Abstract
Polymyxins B and E1 and gramicidin S are bacterium-derived cationic antimicrobial peptides. The polymyxins were more potent than gramicidin S against Pseudomonas aeruginosa, with MICs of 0.125 to 0.25 and 8 μg/ml, respectively. These peptides differed in their affinities for binding to lipopolysaccharide, but all were able to permeabilize the outer membrane of wild-type P. aeruginosa PAO1 strain H103, suggesting differences in their mechanisms of self-promoted uptake. Gramicidin S caused rapid depolarization of the bacterial cytoplasmic membrane at concentrations at which no killing was observed within 30 min, whereas, conversely, the concentrations of the polymyxins that resulted in rapid killing resulted in minimal depolarization. These data indicate that the depolarization of the cytoplasmic membrane by these peptides did not correlate with bacterial cell lethality.
Gramicidin S and the polymyxins are nonribosomally produced peptides obtained from bacteria. They have achieved widespread usage as topical agents (4). Gramicidin S is a dibasic cyclic decapeptide with a two-stranded antiparallel β-sheet structure with the strands interconnected by two type II β-turns (4, 6). The polymyxins, including polymyxin B and colistin (a mixture of polymyxins E1 and E2), are a family of closely related pentabasic peptide antibiotics containing a cycloheptapeptide ring with a C-8 or C-9 fatty acid attached through an amide bond (17). Both classes of antibiotics are active against many gram-negative and a few gram-positive microorganisms (13, 15). Colymycin M, a derivative of colistin with the positive charges neutralized by methane sulfonate, has been developed as an anti-Pseudomonas aeruginosa prodrug that is being used in aerosol formulations to treat patients with cystic fibrosis (8). Although these antibiotics were discovered more than 50 years ago, their mode of action is still not precisely known. A number of studies have suggested that polymyxins (4, 18), related octapeptins (14), and gramicidin S (6) can act on bacterial cytoplasmic membranes, although many of these studies, especially those performed with the polymyxins and octapeptins, were done at high multiples of the MIC. Polymyxin B was also shown to have pleiotropic effects on respiration, uptake, and efflux of α-methylglucopyranoside and RNA and DNA synthesis (13, 20).
Gramicidin S and the polymyxins share two unique features with the antimicrobial peptides from animals, plants, and insects in that they are polycationic and amphipathic (4, 5). Such characteristics have been thought to contribute to the mechanism of killing of gram-negative bacteria by cationic antimicrobial peptides by promoting the initial interaction with the negatively charged surface molecule lipopolysaccharide (LPS), leading to self-promoted uptake across the outer membrane and subsequently promoting the interaction with and insertion into the negatively charged cytoplasmic membrane of bacteria (4, 20). This can lead to membrane perturbation and probably the translocation of the peptide across the membrane (21). The actual mechanism of action is not yet fully understood but has been proposed variously to involve cell lysis, breakdown of the cytoplasmic membrane barrier, or interaction with a cytoplasmic target such as DNA (4). Evidence arguing against cell lysis or membrane breakdown as the sole mechanism of action has been presented elsewhere (20, 21).
In the killing of gram-negative bacteria, a cationic antimicrobial peptide must interact with both bacterial cell envelope membranes. In the study described in this report, we studied the abilities of bacterium-derived antimicrobial peptides to bind to LPS, to depolarize both the outer and cytoplasmic membranes, to interact with lipid monolayers, and to kill a wild-type P. aeruginosa PAO1 strain, strain H103. To explore the hypothesis that permeabilization of the cytoplasmic membrane is responsible for killing, we monitored cell viability and cytoplasmic membrane permeabilization at the same time. Our data strongly suggest that cytoplasmic membrane depolarization did not correlate well with bacterial cell lethality.
MATERIALS AND METHODS
Strains and reagents.
P. aeruginosa PAO1 strain H103 (3) was grown in Mueller-Hinton (MH) broth (Difco Laboratories, Detroit, Mich.) at 37°C unless otherwise indicated. Polymyxin E1 and colymycin M were provided by Pathogenesis Corp. (Seattle, Wash.), whereas gramicidin S and polymyxin B were purchased from Sigma Chemical Co., (St. Louis, Mo.). The LPS of P. aeruginosa H103 was isolated as described by Moore et al. (11). 1-N-Phenylnaphthylamine (NPN), carbonyl cyanide-m-chlorophenylhydrazone, and Re LPS from Salmonella enterica serovar Minnesota R595 (Re mutant) were purchased from Sigma Chemical Co. Dansyl polymyxin B was synthesized as described previously (11). The lipids 1-pamitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-ethanolamine (POPE), and phosphatidylglycerol from egg yolk (egg-PG) were purchased from Avanti Polar Lipids Inc. (Alabaster, Ala.). The fluorescent dye 3,3-dipropylthiacarbocyanine (diSC35) was purchased from Molecular Probes (Eugene, Oreg.)
MIC assay.
The MICs of the peptides for a range of microorganisms was determined by the modified broth microdilution method in MH medium in polypropylene microtiter plates (19). The MIC was determined as the lowest peptide concentration at which growth was completely inhibited after overnight incubation of the plates at 37°C. MICs were determined three times on different occasions, and the median values are shown. The minimal bactericidal concentration (MBC) was taken as the lowest concentration of peptide that produced less than 10−3 survivors.
Dansyl polymyxin B displacement assay.
The relative binding affinity of each peptide for LPS was determined by the dansyl polymyxin B displacement assay of Moore et al. (11) with LPS isolated from P. aeruginosa H103. Maximal displacement of LPS was expressed as a percentage in which 100% displacement of dansyl polymyxin B was taken as the displacement observed with polymyxin B. IC50 was defined as the concentration that led to half-maximal displacement of dansyl polymyxin B.
Membrane permeabilization assay.
The outer membrane permeabilization activity of the peptide variants was determined by the NPN uptake assay of Loh et al. (9) with intact cells of P. aeruginosa H103. The concentration of peptide that led to a 50% maximal increase in NPN uptake (PC50) was recorded.
The cytoplasmic membrane depolarization activities of the peptides were determined with the membrane potential-sensitive dye diSC35 (16) and P. aeruginosa H103. Bacterial cells in the mid-logarithmic phase were centrifuged, washed in 5 mM HEPES (pH 7.8), and resuspended in the same buffer to an optical density at 600 nm of 0.05. The cells were first treated with 0.2 mM EDTA (pH 8.0) in order to permeabilize the outer membrane to allow dye uptake, and then a stock solution of diSC35 was added to a final concentration of 0.4 μM and quenching was allowed to occur at room temperature for 20 to 30 min. KCl was then added to the cell suspension to a final concentration of 100 mM to equilibrate the cytoplasmic and external K+ concentrations. A 2-ml cell suspension was placed in a 1-cm cuvette, and the desired concentration of the peptide to be tested was added. Changes in fluorescence due to the disruption of the membrane potential gradient (Δψ) across the cytoplasmic membrane were continuously recorded with a Perkin-Elmer model 650-10S spectrofluorimeter at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. At regular intervals, the surviving cells were plated on MH agar plates, and the plates were incubated at 37°C overnight to assess the residual numbers of CFU.
Langmuir monolayer assay.
Lipid monolayers were formed by applying the appropriate lipids dissolved in hexane or chloroform onto water contained in a circular Teflon trough (diameter, 4.5 cm; total volume, 11.5 ml). Monolayers were allowed to equilibrate until a stable surface pressure was obtained (drift in surface pressure [Δπ], <0.2 mN/m). A small port in the side of the trough enabled injection of reagents into the subphase without disruption of the monolayer. The subphase was gently mixed with a magnetic stir bar at 45 rpm. Surface pressure measurements were obtained by using the Whilhelmy plate method (10). The plate was cleaned with methanol three times and was thoroughly rinsed with double-distilled water prior to each surface pressure measurement. The experiments were run at 23°C.
An LPS monolayer film on the air-water interface was obtained by applying S. enterica serovar Minnesota Re LPS (0.5 mg/ml in chloroform-methanol-H2O [17/7/1; vol/vol]) (2) onto buffer alone (5 mM HEPES, 150 mM NaCl) or in the presence of either 2 or 5 mM MgCl2. The monolayer was spread to achieve an initial pressure of 18 ± 1 mN/m and was allowed to stabilize for 5 min before addition of peptide. Peptides were injected to a final concentration of 0.8 μg/ml for polymyxin B, polymyxin E1, and colymycin M and 0.32 μg/ml for gramicidin S into the subphase without disruption of the monolayer.
RESULTS AND DISCUSSIONS
Antimicrobial activity.
The characteristics of the peptides included in this study are listed in Table 1. The two polymyxins were highly active against strain H103, with polymyxin B and E1 MICs of 0.125 and 0.25 μg/ml, respectively (Table 2), whereas gramicidin S and colymycin M were substantially less active, with MICs of 8 and 4 μg/ml, respectively. Direct plating of samples from each well in which bacterial cell growth was inhibited by peptides permitted an assessment of the MBC. For polymycin B, gramicidin S, and colymycin M, the MIC and MBC were the same concentration. For polymyxin E1, only 2 logarithms of killing occurred at the MIC and the MBC was twofold higher than the MIC. Thus, all of these peptides were bactericidal. The MICs for Escherichia coli were found to be within twofold of the MICs for P. aeruginosa reported in Table 2.
TABLE 1.
Properties of antimicrobial peptides included in this study
| Peptide | Sequence and structurea | Charge |
|---|---|---|
| Gramicidin S | Cyclic (LOVPFdLOVPFd) | +2 |
| Polymyxin B | Cyclized isooctanoyl BTBB(BFdLBBT) | +6 |
| Polymyxin E1 | Cyclized isooctanoyl BTBB(BLdLBBT) | +6 |
| Colymycin M | Methane sulphonate derivative of polymyxin E | 0 |
One-letter amino acid code, with the following additions: bold-face indicates residues that are positively charged at neutral pH; parentheses represent amino acids that are cyclized; a superscript d represents the d-enantiomer (all other amino acids are of the l-form); O, ornothine; B, diaminobutyrate.
TABLE 2.
MICs, relative binding of peptides to LPS isolated from membrane of P. aeruginosa H103, and ability to permeabilize the outer membrane of P. aeruginosa H103
| Peptide antibiotic | MIC (μg/ml) | LPS binding (IC50 [μg/ml] for dansyl polymyxin B displacement) | Maximal dansyl polymyxin B displacement (%) | Outer membrane permeabilization to NPN (PC50 [μg/ml]) |
|---|---|---|---|---|
| Polymyxin B | 0.125 | 9.6 | 80 | 0.65 |
| Polymyxin E1 | 0.25 | 12.0 | 72 | 0.76 |
| Gramicidin S | 8.0 | 50 | 70 | 5 |
| Colymycin M | 4.0 | >300a | 0 | >300 |
No significant displacement of dansyl polymyxin B was observed at all concentrations tested.
Ability to bind to LPS and interaction with LPS monolayers.
A number of studies have shown that cationic peptides are taken up by the self-promoted uptake route, for which the initial step involves an electrostatic interaction with Mg2+ binding sites on LPS molecules (11). Thus, the ability of each peptide to bind to P. aeruginosa LPS was determined by the dansyl polymyxin B displacement assay (11). The polymyxins showed relatively high affinities for purified LPS, as judged by their low IC50s, whereas gramicidin S bound to LPS somewhat more weakly and colymycin M showed no significant displacement of dansyl polymyxin B at concentrations up to 300 μg/ml (Table 2).
Interaction of peptides with LPS monolayers.
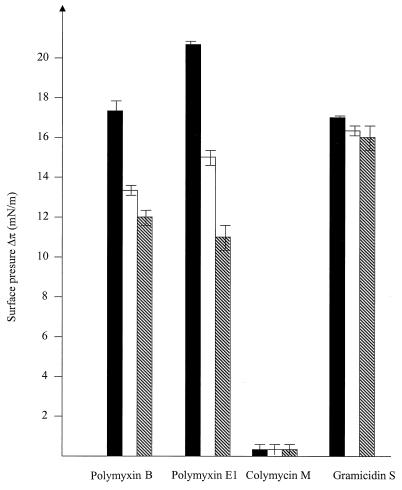
Both polymyxin B and polymyxin E1 induced an increase in surface pressure (Δπ) (Fig. 1), indicating that these peptides penetrated the LPS monolayer. The surface pressure increases mediated by the polymyxins decreased as the external Mg2+ concentration increased, consistent with the ability of divalent cations to competitively inhibit polymyxin binding to LPS (11). Gramicidin S at concentrations above 0.5 μg/ml tended to disrupt the LPS monolayers within seconds. Therefore, the Δπ value was measured with a 0.32-μg/ml concentration. As shown in Fig. 1, it penetrated the LPS monolayers rapidly, resulting in a Δπ of 17 mN/m, which was comparable to that induced by the polymyxins at a concentration of 1 μg/ml. This effect was not significantly reduced by the addition of Mg2+. Colymycin M, on the other hand, showed no interaction with the LPS monolayers (Fig. 1).
FIG. 1.
Influence of peptide addition to the aqueous subphase bathing the LPS monolayers on the surface pressure, as measured in a Langmuir balance. Peptide (0.8 μg/ml [0.32 μg/ml in the case of gramicidin S]) was added to the aqueous subphase bathing the S. enterica serovar Minnesota Re LPS monolayers. The Mg2+ concentration of the subphase was varied from 0 (filled bars) to 2 mM (open bars) to 5 mM (hatched bars). The results shown are the averages of two independent experiments.
Interaction with the outer and cytoplasmic membranes of P. aeruginosa H103.
The ability of each peptide to permeabilize the outer membrane of P. aeruginosa H103 was determined with intact cells by the NPN uptake assay (9). NPN is a small hydrophobic molecule that is excluded by the intact bacterial outer membrane but that exhibits increased fluorescence after partitioning into disrupted outer membranes. Thus, an increase in fluorescence in the presence of a peptide indicates the ability of the peptide to permeabilize the bacterial outer membrane. As shown in Table 1, both polymyxins promoted NPN uptake across the outer membrane of P. aeruginosa H103 to similar extents, with PC50s of about a 0.7 μg/ml. Gramicidin S had a higher PC50 of 5 μg/ml, indicating a weaker ability to permeabilize the outer membrane than polymyxins. Colymycin M, however, mediated NPN uptake only at extremely high concentrations (PC50, >300 μg/ml), and the fluorescence intensity never reached that observed with the polymyxins (data not shown).
Direct and spectrophotometric observation of cells indicated that neither gramicidin S nor the polymyxins were bacteriolytic at modest concentrations above the MBC. Thus, we assessed their abilities to permeabilize the cytoplasmic membrane to ions as a function of time and residual cell viability. A major component of the energy-generating mechanisms of bacteria involves the establishment of a trans-cytoplasmic membrane proton motive force, the largest component of which is an electrical potential gradient, ΔΨ, of −140 mV. This gradient was assessed with a membrane potential-sensitive fluorescent probe, diSC35 (16). Providing it can cross the outer membrane, diSC35 is taken up by bacterial cells according to the magnitude of the electrical potential gradient of the cytoplasmic membrane and becomes concentrated in the cytoplasmic membrane, where it self-quenches its own fluorescence. Any compound that permeabilizes the cytoplasmic membrane and thus depolarizes the ΔΨ will lead to the release of diSC35 and a consequent increase in fluorescence. The diSC35 assay was used previously in conjunction with an outer membrane hyperpermeable mutant of E. coli, mutant DC2, to assess the interactions of cationic peptides with the cytoplasmic membrane (20). We were able to establish this assay with wild-type P. aeruginosa H103 by permeabilizing the outer membrane with 0.2 mM EDTA (pH 8.0). At this concentration, EDTA did not interfere with the diSC35 fluorescence or influence the MICs of the peptides included in this study (data not shown).
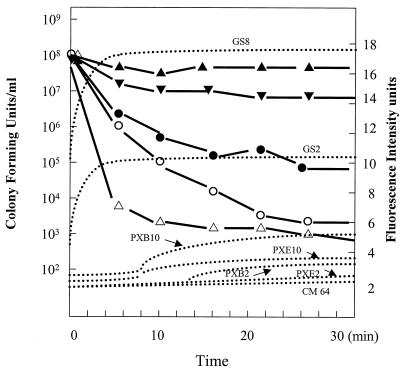
We also assessed cell viability by sampling bacteria at various time intervals during the diSC35 assay and plating for CFU determination to monitor bacterial cell death. Gramicidin S, at its MIC of 8 μg/ml, caused rapid depolarization of the cytoplasmic membrane, resulting in more than 90% maximal release of diSC35 in 2 to 4 min (Fig. 2). However, killing was slow, with only about a 1-logarithm decrease in cell viability in 30 min. In contrast, at their MICs, neither polymyxin B nor polymyxin E1 showed any detectable level of diSC35 release. Polymyxin B at 2 μg/ml (15-fold the MIC) caused only a trace amount of diSC35 release after a lag time of 12 min; however, more than 4 log units of killing was observed in the first 8 min, during which no indication of cytoplasmic membrane depolarization occurred (Fig. 2). Similarly, polymyxin E1 at 2 μg/ml caused about 2 log units of decrease in cell viability, but no release of diSC35 was detected and only a trace amount of diSC35 release was detected at 10 μg/ml, under which conditions more than 3 log units of killing by polymyxin E1 were observed (Fig. 2). Colymycin M showed no cytoplasmic membrane depolarization within 30 min, even at concentrations greater than 64 μg/ml, and no cell killing was observed after 60 min of treatment with colymycin M at 64 μg/ml.
FIG. 2.
Relationship between cytoplasmic membrane permeabilization, as assessed by the diSC35 assay, and cell killing, as measured by determination of the numbers of CFU at the same time as the permeabilization assay. Dotted curves represent data obtained from the diSC35 assay, and solid curves represent data obtained from killing assays. ●, polymyxin E1 at 2 μg/ml; ○, polymyxin E1 at 10 μg/ml; ▾, gramicidin S at 8 μg/ml; ▵, polymyxin B at 2 μg/ml; ▴, colymycin M at 64 μg/ml. Abbreviations: PXB, polymyxin B; PXE1, polymyxin E1; GS, gramicidin S; CM, colymycin M. The number following the abbreviation is the concentration applied (in micrograms per milliliter).
To ensure that these results were not due to the use of EDTA in the modified assay with P. aeruginosa, control experiments were performed with E. coli. Using the same diSC35 assay previously reported for E. coli DC2 (20), we confirmed that gramicidin S at concentrations at or below the MIC caused maximal cytoplasmic membrane depolarization, whereas polymyxins B and E1 at 4-fold the MIC had no effect within 9 min and at concentrations equal to 25-fold the MICs had a minimal effect on cytoplasmic membrane permeabilization (<10% maximal increase in diSC35 fluorescence in 4 min) (data not shown). Colymycin M at 100-fold the MIC had no effect (data not shown). These results suggested that the peptides exerted the same characteristics, in terms of their ability to permeabilize the E. coli cytoplasmic membrane, observed with P. aeruginosa H103 after EDTA treatment, indicating that EDTA has little effect on this particular characteristic. In addition, another cytoplasmic membrane permeabilization assay was used to assess the abilities of the peptides to promote the uptake of the chromogenic substrate ortho-nitrophenyl-β-d-galactopyranoside (ONPG), in which it could be cleaved by the constitutive β-galactosidase of E. coli ML-35. In that assay, gramicidin S at 1.5- to 2-fold the MIC showed maximal unmasking of cytoplasmic β-galactosidase. In contrast, polymyxins B and E1 at 4-fold the MICs had no effect, and even at 200-fold the MICs they caused a minimal permeabilization of the cytoplasmic membrane to ONPG (i.e., the rate of ONPG hydrolysis by cytoplasmic β-galactosidase was 150-fold lower than that caused by 1.5-fold the MIC of gramicidin S).
Interaction with lipid monolayers.
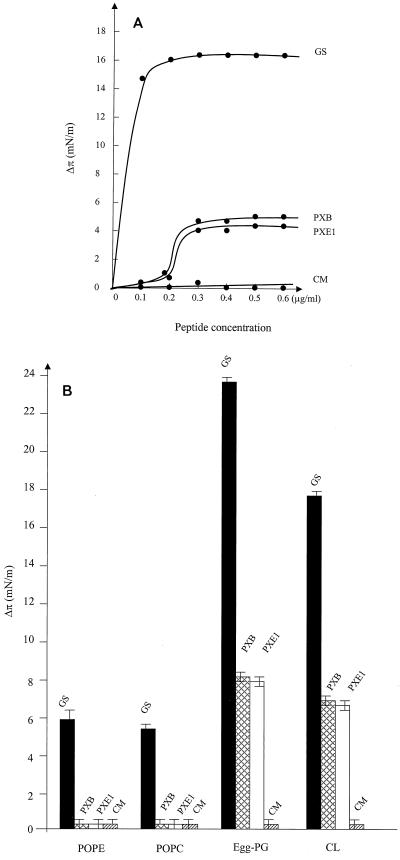
The use of lipid monolayers (10) created at an air-water interface with a Langmuir balance apparatus is a simple means of mimicking biological membranes. Such monolayers provide a powerful tool for the assessment of membrane insertion. A molecule that interacts only with the head groups of a given lipid monolayer will not increase the surface pressure of the monolayer. Thus, when a protein or peptide is injected into the aqueous subphase bathing the monolayer, the corresponding surface pressure change (Δπ) can be interpreted as a result of protein or peptide insertion into the fatty acid chains of the monolayer. To further assess the interactions of these peptides with membranes, we made monolayers with phosphatidylethanolamine (PE)-egg phosphatidylglycerol (PG)-cardiolipin (CL) at a ratio of 78:4.7:14.7 (Avanti Polar Lipids Inc.) to mimic the phospholipid content of the bacterial cytoplasmic membrane and tested the abilities of the peptides to interact with such monolayers (Fig. 3). Gramicidin S caused a large and rapid increase in the monolayer surface pressure even with a single addition of 0.1 μg of peptide per ml into the subphase. The Δπ reached a plateau at about 17 mN/m after the second addition of the same amount of peptide. In contrast, at the same concentration, both polymyxins induced surface pressure increases as a sigmoidal function of the peptide concentration, suggesting cooperativity, and the maximum increase in Δπ was about 5 mN/m for both polymyxin B and polymyxin E1. The use of colymycin M resulted in no detectable change in surface pressure of the same monolayer during the 2-h observation period.
FIG. 3.
Influence of gramicidin S (GS), polymyxin B (PXB), polymyxin E1 (PXE1), or colymycin M (CM) addition to the aqueous subphase on the surface pressure of phospholipid monolayers, as measured in a Langmuir balance. (A) Plot of the surface pressure increase as a function of peptide concentration. Titration of the surface pressure increase was accomplished by adding successive amounts of peptide to the subphase while continuously monitoring the surface pressure of the film. (B) Influence on surface pressure of the addition of 1 μg of a peptide per ml to the aqueous phase of a monolayer made from POPC, POPE, egg-PG, or CL. The results shown are the averages of two independent experiments.
Lipid specificity was monitored by determination of the ability of a peptide to cause maximal surface pressure changes upon injection of 1 μg of peptide per ml into the subphase of pure lipid monolayers. Gramicidin S showed a good ability to insert into both neutral and negatively charged phospholipids, although it seemed to intercalate more efficiently into negatively charged PG or CL monolayers than into neutral phosphatidylcholine (PC) or PE monolayers. Gramicidin S interacted most strongly with PG monolayers. Polymyxins, however, did not cause a surface pressure increase with neutral PC or PE monolayers, indicating their low affinity for neutral lipids, but penetrated to similar extents into negatively charged PG or CL monolayers.
Polymyxins, gramicidin S, and colymycin M are distinctive antimicrobial peptides on the basis of their abilities to interact with phospholipids, LPS, and bacterial cytoplasmic and outer membranes. For example, these peptides displayed somewhat different methods of overcoming the barriers of the outer membranes of gram-negative bacteria. Gramicidin S had a relatively low affinity for binding to isolated LPS but penetrated well into LPS monolayers in a manner that was not strongly inhibitable by Mg2+ ions, and at higher concentrations (0.5 μg/ml) gramicidin S completely disrupted these monolayers. Consistent with its ability to interact with LPS monolayers, gramicidin S at concentrations at about the MIC permeabilized the outer membrane of P. aeruginosa H103. In contrast, polymyxins bound strongly to LPS. Although they penetrated LPS monolayers to a similar extent as gramicidin S, Mg2+ ions were clearly antagonistic, consistent with the antagonism of polymyxin action by Mg2+ ions (9, 11, 12, 13, 15). Hancock and Chapple (4) recently suggested that there are two separate mechanisms of self-promoted uptake: a classical mechanism such as that used by the polymyxins and a variant mechanism involving weak LPS binding and reasonable permeabilization, as is apparently used by gramicidin S (and also, for example, by the bactenecins [19]). Polymyxins, which have a strong positive charge and a hydrophobic acyl chain, have a high binding affinity for LPS molecules and permeabilize the outer membrane by disrupting the negatively charged (surface) head groups through displacement of divalent cations from their binding sites on LPS. In contrast, gramicidin S, which is more weakly positively charged and amphipathic, presumably binds diffusely to the negatively charged surface of the outer membrane and inserts into and passes across the outer membrane by using hydrophobic interactions. By examination of the higher MICs and weaker permeabilizing activity of gramicidin S, the classical mechanism of self-promoted uptake appears to be considerably more efficient. We are unable to explain, on the basis of the current data, how the neutral lipopeptide colymycin M is taken up across either membrane, unless this colistin methane sulfonate acts as a slow-release prodrug for polymyxin E1 and polymyxin E2.
We also examined uptake across the cytoplasmic membrane. As evident from lipid monolayer assays, the polymyxins interacted only with negatively charged lipids, which represent about 20% of the cytoplasmic membrane phospholipids, while gramicidin S interacted well with both neutral and anionic phospholipids. The differences in the lipid affinities of these peptides seemed to correlate well with their abilities to depolarize bacterial cytoplasmic membranes. It has been proposed and is widely cited that the sole target for killing of bacteria by antimicrobial cationic peptides (4, 7), including gramicidin S and the polymyxins (4, 5, 18), is the cytoplasmic membrane. We have previously presented studies that argue against this hypothesis for cationic antimicrobial peptides (20, 21), and the data in this paper extend this to the bacterium-derived antimicrobial peptides. Thus, there was little correlation between membrane permeabilization and bacterial cell death. For example, the concentration required for gramicidin S to cause a 50% maximal release of diSC35 (i.e., to decrease the membrane potential by half) was about 2 μg/ml (one-quarter the MIC). However, at such concentrations, there were no differences in the viabilities of peptide-treated and nontreated cells during a 30-min observation period. In contrast, the use of polymyxin B at 2 μg/ml (16-fold the MIC) resulted in a greater than 4-logarithm decrease in the numbers of CFU within 6 min, but only a 5% maximal release of diSC35 from the cells was detected. This is consistent with data obtained from diSC35 and cytoplasmic β-galactosidase unmasking assays of E. coli cytoplasmic membrane permeability in response to the same peptides. Thus, it appears that polymyxin-mediated cell death takes place prior to cytoplasmic membrane depolarization, whereas for gramicidin S the situation is reversed. We believe that interaction with membranes must be part of the action of such antimicrobial peptides but is not per se the lethal event. This is consistent with the observation that polymyxin B and polymyxin B nonapeptide cause rather similar perturbations of the E. coli cytoplasmic membrane (at high concentrations), even though the latter does not demonstrate antibiotic activity (1). Rather, given the large number of functions of bacteria that are simultaneously inhibited by polymyxins (18) and other cationic antimicrobial peptides (7), we favor a multihit hypothesis, in which there are multiple potential anionic targets, some of which are cytoplasmic.
ACKNOWLEDGMENTS
This work was supported by funding from the Canadian Bacterial Diseases Network and Canadian Cystic Fibrosis Foundation's SPARx program to R.E.W.H., who is also the recipient of Medical Research Council of Canada Distinguished Scientist Award.
REFERENCES
- 1.Dixon R A, Chopra I. Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J Antimicrob Chemother. 1986;18:557–563. doi: 10.1093/jac/18.5.557. [DOI] [PubMed] [Google Scholar]
- 2.Fried V A, Rothfield L I. Interactions between lipopolysaccharide and phosphatidylethanolamine in molecular monolayers. Biochim Biophys Acta. 1978;514:69–82. doi: 10.1016/0005-2736(78)90077-9. [DOI] [PubMed] [Google Scholar]
- 3.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock R E W, Chapple D S. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izumiya N, Kato T, Aoyaga H, Waki M, Kondo M. Synthetic aspects of biologically active cyclic peptides: gramicidin S and tyrocidines. New York, N.Y: Helsted Press; 1979. pp. 49–89. [Google Scholar]
- 6.Katsu T, Kobayashi H, Hirota T, Fujita Y, Sato K, Nagai U. Structure-activity relationship of gramicidin S analogues on membrane permeability. Biochim Biophys Acta. 1987;899:159–170. doi: 10.1016/0005-2736(87)90396-8. [DOI] [PubMed] [Google Scholar]
- 7.Lehrer R I, Barton A, Daher K A, Harwig S S, Ganz T, Selsted M E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Investig. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Littlewood J M, Miller M G, Ghoneim A T, Ramsden C H. Nebulised colymycin for early Pseudomonas colonisation in cystic fibrosis. Lancet. 1985;i:865. doi: 10.1016/s0140-6736(85)92222-6. [DOI] [PubMed] [Google Scholar]
- 9.Loh B, Grant C, Hancock R E W. Use of the fluorescent probe 1-N- phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer L D, Nelsestuen G L, Brockman H L. Prothrombin association with phospholipid monolayers. Biochemistry. 1983;22:316–321. doi: 10.1021/bi00271a013. [DOI] [PubMed] [Google Scholar]
- 11.Moore R A, Bates N C, Hancock R E W. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob Agents Chemother. 1986;29:496–500. doi: 10.1128/aac.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima K, Kawamata J. Structure-activity relationship of colistins. Chem Pharm Bull. 1967;15:1219–1224. doi: 10.1248/cpb.15.1219. [DOI] [PubMed] [Google Scholar]
- 13.Newton B A. The properties and mode of action of the polymyxins. Bacteriol Rev. 1956;20:14–27. doi: 10.1128/br.20.1.14-27.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal K S, Ferguson R A, Storm D R. Mechanism of action of EM49, membrane-active peptide antibiotic. Antimicrob Agents Chemother. 1977;12:665–672. doi: 10.1128/aac.12.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schinder P R G, Teuber M. Action of polymyxin B on bacterial membranes: morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli B. Antimicrob Agents Chemother. 1975;8:95–104. doi: 10.1128/aac.8.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims P J, Waggoner A S, Wang C-H, Hoffman J F. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidyl choline vesicles. Biochemistry. 1974;13:3315–3330. doi: 10.1021/bi00713a022. [DOI] [PubMed] [Google Scholar]
- 17.Storm D R, Rosental K S, Swanson P E. Polymyxin and related peptide antibiotics. Annu Rev Biochem. 1977;46:723–763. doi: 10.1146/annurev.bi.46.070177.003451. [DOI] [PubMed] [Google Scholar]
- 18.Teuber M. Action of polymyxin B on bacterial membranes. III. Differential inhibition of cellular functions in Salmonella typhimurium. Arch Microbiol. 1974;100:131–144. [Google Scholar]
- 19.Wu M, Hancock R E W. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J Biol Chem. 1999;274:29–35. doi: 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- 20.Wu M, Maier E, Benz R, Hancock R E W. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Benz R, Hancock R E W. Influence of proline residues on the antimicrobial and synergistic activities of α-helical peptides. Biochemistry. 1999;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]