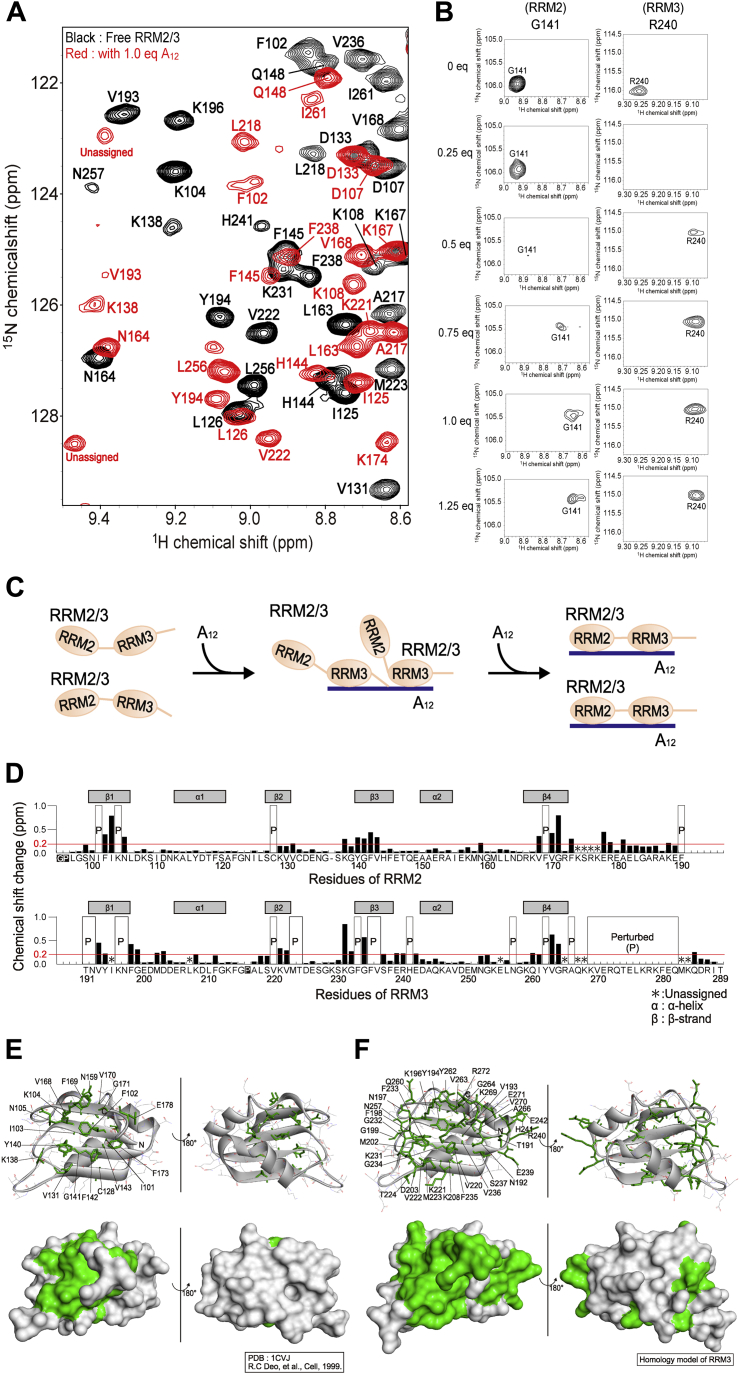
To identify the residues of RRM2/3 involved in the interactions with poly(A) and Paip2A, we analyzed the NMR spectral changes in RRM2/3 after adding A12 or Paip2A(25–83). We assigned 177 backbone NMR resonances (94%) of RRM2/3 of the 189 theoretically observable resonances by analyzing the number of triple resonance experiments using uniformly 13C, 15N-labeled RRM2/3 with reference to the backbone assignments of RRM2 and RRM3 that were independently established (Fig. S8).
Next, A12 titration experiments were performed by monitoring the 1H–15N HSQC spectra of uniformly 15N-labeled RRM2/3 (Fig. 7A). The NMR spectral changes during the titrations differed between signals from the RRM2 and RRM3 regions (Figs. 7B, S9, A and B). As shown in Fig. S9A, when 0.25 equivalents of A12 were added, the intensity was significantly reduced predominantly for RRM3 residues (Fig. S9A): 20 signals for RRM3 residues (V193, K196–G199, A217–S219, M223, E226, G232, F235, V236, R240, H241, N257, I261, V263, G264, and A266), and only four RRM2 residues (I103, K138, G171, and E189). After adding 0.25 to 0.5 equivalents of A12, the intensity of 26 signals from RRM2 was reduced (N100–F102, K104, N105, C128, V130, V131, E134, G139–V143, E152, K157, N159, V168–V170, E182, G184–A187, and F190) and 24 from RRM3 (N192, Y194, M202–D204, F211, V220, V222, T224, S230, K231, G234, F238, E242–A244, G253, L256, I261, V270, E271, and Q273–E275) (Fig. S9B). When 0.75 to 1.0 equivalents of A12 were added, several signals showed different chemical shifts (Fig. S10, D and E); finally, the changes were saturated upon addition of 1.25 equivalents of A12 (Figs. S10F and S11). Assuming the poly(A)-binding affinities of the RRM2 and RRM3 regions of RRM2/3 as those of the isolated RRM2 (Kd = 200 μM) and RRM3 (Kd = 4.7 μM), respectively, this spectral change reflects differences in the affinities between RRM2 and RRM3. In the presence of 0.25 equivalents of A12, most A12 molecules bind to two RRM2/3 molecules, with RRM3 region in RRM2/3 directly binding to poly(A) and leaving the RRM2 region in the A12-unbound state (Fig. 7C). As the Kd value of RRM2/3 and A12 is 1.9 nM, the complex should be sufficiently stable. However, the spectral changes indicate that the RRM2 region in A12-bound RRM2/3 is replaced by the RRM3 region of another RRM2/3 molecule.
Figure 7.
Chemical shift changes in uniformly15N-labeled RRM2/3 after adding A12.A, overlay of the 1H–15N HSQC spectra of 15N-labeled RRM2/3 in the absence (black) or the presence (red) of 1.0 equivalents of A12. B, chemical shift changes of signals derived from RRM2 and RRM3 from 0 to 1.25 equivalents of A12 titrations. The G141 and R240 are residues in RRM2 and RRM3, respectively. C, schematic drawing of the interactions between RRM2/3 and A12. In the presence of excessive RRM2/3 compared with A12, the RRM3 of RRM2/3 binds to A12 leaving RRM2 unbound (center). In the presence of equimolar A12, both RRM2 and RRM3 in one molecule of RRM2/3 simultaneously bind to A12 (right). D, plot showing the chemical shift changes after adding 1.25 equivalents of A12. The upper bar graph shows the chemical shift changes in the signals derived from RRM2, and the lower bar graph shows those from RRM3. The residues of the upper and lower graphs are arranged based on alignments between RRM2 and RRM3, and a secondary structure of these domains is shown in the graph. The corresponding signals in the A12-bound state were assigned by a series of triple resonance experiments, and their chemical shift changes are plotted in D. Some signals were perturbed after adding A12, and their corresponding signals in the A12-bound state could not be assigned, as the triple resonance signals were not observed. These residues are indicated as “Perturbed (P)” in D. Chemical shift changes were calculated using the following equation: , where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively. The symbols α and β in the graph indicate an α-helix and a β-strand, respectively, as observed in the crystal structure of the RRM1/2–poly(A) complex (Protein Data Bank code: 1CVJ (16)). The red lines at the 0.2 ppm represent the thresholds of residues mapped in Figure 7, E and F. E, RRM2 residues showing large chemical shift changes (>0.2 ppm) and labeled as “Perturbed” were mapped in green onto the crystal structure of RRM2 (Protein Data Bank code: 1CVJ (16)). The upper and lower parts are the ribbon model and surface drawing, respectively. F, RRM3 residues showing large chemical shift changes (>0.2 ppm) and labeled as “Perturbed” were mapped in green onto the homology model of RRM3. HSQC, heteronuclear single quantum coherence; RRM, RNA recognition motif.
To investigate the CSP of RRM2/3 after adding A12, we established backbone NMR assignments for RRM2/3 in the A12-bound state based on a series of triple resonance experiments with reference to the assignments of RRM2 and RRM3 in complex with A7, where 152 backbone resonances (96.7%, 65.3% of the theoretically observed RRM2 and RRM3 resonances, respectively) were successfully assigned (Fig. S12). The CSPs are shown in Figure 7D. The resonances with the CSP were more than 0.20 ppm, and those that underwent chemical shift changes in a slow exchange regime were regarded to be from perturbed residues, as follows: I101–N105, C128, V131, K138, Y140–V143, N159, V168–G171, F173, E178, R179, A181, K188, and F190 for RRM2, and T191–Y194, K196–G199, M202, D203, K208, V220–T224, K231–S237, E239–E242, N257, Q260, Y262–G264, A266, K269–Q282, and Q285 for RRM3. These residues mostly corresponded to the RRM2 and RRM3 regions of RRM2/3 (Fig. 7D). We mapped the perturbed residues on the RRM2 and RRM3 structures, obtained from the crystal structure of the RRM1/2-poly(A) complex (Protein Data Bank code: 1CVJ (16)), and modeled them using the structure of the highly homologous RRM2. The results suggested that the perturbed residues were localized at and around the poly(A)-binding interface (Fig. 7, E and F).
Furthermore, we performed 15N relaxation experiments to investigate the role of the linker between the two RRMs. The spin–lattice relaxation time (T1), spin–spin relaxation time (T2), and rotational correlation time (τc) values are summarized in Tables S1 and S2, Figs. S13 and S14, for RRM2/3 in the absence or the presence of A12. In the absence of A12, the τc values for RRM2 and RRM3 were 10.8 and 10.2 ns, respectively, which correspond to the typical τc values of 8 to 10 ns often observed for independently tumbling RRM or a single RRM, and were substantially lower than the τc values typically obtained for tandem interacting RRMs (15–17 ns) (20, 21), indicating that RRM2 and RRM3 in RRM2/3 tumble independently. Consistently, spectral overlay of RRM2 (residues 100–190) and RRM3 (residues 191–289) reproduced the spectrum of RRM2/3 (residues 100–289) well overall (Fig. S15 and Supporting Information). The mean value of τc for the linker region, 8.6 ns, was smaller than those of the RRM2 and RRM3 regions, suggesting that the linker region is highly mobile and flexible.
In the presence of A12, the mean τc values for RRM2 and RRM3 in the RRM2/3–A12 complex were 15.3 and 16.3 ns, respectively, corresponding to the typical τc values of 15 to 17 ns typically observed for tandem interacting RRMs (15–17 ns) (20, 21). The mean τc of the linker region is 14.3 ns, which corresponds to those for RRM2 and RRM3 within the error range. Thus, the mobility of the linker decreased upon A12 binding.
Paip2A binds to the poly(A)-binding interface of RRM2 and RRM3
We observed several 1H–15N HSQC spectra from uniformly 15N-labeled RRM2/3 after sequential addition of unlabeled Paip2A(25–83), which exhibited large spectral changes (Figs. 8A and S16), where most signals shifted in a slow exchange regime (Fig. 8B). Although we attempted to assign the Paip2A(25–83)-bound RRM2/3 resonances using triple resonance experiments, sequential assignments could not be established because of severe line broadening. There were 79 unassigned 1H–15N HSQC signals of RRM2/3 in complex with Paip2A(25–83), 45 of which provided Cα signals and none of which provided Cβ signals.
Figure 8.
Chemical shift changes in uniformly15N-labeled RRM2/3 after adding Paip2A(25–83). A, overlay of 1H–15N HSQC spectra of 15N-labeled RRM2/3 in the absence (black) and the presence (red) of 1.25 equivalents of Paip2A(25–83). B, change in A150 signal of RRM2/3 upon titration of Paip2A(25–83). C, the upper bar graph shows the chemical shift changes in signals derived from RRM2, and the lower bar graph shows those from RRM3. Chemical shift changes were calculated using the following equation: , where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively. The residues of the upper and lower graphs are arranged based on alignment between RRM2 and RRM3, and a secondary structure of these domains is shown in the graph. Signals with green labels were perturbed after adding Paip2A(25–83), and their corresponding signals in the Paip2A(25–83)-bound state could not be assigned because of broadening of the triple resonance signals. These residues are indicated as “Perturbed (P)” in C. The symbols α and β in the graph indicate an α-helix and a β-strand, respectively, as observed in the crystal structure of the RRM1/2–poly(A) complex (Protein Data Bank code: 1CVJ (16)). D, RRM2 residues showing large chemical shift changes labeled as “Perturbed” and I101 (>0.5 ppm) are mapped in green on the crystal structure of RRM2 (Protein Data Bank code: 1CVJ (16)). The upper and lower parts are the ribbon model and surface drawing, respectively. The square labels indicate that the residues interact with poly(A) directly in the crystal structure. E, RRM3 residues showing large chemical shift changes labeled as “Perturbed” were mapped in green onto the homology model of RRM3. The squares indicate the residues that are homologous to the residues of RRM2 that interact with poly(A) directly in the crystal structure. HSQC, heteronuclear single quantum coherence; Paip2A, PABP-interacting protein 2A; RRM, RNA recognition motif.
Although RRM2/3 resonances in the Paip2A(25–83)-bound state could not be assigned, the signals that shifted in a slow exchange regime were treated as having significant CSPs. These changes are summarized in Figure 8B. The significantly perturbed residues in RRM2 were I101–K108, F119, L126–E134, K138, Y140–E146, F169–F173, E178, R179, and R186–F190 and those in RRM3 are T191–Y194, K196–F198, L218–E226, F233–H241, Y262–G264, A266, and K269–E281 (Fig. 8C).
These affected residues were mapped onto the crystal structure of RRM2 and a homology model of RRM3 (Fig. 8, D and E). The results clearly indicated that these residues were localized on the β-sheet surface, on the opposite side of the two α-helices (Fig. 8, D and E), which largely overlapped with the poly(A)-binding surface (Fig. S17). Particularly, the residues whose labels are surrounded by squares (F102, K104, N105, S127, and F142) directly interact with poly(A) (Fig. 8D). These results suggest that Paip2A can compete against poly(A) on identical surfaces of RRM2 and RRM3 of PABPC1.
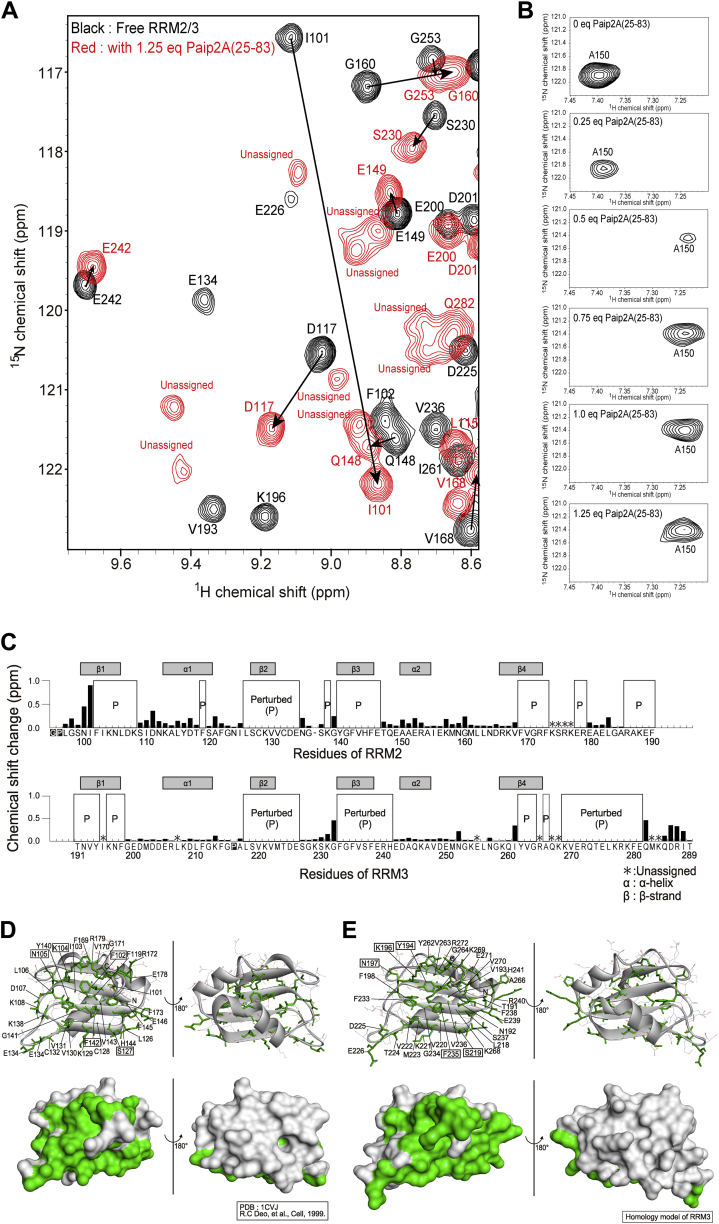
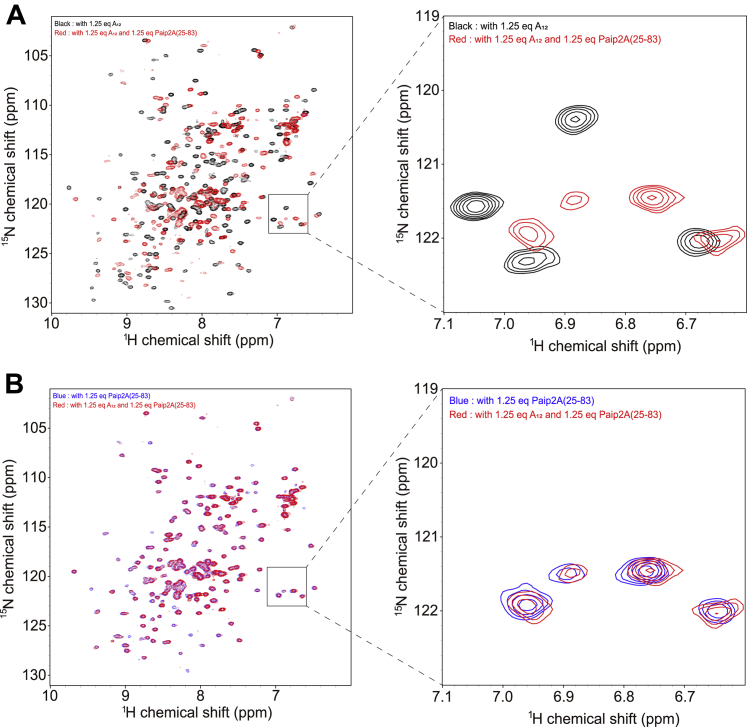
Finally, we conducted Paip2A(25–83) titrations with uniformly 1H, 15N-labeled RRM2/3 in complex with A12 by monitoring the 1H–15N HSQC spectra of RRM2/3. Figure 9 clearly shows that the HSQC spectrum of RRM2/3 in the A12-bound state (Fig. 9A, black) changed completely compared with that of the Paip2A(25–83)-bound state (Fig. 9B, blue, red) when equimolar Paip2A(25–83) was added. This indicates that Paip2A(25–83) competitively binds to RRM2/3 and dissociates A12 from RRM2/3, although the binding affinities of Paip2A(25–83) and A12 for RRM2/3 were comparable (Kd = 1.3–1.9 nM, Tables 1 and 2).
Figure 9.
1H–15N HSQC spectra of the RRM2/3 in the presence of equimolar A12and Paip2A(25–83). A, overlay of the 1H–15N HSQC spectra of 15N-labeled RRM2/3 in the presence of 1.25 equivalents of A12 (black) and 1.25 equivalents of both A12 and Paip2A(25–83) (red). B, overlay of the 1H–15N HSQC spectra of 15N-labeled RRM2/3 in the presence of 1.25 equivalents of Paip2A(25–83) (blue) and presence of 1.25 equivalents of both A12 and Paip2A(25–83) (red). HSQC, heteronuclear single quantum coherence; Paip2A, PABP-interacting protein 2A; RRM, RNA recognition motif.
Discussion
Among the several identified PABPC1-interacting factors, a translational repression factor, Paip2, has attracted attention because it represses the proliferation of cells and viruses (9, 12, 13) through its ability to dissociate PABPC1 from poly(A). However, how Paip2 efficiently dissociates PABPC1 from poly(A) remains unclear, as the Kd values are comparable between the Paip2A(FL)–PABPC1 and poly(A)–PABPC1 interactions.
This investigation, in agreement with previous findings (17), indicates that Paip2A primarily interacts with the RRM2–RRM3 region of PABPC1. The minimal interacting region of`Paip2A is residues 26 to 83, which was confirmed by spectral overlay of RRM2/3 in complex with Paip2A(FL) and Paip2A(25–83) (Fig. S18).
However, the contribution of each RRM region of human PABPC1 to poly(A) binding has not been well characterized. A previous study of Xenopus laevis PABPC1 reported that RRM4 had the highest poly(A)-binding affinity, followed by RRM2 and RRM3, whereas RRM1 had almost no binding affinity (22). Our quantitative ITC analyses of RRMs from human PABPC1 revealed that RRM3 and RRM2 had the highest and second highest binding affinities for A7 (Kd = 4.7 and 200 μM, respectively), whereas no binding was detected for RRM1 and RRM4 at concentrations of 98 and 260 μM, respectively. Although the amino acid sequence of each RRM is very similar (Fig. S19), there were significant differences in their affinity for poly(A). Notably, RRM1 was observed in the poly(A)-bound state in the crystal structure of RRM1/2 of human PABPC1 (16). The poly(A) binding of RRM1 in the crystal structure is likely related to the support by the linker that tethers to poly(A)-bound RRM2, although the poly(A)-binding affinity of RRM1 was much lower than that of RRM2.
The binding affinity of RRM2/3 for A12, which is a deletion mutant of PABPC1 corresponding to the region from RRM2 to RRM3, showed a binding affinity for A12 (Kd = 1.3 nM) that was comparable to the Kd of the PABPC1–A24 interaction (0.69 nM) in our previous report (15). These results clearly indicate that the RRM2–RRM3 region of PABPC1 plays the most important role in binding not only to Paip2A but also to poly(A).
NMR relaxation experiments revealed that the two RRM regions in RRM2/3 tumble independently, with the flexible linker in the absence of A12 (Table S1), whereas the tumbling motion of the RRMs and linker is suppressed by A12 binding. As the linker region exhibited CSPs upon binding to A12, the linker is likely involved in the interactions with A12. However, as the linker region (residues 173–190) is in the isolated RRM2 protein (residues 100–190), the 1000-fold higher affinity of RRM2/3 compared with that of isolated RRM was not related to the linker interactions but rather to “the tethering effect,” that is, an increase in the local concentration of one RRM around poly(A) upon binding of another RRM to poly(A), as previously reported (16).
The two RRM regions appear to contribute to the poly(A) binding independently, as the Kd values satisfy the following relationship:
The tethering effect by the linker and repeating feature of poly(A) likely led to the extremely high affinity of RRM2/3 as compared with that of isolated RRM.
The result of NMR titration experiment for RRM2/3-A12 suggested that RRM2/3 binds to poly(A) on the surface of the β-sheets of RRM2 and RRM3. This binding mode is consistent with the poly(A)-binding region of RRM1/2 of PABPC1 (16). It has also been reported that many other nucleic acid–binding proteins such as TAR DNA-binding protein 43 (23) and Musashi-1 (24, 25) bind to RNA on these surfaces of β-sheets of RRMs. The poly(A) binding of RRM2/3 in our study was also a typical RNA-binding mode of nucleic acid–binding proteins.
In contrast, Paip2A also interacts with the RRM2–RRM3 region of PABPC1, wherein the Paip2A-binding sites on the RRMs overlap with the poly(A)-binding site (Figs. 7, E and F, 8, D and E), strongly suggesting that Paip2A competitively dissociates poly(A) from PABPC1. However, the roles of RRM2 and RRM3 in Paip2A binding differ from those in poly(A) binding. Although Paip2A(25–83) had almost the same binding affinities for RRM2 and RRM3 with Kd values of 4.0 and 1.3 μM, respectively, the Kd of 1.9 nM for the Paip2A(25–83)–RRM2/3 interaction was 1000-fold larger than the product of the Kd values for the Paip2A(25–83)–RRM2 interaction and Paip2A(25–83)–RRM3 interaction. These results indicate that the interactions between isolated RRM domains and Paip2A were not reproduced in the interactions between RRM2/3 and Paip2A, which was also reflected in the NMR observations (Fig. S7). As mentioned previously, poly(A) and Paip2A bind to the same interfaces of the RRM2–RRM3 region of PABPC1 with comparable apparent Kd values. However, our NMR data clearly indicate that Paip2A(25–83) dissociates RRM2/3 from A12, forming the Paip2A(25–83)–RRM2/3 complex (Fig. 9).
To determine how Paip2A facilitated efficient dissociation of PABPC1 from poly(A), we focused on the 40-fold lower binding affinity of RRM2 for poly(A) compared with RRM3. The NMR titration experiments between uniformly 15N-labeled RRM2/3 and A12 indicated that the RRM3 region preferentially bound to poly(A) in the presence of RRM2/3 in excess of A12 (Fig. 7, B and C, S9A). Thus, even if the RRM2 region of RRM2/3 (Kd = 200 μM for RRM2-A7) is tethered to poly(A) via the poly(A)-bound RRM3 region (Kd = 4.7 μM for RRM3-A7), which presumably enhances the RRM2–poly(A) interaction, the RRM3 region of other RRM2/3 molecules inhibits the RRM2–poly(A) interaction. This strongly suggests that the interaction of the RRM2 region of PABPC1 with poly(A) can be inhibited by Paip2A because Paip2A possesses an RRM2-binding affinity (Kd = 4 μM for RRM2–Paip2A(25–83)), which is comparable to the affinity of the RRM3–poly(A) interaction.
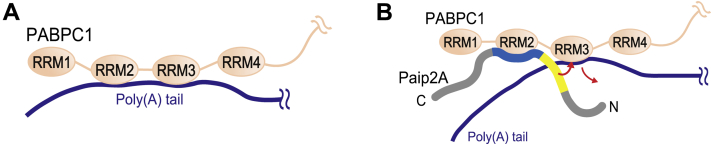
Based on these ITC and NMR data, we propose the following mechanism by which Paip2A dissociates PABPC1 from poly(A): in the poly(A)-bound PABPC1, the RRM2–RRM3 region mainly contributes to poly(A) association (Fig. 10A). As the affinity of the RRM2 region for poly(A) (Kd = 200 μM) was 50-fold weaker than that for Paip2A(25–83) (Kd = 4.0 μM), the RRM2 region transfers from poly(A) to Paip2A when Paip2A approaches, where the RRM3-binding residues of Paip2A are only partly available for RRM3 binding (Fig. 10B). Once Paip2A binds to the RRM2 region of poly(A)-bound PABPC1, which binds to poly(A) mostly through the RRM3 region, Paip2A removes RRM3 from poly(A) because of the 3.6-fold higher affinity of RRM3 for Paip2A(25–83) (Kd = 1.3 μM), as compared with that for poly(A) (Kd = 4.7 μM), as well as the effect of tethering to the RRM2 region (Fig. 10B), resulting in dissociation of PABPC1 from poly(A).
Figure 10.
Schematic representation of the dissociation of PABPC1 from poly(A) by Paip2A. A, in the poly(A)-bound PABPC1, the RRM2–RRM3 region mainly contributes to the poly(A) association. B, as the affinity of the RRM2 region for poly(A) (Kd = 200 μM, Table 1) is 50-fold weaker than that for Paip2A (Kd = 4.0 μM, Table 2), the RRM2 region transfers from poly(A) to RRM2-binding region of Paip2A (blue) when Paip2A approaches sites where the RRM3-binding region of Paip2A (yellow) are partly available for RRM3 binding. Next, Paip2A removes RRM3 from poly(A) because of the threefold higher affinity of RRM3 for Paip2A (Kd = 1.3 μM, Table 2) compared with that for poly(A) (Kd = 4.7 μM, Table 1). PABPC1, poly(A)-binding protein C1; Paip2A, PABP-interacting protein 2A; RRM, RNA recognition motif.
Here, we proposed a mechanism for how Paip2 competitively dissociates PABPC1 from poly(A), although Paip2A and poly(A) possess comparable affinities for PABPC1. A similar molecular mechanism was previously reported for the competition between hypoxia-inducible factor 1α (HIF-1α) and CBP/p300 interacting transactivator with ED-rich tail 2 (CITED2) for the TAZ1 domain of CBP, where TAZ1-bound HIF-1α was effectively replaced by CITED2 (26, 27). Despite the comparable affinities of HIF-1α and CITED2 for TAZ1 (Kd = 10 nM), the NMR spectrum of 15N-labeled TAZ1 with the equivalent molar amount of HIF-1α and CITED2 is consistent with that of binary complex of 15N-labeled TAZ1–CITED2, indicating that TAZ1 binds preferentially to CITED2 compared with HIF-1α. They found that αA (αA-helix) of HIF-1α showed high motility even when HIF-1α formed a complex with TAZ1, and αA of CITED2 showed less motility when CITED2 formed a complex with TAZ1, suggesting that αA of CITED2 has a higher binding affinity for TAZ1 compared with HIF-1α and replaces HIF-1α bound to TAZ1. Furthermore, the LPQL region of HIF-1α bound to TAZ1 is replaced by the LPEL region of CITED2, followed by αB and αC of HIF-1α being replaced by CITED2.
We considered that the dissociation of PABPC1 from poly(A) by Paip2A is achieved via a two-step mechanism in which Paip2A initially binds to RRM2, which showed 50-fold lower binding affinity for poly(A) than for Paip2A, to form a transient ternary complex; next, Paip2A displaces poly(A) from RRM3 by binding to RRM3. Our results extend the efficient replacement despite the comparable affinities through the protein versus protein competition to that through the protein versus nucleic acid competition.
We found that the binding of RRM to poly(A) was competitively inhibited by Paip2A; accordingly, the interaction between Paip2A and RRM2 may be the starting point for dissociation of PABPC1 from poly(A). However, it remains unclear why the binding affinities of RRM2 and RRM3 were 40-fold different despite their high level of homology and ability to recognize Paip2A. Three-dimensional structural analysis at the atomic level of the RRM2/3–poly(A) complex and RRM2–Paip2A and RRM3–Paip2A complexes may further contribute to the understanding of the molecular recognition of PABPC1 and aid in the development of anticancer and/or antiviral drugs.
Experimental procedures
Protein expression and purification
The DNA sequences encoding FL human PABPC1 (residues 1–636) and the mutants RRM1/2/3/4 (residues 1–370), RRM1 (residues 1–99), and RRM4 (residues 290–371) were cloned into the pET-42b(+) vector (Novagen). Mutants of human PABPC1 (RRM2/3 [residues 100–289], RRM2 [residues 100–190], and RRM3 [residues 191–289] were cloned into the pGEX-6p-1 vector (Cytiva). The DNA sequences encoding FL human Paip2A (residues 1–127) and the mutant Paip2A(25–83) were also cloned into a pGEX-6p-1 vector (Cytiva). The glutathione-S-transferase-fusion proteins were expressed in Escherichia coli cells and purified using a Glutathione-Sepharose 4B column (Cytiva), followed by digestion with factor Xa (Novagen) or PreScission Protease (Cytiva). The cleaved glutathione-S-transferase and noncleaved fusion proteins were removed using a Glutathione-Sepharose 4B column. The degradation products were removed using a cation exchange column (HiTrap SP HP for PABPC1 and RRM1/2/3/4 or MONO S for RRM2/3 RRM1, RRM2, RRM3, and RRM4; Cytiva) or an anion exchange column (RESOURCE Q for Paip2A and Paip2A(25–83); Cytiva), followed by gel filtration using HiLoad 16/600 Superdex 75 pg (Cytiva) for PABPC1, RRM1/2/3/4, Paip2A, and Paip2A(25–83). The molecular weight of Paip2A was confirmed via MALDI-TOF mass spectrometry using an AXIMA-CFR Plus mass spectrometer (Shimadzu). Uniformly 15N or 13C, 15N-labeled proteins for NMR experiments were expressed by growing E. coli host cells in M9 minimal medium containing 15NH4Cl or 15NH4Cl and 13C6-glucose. 2H, 15N-labeled proteins were expressed by growing E. coli host cells in M9 minimal medium in 2H2O containing 15NH4Cl. 2H, 13C, 15N-labeled proteins were expressed by growing E. coli host cells in M9 minimal medium in 2H2O containing 15NH4Cl and 13C6-glucose. These isotopically labeled proteins were purified using a similar procedure as used for unlabeled proteins.
SPR analysis
All experiments were conducted at 298 K using a BiacoreX 100 (Cytiva). The running buffer contained 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, 5 mM MgSO4, 1 mM DTT, and 5% (v/v) glycerol at pH 7.4. The 5′-biotinylated A24 was immobilized at a flow rate of 30 μl/min on a streptavidin-coated sensor chip. The purified FL PABPC1, RRM1/2/3/4, and Paip2A(FL) were dialyzed against the running buffer. Next, 128 nM PABPC1 or RRM1/2/3/4 was injected at a flow rate of 5 μl/min for 180 s, and the running buffer flowed over the sensor chip at the same flow rate for 300 s, followed by injection of 1, 10, 100, or 1000 nM Paip2A at the same flow rate for 180 s. The surface was regenerated using 0.05% SDS solution. All samples flowed over the nonimmobilized surface of the flow cells, and the responses were used as a reference.
NMR analysis
Data were collected on Bruker Avance 500, 600, or 800 spectrometer (Billerica) using triple-resonance experiments or cryogenic probes. All spectra were processed using Bruker TopSpin 4.0.8 software, and data were analyzed using Sparky (T.D. Goddard and D.G. Kneller, Sparky 3, University of California).
Titrations of 2H, 15N-labeled Paip2A(FL) with unlabeled RRM1/2/3/4, 2H, 15N-labeled Paip2A(25–83) with unlabeled RRM1/2/3/4, and RRM2/3 were monitored using a 1H–15N TROSY spectrum, and 15N-labeled Paip2A(25–83) with unlabeled RRM2 and RRM3 were monitored using the 1H–15N HSQC spectrum. Analysis of the CSPs of 2H, 13C, 15N-labeled Paip2A(25–83) with unlabeled RRM2/3 was monitored using a 1H–15N TROSY spectrum. All aforementioned experiments were performed at 298 K in buffer containing 18 mM KaH2PO4 (pH 6.0), 90 mM NaCl, and 10% 2H2O. The titrations of 15N-labeled RRM2/3 with unlabeled Paip2A(25–83) and unlabeled A12 were monitored via the 1H–15N HSQC spectrum at 303 K in buffer containing 18 mM NaH2PO4 (pH 6.5), 135 mM NaCl, and 10% 2H2O.
Chemical shift changes were calculated using the following equation:
| (1) |
where Δ1H and Δ15N are the chemical shift changes in the 1H and 15N dimensions, respectively.
Sequential assignments of the backbone NMR resonances for Paip2A(FL) were achieved using HNCACB, CBCA(CO)NH, HN(CA)CO, HNCO, HNCA, and HN(CO)CA experiments, whereas those for Paip2A(25–83) were achieved using 15N-edited TOCSY–HSQC and 15N-edited NOESY–HSQC experiments. Sequential assignments of the backbone NMR resonances for Paip2A(25–83) bound to RRM2/3 were achieved using TROSY-type HNCA and HN(CO)CA experiments and the 2D version (13C–1H plane) of HN(CA)CB and HN(COCA)CB experiments to obtain 13Cβ information. All aforementioned experiments were performed at 298 K in buffer containing 18 mM KaH2PO4 (pH 6.0), 90 mM NaCl, and 10% 2H2O. The backbone NMR resonances of RRM2 and RRM3 were sequentially assigned using HNCACB, CBCA(CO)NH, CC(CO)NH, 15N-edited NOESY–HSQC, and 15N-edited NOESY–HSQC. The backbone NMR resonances of RRM2/3 and those in complex with A12 were sequentially assigned using HNCACB, CBCA(CO)NH, HNCA, and HN(CO)CA. We attempted to determine whether the backbones of the NMR resonances of RRM2/3 were in complexes with Paip2A(25–83), by analyzing the HNCA and HN(CO)CA spectra, and line broadening of several signals was found to preclude the sequential assignments. All aforementioned experiments were performed at 303 K in buffer containing 18 mM NaH2PO4 (pH 6.5), 135 mM NaCl, and 10% 2H2O.
15N relaxation experiments
The rotational correlation time (τc) of RRM2/3 in the absence of A12 and presence of A12 was calculated using following equation (28):
where τc [ns] is rotational correlation time, ωN [1/s] is the Larmor frequency of 15N, T1 [ms] is the 15N spin–lattice relaxation time, and T2 [ms] is the 15N spin–spin relaxation time. In addition, ωN was calculated using the following equation:
where γN (= −27.126188 × 106 [s−1 T−1]) is the gyromagnetic ratio of 15N and B0 (= 14.09 T) is external magnetic field in the case of using a machine with a resonance frequency of 1H of 600 MHz. As γN is a negative value, ωN is also a negative value. Thus, in the calculation of τc, we used the absolute value of ωN.
The experiments were performed at 303 K in buffer containing 18 mM NaH2PO4 (pH 6.5), 135 mM NaCl, and 10% 2H2O. The spectra were processed using Bruker TopSpin 4.0.8 software, and data were analyzed using Sparky.
ITC
Binding of Paip2A(25–83) or poly(A) to each RRM was measured using ITC (29) and MicroCal PEAQ-ITC (Malvern Instruments). All samples were prepared in buffer containing 10 mM Na2HPO4 (pH 7.4), 1.8 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl. The experiments were performed at 25 °C. RRM1 (9.2 μM), RRM2 (211 μM), RRM3 (50 μM), RRM4 (26 μM), and RRM2/3 (50 μM) were titrated with poly(A) (A7) (98 μM, 2.3 mM, 570 μM, 260 μM, and 459 μM), respectively. Poly(A) (A12) (349, 27, and 0.99 μM) was titrated with RRM2 (8.1 mM), RRM3 (525 μM), and RRM2/3 (10.3 μM). Paip2A(25–83) (5, 20, 20, 5, and 0.95 μM) was titrated with RRM1 (50 μM), RRM2 (211 μM), RRM3 (200 μM), RRM4 (50 μM), and RRM2/3 (9.7 μM), respectively. The heats of the injectant dilution were determined by titration of the injectant into the buffer, followed by subtraction from the row titration data before analysis using MicroCal PEAQ-ITC Analysis Software (Malvern). A “One Set of Sites” model was applied for curve fitting. Thermodynamic parameters with error values were calculated from the fitted data.
Quantitation of poly(A)
The molar concentrations of poly(A) were evaluated by measuring the UV absorption at 260 nm based on Lambert–Beer’s law:
| (2) |
where c(An) is the molar concentration of An (poly(A) consisting of n bases), Abs260 is the absorption of UV at a wavelength of 260 nm, εn is the molar extinction coefficient value, and l is the optical path length of 1 cm. εn was calculated using the nearest neighbor method, ε7 was 87,100 (l/mol cm), ε12 was 146,900 (l/mol cm), and ε24 was 290,420 (l/mol cm).
Construction of comparative model for RRM3
A homology model for RRM3 was constructed using MODELLER (University of California, San Francisco, Accelrys) (30). The amino acid sequence of PABPC1 (95–179) corresponding to the RRM2 sequence was adopted as a template sequence, whereas the amino acid sequence of PABPC1 (187–272) corresponding to the RRM3 sequence was adopted as the target. The crystal structure of RRM1/2–poly(A) (Protein Data Bank code: 1CVJ (16)) was adopted as the atomic coordinates for the template.
Data availability
The 1H, 13C, and 15N chemical shift assignments for Paip2A, Paip2A(25–83), RRM2, RRM3, RRM2/3, RRM2/3-A12, RRM2/3-Paip2A(25–83) have been deposited in the Biological Magnetic Resonance Bank (ID: 26314 for Paip2A, 26315 for Paip2A(25–83), 26323 for Paip2A(25–83) when in complex with RRM2/3, 26316 for RRM2, 26317 for RRM3, 26318 for RRM2/3, 26319 for RRM2/3 when in complex with A12, and 26320 for RRM2/3 when in complex with Paip2A(25–83), respectively).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Editage (www.editage.com) for English language editing. We also thank the Graduate School of Pharmaceutical Science, the University of Tokyo for the NMR machine time.
Author contributions
T. S., M. Y., R. S., N. H., S.-i. H., and M. O. methodology; T. S., M. Y., R. S., Y. I., S.-i. H., I. S., and M. O. formal analysis; T. S., M. Y., R. S., Y. I., N. H., S. I., and M. O. investigation; T. S., M. Y., R. S., Y. I., S.-i. H., I. S., and M. O. writing–original draft.
Funding and additional information
This research was supported in part by the Japan Society for the Promotion of Science KAKENHI grant (grant numbers: JP17H03978 and JP19H04973 [to M. O.]), Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from the Japan Agency for Medical Research and Development (AMED) under grant numbers JP21 AM0101071 (support number: 1392), a grant from The Vehicle Racing Commemorative Foundation (to M. O.), Takeda Science Foundation (to M. Y. and M. O.), grant for Basic Science Research Projects from the Sumitomo Foundation (to M. Y.), and Sato Pharmaceutical Research Grant (to T. S).
Edited by Karin Musier-Forsyth
Supporting information
References
- 1.Hocine S., Singer R.H., Grünwald D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Svitkin Y.V., Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- 3.Melo E.O., Dhalia R., De Sa C.M., Standart N., De Melo Neto O.P. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: Role in cooperative binding to poly(A) and efficient cap distal translational repression. J. Biol. Chem. 2003;278:46357–46368. doi: 10.1074/jbc.M307624200. [DOI] [PubMed] [Google Scholar]
- 4.Baer B.W., Kornberg R.D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safaee N., Kozlov G., Noronha A.M., Xie J., Wilds C.J., Gehring K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell. 2012;48:375–386. doi: 10.1016/j.molcel.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino S.I., Imai M., Kobayashi T., Uchida N., Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 7.Chorghade S., Seimetz J., Emmons R., Yang J., Bresson S.M., de Lisio M., Parise G., Conrad N.K., Kalsotra A. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. Elife. 2017;6 doi: 10.7554/eLife.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Zhou W., Velculescu V.E., Kern S.E., Hruban R.H., Hamilton S.R., Vogelstein B., Kinzler K.W. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 9.Polacek C., Friebe P., Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 2009;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 10.Perez C., McKinney C., Chulunbaatar U., Mohr I. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J. Virol. 2011;85:156–164. doi: 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaleghpour K., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld A.B. Suppression of cellular transformation by poly (A) binding protein interacting protein 2 (Paip2) PLoS One. 2011;6:2–6. doi: 10.1371/journal.pone.0025116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney C., Yu D., Mohr I. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev. 2013;27:1809–1820. doi: 10.1101/gad.221341.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliseeva I.A., Lyabin D.N., Ovchinnikov L.P. Poly(A)-binding proteins: Structure, domain organization, and activity regulation. Biochem. 2013;78:1377–1391. doi: 10.1134/S0006297913130014. [DOI] [PubMed] [Google Scholar]
- 15.Sawazaki R., Imai S., Yokogawa M., Hosoda N., Hoshino S.I., Mio M., Mio K., Shimada I., Osawa M. Characterization of the multimeric structure of poly(A)-binding protein on a poly(A) tail. Sci. Rep. 2018;8:1455. doi: 10.1038/s41598-018-19659-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 17.Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y.V., Imataka H., O’Connor-McCourt M., Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlov G., Ménade M., Rosenauer A., Nguyen L., Gehring K. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J. Mol. Biol. 2010;397:397–407. doi: 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 19.Wishart D.S., Sykes B.D. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez C., Schubert M., Duss O., Ravindranathan S., Allain F.H.T. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011;58:1–61. doi: 10.1016/j.pnmrs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Barraud P., Allain F.H.T. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: How the relative orientation between RRMs influences the nucleic acid binding topology. J. Biomol. NMR. 2013;55:119–138. doi: 10.1007/s10858-012-9696-4. [DOI] [PubMed] [Google Scholar]
- 22.Nietfeld W., Mentzel H., Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukavsky P.J., Daujotyte D., Tollervey J.R., Ule J., Stuani C., Buratti E., Baralle F.E., Damberger F.F., Allain F.H.T. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 24.Ohyama T., Nagata T., Tsuda K., Kobayashi N., Imai T., Okano H., Yamazaki T., Katahira M. Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2012;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwaoka R., Nagata T., Tsuda K., Imai T., Okano H., Kobayashi N., Katahira M. Structural insight into the recognition of r(UAG) by Musashi-1 RBD2, and construction of a model of Musashi-1 RBD1-2 bound to the minimum target RNA. Molecules. 2017;22:1207. doi: 10.3390/molecules22071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berlow R.B., Dyson H.J., Wright P.E. Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature. 2017;543:447–451. doi: 10.1038/nature21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berlow R.B., Dyson H.J., Wright P.E. Multivalency enables unidirectional switch-like competition between intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2117338119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gryk M.R., Abseher R., Simon B., Nilges M., Oschkinat H. Heteronuclear relaxation study of the PH domain of β-spectrin: Restriction of loop motions upon binding inositol trisphosphate. J. Mol. Biol. 1998;280:879–896. doi: 10.1006/jmbi.1998.1731. [DOI] [PubMed] [Google Scholar]
- 29.Wiseman T., Williston S., Brandts J.F., Lin L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 30.Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 1H, 13C, and 15N chemical shift assignments for Paip2A, Paip2A(25–83), RRM2, RRM3, RRM2/3, RRM2/3-A12, RRM2/3-Paip2A(25–83) have been deposited in the Biological Magnetic Resonance Bank (ID: 26314 for Paip2A, 26315 for Paip2A(25–83), 26323 for Paip2A(25–83) when in complex with RRM2/3, 26316 for RRM2, 26317 for RRM3, 26318 for RRM2/3, 26319 for RRM2/3 when in complex with A12, and 26320 for RRM2/3 when in complex with Paip2A(25–83), respectively).