Abstract
The glycoprotein alpha-1-antichymotrypsin (AACT), a serine protease inhibitor, is mainly synthesized in the liver and then secreted into the blood and is involved in the acute phase response, inflammation, and proteolysis. The dysregulation of AACT and its glycosylation levels are associated with tumor progression and recurrence, and could be used as a biomarker for tumor monitoring. In this review, we summarized the expression level, glycosylation modification, and biological characteristics of AACT during inflammation, neurodegenerative or other elderly diseases, and tumorigenesis, as well as, focused on the biological roles of AACT in cancer. The aberrant expression of AACT in cancer might be due to genetic alterations and/or immune by bioinformatics analysis. Moreover, AACT may serve as a diagnostic or prognostic biomarker or therapeutic target in tumors. Furthermore, we found that the expression of AACT was associated with the overall survival of patients with human cancers. Decreased AACT expression was associated with poor survival in patients with liver cancer, increased AACT expression was associated with shorter survival in patients with pancreatic cancer, and decreased AACT expression was associated with shorter survival in patients with early lung cancer. The review confirmed the key roles of AACT in tumorigenesis, suggesting that the glycoprotein AACT may serve as a biomarker for tumor diagnosis and prognosis, and could be a potential therapeutic target for human diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-022-02572-4.
Keywords: Alpha-1-antichymotrypsin, Inflammation, Cancer, Biomarker, Diagnosis, Prognosis, Therapeutic target
Introduction
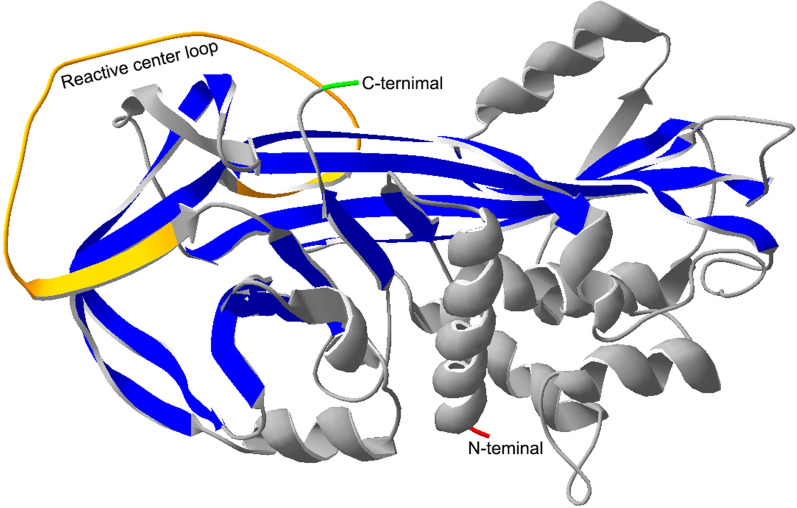
Alpha-1-antichymotrypsin (AACT), namely serpin family A member 3 (SERPINA3), is encoded by the serpin A3 gene and has 423 amino acids with a molecular weight of 47.651 kDa. We have remodeled the structure of AACT with the SWISS-MODEL WORKSPACE (http://swissmodel.expasy.org) [1–3] and SPDBV software (http://swissmodel.expasy.org/), which is based on Protein Data Bank files (PDB ID: 1qmn.1 A) and has 99.75% identity with 1qmn.1 A. The 3D structure of AACT is shown in Fig. 1. The protein structure of AACT consists of an α -helix, β -folded sheets and a reaction center loop (RCL) [4]. AACT is mainly synthesized in the liver and then secreted into the blood. Some reports have shown that AACT is also expressed in other organs, such as the brain and aorta [5, 6], and is also secreted in astrocytes [7]. As a serine protease inhibitor, AACT can inhibit neutrophil cathepsin G (CTSG) and mast cell chymase and protects cells or tissues from damage caused by proteolysis after inflammation [8], which is essential to maintain intracellular homeostasis and extracellular matrix reconstruction. The function of AACT is mainly involved in the acute phase response [9], inflammation [10], and proteolysis [11] and could be used as a biomarker for the diagnosis and prognosis of diseases [12], including liver cancer [13], pancreatic cancer [14, 15], lung cancer [16, 17], ovarian cancer [18], and diffuse large B-cell lymphoma [19].
Fig. 1.
The 3D structure of AACT. The color red represents the N-terminal amino acid, green represents the C-terminal amino acid, yellow represents the reactive center loop, gray represents α-helixes and blue represents β-fold sheets. The amino acid sequences were derived from UniProt (https://www.uniprot.org/uniprot/P01011), and the 3D structure was constructed by the SWISS-MODEL WORKSPACE (http://swissmodel.expasy.org)
Accumulating evidence has demonstrated a crucial role of protein glycosylation in biological processes, and abnormal glycosylation is tightly associated with tumorigenesis. The glycan sites, different types of glycosylation and complex sugar chain structure of AACT affect its structure and biological functions [20, 21] and are involved in the occurrence and development of diseases; for example, the changes in glycoform profiles in AACT reflected the physiological state of patients with sepsis [22]. Data from the UniProt and GeneCards databases show that AACT has six N-linked N-acetylglucosamine (GlcNAc) modification sites [23] (33, 93, 106, 127, 186 and 271), but their N-glycan structures are not clear. Santamaria et al. reported that the N-linked glycosylation modification of AACT in the nucleus has less complex form than secreted AACT, and perhaps the protease inhibitory activity of AACT helps some proteins escape the degradation of protease and transport into the nucleus [24].
However, the expression changes, glycan modification, and biological functions of AACT remain unclear during disease development. In our latest reports, we found that AACT is significantly decreased in the sera of patients with stage I NSCLC and reported that GlcNAc-modified AACT could be a novel biomarker for the early diagnosis of NSCLC [16, 17]. In this review, we have mainly focused on the differential expression and/or modifications of ACCT in disease to better understand its biological functions, especially clarifying the biological significance of AACT as a novel biomarker for diagnosis, prognosis, and therapy prediction in cancer.
The biological significance of AACT in inflammatory diseases
AACT is also an acute-phase protein (APP) whose expression is changed during the inflammatory response and is involved in monitoring inflammatory diseases [25, 26]. In the cerebrospinal fluid (CSF) of patients with fibromyalgia, the expression of AACT is decreased and is involved in the inflammatory response and signal transduction [5]. Kroksveen et al. identified differentially expressed proteins using cerebrospinal fluid from the brain by iTRAQ labeling and proteomics and found that AACT could be used as a diagnostic or prognostic indicator for multiple sclerosis [27]. In heart tissue, the expression of AACT is directly regulated by miR-37, and elevated AACT and miR-37 play an important role in the pathology of patients with chronic heart failure (HF), thereby providing a potential therapeutic target [28]. Brioschi et al. reported that AACT with three other abundant plasma proteins in patients with HF, namely, neuropilin-2, beta 2 microglobulin, and complement component C9, could discriminate patients with HF from healthy subjects with sufficient accuracy [29] and could be used as a diagnostic biomarker. AACT is significantly increased in patients with aortic stenosis, which is important in the pathophysiology of this disease, and AACT may serve as a biomarker for the early diagnosis of aortic stenosis [6]. The level of AACT in serum is generally increased in patients with active tuberculosis [30]. Sun et al. screened new plasma markers to distinguish pulmonary tuberculosis from underlying infection with quantitative proteomics technology, and found that the combination of AACT, α-1-acid glycoprotein (alpha-1-acid glycoprotein 1, AGP1) and cadherin (E-cadherin, CDH1) could distinguish patients with pulmonary tuberculosis and underlying infection, with a sensitivity of 81.2% and specificity of 95.02% [26]. Therefore, AACT can be used as a biomarker for inflammatory diseases.
In addition, the glycosylation level of AACT is also changed in inflammatory diseases. For example, the glycosylation profile of AACT could be used as an indicator of physiological inflammatory status [31]. The glycan structure of the APP is dynamically modified by circulating glycosidase or glycosyltransferase in the inflammatory environment. AACT is highly glycosylated in acute inflammatory reactions, and the glycan form of fucosylated modification of SLex was significantly increased [32, 33]. In septic episodes, the alteration of AACT glycoforms such as increased fucosylation and additional branching of N-glycans could be used to monitor sepsis progression, and the individual AACT glycoproteoform profiles are unique, which may be linked with physiological states [22].
The role of AACT in neurodegenerative or related elderly diseases
AACT promotes the secretion of cytokines, including IL-6 and tumor necrosis factor-alpha (TNF-α), by regulating the NF-κB signaling pathway to be involved in nervous system diseases [34]. We analyzed the proteins that interacted with AACT by STRING, which found that AACT interacted with IL-6 (Fig. 2). AACT is a major component of nerve fiber nodules, which play an important role in the pathogenesis of Alzheimer’s disease (AD). The protein expression of AACT is higher in the CSF and serum of patients with AD [5], Aβ amyloid plaque deposits with increasing AACT concentration, and AACT can activate c-Jun N-terminal kinase, further mediating the hyperphosphorylation of tau protein and leading to neurofibrillary tangles [35]. Braghin et al. found that AACT induced mRNA expression and released TNF-α in murine microglial cells, promoting NF-κB translocation into the nucleus in AD pathogenesis, which suggested that inhibition of AACT could be useful for the treatment of AD [36]. Moreover, Chen et al. reported that the N-linked glycosylation pattern of alpha-1-antichymotrypsin was altered in AD, which could contribute to elucidating the role of glycosylation in AD progression [37]. Lanni et al. found that the sialic acid levels of AACT were significantly reduced in AD [38]. For other diseases of aging, Liu et al. reported that AACT may serve as a biomarker for diagnosing amnestic mild cognitive impairment (aMCI) [39]. AACT combined with other proteins can be used as predictor of the five-year risk of death in older men through high-throughput serum proteomics identification [40].
Fig. 2.

The protein-protein interaction with AACT. The data were analyzed by STRING (https://string-db.org). DNAJC1: DnaJ homolog subfamily C member 1; KLK3: prostate-specific antigen; A2M: Alpha-2-macroglobulin; GIG25: Serpin peptidase inhibitor; HP: Haptoglobin; CTSG: Cathepsin G; IL6: Interleukin-6; PMVK: Phosphomevalonate kinase; MVK: Mevalonate kinase; MVD: Diphosphomevalonate decarboxylase
The biological significance of AACT in human cancer
The abnormal expression and glycosylation level of AACT in tumors
AACT exhibits low specificity in cancers, and the protein or glycosylation level of AACT is aberrantly expressed in various tumors, and associated with survival and tumor progression, including ovarian cancer [18], colorectal cancer [41], melanoma [42], and endometrial cancer [43], as well as can be used as a biomarker for cancer monitoring. Weiz et al. reported that biantennary and tetraantennary glycoforms containing Lewisx structures in AACT were increased in epithelial ovarian cancer, while the β (1–4) branch and triantennary-branched structures were reduced [18]. The glycoforms containing SLex of AACT and other APPs, such as AGP and haptoglobin, have been elevated in ovarian cancer [44], and may be candidates for monitoring breast cancer progression [45]. In prostate cancer, prostatic specific antigen (PSA) is a useful prostate cancer marker that is complexed with AACT, as shown in Fig. 2, and the PSA-AACT levels were increased in moderately differentiated prostate cancer tissues [46]. Julia et al. also found that the combination of AACT and PSA significantly increased the diagnostic rate of prostate cancer compared to PSA alone, with an AUC value of 0.71 [47].
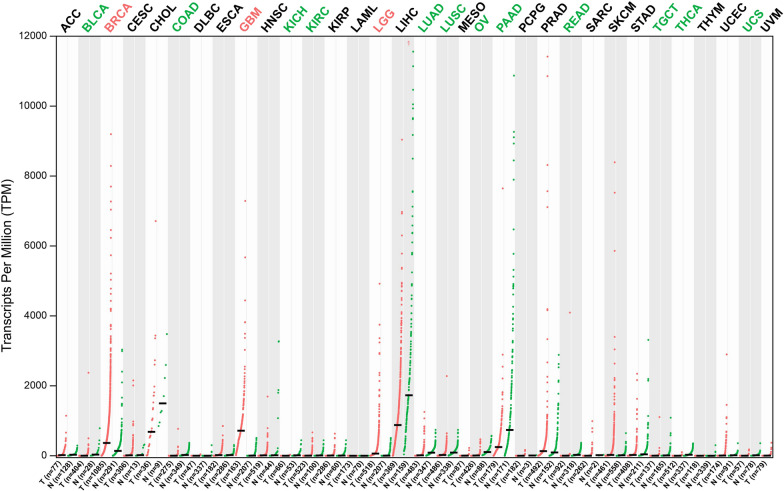
We also used the GEPIA database to test the mRNA expression levels of AACT in different tumor tissues and found that the expression of AACT was significantly differentially expressed in multiple cancer types compared to their normal counterparts (Fig. 3). These cancer types included invasive breast carcinoma (BRCA), glioblastoma multiforme (GBM), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), etc.
Fig. 3.
The gene expression profile across all tumor samples (T) and paired normal tissues (N) (dot plot). Each dot represents the AACT mRNA expression level of the samples. Data were downloaded from the GEPIA database (http://gepia.cancer-pku.cn/detail.php?gene=SERPINA3). ACC: Adrenocortical carcinoma; BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: Cholangio carcinoma; COAD: Colon adenocarcinoma; DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: Esophageal carcinoma; GBM: Glioblastoma multiforme; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LAML: Acute myeloid leukemia; LGG: Brain lower grade glioma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; MESO: Mesothelioma; OV: Ovarian serous cystadenocarcinoma; PAAD: Pancreatic adenocarcinoma; PCPG: Pheochromocytoma and paraganglioma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; SARC: Sarcoma; SKCM: Skin cutaneous melanoma; STAD: Stomach adenocarcinoma; TGCT: Testicular germ cell tumors; THCA: Thyroid carcinoma; THYM: Thymoma; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma; UVM: Uveal melanoma
Why is AACT abnormally expressed in cancer?
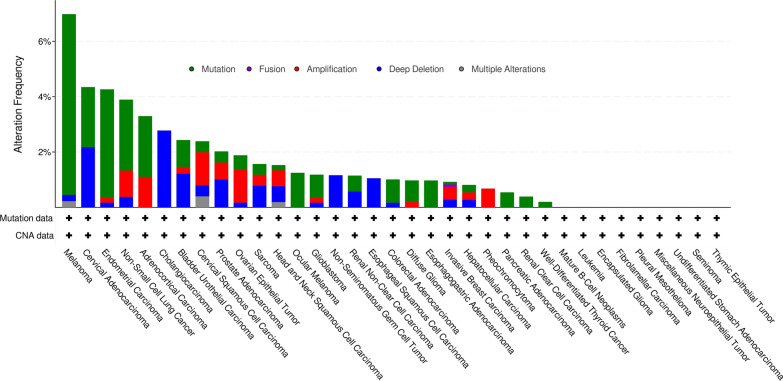
Aberrant AACT expression might be induced through gene mutation, deep deletion, fusion, amplification or multiple alterations in tumors. We performed a statistical analysis of the DNA alteration frequency of AACT in various cancer types by using the cBioPortal database (http://www.cbioportal.org/) (Fig. 4) [48]. Interestingly, we recorded 6.94%, 3.89%, and 0.81% genetic alteration frequencies of AACT in 444 melanoma cases, 1053 NSCLC cases, and 389 liver cancer cases, respectively. The changes in the expression levels of AACT may be regulated by different signaling pathways or immune microenvironments, such as immunosuppression in glioma [49, 50].
Fig. 4.
The gene alterations of AACT in tumors. Summary of the gene alterations for AACT in various cancers. Data were downloaded from cBioPortal (http://www.cbioportal.org/)
Furthermore, we also used the COSMIC database to analyze the mutation distribution of AACT in different tissues (Table 1), which indicated that the mutation rate of AACT was different in tissues and diseases. For example, 3.48% of cases were found to exhibit point mutations in lung adenocarcinoma, and in bladder carcinoma, the point mutation, copy number variation, and hypomethylation were 1.44%, 0.5% and 10.62% of the cases, respectively.
Table 1.
The distribution of mutations across the primary tissue types that are curated by COSMIC database
| Tissue/sub-tissue | Histology | Sub histology | Point mutations (%) | Copy number variation (%) | Hypomethylation (%) | |
|---|---|---|---|---|---|---|
| Gain | Loss | |||||
| Bile duct/gallbladder | Carcinoma | Small cell carcinoma | 5.88 (n = 17) | / | / | / |
| Bile duct/bile duct | Carcinoma | Adenocarcinoma | 3.5 (n = 457) | / | / | / |
| Breast | Carcinoma | / | 0.14 (n = 1429) |
0.18 (n = 1131) |
0.35 (n = 1131) |
/ |
| Central nervous system/cerebellum | Glioma | Astrocytoma Grade IV |
7.14 (n = 14) |
/ | / | / |
| Cervix | Carcinoma | Squamous cell carcinoma |
2.83 (n = 318) |
/ | 0.33 (n = 299) | / |
| Endometrium | Carcinoma | Endometrioid carcinoma |
3.61 (n = 581) |
/ | 0.19 (n = 530) | / |
| Haematopoietic and lymphoid | Lymphoid neoplasm | Follicular lymphoma | 2.33 (n = 43) | / | / | / |
| Kidney | Carcinoma | Papillary renal cell carcinoma | 0.59 (n = 340) | / | 1.05 (n = 286) | / |
| Liver | Carcinoma | Hepatocellular carcinoma | 0.69 (n = 1012) | 0.15 (n = 660) | 0.15 (n = 660) | / |
| Lung | Carcinoma | Adenocarcinoma | 3.48 (n = 1035) | 0.53 (n = 375) | 0.27 (n = 375) | / |
| Lung | Carcinoma | Squamous cell carcinoma | 2.62 (n = 762) | 0.4 (n = 500) | / | / |
| Ovary | Carcinoma | Serous carcinoma | 0.59 (n = 673) | 0.18 (n = 568) | 0.18 (n = 568) | / |
| Oesophagus | Carcinoma | Adenocarcinoma | 0.64 (n = 467) | |||
| Pancreas | Carcinoma | Ductal carcinoma | 0.7 (n = 1281) | |||
| Prostate | Carcinoma | Adenocarcinoma | 0.48 (n = 1462) | / | / | / |
| Skin | Malignant melanoma | / | 8.35 (n = 826) | |||
| Stomach | Carcinoma | Adenocarcinoma | 2.35 (n = 596) | / | / | / |
| Thyroid | / | neoplasm | 6.67 (n = 15) | / | / | / |
| Upper aerodigestive tract/ head neck | Carcinoma | Squamous cell carcinoma | 0.8 (n = 519) | 0.58 (n = 626) | / | 0.6 (n = 496) |
| Urinary tract/Bladder | Carcinoma | / | 1.44 (n = 555) | 0.5 (n = 399) | / | 10.62 (n = 273) |
The data was derived from the COSMIC database (https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=SERPINA3_ENST00000393078), and there are 40,656 unique samples were used in this database, of which 506 unique samples with mutations were distributed in 40 types of tissues. n represents the number of samples
In addition, the abnormal expression of AACT in cancer may be associated with immunity. The immune microenvironment and immune cells are closely related to cancer occurrence, and most tumor-infiltrating immune cells are imbalanced in the development of tumors [51]. We analyzed the correlation between AACT expression and immune cells, including NK cells, CD8+ T cells, macrophages and neutrophils, in 40 types of tumors by Timer 2.0 software. Different immune invasion assessment algorithms have different results, such as the ESTIMATE algorithm [52], XCELL (http://xcell.ucsf.edu/), MCP-counter (http://github.com/ebecht/MCPcounter), and CIBERSORT (https://cibersort.stanford.edu/index.php), and the results are shown in Fig. 5. For example, in cervical squamous cell carcinoma (CESC), AACT has a positive correlation with neutrophils and a negative correlation with CD8+ T cells with the XCELL algorithm. However, the results showed that AACT had no significant correlation with CD8+ T cells and a positive correlation with NK cells, macrophages and neutrophils with the MCP-counter algorithm. However, the detailed relationship between AACT expression and immunity in tumors needs to be further elucidated by experiments.
Fig. 5.
The correlation between AACT expression and immune infiltration in tumors. The correlation between AACT expression and immune infiltration in 40 types of tumors was analyzed by Timer 2.0 software (http://timer.cistrome.org/). The NK cells, neutrophils, macrophages and CD8 + T cells were selected to analyze the correlation with AACT expression in tumors by Spearman’s test. Red represents a significantly positive correlation (p < 0.05), and blue represents a significantly negative correlation (p < 0.05). The depth of color represents the value of the correlation coefficient
As above, it may be the reason to clarify the abnormal expression of AACT in cancer. However, the molecular mechanisms of AACT dysregulation in every tumor still need to be investigated further.
The biological roles of AACT in cancer
AACT as a prognostic biomarker or therapeutic target in glioma
AACT could be a candidate biomarker for the early diagnosis of glioblastoma, which was identified using quantitative proteomics detection [53]. Furthermore, Lara-Velazquez et al. reported that AACT overexpression led to an increase in cell migration, while silencing of AACT decreased cell migration in glioblastoma and increased the survival of mice, which is positively correlated with glioma grade and negatively correlated with the prognosis of patients with GBM [54, 55]. AACT expression regulates the MAPK signaling pathway and matrix metalloproteinase expression, and AACT may serve as a potential immunomodulator and therapeutic target in GBM [49]. Yuan et al. found that AACT expression was involved in immune suppression in glioma, and higher AACT expression was associated with lower CD4+ T cell levels, which indicated that AACT played a key role in the occurrence of glioma [50].
AACT as a diagnostic biomarker or therapeutic target in hepatocellular carcinoma
Alterations in core fucosylation, increased sialylation and glycan branching have been identified in the serum of patients with hepatocellular carcinoma (HCC), and are involved in tumor development [56]. Both the expression levels of AACT and its glycosylation levels were abnormal in the sera of patients with HCC [57]. Ji et al. applied hydrophilic interaction liquid chromatography (HILIC) to enrich N-linked glycopeptides from plasma in patients with liver cancer and then analyzed them by nano-LC/ESI-MS/MS. The N-linked glycopeptide modification sites N106 and N271 of AACT were detected by collision-induced dissociation (CID) and higher energy C trap dissociation (HCD) mass spectrometry, and further identified that the fucosylation modification of N-linked glycopeptides in AACT was mainly tri- and/or tetra-antennary glycan structure [58]. The aleuria aurantia lectin (AAL) was used for enriching the plasma from liver cancer and combined with multiple reaction monitoring (MRM) quantitative proteomic analysis, which found that AACT captured by AAL could significantly distinguish liver cancer from healthy samples, with an AUC of 0.927 and 90.0% sensitivity at a specificity of 83.3%, which suggested that AACT could be a potent biomarker for patients with HCC [13].
Santamaria et al. reported that the protein level of AACT is normally expressed in healthy noncancerous liver tissue and decreased in tissues and cells of liver cancer. The decreased expression of AACT was associated with liver cancer development, while AACT overexpression inhibited the proliferation of liver cancer cells. In vitro studies indicated that AACT inhibited DNA synthesis by inhibiting DNA polymerase activity, and found that AACT was located in the nucleus through binding tightly with chromatin, which promoted chromatin to be in a condensed state and prevented cell proliferation [24]. Zhu et al. found that AACT acts as a tumor suppressor in liver cancer and inhibits the PI3K/AKT/mTOR pathway by activating PTEN, thus inhibiting the development and metastasis of liver cancer. We analyzed the mRNA expression levels of AACT and found that AACT expression was associated with the overall survival of liver cancer in 333 patients, and decreased expression of AACT was associated with poorer survival of patients with liver cancer (Fig. 6A). These reports suggest that AACT can also be used as a target for treatment intervention in liver cancer [59].
Fig. 6.
Analysis of the association between mRNA expression level of AACT and overall survival in tumors. A, B The fragment per kilobase of exon model per million mapped reads (FPKM) data of AACT mRNA expression levels were downloaded from the Human Protein Atlas database (https://www.proteinatlas.org/ENSG00000196136-SERPINA3) (Additional file 1), and overall survival analysis was performed using the Kaplan–Meier method, followed by the log-rank test with SPSS 24.0 software. The FPKM cutoff of AACT mRNA expression in liver cancer was 1.41 (A) and in pancreatic cancer was 0.9 (B). C, D The overall survival analysis was performed using the Kaplan–Meier Plotter database (http://www.kmplot.com/analysis/index.php?p=service&cancer=lung). The parameter settings: Affy ID: 232376_at; Split patients by: Auto select best cutoff; Survival: OS; History: all; grade: all; Gender: all; Smoking history: all; Cox regression: univariate. C. In early-stage lung cancer. Stage: 1; AJCC stage T: 1; AJCC Stage N: 0; AJCC stage M: 0. D. In lung cancer. Stage: all; AJCC stage T: all; AJCC Stage N: all; AJCC stage M: all
AACT as a diagnostic or prognostic biomarker in pancreatic cancer
Pancreatic cancer is an intractable malignancy with a 5-year survival rate lower than 5% [60]. AACT could be used as a biomarker for the diagnosis of pancreatic cancer, and the combination of AACT, thrombospondin-1 (THBS1) and peptides containing single amino acid variants (SAAVs) has shown an improved diagnostic performance in the identification of pancreatic cancer from healthy controls, with an AUC of 0.98, which was demonstrated to improve the survival rate if diagnosed at an early stage [15]. Moreover, Nie et al. also used AAL to extract serum fucosylated glycoproteins with quantitative proteomics, and found that the combination of AACT, THBS1, and haptoglobin (HPT) could differentiate patients with pancreatic cancer from healthy controls, with an AUC of 0.95 [14]. AACT, vimentin, β-catenin, and α-1 antitrypsin can be used as new markers for the diagnosis of solid serous cystadenoma (SCA) of the pancreas, which is a rare type of pancreatic solid tumor [61]. AACT and other markers were used to identify extraordinarily rare pancreatic-type acinar cell carcinoma in the stomach [62].
In addition, Roberts et al. showed that AACT was negatively correlated with the overall survival of patients with pancreatic cancer, and increased AACT expression was associated with shorter survival of patients with pancreatic cancer [63]. The obtained results in this review are in line with previously published data, as shown in Fig. 6B. Moreover, AACT was used for survival prediction of patients with pancreatic cancer following gemcitabine treatment [64]. Together, these data suggest that AACT can be an effective prognostic indicator for advanced pancreatic cancer. However, the mechanism and related functions of AACT in pancreatic cancer still need to be investigated further.
Aberrant glycosylation has been identified in pancreatic cancer [65, 66], and glycosciences.de software (http://www.glycosciences.de/) was used to mine the glycoforms, such as fucosylation, sialyl Lewis A and Lewisx, for the early detection of pancreatic cancer [67–69]. N-glycosylation has a large variation in the metastatic pathogenesis of pancreatic cancer [70]. Currently, the glycosylation of AACT in pancreatic cancer has not been reported.
AACT as a diagnostic or prognostic biomarker in lung cancer
Previous studies have reported that AACT is also synthesized in lung adenocarcinoma cells, and the expression levels of AACT in lung cancer cell lines are associated with tumor progression, especially tumor growth. Zhang et al. reported that AACT in urine can be used as a biomarker for the diagnosis of NSCLC, and the expression level of AACT in the urine of patients with stage IV NSCLC was increased. The expression level of AACT is increased in advanced lung cancer tissues, and the level of AACT was found to be significantly increased in the plasma of patients with metastatic lung cancer [71]. In our previous study, we found that the expression level of the glycoprotein AACT and its GlcNAcylation level were decreased in the serum of patients with early-stage NSCLC, and were especially significantly decreased in stage IA, but AACT and its GlcNAcylation levels were restored and presented an increasing trend in late-stage NSCLC [16]. Perhaps the role of AACT in tumorigenesis and progression depends not only on its expression level but also on the complex regulation of its glycan modification levels. A study reported that the expression level of beta 1,6-N-acetylglucosaminyl transferase (GnT-V) was decreased in 50% of patients with NSCLC, resulting in a decrease in the βl,6 GlcNAc branch of the N-glycan structure, which was associated with a poor prognosis of NSCLC [72]. The decrease in the GlcNAcylation level of AACT in the early occurrence of NSCLC may be related to the downregulated expression of GnT-V. The glycosylation level changes of AACT may be associated with glycosyltransferase, but there are no directly related reports. To date, there are no related reports about the function of AACT in lung cancer.
Our previous studies showed that GlcNAcylated AACT could effectively distinguish stage I from benign samples by lectin-ELISA tests, with an AUC value of 0.932 and 90.9% sensitivity at a specificity of 93.8%. Moreover, GlcNAcylated AACT could also effectively distinguish stage I from tumor-free (healthy and benign) samples, with an AUC value of 0.908 and 90.9% sensitivity at a specificity of 86.2%. Combining the GlcNAcylated AACT and CEA significantly improved the specificity of CEA in the diagnosis of early-stage NSCLC [16]. Furthermore, we also found that combining GlcNAcylated AACT and GlcNAcylated serum paraoxonase/arylesterase l (PONl) can provide better accuracy in distinguishing early NSCLC from tumor-free samples, and the AUC for this combination was as high as 0.940, with 94.4% sensitivity and 90.2% specificity [17]. These results show that GlcNAc modified AACT with high sensitivity and specificity can be used as a novel biomarker for the early diagnosis of NSCLC.
In addition, the correlation between AACT mRNA expression levels and the overall survival of lung cancer was also analyzed by the Kaplan Meier Plotter database. In early-stage lung cancer, the median survival was 69 months, with lower expression of AACT in patients with early lung cancer patients (n = 22) and with shorter survival; the median survival was 129.3 months, with higher expression of AACT in patients with early lung cancer (n = 57) with longer survival (Fig. 6C), which suggested that mRNA expression of AACT was strongly associated with the survival of patients. However, in all patients with lung cancer, the median survival was 76 months, with lower expression of AACT in patients with lung cancer (n = 1386) and with longer survival. The median survival was 56.5 months, with a higher expression of AACT in patients with lung cancer (n = 539) (Fig. 6D), which suggested that AACT could be used as a prognostic marker for therapeutic efficiency and survival evaluation in lung cancer.
Conclusion and overall perspectives
Here, we have provided the updated structure, glycosylation modification, and biological characteristics of AACT during inflammatory and neurodegenerative diseases and tumorigenesis (Table 2). As a serine protease inhibitor and an APP, the protein expression and glycan modification of AACT, such as fucose, sialyl acid, SLex, GlcNAc, etc., is changed in inflammatory conditions and cancers. In addition, we have described the correlation between AACT expression and disease progression, particularly in cancer. The correlation between AACT expression and prognosis in inflammatory or neurodegenerative diseases is different from that found in tumors, which is most likely due to the different regulation of the immune microenvironment, but the underlying molecular mechanism still needs to be explored further. AACT expression is associated with the overall survival of cancer patients; however, the occurrence mechanisms of AACT in other diseases are still unknown and inhibitors that regulate AACT levels have not been reported, so further research on AACT-mediated disease progression and related therapy is still needed. This review provides recent findings that may be useful for designing basic research in investigating glycoprotein AACT as diagnostic and prognostic biomarkers and therapeutic targets in tumors.
Table 2.
List the expression changes and roles of alpha-1-antichymotrypsin in human diseases
| Diseases | AACT derived | Abnormal expression of AACT | Roles | Reference | |
|---|---|---|---|---|---|
| Inflammatory diseases | Fibromyalgia | CSF | ↓ | Involve in the inflammatory response and signal transduction | [5] |
| Multiple sclerosis | CSF | ↑ | Diagnostic or prognostic indicator | [27] | |
| Heart failure | Plasma | ↑ | Therapeutic or diagnostic target | [28, 29] | |
| Aortic stenosis | Plasma | ↑ | Indicator for early diagnosis | [6] | |
| Active tuberculosis | Serum | ↑ | Use for diagnosis | [30] | |
| Neurodegenerative diseases | AD | Serum, CSF | ↑ | Lead to neurofibrillary tangles in AD | [5, 35] |
| aMCI | Serum, CSF | ↑ | Biomarker for diagnosing aMCI | [39] | |
| Cancer | Ovarian cancer | Serum, ascites | ↑ | Biomarker for diagnosis | [18, 73] |
| Prostate cancer | Tissues | ↑ | Increased the rate of prostate diagnosis | [46, 47] | |
| glioblastoma | CSF | ↑ | Therapeutic target in GBM | [49] | |
| HCC | Tissues | ↓ | Tumor suppressor and as a target for treatment intervention | [24, 59] | |
| Pancreatic cancer | Serum | ↑ | Diagnostic or prognostic indicator | [14, 64] | |
| Tissues | ↑ | [61–63] | |||
| Lung cancer | Serum | in early-stage:↓; in late-stage:↑ | Early diagnostic biomarker of NSCLC | [16, 17] | |
| Urine | in late-stage: ↑ | Diagnostic biomarker of NSCLC | [71] | ||
| Colorectal cancer | Tissues | ↓ | Potential biomarker for CRC progression | [74] | |
CSF: cerebrospinal fluid; AD: Alzheimer’s disease syndrome; aMCI: amnestic mild cognitive impairment; GBM: glioblastoma multiforme; HCC: hepatocellular carcinoma; NSCLC: non-small cell lung cancer; CRC: colorectal cancer; ↓: down-regulated. ↑: up-regulated
Supplementary Information
Additional file 1: Table S1. List the samples in liver cancer and pancreatic cancer used in this manuscript.
Acknowledgements
Not applicable.
Abbreviations
- AACT
Alpha-1-antichymotrypsin
- CTSG
Neutrophil cathepsin G
- RCL
Reactive center loop
- GlcNAc
N-acetylglucosamine
- APP
Acute-phase protein
- HF
Heart failure
- AGP1
Alpha-1-acid glycoprotein 1
- CDH1
E-cadherin
- TNF-α
Tumor necrosis factor
- PSA
Prostatic specific antigen
- HILIC
Hydrophilic interaction liquid chromatography
- CID
Collision-induced dissociation
- HCD
Higher energy C trap dissociation
- AAL
Aleuria aurantia Lectin
- MRM
Multiple reaction monitoring
- THBS1
Thrombospondin-1
- SAAVs
Single amino acid variants
- HPT
Haptoglobin
- SCA
Solid serous cystadenoma
- GnT-V
Beta 1,6-N-acetylglucosaminyl transferase
- ACC
Adrenocortical carcinoma
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangio carcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and neck squamous cell carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute myeloid leukemia
- LGG
Brain lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin cutaneous melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- UVM
Uveal melanoma
- CSF
Cerebrospinal fluid
- AD
Alzheimer's disease syndrome
- aMCI
Amnestic mild cognitive impairment
- GBM
Glioblastoma multiforme
- HCC
Hepatocellular carcinoma
- NSCLC
Non-small cell lung cancer
- CRC
Colorectal cancer
Author contributions
YJ designed the project. YJ and WW wrote the manuscript and submitted the final version. QW, YZ and YG performed data collection. YG, KRZ and UR revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 320000908), the Natural Science Foundation of Hubei Science and Technology Department (Grant No. 2020CFB417), Hubei Key Laboratory of Edible Wild Plants Conservation and Utilization (No. EWPL202002). and the Hubei Normal University Scientific Research Foundation for the introduction of talent in 2018 (Grant No. HS2019RC008).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weidong Wang, Email: wangweidong@hbnu.edu.cn.
Yongsheng Gong, Email: gongshuishengone@aliyun.com.
References
- 1.Arnold K, Bordoli L, Kopp J, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 2.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 3.Schwede T, Kopp J, Guex N, et al. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt K, Gardill BR, Kern A, et al. Design of an allosterically modulated doxycycline and doxorubicin drug-binding protein. Proc Natl Acad Sci U S A. 2018;115(22):5744–9. doi: 10.1073/pnas.1716666115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoonsari PE, Ossipova E, Lengqvist J, et al. The human CSF pain proteome. J Proteom. 2019;190:67–76. doi: 10.1016/j.jprot.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Rojas T, Mourino-Alvarez L, Gil-Dones F, et al. A clinical perspective on the utility of alpha 1 antichymotrypsin for the early diagnosis of calcific aortic stenosis. Clin Proteom. 2017;14:12. doi: 10.1186/s12014-017-9147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Xu XF, Zhang RQ, et al. Remarkable increases of alpha1-antichymotrypsin in brain tissues of rodents during prion infection. Prion. 2017;11(5):338–51. doi: 10.1080/19336896.2017.1349590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker C, Belbin O, Kalsheker N, et al. SERPINA3 (aka alpha-1-antichymotrypsin) Front Biosci. 2007;12:2821–35. doi: 10.2741/2275. [DOI] [PubMed] [Google Scholar]
- 9.Pucher B, Sobieska M, Grzegorowski M, et al. The Acute phase proteins reaction in children suffering from pseudocroup. Mediat Inflamm. 2019;2019:6518308. doi: 10.1155/2019/6518308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellocchi C, Ying J, Goldmuntz EA, et al. Large-scale characterization of systemic sclerosis serum protein profile: comparison to peripheral blood cell transcriptome and correlations with skin/lung fibrosis. Arthritis Rheumatol. 2021;73(4):660–70. doi: 10.1002/art.41570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan DBA, Ito J, Peters K, et al. Protein network analysis identifies changes in the level of proteins involved in platelet degranulation, proteolysis and cholesterol metabolism pathways in AECOPD patients. COPD. 2020;17(1):29–33. doi: 10.1080/15412555.2019.1711035. [DOI] [PubMed] [Google Scholar]
- 12.Hou W, Janech MG, Sobolesky PM, et al. Proteomic screening of plasma identifies potential noninvasive biomarkers associated with significant/advanced fibrosis in patients with nonalcoholic fatty liver disease. Biosci Rep. 2020 doi: 10.1042/BSR20190395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn YH, Shin PM, Oh NR, et al. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J Proteom. 2012;75(17):5507–15. doi: 10.1016/j.jprot.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Nie S, Lo A, Wu J, et al. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J Proteome Res. 2014;13(4):1873–84. doi: 10.1021/pr400967x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie S, Yin H, Tan Z, et al. Quantitative analysis of single amino acid variant peptides associated with pancreatic cancer in serum by an isobaric labeling quantitative method. J Proteome Res. 2014;13(12):6058–66. doi: 10.1021/pr500934u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y, Wang J, Ye X, et al. Identification of GlcNAcylated alpha-1-antichymotrypsin as an early biomarker in human non-small-cell lung cancer by quantitative proteomic analysis with two lectins. Br J Cancer. 2016;114(5):532–44. doi: 10.1038/bjc.2015.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y, Yang Y, Su Y, et al. Identification a novel clinical biomarker in early diagnosis of human non-small cell lung cancer. Glycoconj J. 2019;36(1):57–68. doi: 10.1007/s10719-018-09853-z. [DOI] [PubMed] [Google Scholar]
- 18.Weiz S, Wieczorek M, Schwedler C, et al. Acute-phase glycoprotein N-glycome of ovarian cancer patients analyzed by CE-LIF. Electrophoresis. 2016;37(11):1461–7. doi: 10.1002/elps.201500518. [DOI] [PubMed] [Google Scholar]
- 19.Riby J, Mobley J, Zhang J, et al. Serum protein profiling in diffuse large B-cell lymphoma. Proteom Clin Appl. 2016;10(11):1113–21. doi: 10.1002/prca.201600074. [DOI] [PubMed] [Google Scholar]
- 20.Comamala G, Madsen JB, Voglmeir J, et al. Deglycosylation by the Acidic Glycosidase PNGase H(+) enables analysis of N-linked glycoproteins by hydrogen/deuterium exchange mass spectrometry. J Am Soc Mass Spectrom. 2020;31(11):2305–12. doi: 10.1021/jasms.0c00258. [DOI] [PubMed] [Google Scholar]
- 21.Madsen JB, Andersen LM, Dupont DM, et al. An RNA aptamer inhibits a mutation-induced inactivating misfolding of a Serpin. Cell Chem Biol. 2016;23(6):700–8. doi: 10.1016/j.chembiol.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Caval T, Lin YH, Varkila M, et al. Glycoproteoform profiles of individual patients’ plasma alpha-1-antichymotrypsin are unique and extensively remodeled following a septic episode. Front Immunol. 2020;11:608466. doi: 10.3389/fimmu.2020.608466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T, Qian WJ, Gritsenko MA, et al. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–80. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santamaria M, Pardo-Saganta A, Alvarez-Asiain L, et al. Nuclear alpha1-antichymotrypsin promotes chromatin condensation and inhibits proliferation of human hepatocellular carcinoma cells. Gastroenterology. 2013;144(4):818–28. doi: 10.1053/j.gastro.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 25.Okano T, Seike M, Kuribayashi H, et al. Identification of haptoglobin peptide as a novel serum biomarker for lung squamous cell carcinoma by serum proteome and peptidome profiling. Int J Oncol. 2016;48(3):945–52. doi: 10.3892/ijo.2016.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Pan L, Jia H, et al. Label-Free Quantitative Proteomics Identifies Novel Plasma Biomarkers for Distinguishing Pulmonary Tuberculosis and Latent Infection. Front Microbiol. 2018;9:1267. doi: 10.3389/fmicb.2018.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroksveen AC, Aasebo E, Vethe H, et al. Discovery and initial verification of differentially abundant proteins between multiple sclerosis patients and controls using iTRAQ and SID-SRM. J Proteom. 2013;78:312–25. doi: 10.1016/j.jprot.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 28.Lok SI, van Mil A, Bovenschen N, et al. Post-transcriptional regulation of alpha-1-antichymotrypsin by microRNA-137 in chronic heart failure and mechanical support. Circ Heart Fail. 2013;6(4):853–61. doi: 10.1161/CIRCHEARTFAILURE.112.000255. [DOI] [PubMed] [Google Scholar]
- 29.Brioschi M, Gianazza E, Agostoni P, et al. Multiplexed MRM-based proteomics identified multiple biomarkers of disease severity in human heart failure. Int J Mol Sci. 2021 doi: 10.3390/ijms22020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song SH, Han M, Choi YS, et al. Proteomic profiling of serum from patients with tuberculosis. Ann Lab Med. 2014;34(5):345–53. doi: 10.3343/alm.2014.34.5.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobieska M, Gajewska E, Kalmus G, et al. Obesity, physical fitness, and inflammatory markers in Polish children. Med Sci Monit. 2013;19:493–500. doi: 10.12659/MSM.883959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otvos JD, Shalaurova I, Wolak-Dinsmore J, et al. GlycA: a composite nuclear magnetic resonance biomarker of systemic inflammation. Clin Chem. 2015;61(5):714–23. doi: 10.1373/clinchem.2014.232918. [DOI] [PubMed] [Google Scholar]
- 33.Arnold JN, Saldova R, Galligan MC, et al. Novel glycan biomarkers for the detection of lung cancer. J Proteome Res. 2011;10(4):1755–64. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- 34.Alfadda AA, Benabdelkamel H, Masood A, et al. Differences in the plasma proteome of patients with hypothyroidism before and after thyroid hormone replacement: a proteomic analysis. Int J Mol Sci. 2018 doi: 10.3390/ijms19010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi E, Fiorelli T, Norden M, et al. Alpha 1-Antichymotrypsin, an Inflammatory protein overexpressed in the brains of patients with Alzheimer’s Disease, induces tau hyperphosphorylation through c-Jun N-Terminal kinase activation. Int J Alzheimers Dis. 2013;2013:606083. doi: 10.1155/2013/606083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braghin E, Galimberti D, Scarpini E, et al. Alpha1-antichymotrypsin induces TNF-alpha production and NF-kappaB activation in the murine N9 microglial cell line. Neurosci Lett. 2009;467(1):40–2. doi: 10.1016/j.neulet.2009.09.057. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Yu Q, Yu Q, et al. In-depth site-specific analysis of N-glycoproteome in human cerebrospinal fluid and glycosylation landscape changes in alzheimer’s disease. Mol Cell Proteomics. 2021;20:100081. doi: 10.1016/j.mcpro.2021.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ianni M, Manerba M, Di Stefano G, et al. Altered glycosylation profile of purified plasma ACT from Alzheimer’s disease. Immun Ageing. 2010;7(Suppl 1):6. doi: 10.1186/1742-4933-7-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Pan J, Tang K, et al. Alpha 1-antichymotrypsin may be a biomarker for the progression of amnestic mild cognitive impairment. Acta Neurol Belg. 2021;121(2):451–64. doi: 10.1007/s13760-019-01206-3. [DOI] [PubMed] [Google Scholar]
- 40.Orwoll ES, Wiedrick J, Jacobs J, et al. High-throughput serum proteomics for the identification of protein biomarkers of mortality in older men. Aging Cell. 2018 doi: 10.1111/acel.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao LL, Pei XF, Qiao X, et al. SERPINA3 silencing inhibits the migration, invasion, and liver metastasis of colon cancer cells. Dig Dis Sci. 2018;63(9):2309–19. doi: 10.1007/s10620-018-5137-x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Jiang H, Dai D, et al. Alpha 1 antichymotrypsin is aberrantly expressed during melanoma progression and predicts poor survival for patients with metastatic melanoma. Pigment Cell Melanoma Res. 2010;23(4):575–8. doi: 10.1111/j.1755-148X.2010.00715.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhou ML, Chen FS, Mao H. Clinical significance and role of up-regulation of SERPINA3 expression in endometrial cancer. World J Clin Cases. 2019;7(15):1996–2002. doi: 10.12998/wjcc.v7.i15.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17(12):1344–56. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 45.Abd Hamid UM, Royle L, Saldova R, et al. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18(12):1105–18. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- 46.Zhu L, Jaamaa S, Af Hallstrom TM, et al. PSA forms complexes with alpha1-antichymotrypsin in prostate. Prostate. 2013;73(2):219–26. doi: 10.1002/pros.22560. [DOI] [PubMed] [Google Scholar]
- 47.Oto J, Fernandez-Pardo A, Royo M, et al. A predictive model for prostate cancer incorporating PSA molecular forms and age. Sci Rep. 2020;10(1):2463. doi: 10.1038/s41598-020-58836-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu G, Claret FX, Zhou F, et al. Jab1/COPS5 as a novel biomarker for diagnosis, prognosis, therapy prediction and therapeutic tools for human cancer. Front Pharmacol. 2018;9:135. doi: 10.3389/fphar.2018.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lara-Velazquez M, Zarco N, Carrano A, et al. Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of glioblastoma. Neuro Oncol. 2021;23(4):599–610. doi: 10.1093/neuonc/noaa264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan Q, Wang SQ, Zhang GT, et al. Highly expressed of SERPINA3 indicated poor prognosis and involved in immune suppression in glioma. Immun Inflamm Dis. 2021 doi: 10.1002/iid3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghorbaninezhad F, Asadzadeh Z, Masoumi J, et al. Dendritic cell-based cancer immunotherapy in the era of immune checkpoint inhibitors: From bench to bedside. Life Sci. 2022;297:120466. doi: 10.1016/j.lfs.2022.120466. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Du X, Fan G. Identifying potential biomarkers of prognostic value in colorectal cancer via tumor microenvironment data mining. Front Genet. 2021;12:787208. doi: 10.3389/fgene.2021.787208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyauchi E, Furuta T, Ohtsuki S, et al. Identification of blood biomarkers in glioblastoma by SWATH mass spectrometry and quantitative targeted absolute proteomics. PLoS ONE. 2018;13(3):e0193799. doi: 10.1371/journal.pone.0193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norton ES, Da Mesquita S, Guerrero-Cazares H. SERPINA3 in glioblastoma and Alzheimer’s disease. Aging. 2021;13(18):21812–3. doi: 10.18632/aging.203603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nimbalkar VP, Kruthika BS, Sravya P, et al. Differential gene expression in peritumoral brain zone of glioblastoma: role of SERPINA3 in promoting invasion, stemness and radioresistance of glioma cells and association with poor patient prognosis and recurrence. J Neurooncol. 2021;152(1):55–65. doi: 10.1007/s11060-020-03685-4. [DOI] [PubMed] [Google Scholar]
- 56.Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res. 2015;126:257–79. doi: 10.1016/bs.acr.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim KH, Park GW, Jeong JE, et al. Parallel reaction monitoring with multiplex immunoprecipitation of N-glycoproteins in human serum for detection of hepatocellular carcinoma. Anal Bioanal Chem. 2019;411(14):3009–19. doi: 10.1007/s00216-019-01775-5. [DOI] [PubMed] [Google Scholar]
- 58.Ji ES, Hwang H, Park GW, et al. Analysis of fucosylation in liver-secreted N-glycoproteins from human hepatocellular carcinoma plasma using liquid chromatography with tandem mass spectrometry. Anal Bioanal Chem. 2016;408(27):7761–74. doi: 10.1007/s00216-016-9878-0. [DOI] [PubMed] [Google Scholar]
- 59.Zhu H, Liu Q, Tang J, et al. Alpha1-ACT functions as a tumour suppressor in hepatocellular carcinoma by inhibiting the PI3K/AKT/mTOR signalling pathway via activation of PTEN. Cell Physiol Biochem. 2017;41(6):2289–306. doi: 10.1159/000475648. [DOI] [PubMed] [Google Scholar]
- 60.Lin QJ, Yang F, Jin C, et al. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21(26):7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu W, Hong X, Li J, et al. Solid serous cystadenoma of the pancreas: a case report of 2 patients revealing vimentin, beta-catenin, alpha-1 antitrypsin, and alpha-1 antichymotrypsin as new immunohistochemistry staining markers. Med (Baltim) 2015;94(12):e644. doi: 10.1097/MD.0000000000000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yonenaga Y, Kurosawa M, Mise M, et al. Pancreatic-type acinar cell carcinoma of the stomach included in multiple primary carcinomas. Anticancer Res. 2016;36(6):2855–64. [PubMed] [Google Scholar]
- 63.Roberts AS, Campa MJ, Gottlin EB, et al. Identification of potential prognostic biomarkers in patients with untreated, advanced pancreatic cancer from a phase 3 trial (Cancer and Leukemia Group B 80303) Cancer. 2012;118(2):571–8. doi: 10.1002/cncr.26270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsubara J, Ono M, Honda K, et al. Survival prediction for pancreatic cancer patients receiving gemcitabine treatment. Mol Cell Proteomics. 2010;9(4):695–704. doi: 10.1074/mcp.M900234-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drabik A, Bodzon-Kulakowska A, Suder P, et al. Glycosylation changes in serum proteins identify patients with pancreatic cancer. J Proteome Res. 2017;16(4):1436–44. doi: 10.1021/acs.jproteome.6b00775. [DOI] [PubMed] [Google Scholar]
- 66.Pan S, Brentnall TA, Chen R. Glycoproteins and glycoproteomics in pancreatic cancer. World J Gastroenterol. 2016;22(42):9288–99. doi: 10.3748/wjg.v22.i42.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terao N, Takamatsu S, Minehira T, et al. Fucosylation is a common glycosylation type in pancreatic cancer stem cell-like phenotypes. World J Gastroenterol. 2015;21(13):3876–87. doi: 10.3748/wjg.v21.i13.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyoshi E, Kamada Y. Application of glycoscience to the early detection of pancreatic cancer. Cancer Sci. 2016;107(10):1357–62. doi: 10.1111/cas.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munkley J. The glycosylation landscape of pancreatic cancer. Oncol Lett. 2019;17(3):2569–75. doi: 10.3892/ol.2019.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holst S, Belo AI, Giovannetti E, et al. Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation. Sci Rep. 2017;7(1):16623. doi: 10.1038/s41598-017-16811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Li Y, Qiu F, et al. Comparative analysis of the human urinary proteome by 1D SDS-PAGE and chip-HPLC-MS/MS identification of the AACT putative urinary biomarker. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(32):3395–401. doi: 10.1016/j.jchromb.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 72.Lemjabbar-Alaoui H, McKinney A, Yang YW, et al. Glycosylation alterations in lung and brain cancer. Adv Cancer Res. 2015;126:305–44. doi: 10.1016/bs.acr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miyamoto S, Ruhaak LR, Stroble C, et al. Glycoproteomic analysis of malignant ovarian cancer ascites fluid identifies unusual glycopeptides. J Proteome Res. 2016;15(9):3358–76. doi: 10.1021/acs.jproteome.6b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dimberg J, Strom K, Lofgren S, et al. Expression of the serine protease inhibitor serpinA3 in human colorectal adenocarcinomas. Oncol Lett. 2011;2(3):413–8. doi: 10.3892/ol.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List the samples in liver cancer and pancreatic cancer used in this manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.