Summary
Oligo library pools are powerful tools for systematic investigation of genetic and transcriptomic machinery such as promoter function and gene regulation, non-coding RNAs, or RNA modifications. Here, we provide a detailed protocol for cloning DNA oligo pools made up of tens of thousands of different constructs, aiming to preserve the complexity of the pools. This system would be suitable for expression in cell lines and can be followed up by next-generation sequencing analysis.
For complete details on the use and execution of this profile, please refer to Uzonyi et al. (2021).
Subject areas: Genetics, Molecular Biology, Systems biology
Graphical abstract

Highlights
-
•
Restriction-based cloning of DNA pools
-
•
Preservation of complexity of thousands of constructs
-
•
Used to investigate genetic and transcriptomic machineries
-
•
To be expressed in cell lines and follow up by NGS analysis
Oligo library pools are powerful tools for systematic investigation of genetic and transcriptomic machinery such as promoter function and gene regulation, non-coding RNAs, or RNA modifications. Here, we provide a detailed protocol for cloning DNA oligo pools made up of tens of thousands of different constructs, aiming to preserve the complexity of the pools. This system would be suitable for expression in cell lines and can be followed up by next-generation sequencing analysis.
Before you begin
The protocol here describes the specific steps for amplifying and cloning a DNA oligo library pool consisting of 12,000 sequence variants, into the 3′ end of a fluorescent reporter in a plasmid. The plasmid used in this example contains an ampicillin resistance gene, attB sites and a GFP, driven by a mouse SNRPN (Small Nuclear Ribonucleoprotein Polypeptide N) promoter. The full plasmid sequence is available in Table S1. We expect similar efficiency and outcome with any high copy number plasmid.
This protocol is based on the Agilent library cloning protocol developed by the lab of Eran Segal, employing a large scale restriction-ligation approach, allowing adequate representation of the library complexity. The protocol was developed for transformation of yeast (Sharon et al., 2012), and later adapted to work with mammalian systems (Weingarten-Gabbay et al. 2016, 2019; Vainberg Slutskin et al., 2018).We then modified this protocol for oligo library pools from Twist Bioscience, and carried out additional changes (Safra et al., 2017; Uzonyi et al., 2021).
Compared to the original protocol (which was not published as a detailed step-by-step protocol), adaptations were made due to the different starting material (Agilent versus Twist library pool). Furthermore, the scale of several steps have been revised. Due to discontinuation and availability issues, some of the reagents have been replaced. Additional cleaning steps throughout the protocol led to increased efficiency of this updated version.
Note:we have marked all example calculations in italics throughout the protocol.
Planning and ordering of an oligo library pool
Timing: 2–12 weeks
This time-frame is dependent on non-technical factors (such as delivery times) and may vary from one set up to another.
The protocol is preceded by the planning and ordering of the oligo library pool according to the topic of interest (e.g., RNA modifications, non-coding RNAs, promoters, enhancers, gene regulatory elements or other). The following notes should be considered when planning the sequences:
Each sequence should be planned with a DNA barcode with at least 2 nucleotides distance between any two barcodes. This is to ensure that amplification or sequencing errors would not lead to confusion between constructs. A full set of 10 nucleotide long barcodes with a 2 nucleotide distance and no triplets planned with the DNABarcodes R package is available in Table S1 (Buschmann and Bystrykh 2013).
The cloning approach used in this protocol is the highly efficient restriction and ligation approach, to allow maintaining the complexity of the library. Make sure that the used restriction sites are not present elsewhere in the sequences. This can be done by simple manual text search of the final sequence set. Any sequence that is found to contain a used restriction motif should be point mutated or removed.
If relevant, make sure that the sequences don’t contain strong canonical poly-adenylation sites or other sequences that can influence the results. We recommend doing this by simple manual text search followed by replacement or removal of problematic sequences. Sequence constraints can also be applied during the sequence planning step, depending on the specific goal and use.
We recommend designing a subset of at least 100 sequences with two different barcodes (BC), to assess the effect of the barcode sequence on the results, and to measure technical noise.
Please refer to Figure 1 for an example oligo pool plan.
CRITICAL: Always plan the libraries with two different restriction enzymes. Using the same restriction enzyme will lead to some of the sequences being integrated in the wrong direction. In this large-scale cloning, there’s no way to select or adjust for this. Select highly efficient enzymes with low star activity, to ensure efficient cutting at the target location and no off-target cuts. Ideally use 7-or 8 cutters, to reduce the frequency of unwanted cuts in the library pool. Although FastDigest enzymes (Thermo Scientific) are often used in this protocol, it is possible to refer to the NEB list of restriction enzyme motifs as a resource for the selection of appropriate enzymes: https://international.neb.com/tools-and-resources/selection-charts/alphabetized-list-of-recognition-specificities. Enzymes marked HF (high fidelity) have very low star activity.
CRITICAL: When designing the plasmid, make sure that there is no need to gel-purify the plasmid after restriction digestion, meaning that there should be no more than 40 bases between the two restriction sites (to allow for the excised fragment to be eliminated by a PCR purification kit). Adding a gel purification step would dramatically decrease the ligation efficiency.
Figure 1.
Oligo pool planning strategy
RE = restriction enzyme, UTR = 3′ untranslated region, BC = barcode. Upper panel: Constructs consist of a sequence of interest, in this example 101 nt long, followed by a 10 nt barcode, in-between two restriction sites (RE1 and RE2, in this example: SpeI and BstBI). Forward and reverse primer binding sites are present to allow amplification of the pool. For libraries with multiple subsets, different primer pairs are used. The shoulder regions allow equal length of all sequences. Lower panel: plasmid used to clone the library pool to the 3′ end of a GFP reporter. In this example, we used XbaI as RE1 and BstBI as RE2 in the plasmid. Note that XbaI and SpeI lead to compatible cohesive restriction ends.
Preparation or ordering of agar plates and LB medium
Timing: 1 day
-
1.Prepare or order from your institute’s relevant unit the following reagents, to be available for the day of the transformation. A detailed protocol of how to prepare LB-Agar plates is available here: https://www.addgene.org/protocols/pouring-lb-agar-plates/. Pre-poured plates can be ordered from several commercial vendors, such as Sigma #L5542 or SSI diagnostica #99112.
-
a.14 cm LB-Agar 1.3% plates with 200 mg/L Ampicillin, or other suitable antibiotic according to the used plasmid (28 plates for 12,000 variants in this example) (Table 1).
-
b.9 cm LB-Agar plates with 200 mg/L Ampicillin, or other suitable antibiotic according to the used plasmid (2 plates for the one pool in this example).
-
c.350 mL LB (∼10 mL/14 cm plate) (Table 1).
-
d.SOC medium (for the 1:10 and 1:100 dilutions).
-
a.
Table 1.
Reaction numbers and sizes according to variants in the library
| Variants in library/number of reactions | PCR reaction | Ligation reaction | electroporation cuvette | 14 cm plate | LB medium (mL) |
|---|---|---|---|---|---|
| 600–2,000 | 5 | 1 | 1–3 | 12 | 130 |
| 2,000–5,000 | 5 | 2 | 3–5 | 20 | 210 |
| 5,000–15,000 | 5 | 2 | 4–6 | 24 | 250 |
| 15,000–55,000 | 16 | 2 | 7 | 28 | 290 |
LB medium and LB-Agar plates can be stored at 4°C for one month.
Note: The number of 14 cm plates used should be adjusted to the size of the pool (Table 1). Larger pools require more plates to ensure keeping the complexity of the library.
Note: The 9 cm plates are used to count the number of colonies and estimate library complexity, as well as to obtain single colonies that would allow to perform a colony PCR assay to verify single insertions of the library to the vector. Consider preparing a 1:10 and a 1:100 dilution for each pool or subset, and prepare or order plates accordingly.
Note: We are aiming to obtain at least 30–40 colonies per construct. Table 1 (below) can serve as a starting point for adjusting the scale of the experiment to the size of the library. Note, that this is according to the library sizes tested in our lab (600–55,000 variants). In theory, we assume that the largest scale reaction in this table (16 PCR reactions, 7 electroporation cuvettes) can support up to 0.5 million variants.
Note: If an oligo pool includes several library subsets, a few of them can be cloned in parallel, using a single tube of bacteria, as long as they do not each contain a very high number of variants. Since it is not possible to freeze leftover bacteria after they are thawed without substantial loss of transformation efficiency, planning such parallel cloning maximizes the output of the bacteria. This is achieved by dividing the bacteria and the 7 cuvettes between the different subsets, according to the ratios between variant numbers.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. cloni 10G CLASSIC electrocompetent cells with recovery medium | Lucigen Corporation | #LC-60117-2 |
| Chemicals, peptides, and recombinant proteins | ||
| dNTPs | Larova | #DMIX10 |
| Water (DNase free) | Sigma-Aldrich | #W4502-1L |
| TAE electrophoresis buffer (x50) | Biological Industries | #01-870-1A |
| Tris-EDTA buffer solution (x100) | Sigma-Aldrich | #T9285-100ML |
| Herculase II Fusion DNA polymerase | Agilent | #600679 |
| FD buffer | Thermo Fisher Scientific | #B64 |
| XbaI | Fermentas | #ER0681 |
| BstBI | NEB | #R0519S |
| BcuI (SpeI) | Thermo Fisher Scientific | #FD1254 |
| FastAP | Thermo Fisher Scientific | #EF0651 |
| GlycoBlue | Thermo Fisher Scientific | #AM9516 |
| FastLink Ligase | Lucigen | #LK0750H |
| Dream Green polymerase | Thermo Fisher Scientific | #EP0702 |
| GelStar dye | Lonza | #50535 |
| Ethidium Bromide | Vita Scientific | #E406-5ML |
| Multi-purpose Agarose | HydraGen | #R9012LE-500g |
| 5× Loading dye | QIAGEN | #239901 |
| Critical commercial assays | ||
| QiaQuick PCR purification column | QIAGEN | #28104 |
| 2% e-gel EX agarose gel | Thermo Fisher Scientific | #G401002 |
| Midi GeBAFlex electroelution tubes | Gene Bio-Application Ltd | #D010-100 |
| Supporting tray for 1–4 Midi GeBAflex tubes | Gene Bio-Application Ltd | #T001 |
| Qubit dsDNA HS assay | Thermo Fisher Scientific | #Q328541 |
| Zymo DNA Clean up and Concentration | Zymo Research | #D4003T |
| Gene Pulser Cuvettes | Bio-Rad Laboratories | #165-2089 |
| Cell lifter | Biologix | #70-2180 |
| NucleoBond Xtra Maxiprep kit | MACHEREY-NAGEL | #740414.10 |
| Oligonucleotides | ||
| Forward primer for pool amplification: ATGGGGTT CGGTATGCGC |
Integrated DNA Technologies | N/A |
| Reverse primer for pool amplification: ATCGTCTC GGGGAGCCTT |
Integrated DNA Technologies | N/A |
| Colony PCR forward: GATCACTCTCGGCATGGA CGAG |
Integrated DNA Technologies | N/A |
| Colony PCR reverse: CTTCCAGGGTCAAGGAAG GCAC |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Oligo library pool | Twist Bioscience | N/A |
| Plasmid | this study (Table S1) | N/A |
| Other | ||
| UV transilluminator | MaestroGen | #MLB-21 |
| PowerPac Basic Power Supply | Bio-Rad Laboratories | #1645050 |
| Owl EasyCast Mini Gel Electrophoresis system (used for electroelution) | Thermo Fisher Scientific | #B1A |
| MicroCL 17R Microcentrifuge | Thermo Fisher Scientific | # 75002499 |
| Heraeus Megafuge 16R | Thermo Fisher Scientific | # 75004230 |
| Nanodrop | Thermo Fisher Scientific | #ND-ONE-W |
| Qubit 3.0 Fluorometer | Life technologies | #Q33216 |
Materials and equipment
Amplification of library pool: We recommend using Herculase II Fusion DNA polymerase, as this enzyme is known to easily amplify low abundance, high GC, and other difficult targets with robust yields.
Alternatives: We assume that other enzymes could also work well, such as Q5 High-Fidelity DNA polymerase (NEB) and Kapa HiFi (Kapa Biosystems).
Centrifuges: for all microtube centrifugations, the MicroCL 17R Microcentrifuge (Thermo Scientific) was used with the 24 × 1.5/2.0 mL Rotor with ClickSeal™ Biocontainment Lid (Thermo Scientific).
The Heraeus Megafuge 16R (Thermo Scientific) was used for centrifugation of 50 mL tubes with the TX-400 4 × 400 mL Swinging Bucket Rotor (Thermo Scientific).
Alternatives: Any centrifuge with similar speed and appropriate tube holder is equally applicable.
Electroporator set up: On BioRad Micropulser electroporator we select the EcI (bacteria) program. This program gives a 1.8 kV pulse. If the ligation reaction is cleaned up properly from salts and there are no air bubbles, the time constant should be over 4 ms. A time constant lower than this value indicates a high level of salt in the solution and makes the electroporation inefficient. See troubleshooting 5 for potential solutions. In this example, we had time constant values of 5.7–5.9 ms.
Electroporation of bacteria: We recommend using E. cloni 10G CLASSIC electrocompetent cells. Chemical competent bacteria give significantly lower yield and are less recommended for cloning of libraries.
Alternatives: please refer to the manufacturer’s instructions to find alternative electro competent cells with similar genotype functions: https://www.lucigen.com/docs/slide-decks/Lucigen-Webinar-Competent-Cells-101-13July2016.pdf.
Step-by-step method details
Amplification of library pool
Timing: 3 h
This step describes the resuspension, amplification, quality control and clean-up of the library pool or library subsets.
Note: Library pools can contain several subsets planned with different primer pairs to allow amplification and downstream applications separately.
-
1.Resuspend the oligo library pool as follows:
-
a.Spin down the tube for 2 min at 18.000 g at 18°C–25°C (MicroCL 17R Microcentrifuge or similar).
-
b.Add 200 μL TE (Tris-EDTA, pH 8) and vortex for 30 s at medium speed.
-
c.Incubate at 65°C for 10 min.
-
d.Divide to aliquots and freeze at −20°C.
-
a.
Note: We use 200 μL TE for libraries ranging between ∼0.4–0.8 pmol (for example, a 212 ng library of 200 nt length). For drastically different library sizes, TE volume should be adjusted. We recommend using 25–50 μL TE for each 0.1 pmol of the library.
-
2.Set up adequate PCR conditions on one pool or subset before amplifying the samples on a large scale.Note: We recommend trying different dilutions and/or cycle numbers, to find the minimal necessary amplification for adequate DNA amount. Over-amplification can lead to the accumulation of mutations and reduce library complexity, while under-amplification leads to inefficient cloning due to low amounts of DNA. Using highly concentrated DNA can lead to unspecific amplification. For example, try 10 and 14 cycles after a 1:10 or 1:50 dilution of the pool for a single reaction, before proceeding with the large-scale amplification.Note: If a subset makes up a smaller part of a pool (e.g. 20%) adjust the dilutions accordingly, in fractions of 10% or smaller it is worth examining the use of an undiluted sample.Note: The forward and reverse primers used here (See the key resources table for the sequence) are not specific to this plasmid or experiment and can be added to any library and used for amplification. It is possible to design and use any other primer, but we recommend testing if the designed primer works efficiently on a single sequence before ordering the library pool. For primer design, we recommend using primer3 (Koressaar and Remm 2007; Untergasser et al., 2012): https://bioinfo.ut.ee/primer3-0.4.0/.Note: See the materials and equipment section for the rationale and alternatives of polymerase selection.
-
a.Prepare PCR reaction mixture for one sample or subset.
Reagent Amount (μL) Buffer (5×) 10 dNTPs (2.5 mM each) 5 Primer Forward (20 uM) 2.5 Primer Reverse (20 uM) 2.5 Herculase polymerase 1 Diluted DNA (e.g., 1:10 or 1:50) 5 DNase free double distilled water 24 Total 50 -
b.Run PCR.
PCR cycling conditions
Steps Temperature Time Cycles Initial Denaturation 95°C 1 min 1 Denaturation 95°C 20 s 10–14 cycles Annealing and extension 68°C 1 min Final extension 68°C 4 min 1 Hold 4°C Forever 1
-
a.
-
3.
Run 5 μL of the PCR product on a 2% agarose e-gel. Select the sample that gives a strong single band of the expected size with the minimum number of cycles and low background (Figure 2A). See troubleshooting 1 for potential solutions if no band is visible at this point.
Note: Throughout the protocol, a regular 2% agarose gel can be used instead of the e-gel. As the e-gel is very sensitive, using a regular agarose gel requires loading 3–6 times higher amounts of DNA than the e-gel.
-
4.Amplify library pool or subsets.
-
a.Re-run PCR with the optimized conditions. For each pool or subset, prepare 5 reactions (or more, according to Table 1) to minimize biases and allow equal amplification of all sequences.
-
b.Pool PCR reactions for each set (5∗50 μL = 250 μL total volume).
-
c.Run 5–20 μL (depending on the strength of the band in the trial PCR) of the 250 μL on a 2% agarose e-gel. Proceed if the band strength and size are as expected (Figure 2B).
-
a.
-
5.Clean up PCR products on QIAQuick PCR purification columns from Qiagen, according to the manufacturer’s instructions: https://www.qiagen.com/cn/resources/download.aspx?id=e0fab087-ea52-4c16-b79f-c224bf760c39&lang=en
-
a.At the end elute DNA in 30 μL DNase free water, then re-elute in another fresh 20 μL (total volume 50 μL) to ensure collecting all of the DNA.
-
b.Measure the concentration on Nanodrop.
-
a.
Note: The expected DNA concentration at this point is 10–50 ng/μL. In this case, we had 42.2 ng/μL.
Pause Point: The digested DNA can be stored at −20°C.
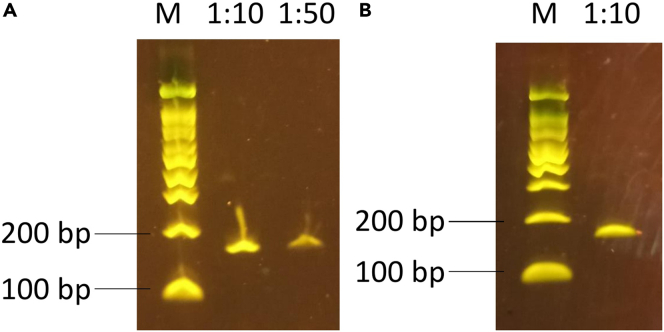
Figure 2.
Amplification of an oligo pool
(A) PCR product with 10-fold or 50-fold dilution of the ssDNA pool and 14 cycles. M: marker. As the 1:10 dilution did not have more background than the 1:50 dilution, it was selected for the large-scale amplification.
(B) PCR product of 5 reactions of oligo pool amplification with a 1:10 dilution. The expected fragment size is 159 nt and there is no background.
Restriction digestion of the library and plasmid
Timing: 1.5 day
This step explains how to perform the restriction digestion and subsequent clean-up of the plasmid and the library pool.
CRUCIAL: Make sure to check the conditions of the enzymes of choice.
-
6.Digest the plasmid and treat with phosphatase to prevent plasmid self-ligation:Note: Digest 2 μg of a ∼4000 nt plasmid. Use more DNA of larger plasmid (e.g. 4 μg for 8000 nt), aiming for ∼0.6–0.7 pmol plasmid.
-
a.Prepare the following mixture (for each sample):
Reagent Amount (μL) Buffer FD (10×) 6 Plasmid DNA (e.g., 2 μg) X Restriction enzyme 1 3 Restriction enzyme 2 3 DNase free double distilled water 48-x Total 60 -
b.Digest for 2 h at appropriate temperature (e.g., 37°C). We recommend using a PCR machine for the restriction digestion steps, as it has a more stable temperature then heat blocks or water baths.
-
c.Add to the reaction:
Reagent Amount (μL) Buffer FD (10×) 3 FastAP Alkaline Phosphatase 3 DDW 24 Total with previous 90 -
d.Incubate at 37°C for 30 min.
-
e.Heat inactivate at 65°C for 20 min.
-
f.Run 5 μL of the cut product and 200 ng of undigested plasmid on an agarose e-gel to check if the plasmid is cut well. See Figure 3A for a scheme of the expected gel pattern of the uncut, single cut, and double cut vectors. If the digestion was inefficient, refer to Troubleshooting 2.
-
g.Purify the digested plasmid on QIAGEN QIAQuick PCR purification columns, according to the manufacturer’s instructions (same as above).
-
h.Elute in 30 μL Dnase free water at the final step.Note: The expected DNA concentration at this point is ∼50 ng/μL (∼70% recovery). Here, we obtained 54.8 ng/μL.
-
a.
-
7.Digest the oligo pool with appropriate restriction enzymes (same as the ones used for digesting the plasmid):
-
a.Dilute the DNA to 10 ng/μL.
-
b.Three parallel reactions were set up to increase the efficiency of restriction enzyme digestion and allow the digestion of a high amount of DNA. Prepare a pool for 3 reactions based on the following (per tube):
Reagent Amount (μL) Buffer FD (10×) 4 DNA (10 ng/μL) 18 Restriction enzyme 1 1 Restriction enzyme 2 1 DDW 16 Total 40 -
c.Digest for 2 h at the temperature required by the enzymes of choice.Note: In this example, we used the restriction enzymes SpeI and BstBI. As they work at different temperatures, we performed the restriction in two steps, first with SpeI at 37°C, then with BstBI at 65°C, for 2 hours each. SpeI was added at time point 0, BstBI was added two hours later. Make sure to check the conditions of the enzymes of choice.
-
d.Heat inactivation according to the requirements of the selected restriction enzymes. Here, we used 20 min of heat inactivation at 80°C.
 Pause Point: The digested DNA can be stored at −20°C.
Pause Point: The digested DNA can be stored at −20°C.
-
a.
-
8.Gel cleanup of the cut fragments:
-
a.Prepare a long (min 10 cm) 2% agarose gel with GelStar dye (1:10000) from Lonza.Note: GelStar is more sensitive than EtBr, hence allows the use of smaller amounts to avoid having any remains of the dye in the sample after cleanup. The gel should be long to allow a very good separation of small bands.
-
b.Mix the samples (120 μL) with 28 μL glycerol (30%) and 2 μL Qiagen 5× loading dye.Note: We want to use as little of the loading buffer as possible, to avoid residual dye in the DNA sample. The additional transparent glycerol ensures that the sample sinks to the bottom of the well after loading.
-
c.Load the gel.Note: In case of having multiple pools or subsets, leave a blank well in between any two samples to avoid cross-contamination.
-
d.Run the gel at constant 125 V in 1× TAE buffer for at least 60 min or until full separation of bands (Figure 3B). If the library digestion is insufficient, see troubleshooting 2.
-
e.Cut the band from the gel under a UV lamp (302 and 365 nm) and place it into a 1.5 mL microcentrifuge tube.Note: Due to the low amount of dye, the bands may be dim and hard to see. Increase UV strength if the bands are not visible, or cut according to the expected band size.Note: Do not expose the gel to the UV more than necessary to avoid DNA damage. Only turn on the UV light for excising the band, and perform the cutting as quickly as possible.
-
a.
-
9.Extract the gel fragments using electroelution with the GeBAflex Midi electroelution tubes:Note: Cleanup with chaotropic salt containing gel extraction kits can potentially create sequence biases (Guo and Zhang 2013; Han et al., 2019; Levo et al., 2015), while the electroelution should allow high yield and shouldn’t be affected by the nucleotide composition.If using an electroelution apparatus is not an option, it is possible to use a chemical elution kit, although some bias may be expected.
-
a.Prepare fresh 1× TAE buffer. Wash a small gel tank with water and fill it with TAE.
-
b.Fill the GeBAflex tube with 800 μL dH2O and incubate at RT for 5 min.
 CRITICAL: Check carefully that no water is leaking from the tube. Leakage may lead to loss of the entire sample.
CRITICAL: Check carefully that no water is leaking from the tube. Leakage may lead to loss of the entire sample. -
c.Carefully remove all the water by shaking or pipetting.
-
d.Insert the gel slice into the tube by flipping the microcentrifuge tube onto the GeBAflex tube.
 CRITICAL: The maximum size of gel slice per GeBAflex tube is 1 cm ∗ 0.5 cm. Don’t fill the tube with several gel slices, for large gel slices use more than one tube.
CRITICAL: The maximum size of gel slice per GeBAflex tube is 1 cm ∗ 0.5 cm. Don’t fill the tube with several gel slices, for large gel slices use more than one tube. -
e.Add 700 μL dH20 and close the tube carefully to avoid bubbles. For larger gel slices use smaller amounts of water.
-
f.Place the GeBAflex tubes directly (without gel casting tray) into a small gel tank (e.g., Owl EasyCast Mini Gel Electrophoresis system or similar).
 CRITICAL: The arrow on the plug of the tube is pointed upwards (to ensure that the current flows through the membrane, Figure 3C).
CRITICAL: The arrow on the plug of the tube is pointed upwards (to ensure that the current flows through the membrane, Figure 3C). CRITICAL: The GeBAflex tubes with the tray need to be fully immersed in TAE. The two membranes of the GeBAflex-tube must be in parallel to the electric field to permit the electric current to pass through the tube (Figure 3C).
CRITICAL: The GeBAflex tubes with the tray need to be fully immersed in TAE. The two membranes of the GeBAflex-tube must be in parallel to the electric field to permit the electric current to pass through the tube (Figure 3C). -
g.Elute DNA for 1 h at constant 125 V, and then for an additional 3 min with reverse current (Bio-Rad PowerPac Basic Power or similar). The short reverse current step detaches the DNA from the dialysis tubing.Note: The time and voltage parameters should be calibrated for each band size, gel percentage and for the thickness of the gel slice, according to the manufacturer’s recommendation: https://www.geba.org/wp-content/uploads/2019/02/GeBAflex-Tube-Electroelution-handbook-Protein-DNA-and-RNA-Extraction.pdf.Note: For reverse current, plug the + (red) electrode into the black socket, and the – (black) electrode in the red one. Change back the set up after the DNA elution to avoid the next user accidentally losing their sample.
-
h.Open the tube carefully to avoid losing any of the liquid.
-
i.Pipette the solution up and down 5 times and transfer it into a clean 1.5 mL tube.
-
j.Centrifuge the tube for 1 min at maximum speed.
-
k.Transfer the solution into a clean 1.5 mL tube. Discard the pellet.
-
l.Measure and record the volume of the liquid with micropipette.
-
a.
-
10.Ethanol precipitation of the DNA
-
a.To each volume × of the liquid, add 0.1× volume of 3M NaOAc, 0.005× of glycogen and of 1× isopropanol. In this example, we added 640 μL of isopropanol, 64 μL of NaOAc and 3.2 μL of glycogen to 640 μL of eluate.
-
b.Invert the tube a few times to mix thoroughly.
-
c.Incubate the sample in −20°C for at least 12 h to ensure sufficient DNA precipitation.
-
d.Prepare 70% ethanol for the next day and store in −20°C.
-
e.Continue the next day:
-
f.Cool a microcentrifuge to 4°C (Thermo Fisher MicroCL 17R Microcentrifuge or similar).
-
g.Centrifuge the sample in a cold centrifuge for 5 min at maximum speed.
-
h.Remove the supernatant without disturbing the pellet using a micropipette.
-
i.Add 1 mL ethanol (pre-chilled at −20°C) 70% ethanol, centrifuge for 5 min, remove liquid.
-
j.Air dry pellet on ice in a chemical hood for 5 min with open tube cap.
-
k.Repeat the washing step (i & j), including the 5 min drying step 2 more times.Note: The washing steps are required in order to get rid of the salts that will damage the ligation reaction.
-
l.Dry the pellet in a chemical hood, on ice for about 20 min. See troubleshooting 3 for potential solutions if no pellet is visible at this point.
-
m.Resuspend the pellet in 13 μL DNase free water. Combine pellets of the same sample into the same tube with this volume (in case the electroelution was done in multiple tubes).
-
n.Measure the concentration with Qubit: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/Qubit_dsDNA_HS_Assay_UG.pdf.Note: The concentration should be ∼1 ng/μL. Here, we had 1.3 ng/μL.
-
a.
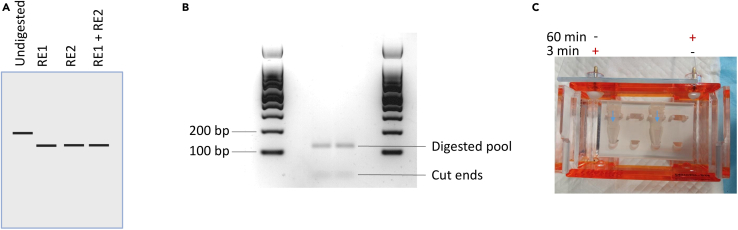
Figure 3.
Restriction digestion and electroelution
(A) Schematic of plasmid digestion. RE = restriction enzyme. The digested bands run faster than the circular plasmid. There is no significant difference in the pattern of the single and double digestions. B: Oligo pool digested with SpeI and BstBI. Cut library has a size of 123 nt.
(C) Electroelution set up. The first 60 min forward current is used, followed by 3 min reverse current. The transparent arrows on the tubes are marked in blue for better visibility.
Ligation and transformation
Timing: 1 day
This step describes the ligation reaction of the pool into the plasmid, followed by clean up and the transformation of electrocompetent bacteria. The transformation is followed by plating the cells on large agar plates for 8–16 h incubation, as well as serial dilutions to estimate the library complexity.
-
11.Preparations:
-
a.Dry the agar plates in a 37°C incubator, for 1–2 h, placing the plate slightly tilted over its lid to dry. In case significant amount of water is present in the plates due to condensed humidity, carefully tap the plates over absorbent paper to dry them before placing in the incubator.
-
b.Take out the recovery medium (supplied with the bacteria) from −80°C to thaw.
-
a.
-
12.Ligation:
-
a.Prepare ligation mix in a PCR tube. For an example of 123 nt insert and 4213 nt plasmid:Note: We are using 150 ng of plasmid DNA, and calculate the amount of insert according to a 1:1 plasmid:insert molar ratio. This minimizes the amount of multiple insertions while maximizing ligation efficiency. With this ratio, we get about 2% of the transformants with multiple inserts.Note: Calculate the required amount (ng) of insert as follows: 150/(size of plasmid/size of insert).Note: For libraries with many variants, we recommend the preparation of two ligation reactions, according to Table 1.
Reagent Amount Buffer (10×) 1 Library DNA 150/(4213/123) = 4.39 ng (3.4 μL of 1.3 ng/μL) Plasmid DNA 150 ng (2.7 μL of 54.8 ng/μL) Ligase Lucigen Fast Link 1 ATP 1 DNase free double distilled water Up to 10 μL (here 0.9 μL) Total 10 μL -
b.Let it stand at 18°C–25°C for 30 min.
-
c.Heat inactivate the ligase at 70°C for 15 min.
-
d.Put ligation on ice for 2 min and proceed directly to clean up.
-
e.Merge the ligations together if you use multiple reactions.
-
a.
-
13.Clean-up of ligation: clean ligation product on Zymo DNA Clean up and Concentration columns according to the manufacturer's instructions: https://files.zymoresearch.com/protocols/_D4003T_D4003_D4004_D4013_D4014_DNA_Clean_Concentrator_-5_ver_1_2_1_LKN-SW__1.pdf. Alternative kits that allow elution in a 15 μL volume can be used as well.Note: 2 μL ligation product is used for each transformation cuvette. Total elution volume should be adjusted to the number of electroporation cuvettes. Here, we used a total 15 μL for 7 cuvettes. (Table 1).
-
a.Elute in 7.5 μL water.
-
b.Repeat the elution step on top with another, fresh 7.5 μL of water. The total volume will be 15 μL.Note: Cleaning the ligation product radically increases transformation efficiency. In case multiple ligation reactions were used, it’s necessary to concentrate the ligation product. Keep the total elution volume at 15 μL even for multiple ligation reactions to get a concentrated product.
-
c.Keep the ligation product on ice throughout the next steps.
-
d.Proceed directly with the transformation.
-
a.
-
14.Transformation of E. cloni cells:
 CRITICAL: For adequate transformation efficiency, all reagents should be kept cold. The electroporation is performed in a cold room (4°C).
CRITICAL: For adequate transformation efficiency, all reagents should be kept cold. The electroporation is performed in a cold room (4°C).-
a.Move pipettes and tips into the cold room at least 30 min before use to cool down. Carry the electroporator to the cold room as well.Note: The number of electroporation reactions per sample should increase with library size (see Table 1)
-
b.Place an adequate number of electroporation cuvettes (0.1 cm gap) and the same number of 1.5 mL tubes on ice.
-
c.Divide 975 μL of the recovery medium to the same number of 15 mL tubes for the recovery.
-
d.Thaw on ice an ampule of E. cloni cells.Note: One vial of E. cloni cells is sufficient for 7 electroporation cuvettes (Table 1).
 CRITICAL: The cells are very sensitive. Do not warm or vortex. For additional notes on handling the bacteria, see troubleshooting 4.
CRITICAL: The cells are very sensitive. Do not warm or vortex. For additional notes on handling the bacteria, see troubleshooting 4. -
e.Divide 23 μL of bacteria to each pre-chilled microcentrifuge tube.
-
f.Add 2 μL of the ligation mixture to each tube.
-
g.Stir briefly with a pipette tip.
 CRITICAL: Don’t pipette up and down to mix. This could introduce air bubbles and warm the cells.
CRITICAL: Don’t pipette up and down to mix. This could introduce air bubbles and warm the cells. -
h.Carefully pipette the entire mix into the pre-chilled cuvettes. Avoid generating air bubbles.
-
i.Electroporate the cells with BioRad Micropulser electroporator using the EcI program (1.8 kV pulse). For the detailed set up of the electroporator, see the materials and equipment section. If the time constant is low, or the machine shows arc error, see troubleshooting 5.Note: The time constant should be over 4 ms. In this experiment, we had time constants in the range of 5.7–5.9 ms. For further notes on the time constant, see the materials and equipment section, or troubleshooting 5.
-
j.As quickly as possible, add the recovery medium to the cuvette. Pipette three times to resuspend the cells and put it back to the 15 mL tube.
-
k.Transfer the tubes in a shaking, 37°C incubator for a 1 h recovery.
-
l.Merge together all the reactions into a 15 mL tube.Note: We prepare 1:10 and 1:100 dilutions and spread it on the 9 cm agar plates, to estimate the total number of colonies and to pick colonies for colony PCR.
-
m.Take out 20 μL and dilute 1:10 with SOC medium. From the 1:10 dilution, dilute 20 μL 1:10, to obtain a 1:100 dilution. Seed 50 μL of both dilution to a small agar plate. (These will be used for colony counting and colony PCR the next day.)
-
n.Spread the remaining bacteria on the calculated number of large agar plates (∼240 μL/plate). Light a Bunsen burner and place on the working station to ensure a sterile environment. Pipette ∼240 μL on a plate and spread it with a bacterial cell spreader, plating beads, or any preferred method. Repeat until the entire liquid volume is used.
-
o.Incubate the plates upside-down for 8–16 h at 37°C.Note: We prepare the DNA directly from large agar plates instead of using a liquid medium. This allows independent growth of every colony, while in liquid medium the bacteria are competing for resources, which could lead to depletion of certain sequences, if the fitness of bacteria transformed with every construct isn’t equal.
-
a.
Colony PCR and plasmid preparation
Timing: 1 day
This step explains how to calculate the number of colonies per construct, analyze the library quality and prepare the DNA for subsequent transfection.
-
15.Estimate the number of colonies per construct:
-
a.Count the colonies on the dilution plates.
-
b.Calculate the number of colonies per construct.
-
a.
Note: Multiply the number of colonies on the 1:100 dilution by 480 (if seeding 240 μL per plate) to get the number of colonies on a large (14 cm) plate (Figure 4B).
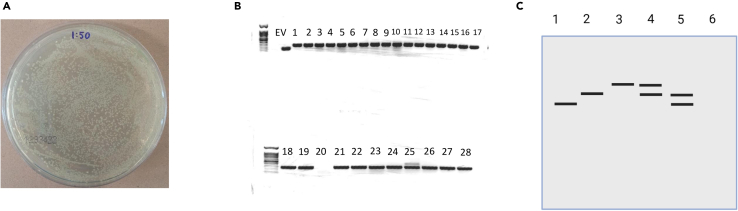
Figure 4.
Counting and quality control of the colonies
(A) Colonies on the 1:50 dilution plate. Approximately 400 colonies in ⅛ of the plate means 3200 colonies on the dilution plate, and ∼18.5 million colonies in total.
(B) Colony PCR of 28 picked colonies. EV is the empty vector control, showing a 154 nt band. For colony #20, the picking or PCR failed (we don’t see the band for the empty vector either). Colony #25 was likely transformed with two plasmid, one with a single insertion, and one with a double insertion. The remaining 26 colonies have correct single integration of 277 nt.
(C) Possible scenarios of band patterns in colony PCR. 1: Empty vector, also used as negative control. 2: Colony with proper single integration. 3: Colony with double integration. 4: Colony that likely received two plasmids, one with a single integration, and one with a double integration. 5: Colony that likely received two plasmids, one with a single integration, and one with an empty vector that closed on itself. 6: Colony picking or PCR failed, no band.
In this example (Figure 4A), we used a 1:10 and 1:50 dilution. While counting and colony picking was possible, for better accuracy and easier handling we recommend 1:10 and 1:100 dilution in this protocol. As we had some additional plates and used a total of 28 plates, the seeded volume per plate in this example is 200 μL.
In this example, we had ∼400 colonies in ⅛ of a 1:50 dilution plate. On the entire 1:50 dilution plate, we had 400∗8 =3200 colonies. On a single 14 cm plate, that gives 3200∗200=640000 colonies, and on the total 29 plates, approximately 18.5 million colonies. 18.5 million colonies for the 12000 constructs gives ∼1500 colonies per construct. If the number of colonies is too low, see troubleshooting 4.
-
16.Maxi prep of library DNA
-
a.Scrape the bacteria off all plates with a cell lifter into an adequate sized bottle, to mix all the bacteria together.
-
b.Divide the bacteria into 50 mL tubes.
-
c.Take out 10% of the bacteria into a separate tube for freezing glycerol stocks.
-
d.Centrifuge the bacteria both for the maxi prep and for the glycerol stock at 5,000×g for 15 min in 4°C (Heraeus Megafuge 16R or similar).
-
e.For the glycerol stocks, resuspend the bacteria in 6 mL LB. Aliquot 1.2 mL of the bacteria into each of 5 freezing tubes, and to each add 0.4 mL 80% glycerol. Vortex and transfer to a cryotube.
-
f.Put directly to −80°C.
-
g.Freeze bacterial pellets for the maxi prep at −20°C.
-
a.
 Pause point: Bacteria pellet for the maxi prep can be stored at −20°C for a few days.
Pause point: Bacteria pellet for the maxi prep can be stored at −20°C for a few days.
-
17.Colony PCR
-
a.Prepare PCR mix for about 30 colonies. Reagents per sample:
Reagent Amount (μL) Dream Green Buffer (10×) 3 dNTPs (2 mM each) 3 Primer Forward 1.2 Primer Reverse 1.2 Dream Enzyme 0.3 DNase free double distilled water 1.3 Total 10 -
b.Divide the mix into PCR strips.
-
c.With a clean pipette tip, pick a colony from the 9 cm agar plate (with the appropriate dilution for single colony picking) and place the pipette into a cell in the PCR strip.
-
d.Continue until all wells have a tip.
-
e.Remove tips.
-
f.Run the PCR.
PCR cycling conditions
Steps Temperature Time Cycles Initial Denaturation 95°C 10 min 1 Denaturation 95°C 30 s 32 cycles Annealing 56°C 30 s Extension 72°C 1 min Final extension 72°C 10 min 1 Hold 10°C Forever -
g.Run samples on a 1% ethidium bromide agarose gel. If the insert size is less than 50 nucleotides, use a 2% agarose gel instead of the 1%, to enable the detection of double integrations appearing as a double band of similar sizes.
-
h.Analyze the ratio of good colonies to empty vectors or multiple insertions. Colonies with no insertion have the same band size as the empty vector control. Colonies with multiple insertions have larger band size. See Figure 4C for a schematic depicting the most likely integration patterns and representative gel patterns. If you have a high ratio of colonies with no insertions, or multiple insertions, see troubleshooting 6.
-
a.
-
18.
Perform DNA maxi prep according to the manufacturer’s instructions: https://www.takarabio.com/resourcedocument/x32861. In this example, the yield of the maxi prep was ∼ 600 μg DNA.
Expected outcomes
We have used this protocol to clone oligo pools in the range of ∼2000 sequences with 100% of the planned barcodes detected (Uzonyi et al., 2021)), up to 55,000 sequences, with over 90% detection rate.
Limitations
It is crucial to perform each step on a large scale and with high efficiency, in order to keep the complexity of the pool. The protocol is sensitive to ineffective or expired reagents and to material loss due to experimenter error. Sequence heterogeneity that affects PCR efficiency or bacterial DNA production can also reduce library complexity. Therefore, the method is not suitable for highly repetitive sequences, or sequences forming strong secondary structures.
Due to the large-scale of certain steps, parts of the protocol are repetitive and work-intensive.
Troubleshooting
Problem 1 (related to point 3)
No band or weak band after library amplification.
Potential solution
Increase the concentration of input DNA (lower dilution) or add more cycles (up to 18 cycles total). Make sure to plan the libraries with previously tested, good primer binding sites.
Problem 2 (related to point 6)
Library or plasmid is not sufficiently digested. See Figure 3A for a schematic of the expected digestion patter.
Potential solution
Increase the amount of restriction enzymes, and/or increase the duration of the restriction reaction. Make sure that the enzyme is not expired, and previously tested to be efficient.
Problem 3 (related to point 10)
No pellet is visible after the ethanol precipitation of the library.
Potential solution
Place the tube to the microcentrifuge with the closed side facing outside, to know the expected location of the pellet. You can try to use stained glycogen (e.g., GlycoBlue) for better visibility.
If the pellet is still not visible, it was likely lost in the electroelution step. Repeat the amplification and restriction digestion, and make sure that the electroelution tubes are not leaking. Open and close the electroelution tubes carefully to avoid material loss and air bubbles.
Problem 4 (related to point 14)
The number of colonies per construct is low.
Potential solution
Make sure to start the ligation with clean plasmid DNA and library DNA. Clean up and concentrate the ligation product. Make sure to handle the bacteria carefully. Thaw them on ice and do not warm or vortex. Perform all steps of the transformation on ice or in a cold room. Do not refreeze vials of bacteria.
Problem 5 (related to point 14)
Machine shows arc error or the time constant is low.
Potential solution
This is likely due to high salt concentration or air bubbles in the solution. Perform a cleanup with extensive wash steps after ligation. Don’t use more than 2 μL of ligation reaction per cuvette. Avoid introducing air bubbles.
The time constant should be over 4 ms.
Problem 6 (related to point 17)
The colony PCR shows a high ratio of empty vectors or plasmids with multiple insertions.
Potential solution
If you have a high ratio of empty vectors, make sure that the digestion of the vector is close to 100%. Always use a pair of restriction enzymes that can’t re-ligate with each other. Make sure that the FastAP enzyme is not expired. The ratio of insert:vector can be mildly increased.
In case of a high ratio of multiple insertions, decrease insert:vector ratio. Instead of the 1:1 ratio recommended in this protocol, try 0.75:1. Re-measure the concentration before repeating the ligation to make sure that the ratio is correct.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Schraga Schwartz (schwartz@weizmann.ac.il).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank Dr. Yael Kalma, Dr. Eilon Sharon, Dr. Shira Weingarten-Gabbay, and Professor Eran Segal, who developed the Agilent oligo pool cloning protocol, for their permission to publish this revised and updated version. We thank Dr. Yoach Rais for the kind donation of the plasmid. This project has received funding from the Israel Science Foundation (543165), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 714023), and the Estate of Emile Mimran. S.S. is the incumbent of the Robert Edward and Roselyn Rich Manson Career Development Chair in Perpetuity.
Author contributions
A.U. tested the protocol and wrote this manuscript with input from all authors. R.N. improved and tested the protocol. S.S. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.101103.
Contributor Information
Anna Uzonyi, Email: anna.uzonyi@weizmann.ac.il.
Schraga Schwartz, Email: schwartz@weizmann.ac.il.
Supplemental information
Data and code availability
This study did not generate/analyze datasets or code.
References
- Buschmann T., Bystrykh L.V. Levenshtein error-correcting barcodes for multiplexed DNA sequencing. BMC Bioinformatics. 2013;14:272. doi: 10.1186/1471-2105-14-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Zhang T. Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl. Microbiol. Biotechnol. 2013;97:4607–4616. doi: 10.1007/s00253-012-4244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Sun J., Lv A., Wang A. Biases from different DNA extraction methods in intestine microbiome research based on 16S rDNA sequencing: a case in the koi carp, Cyprinus Carpio Var. Koi. MicrobiologyOpen. 2019;8:e00626. doi: 10.1002/mbo3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Levo M., Zalckvar E., Sharon E., Machado A.C.D., Kalma Y., Lotam-Pompan M., Weinberger A., Yakhini Z., Rohs R., Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015;25:1018–1029. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M., Nir R., Farouq D., Slutskin I.V., Schwartz S. TRUB1 is the predominant pseudouridine synthase acting on mammalian MRNA via a predictable and conserved code. Genome Res. 2017;27:393–406. doi: 10.1101/gr.207613.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E., Kalma Y., Sharp A., Raveh-Sadka T., Levo M., Zeevi D., Keren L., Yakhini Z., Weinberger A., Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzonyi A., Nir R., Shliefer O., Stern-Ginossar N., Antebi Y., Stelzer Y., Levanon E.Y., Schwartz S. Deciphering the principles of the RNA editing code via large-scale systematic probing. Mol. Cell. 2021 doi: 10.1016/j.molcel.2021.03.024. [DOI] [PubMed] [Google Scholar]
- Vainberg Slutskin I., Weingarten-Gabbay S., Nir R., Weinberger A., Segal E. Unraveling the determinants of microRNA mediated regulation using a massively parallel reporter assay. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-02980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S., Elias-Kirma S., Nir R., Gritsenko A.A., Stern-Ginossar N., Yakhini Z., Weinberger A., Segal E. Comparative genetics. systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351 doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S., Nir R., Lubliner S., Sharon E., Kalma Y., Weinberger A., Segal E. Systematic interrogation of human promoters. Genome Res. 2019;29:171–183. doi: 10.1101/gr.236075.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets or code.