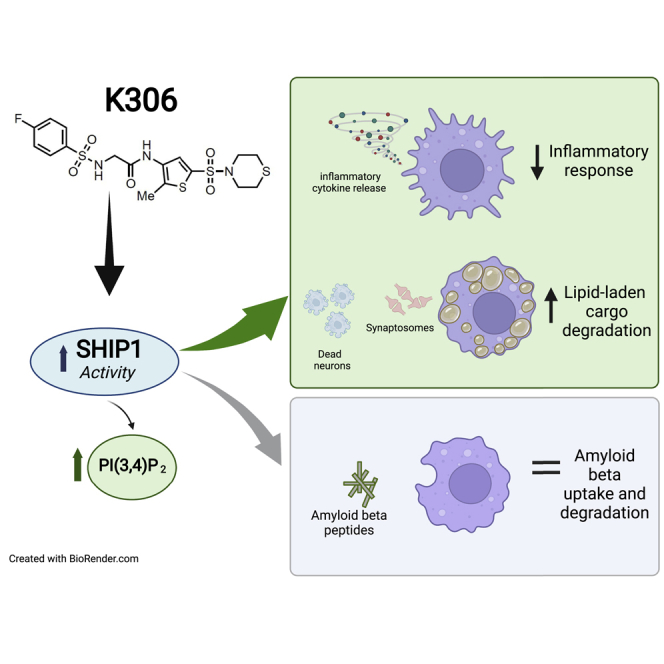
Summary
Here, we describe the use of artificial intelligence to identify novel agonists of the SH2-containing 5′ inositol phosphatase 1 (SHIP1). One of the compounds, K306, represents the most potent agonist identified to date. We find that K306 exhibits selectivity for SHIP1 vs. the paralog enzyme SHIP2, and this activation does not require the C2 domain of SHIP1 which other known SHIP1 agonists require. Thus, K306 represents a new class of SHIP1 agonists with a novel mode of agonism. Importantly, we find that K306 can suppress induction of inflammatory cytokines and iNOS in macrophages or microglia, but not by their SHIP1-deficient counterparts. K306 also reduces TNF-α production in vivo in an LPS-induced endotoxemia assay. Finally, we show that K306 enhances phagolysosomal degradation of synaptosomes and dead neurons by microglia revealing a novel function for SHIP1 that might be exploited therapeutically in dementia.
Subject areas: Health sciences, Biochemistry, Biochemical mechanism, Cellular neuroscience, Artificial intelligence
Graphical abstract
Highlights
-
•
Discovery of a potent SHIP1 selective agonist (K306) via artificial intelligence
-
•
SHIP1 agonism via K306 is independent of the C2 domain and increases PI(3,4)P2 levels
-
•
K306 reduces IL-6, TNF-α, and iNOS induction in microglia and macrophages
-
•
K306 promotes phagocytic degradation of lipid-laden but not protein cargo in microglia
Health sciences; Biochemistry; Biochemical mechanism; Cellular neuroscience; Artificial intelligence
Introduction
Over the last decade, SHIP1 has emerged as a therapeutic target due to its essential role in immune cells, but also importantly for survival of certain cancers (Kerr, 2011). Indeed, a SHIP1-selective inhibitor has been shown to be an effective promoter of immune responses to tumor cells, a chemotherapeutic for B lymphoid cancers, hematopoietic stem cell (HSC) mobilization, and engraftment of autologous and allogeneic HSC in murine models of disease or transplantation (Chen et al., 2015; Ecker et al., 2021; Fernandes et al., 2015; Fuhler et al., 2012; Gumbleton et al., 2017). Pan-SHIP1/2 inhibitors have also been identified and these compounds have shown in vivo efficacy in reversing diet-induced obesity and metabolic syndrome (Srivastava et al., 2016). More recently, pan-SHIP1/2 inhibitors have emerged as a class of small molecules that can increase phagocytic function of microglia in the CNS suggesting potential applications in dementias like Alzheimer disease (Pedicone et al., 2019). The identification of the SHIP1 C2 domain that binds the SHIP1 product, PI(3,4)P2, to increase its catalytic activity in a feed-forward manner, enabled the identification of pelorol-based SHIP1 agonists. These first-in-class SHIP1 agonists (AQX-MN100, -16A) all based on the pelorol scaffold were found to be effective at reducing proinflammatory functions of macrophages in vitro and in vivo (Chamberlain et al., 2020; Ong et al., 2007). Unfortunately, pharmaceutical development and testing of a novel SHIP1 agonist, AQX-1125, was halted after failing to demonstrate efficacy in a Phase III trial for the treatment of inflammatory bladder disease (Nickel et al., 2019). However, a recent re-examination of AQX-1125 failed to show significant binding to recombinant SHIP1 in vitro (Chamberlain et al., 2020) suggesting that failure in clinical testing was not necessarily that of SHIP1 agonism but rather the poor potency of the compound that was chosen to be pursued clinically. Like SHIP1, SHIP2 has recently been shown to be able to bind both its substrate PI(3,4,5)P3 and its product PI(3,4)P2 through the C2 domain (Le Coq et al., 2017, 2021). Binding of either inositol phospholipid species increases SHIP2 enzymatic activity with PRR domain binding enabling a 10- to 15-fold increase in activity (Le Coq et al., 2021). Thus, small molecule SHIP2 agonists are very likely to be developed that could have a role in certain cancers where SHIP2 is a tumor suppressor, but also potentially in diabetes, metabolic syndrome, and hypertension due to SHIP2’s role in insulin signaling (Clement et al., 2001; Kagawa et al., 2005; Kaisaki et al., 2004; Sleeman et al., 2005), as supported by associations of SNPs in the SHIP2 gene (INPPL1) in such diseases in both European and Asian populations (Ghosh et al., 2000; Panhuysen et al., 2003; Xu et al., 1999). Interestingly, one of the SNPs found in SHIP2 creates an amino acid change (L632I) in the catalytic domain that reduces its enzymatic activity (Kagawa et al., 2005).
The recent identification of single nucleotide polymorphisms (SNPs) in the INPP5D (SHIP1) gene linked to Alzheimer disease (AD) risk, including the rs35349669 (rs669) SNP which is strongly associated with AD (Ruiz et al., 2014; Zhang et al., 2016), has led to increased interest in targeting SHIP1 in Alzheimer disease. Currently, there is a focus on SHIP1 inhibition approaches as a means to increase microglial functions promoted by the SHIP1-regulated TREM2 receptor to decrease amyloid burden in the CNS (Kerr et al., 2020). Indeed, our recent analysis showed that pan-SHIP1/2 inhibitory compounds promote the phagocytic function of microglia both in vitro and in vivo, including phagocytosis of pathogenic Aβ42 (Pedicone et al., 2019). However, reduced SHIP1 expression was recently shown to be part of a seven gene AD risk cassette in a murine model of AD (Sala Frigerio et al., 2019), suggesting that SHIP1 agonism might also prove to be an effective therapeutic strategy in AD. The answer is likely more complex in that SHIP1 might be antagonized or agonized depending upon the stage of disease in AD, with SHIP1 antagonism advocated in early-stage disease to enhance microglial homeostatic function with agonism needed in late-stage disease to reduce the damage caused by dysfunctional microglia.
Agonism of SHIP1 has also been advocated for the treatment of mucosal inflammatory diseases as SHIP1 null mice demonstrate consolidation of their lungs (Helgason et al., 1998; Paraiso et al., 2007), but also in inflammatory bowel diseases (IBD) like Crohn disease (CD) as SHIP1 null adult mice develop a profound ileitis that closely resembles human Crohn disease (Kerr et al., 2011). In fact, a significant subset of patients with IBD (15%) are SHIP1-deficient at the protein level (Fernandes et al., 2018; Somasundaram et al., 2017). The SHIP1-deficient subset of patients with IBD was subsequently found to have more severe disease that requires surgical resection(s) or the use of biologics for treatment with respect to other patients with IBD who are SHIP1-sufficient (Fernandes et al., 2018). Interestingly, these patients still retain a low level of SHIP1 protein expression (<10%) relative to healthy controls, suggesting SHIP1 agonism may be a feasible treatment strategy as the protein target is not completely absent. Consistent with this, Chamberlain et al. have recently shown that AQX-MN100, a pelorol-type SHIP1 agonist, is effective at treatment in a murine model of colitis (Chamberlain et al., 2020).
We show here that a virtual high-throughput screen of the SHIP1 active site guided by an artificial intelligence-based algorithm is effective in the identification of novel SHIP1 agonists. Moreover, one of these agonists, K306, has a distinct mode of action that does not involve the C2 domain of SHIP1 as has been shown for the pelorol derivatives (Ong et al., 2007). We show that the newly identified SHIP1 agonist K306 can reduce inflammatory responses by both macrophages and microglia. Finally, studies with K306 enabled the discovery of an unappreciated role for SHIP1 in promoting degradation of lipid-laden phagocytic cargo by microglia.
Results
Identification of new SHIP1 agonists via an innovative computational screening strategy
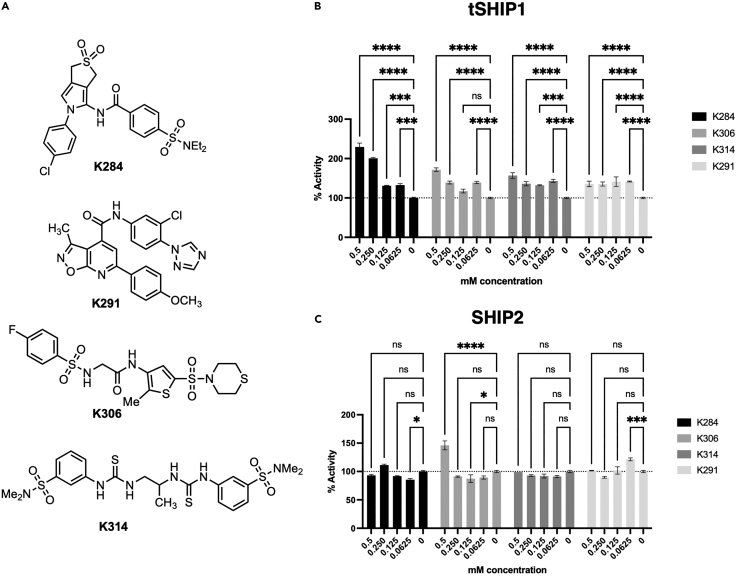
Until recently, there have been no X-ray structures of SHIP1 (Bradshaw et al., 2019, Released: 2019-01-16), although, several crystal structures of portions of the highly homologous paralog enzyme, SHIP2, have been disclosed (Le Coq et al., 2017; Mills et al., 2012; Tresaugues et al., 2014). To gain insight into the SHIP1 structure, the sequence for SHIP1 was threaded into these SHIP2 structures, creating models of the SHIP1 phosphatase domain. A virtual screen against the phosphatase active site was then conducted using a scoring algorithm based on a convolutional neural network (Wallach et al., 2015). This yielded 76 structures that were predicted to be SHIP1 antagonists. Indeed, screening of these compounds identified several novel SHIP1 inhibitors (data not shown). However, several compounds (K284, K291, K314, and K306) (Figure 1A) were surprisingly found to agonize SHIP1 activity using the malachite green assay and the SHIP1/2 substrate PI(3,4,5)P3 (Figure 1B). All showed potent agonistic activity for SHIP1 (Figure 1B). Both K284 and K314 failed to significantly agonize the other SHIP paralog, SHIP2, at any concentration tested, while K306 agonized SHIP2 activity, but only at the highest concentration tested (0.5mM) (Figure 1C). Interestingly, K306 significantly inhibited SHIP2 at 125μM, but not at the other concentrations tested (Figure 1C). K291 also showed significant agonism of SHIP2, but interestingly only at the lowest concentration tested (62.5μM) (Figure 1C). Taken together, this in silico screen led to the identification of four novel chemical classes that can selectively agonize SHIP1 enzyme activity in vitro.
Figure 1.
Novel SHIP1 agonists and their relative activity on SHIP1 and SHIP2
(A) Structures of novel SHIP1 agonists. Percentage increase of phosphatase activity for (B) SHIP1 and (C) SHIP2 enzyme in the malachite green assay. The assay was performed at the indicated mM concentration with the indicated agonist or its vehicle control (0) with 100μM PI(3,4,5)P3-diC8. (Data are representative of 2 independent experiments. Bars indicate mean with ±SEM. The significance of agonism (or inhibition) for each compound vs. vehicle was assessed for all concentration tested via a two-way ANOVA. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05).
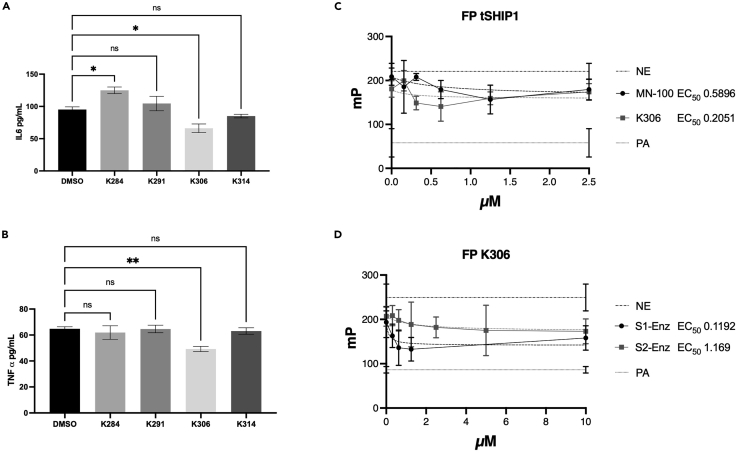
To further discriminate which of these novel SHIP1 agonists might have useful biological activity, we examined their ability to suppress LPS-induced production of TNF-α and IL-6 by BV2 microglial cells. SHIP1 is known to limit production of these inflammatory cytokines in LPS-stimulated macrophages, and this can be partially repressed by pelorol SHIP1 agonists like AQX-MN100 (Ong et al., 2007). Thus, we reasoned that, if these agonists can cross a cell membrane, they might also suppress inflammatory cytokine production by microglia. Testing of all 4 compounds on LPS-stimulated BV2 cells revealed that only K306 had activity consistent with SHIP1 agonism as it reduced both TNF-α (Figure 2B) and IL-6 production (Figure 2A). Intriguingly, K284 increased IL-6 (Figure 2A), but not TNF-α production (Figure 2B) by LPS-stimulated BV2 cells suggesting it might have some SHIP1 inhibitory activity in cells. Based on these cell-based assays, we then selected K306 for further enzymatic and biological testing, including comparison with the known and potent SHIP1 agonist, the pelorol AQX-MN100 (Ong et al., 2007). To accomplish this, we utilized a more sensitive assay for SHIP1 agonism than the malachite green assay, a fluorescent polarization assay (FP) for SHIP1 activity that we have described previously (Brooks et al., 2010). The activity of K306 and AQX-MN100 on purified recombinant SHIP1 (tSHIP1) or on comparable recombinant forms of SHIP1 (S1-Enz) and SHIP2 (S2-Enz) (see Figure 3A for SHIP gene constructs) was measured by incubating dilutions of K306 or AQX-MN100 with either enzyme for 20 min at 37°C followed by inactivation of the enzyme at 95°C for 3 min. Enzymatically generated PI(3,4)P2 from PI(3,4,5)P3 was measured using the 5′ PtdIns(3,4,5)P3 phosphatase activity fluorescence polarization assay (Echelon Biosciences), by adding PI(3,4)P2 detector and fluorescent PI(3,4)P2 probe to replicates of the enzymatic reactions. Polarization (mP) values decrease as probe binding to the PI(3,4)P2 detector is displaced by PI(3,4)P2 produced by SHIP enzymatic activity and thus the amount of unbound fluorescent probe in the mixture increases. The FP measurements confirmed that K306 is a potent SHIP1 agonist that has greater potency than AQX-MN100 with significant SHIP1 agonism detected into the nanomolar range (EC50 of 0.2051μM for K306 vs. 0.5896μM for AQX-MN100) (Figure 2C). Comparisons of K306 agonism on recombinant SHIP1 and SHIP2 (S1-Enz and S2-Enz, respectively) showed it has approximately 10-fold higher selectivity for SHIP1 agonism vs. SHIP2 (EC50 of 0.1192μM, SHIP1 vs. 1.169μM, SHIP2) (Figure 2D). Thus, K306 is a potent SHIP1-selective agonist that appears, to the best of our knowledge, to have the strongest agonistic activity of any small molecule SHIP agonist identified to date.
Figure 2.
K306 has biological activity consistent with SHIP1 agonistic activity and is a more potent agonist than the pelorol AQX-MN100
(A) IL-6 and (B)TNF-α production by BV2 microglia cells stimulated with LPS for 6 h or 2 h, respectively, as measured from supernatants by ELISA. (Representative results of two independent comparisons of all potential agonists at 5μM). All cells were treated with agonists or vehicle control 1 h before LPS challenge. Bars indicate mean ±SEM. Statistical analysis was performed with one-way ANOVA with Dunnett correction for multiple comparisons versus control (DMSO), ∗∗p < 0.01, ∗p < 0.05). 5′ fluorescence polarization Assay (FP) to measure PtdIns(3,4,5)P3 5′ phosphatase activity on (C) tSHIP1 with K306 and MN-100 and (D) SHIP1-Enzyme (S1-Enz) and SHIP2-Enzyme (S2-Enz) with K306. EC50 of K306 and MN-100 on tSHIP1 and the EC50 of K306 on S1-Enz and S2-Enz were calculated by incubating dilutions of K306 or MN-100 with either enzyme for 20 min at 37°C. PI(3,4)P2 generated by the SHIP enzymes is then measured using the FP assay (Echelon Biosciences). Control reactions include probe alone (PA), where all the probe is free rotating and thus not polarized and no-enzyme (NE) control, where the probe is not displaced from the detector by enzymatically generated PI(3,4)P2 and highly polarized. Shown is one of 2 independent experiments with 6 replicate wells/compound concentration. EC50 was calculated with Prizm version 9.3.1, using Nonlinear regression [Agonist] vs. response (three parameters), using Robust Fitting method, Medium Convergence Criteria, No Weighting and Considering every Y replicate as an individual point.
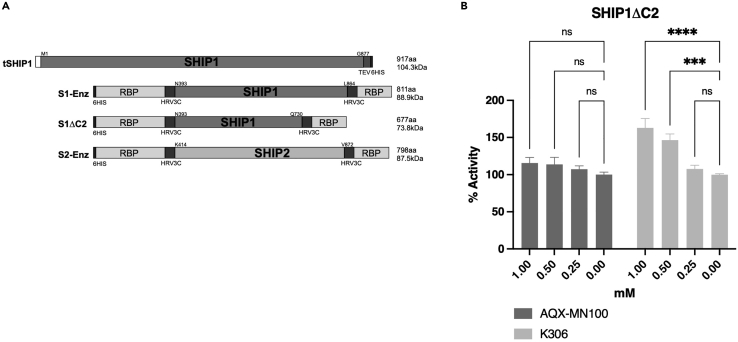
Figure 3.
K306 agonism does not require the C2 domain to agonize SHIP1
(A) Structure of the tSHIP1, SHIP1-Enzyme (S1-Enz), SHIP1ΔC2 (S1ΔC2), and SHIP2-Enzyme (S2-Enz) constructs.
(B) Malachite Green Phosphatase Release assay measurements of K306 or AQX-MN100 agonism on purified SHIP1ΔC2. Data in (B) are representative of three independent experiments. Bars indicate mean ±SEM. The significance of agonism for each compound vs. vehicle was assessed for all concentration tested via a two-way ANOVA∗∗∗∗p < 0.0001, ∗∗∗p < 0.001).
K306 has a novel mechanism of action relative to pelorol-based SHIP1 agonists
Others have shown the C2 domain of SHIP1 to be important for agonistic function, in particular Ong et al. reported that the C2 domain was essential for SHIP1 agonism by the pelorols AQX-MN100 and AQX-16A (Ong et al., 2007). Thus, we examined the dependency of K306 agonistic activity on recombinant SHIP1 that lacks the C2 domain, SHIP1ΔC2 (Figure 3A). When we compared the ability of AQX-MN100 and K306 to agonize SHIP1ΔC2 enzyme activity (Figure 3B), the novel agonist K306 retained significant agonistic activity for SHIP1ΔC2, while AQX-MN100 showed no agonistic activity on SHIP1ΔC2 (Figure 3B), consistent with previous findings (Ong et al., 2007). Thus, K306 agonism of SHIP1 is not dependent on the C2 domain required for pelorol-based SHIP1 agonists (e.g., AQX-MN100), indicating K306 has a different molecular mechanism for agonizing SHIP1 vs. pelorol-based agonists.
K306 acts specifically on SHIP1 to dampen inflammatory cytokine production by myeloid-lineage cells in vitro and in vivo
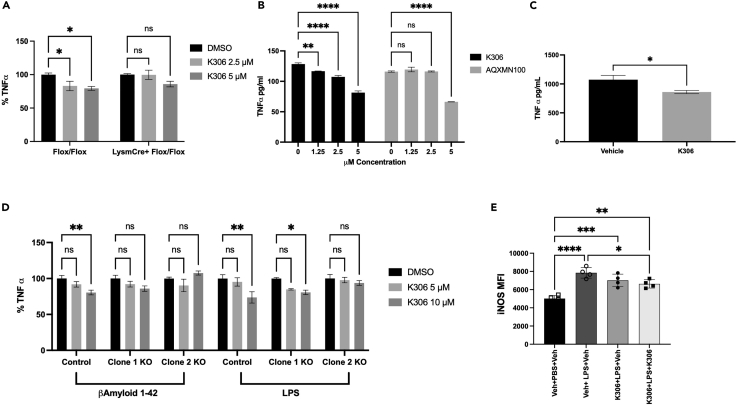
Previous studies of the pelorol-based agonists AQX-MN100 and AQX-16A used bone-marrow-derived macrophages (BMDM) prepared from SHIP1 knockout and WT mice to validate that these compounds specifically act on SHIP1 to suppress inflammatory cytokine production following an LPS challenge (Ong et al., 2007). We adopted a similar approach to validate target specificity of K306 using BMDM derived from either LysMCreSHIP1flox/flox mice or SHIP1-sufficient SHIP1flox/flox control mice that we have previously used to demonstrate that SHIP1 controls macrophage activation and training by PAMP ligands like LPS (Saz-Leal et al., 2018). In Figure 4A, we show that K306 significantly reduced LPS-induced TNF-α production (vs. DMSO controls) after 6 h (6h) stimulation of SHIP1-sufficient SHIP1flox/flox BMDM, but not with LPS-stimulated SHIP1-deficient LysMCreSHIP1flox/flox BMDM. The selective reduction of LPS-induced TNF-α production by SHIP1-sufficient BMDM indicates K306 is acting on SHIP1 to repress LPS stimulated TNF-α production. To assess the relative potency of K306 vs. the pelorol AQX-MN100, we compared their ability to repress TNF-α production by WT BMDM from WT C57BL6 mice stimulated with LPS for 1h. In this setting, K306 significantly reduced TNF-α production at all concentrations tested in SHIP1-sufficient SHIP1flox/flox BMDM as compared to vehicle, while MN100 significantly decreased TNF-α production only at the highest dose (5μM) as compared to vehicle (Figure 4B). Based on these findings with primary BMDM in vitro, we then evaluated whether K306 had sufficient potency to act on macrophages in vivo using an LPS-induced septicemia model that was used to validate in vivo efficacy of AQX-MN100 (Ong et al., 2007). To accomplish this, we dosed mice twice with K306 (20 mg/kg, i.p.), 30 min before and 30 min after LPS challenge (2 mg/kg). We collected the blood via intracardiac puncture 1h after LPS stimulation and analyzed TNF-α levels in the serum via ELISA. We observed a significant reduction of TNF-α in the serum of K306-treated mice versus vehicle controls (Figure 4C). Therefore, K306 has sufficient stability and potency to reduce LPS-induced inflammatory cytokine production in vivo suggesting a possible application of this compound in dampening cytokine storms driven by bacterial or viral infections. Macrophages are not the only terminally differentiated myeloid cells that can respond to inflammatory stressors to produce inflammatory mediators. Brain-resident microglia also do this leading to neurotoxic effects in various dementias (Brown and Vilalta, 2015; Colonna and Butovsky, 2017; Hansen et al., 2018; St-Pierre et al., 2020; Ulland et al., 2015; Zhang et al., 2011). In fact, microglia can respond to both LPS and Aβ42 peptides to produce TNF-α (Hughes et al., 2020). We found that K306 reduces both TNF-α and IL-6 production by BV2 microglia cells stimulated with LPS (Figures 2A and 2B), and thus we sought to determine whether this might also be the case for Aβ42 stimulation. Aβ42 is a ligand for the PAMP receptor TREM2 (Wang et al., 2015) and SHIP1 opposes TREM2 distal signaling (Peng et al., 2010). Hence, we used parental BV2 cells and two clones that have been gene edited by CRISPR/Cas9 to have reduced SHIP1. We confirmed by Western blot, flow cytometry and immunofluorescence that the two gene-edited clones had lower or lacked expression of SHIP1 relative to parental BV2 cells (Figures S1A–S1C). Specifically, Clone 1 has reduced SHIP1 expression as compared to parental BV2 cells, while Clone 2 lacks SHIP1 expression (Figures S1A–S1C). Next, we functionally validated the biological activity of K306 as a selective SHIP1 agonist, by measuring the level of the direct SHIP1 product PI(3,4)P2 in parental and Clone 2 BV2 cells. While, as expected, K306 treatment induced a significant increase in PI(3,4)P2 in parental cells, this was not the case in Clone 2, which lacks SHIP1 expression, further confirming that the agonist activity of K306 is exerted through SHIP1 (Figures S1D-S1F). When testing cytokine production, we found that K306 was able to reduce Aβ42-stimulated TNF-α production by parental (SHIP1 sufficient) BV2 cells, but not by either of the gene-edited BV2 clones that lack or have lower SHIP1 expression, including Clone 1 that still retains some SHIP1 expression (Figure 4D). In LPS-stimulated BV2 cells, we did find, however, that both parental BV2 cells and Clone 1 cells which have reduced SHIP1 expression showed reduced TNF-α induction when treated with K306, while Clone 2 cells that lack SHIP1 expression were unresponsive to K306 implying different activating receptors (e.g., TREM2 vs. TLR4) found on microglia have different sensitivities to SHIP1 agonism (Figure 4D). We further tested the ability of K306 to reduce inflammatory gene expression in microglia by examining its ability to impact LPS induction of iNOS expression. We found that K306 also reduces LPS-mediated induction of iNOS in BV2 cells stimulated with LPS, but this requires addition of K306 before and shortly after LPS stimulation (Figures 4E and S2). Importantly, these findings are the first to demonstrate that SHIP1 agonism can significantly reduce production of inflammatory cytokines and an enzyme that produces NO in microglia.
Figure 4.
K306 reduces TNF-α production in vitro and in vivo
ELISA quantitation of TNF-α production by (A) BMDM from LysMCre+SHIP1flox/flox or SHIP1flox/flox mice after 6 h of LPS stimulation, reported as percentage of TNF-α production compared to DMSO-treated BMDM of the same genotype (B) WT BMDM treated with K306 or MN100 for 1 h and incubated with LPS for 1 h. TNF-α was quantified as pg/mL in the cellular supernatant and normalized to the number of the cells present after stimulation.
(C) Mice were treated with K306 (20 mg/kg) or vehicle at 30 min prior to and 30 min after LPS challenge (2 mg/kg). Blood was collected 1 h after LPS challenge and TNF-α levels were measured in the serum with ELISA. Data are representative of 2 independent experiments, N = 5.
(D) BV2 clones were stimulated with Aβ42 (100 ng/mL) for 3 h or 10 ng/mL LPS for 1 h, as indicated. On the y axes is reported % of TNF-α induction relative to the vehicle control for each clone to enable a comparison of % suppression of TNF-α induction in these clones at the indicated K306 concentration. All cells were pretreated with the agonist or vehicle control for 1 h prior to Aβ42 or LPS stimulation. Data are representative of three independent experiments. Bars indicate Mean ± SEM; statistical tests: two-way ANOVA with Tukey correction for multiple comparisons (A, B,E) or unpaired Students T test (C) ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05 (E) K306 reduces induction of iNOS in microglia. Mean fluorescence intensity (MFI) for iNOS staining of vehicle or K306 LPS-stimulated BV2 cells. BV2 cells were plated and treated for 1 h with Veh (0.25%DMSO) or K306 (10μM in Veh) prior to addition of LPS (100 ng/mL). Unstimulated BV2 cells treated with Veh are shown as an additional control to assess LPS induction of iNOS (lane 1). After a 1 h incubation with LPS, Veh (0.25% DMSO) or K306 (10μM in Veh) was again added to the indicated samples and incubation was continued for 16 h (Final DMSO concentration in each well was 0.5% DMSO and 0μM (lane 1 and 2), 10μM (lane 3), or 20μM K306 (lane 4). Cells were harvested and expression of iNOS was analyzed by intracellular spectral flow cytometry. A one-way ANOVA with Tukey’s multiple comparison test was used to compare all-pairs of columns. Shown is one representative experiment of 4 independent experiments with 4 replicate wells/condition.
SHIP1 agonism via K306 selectively promotes degradation of membrane-bound phagocytic cargoes in microglia
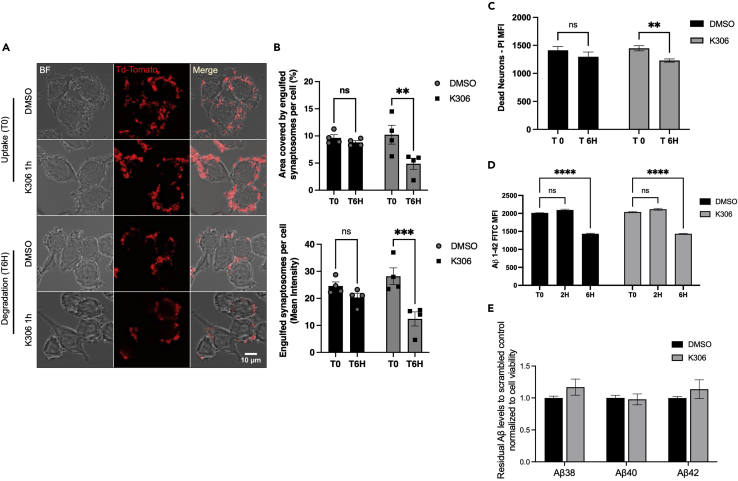
Microglia, in addition to taking up proteinaceous debris for degradation, also phagocytose dead or dying neurons as well as synapses (Hume et al., 1983). These are essential microglial homeostatic functions that are necessary for the development and maintenance of normal cognitive abilities (Paolicelli et al., 2017; Yanuck, 2019; Zhan et al., 2014). We previously found that pan-SHIP1/2 inhibitory compounds selectively enhanced microglial phagocytic uptake, or engulfment, of Aβ42 peptide, and also dead neurons (Pedicone et al., 2019). We then considered that SHIP1 agonism might reduce microglial phagocytic functions. We therefore examined engulfment and degradation of different phagocytic cargoes by BV2 microglial cells treated with K306 (or vehicle) using three different physiologically relevant targets of microglia—namely synaptosomes, dead neurons, and Aβ42. We analyzed the initial engulfment of synaptosomes labeled with TdTomato by confocal microscopy and compared their intracellular levels at 0 and 6 h after initial uptake to study cargo degradation (Figures 5A and 5B). While K306 treatment did not reduce the initial uptake of synaptosomes, it did surprisingly increase the rate of degradation of the synaptosomal cargo. Efficient internalization and degradation in this assay were validated by using 3D reconstruction of cells and engulfed synaptosomes (Figure S3A) and by time-lapse light sheet microscopy (Figure S3F and Video S1). Furthermore, synaptosomes were conjugated with the pH-sensitive dye pHrodo, to confirm intracellular trafficking into acidic compartments (Figures S3G–S3I). We then assessed whether this was also the case for another lipid-laden cargo. Indeed, K306 did not promote a significant increase in the initial engulfment of dead neurons but did increase their rate of degradation by BV2 microglia (Figure 5C). Interestingly, this was not the case for the peptide cargo Aβ42, as K306 neither enhanced nor decreased its initial uptake by microglia or its degradation when the latter was analyzed 2 and 6 h after initial engulfment (Figure 5D). To further investigate the effects of K306 on amyloid clearance by microglia, we used a previously established paradigm in which BV2 cells are exposed to conditioned medium from HeLa cells overexpressing the human amyloid precursor protein carrying the Swedish mutation (swAPP) (Paolicelli et al., 2017). In this assay, the residual amyloid levels are measured by a multiplex immunoassay, which allows simultaneous detection of different Aβ species (Aβ38, Aβ40, and Aβ42). Consistent with Aβ42−FITC experiments, K306 treatment had no effect on microglial clearance of amyloid, regardless the Aβ species (Figure 5E), indicating that K306 does not modulate uptake/degradation of amyloid peptides. The above analysis of K306 in microglial biology reveals a novel function for SHIP1 in promoting phagolysosomal degradation of large membrane-bound cargoes, such as synaptosomes and dead neurons.
Figure 5.
K306 selectively increases microglial degradation of membrane-containing phagolysosomal cargo
BV2 pretreated with 10μM K306 for 1 h was incubated with the indicated cargo for 1 h and degradation of the cargo was detected by either fluorescence microscopy (A) or flow cytometry (C and D).
(A) Image of TdTomato-labeled synaptosomes phagocytosed by BV2 cells at time 0 (T0) for uptake and 6 h (T6H) after uptake for DMSO control or K306 treatment.
(B) Quantification of area covered by synaptosomes and mean intensity for synaptosome per cell.
(C) Dead neuron uptake (T0) and degradation (T6H) evaluated by flow cytometry as mean fluorescence intensity of PI-labeled engulfed dead neurons for BV2 cells treated with DMSO or K306 (10μM).
(D) Flow cytometry detection of Aβ42-FITC uptake (T0) and degradation at 2 and 6 h (T2H, T6H). (Data are representative of four independent experiments in (A and B) or three independent experiments in (C, D). Bars indicate mean ±SEM. The significance of engulfment or degradation for K306 vs. vehicle was assessed by unpaired Student’s T-test (C) or via a two-way ANOVA(B, D) with Tukey correction for multiple comparisons. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05).
(E) Residual Aβ38, Aβ40, and Aβ42 levels from HeLa swAPP-conditioned medium, after 6 h incubation with BV2 cells treated with K306 or DMSO control, normalized to DMSO and to cell viability. Bars indicate mean ± SEM from three independent experiments.
Discussion
Here, we have shown that an artificial intelligence-guided virtual screening approach can be used to identify novel SHIP1 agonists. All the agonists identified through this in silico approach showed selective agonism of SHIP1 vs. its paralog SHIP2 with most not agonizing SHIP2 to any significant degree. Most of the initial SHIP1 agonists identified did not have biological activity, which might be due to a lack of cell membrane permeability.
The SHIP1 agonist K306 displayed biological activity in a cell-based assay enhancing SHIP1 phosphatase activity, as supported by the significant increase in PI(3,4)P2 levels observed in BV2 cells upon treatment. K306 was also able to significantly suppress production of inflammatory cytokines (TNF-α, IL-6) in both primary macrophages and BV2 microglial cells. Suppression of inflammatory cytokine production by K306 was found to be dependent on SHIP1 expression in both cell types, demonstrating K306 anti-inflammatory activity is mediated through agonism of SHIP1. Surprisingly K306 also enhanced phagocytic degradation of synaptosomes and dead neurons by microglia, but not that of Aβ42, a proteinacious cargo. These findings reveal a novel function for SHIP1 in microglial biology. Moreover, we find that K306 has a new mode of agonism for SHIP1 and interestingly it may be the most potent small molecule agonist of SHIP1 identified to date. Furthermore, we show that K306 can suppress LPS-induced TNF-α production in vivo, suggesting it, or an analog of it, might have therapeutic efficacy in inflammatory diseases, and perhaps also in diseases involving microglial dysfunction due to their inability to process large, lipid-bearing phagocytic cargoes as has recently been proposed in aging and dementia (Marschallinger et al., 2020).
Our screen identified four novel classes of small molecules that can increase SHIP1 enzyme activity in vitro and all four agonists (K284, K291, K306, and K314) showed selectivity for SHIP1 vs. SHIP2. Somewhat disappointingly, only one of the four, K306, appears to have biological activity consistent with a SHIP1 agonist on cells, although we did find that K284 significantly increases LPS-induced IL-6 production by BV2 cells, but surprisingly not that of TNF-α. As SHIP1 limits both TNF-α and IL-6 production by macrophages, the selective super-induction of TNF-α mediated by K284 is not consistent with it being a SHIP1 inhibitor. Further analysis is required to confirm this selective induction is specific to BV2 cells or perhaps an off-target effect of K284. We speculate that both K291 and K314 are not able to pass through the plasma membrane lipid bilayer to access SHIP1 in its cytosolic location, but further studies are required to confirm these molecules are indeed unable to enter the cytosol. Should this prove to be the case, it is feasible that K291 and K314 can be salvaged as biologically useful SHIP1 agonists by modifying them to increase their lipophilicity to improve cell permeability.
By combining protein engineering with analysis of K306 and AQX-MN100 agonism, we were also able to demonstrate that K306 exerts its activating effects on SHIP1 via a novel mechanism at the molecular level. K306 clearly does not require the C2 domain that is adjacent to its enzyme domain to increase SHIP1 phosphatase activity. This suggests it does not act as a PI(3,4)P2 mimic as the C2 domain mediates physiological agonism of SHIP1 in cells through binding of its PI(3,4)P2 product in a feed-forward fashion (Ong et al., 2007). Future mutational analyses and structural modeling should reveal more information regarding the K306 binding site on the SHIP1 protein, although NMR and X-ray crystallographic studies are likely needed to provide a complete understanding of the K306 mode of agonism.
There are several SHIP1 signaling roles, that might limit inflammatory cytokine production, that might now be explored for K306, in comparison with other SHIP1 agonists like AQX-MN100. For instance, how K306 reduces inflammatory cytokine production should be explored, including signaling pathways that lead to activation of NF-κB, NFAT, or STAT1. In addition, the ability of K306 to reduce production of other proinflammatory mediators, ROS and NO, by macrophages might also be examined as we find that it can reduce induction of iNOS. It will also be interesting to determine if K306 influences SHIP1 signaling pathways where it has a non-enzymatic function. SHIP1 plays a masking role for the intracellular portions of receptors to limit their signaling as shown for the SLAM family receptor 2B4 (Wahle et al., 2007), TREM2 (Peng et al., 2010), and CD95/Fas (Park et al., 2014). We show here that K306 reduced Aβ42-mediated induction of TNF-α by microglia. This suggests K306 might enhance the stability of SHIP1’s association with DAP10, the co-receptor for TREM2 in microglia and macrophages (Peng et al., 2010). TREM2 is thought to be the major microglial sensor for Aβ42 (Wang et al., 2015).
It is particularly exciting that the discovery of a novel SHIP1 agonist has led to identification of a previously unknown biological function for SHIP1—promoting the degradation of large, lipid-laden cargo by microglial cells. Whether this is an enhancement of conventional phagocytic flux via Rab5 or Rab7, improved autophagic flux into the phagolysosomal compartment via Atg7 or possibly by promoting flux into the recently identified gastrosome (Villani et al., 2019) remains to be determined. Improved gastrosomal flux is a distinct possibility as K306 selectively enhances degradation of large lipid-bearing cargo, and not a proteinaceous cargo like Aβ42. Further studies of K306 in mutant microglia that selectively impair each of these phagocytic processes are now warranted. In addition to defining the cell biological processes and their molecular components that are promoted by K306 to increase degradation of dead cells and synaptosomes, it will also be interesting to examine the physiological and pathological processes that K306, or an analog, might have in aging and diseases impacted by microglial dysfunction. For instance, lipid-droplet-associated microglia (LDAM) are enriched in both aging and in dementias like Alzheimer and also acquire proinflammatory function (Marschallinger et al., 2020). That K306 can both reduce production of inflammatory cytokines by microglia in response to beta-amyloid, while also improving their ability to process lipid-laden cargo, suggests this is a valid possibility. In addition, lipid-laden macrophages (e.g., foam cells) that are unable to recycle, and efflux cholesterol properly contribute to atherosclerotic disease (McLaren et al., 2011; Yu et al., 2013). Thus, the potential for K306 to re-balance cholesterol uptake, processing, and efflux in such macrophages might also be explored in atherosclerosis models.
In addition to the above important biochemical, cell signaling and pathology questions concerning the utility of K306, an assessment of its potential to abrogate inflammatory disease in the lungs and gut seems warranted. This is important because the failure of AQX-1125 to modulate these diseases, including COPD, has been viewed as a clinical failure for SHIP1 agonism approaches. However, Chamberlain et al. have recently found that AQX-1125 exhibits negligible binding for SHIP1 suggesting this may not have been an ideal small molecule to advance clinically (Chamberlain et al., 2020). The relative water insolubility of pelorols like AQX-MN100, -16A, and −435 may in turn limit their clinical testing (Lemm et al., 2020). K306 is more water-soluble than the pelorols (Table S1), has greater potency, and thus it, or possibly an analog, might now be explored to allow a proper test of the true potential for SHIP1 agonism to abrogate diseases like COPD and IBD. That a subset of patients with IBD is SHIP1-deficient, but still retains some SHIP1 activity further supports the necessity of testing novel potent SHIP1 agonists in IBD and perhaps other inflammatory diseases, including Alzheimer disease.
Limitations of the study
As with any study describing a novel small molecule that impacts the function of a key signaling molecule like SHIP1, there is always the possibility that it also has undesired effects, by interacting with some other protein in the cell. However, we feel that we have mitigated this possibility to a significant degree by testing the novel SHIP1 agonist in SHIP1 null macrophages and microglial cells. This analysis did not reveal an effect of the novel SHIP1 agonist on inflammatory mediators when SHIP1 was absent, but nonetheless off-target effects could subsequently be found to impact other cellular functions that we have not anticipated. We also should point out that K306 represents a novel chemical class of SHIP1 agonists that contains a sulfur atom and potentially other chemical groups that are subject to oxidation by oxidases elaborated by various cell types in the body, or even in microglia and macrophages themselves. This may have reduced some of the cellular or biological potency of this first-generation molecule (K306) as it may be subject to rapid degradation in cells. Some of our data on the effect of K306 on iNOS induction in microglia already suggest this possibility. Thus, over time, it will be important to test analogs of K306 that retain agonistic activity but where the sulfur atom, or other groups, have been replaced to render this novel class of agonists more resistant to cellular oxidation pathways. This might result in more efficacious analogs that could significantly increase the utility of this novel class of SHIP1 agonists in inflammatory diseases and potentially also dementias like Alzheimer Disease. It should also be pointed out that we did not see increased degradation of Aβ42 peptide in vitro in microglia treated with K306. It merits mention that it is currently thought that most neurotoxic Aβ42 deposits are associated with lipids. Thus, it will be interesting to see if Aβ42 degradation by microglia is enhanced by K306 treatment when Aβ42:lipid compositions are tested, or perhaps in vivo with plaque-associated Aβ42 deposits in amyloidogenic mouse models of AD.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SHIP-1 Antibody (P1C1) | Santa Cruz Biotechnology | Cat# Sc-8425 |
| Santa Cruz Biotechnology | Cat# Sc-13119 | |
| Anti-β-Actin Antibody (C4) | Santa Cruz Biotechnology | Cat# Sc-47778 |
| m-IgGκ BP-HRP | Santa Cruz Biotechnology | Cat# Sc-516102 |
| TruStain FcX (anti-mouse CD16/32, Clone 93) | Biolegend | Cat# 101320 |
| SHIP1-PE (Clone P1C1) | Biolegend | Cat # 656604 |
| APC/Cyanine7 anti-mouse CD45 Antibody | Biolegend | Cat# 103116 |
| CD45-PerCP (Clone 30-F11) | Biolegend | Cat# 557235 |
| CD11b-APC_Cyanine7 (Clone M1/70) | BD Biosciences | Cat# 557657 |
| iNOS-PE_eFlour610 (CloneCxNFT) | ThermoFisher Scientific | Cat #61-5920-80 |
| Anti-PtdIns(3,4)P2 | Echelon Biosciences | Cat# Z-P034 |
| Phalloidin CruzFluor, 647 Conjugate | Santa Cruz Biotechnology | Cat#sc-363797 |
| Alexa Fluor Plus 555 Goat anti-Mouse IgG (H+L) | Thermofisher Scientific | Cat# A32727 |
| Bacterial and virus strains | ||
| DH 5 alpha competent E.coli | New England Biolabs | Cat# C2987H |
| BL2I (DE3) competent E.coli | New England Biolabs | Cat# C2527H |
| Chemicals, peptides, and recombinant proteins | ||
| beta-Amyloid Peptide (1-42) Human | Abcam | Cat#ab120301 |
| Lipopolysaccharides from, E coli 0111:B4 (LPS) | Sigma-Aldrich | Cat# L2630 |
| Beta-Amyloid (1-42), HiLyte™ Fluor 488-labeled Human | Anaspec | Cat# AS-60479-01 |
| SHIP2 Lipid Phosphatase Enzyme | Echelon Biosciences | Cat # E-1000-2 |
| BugBuster HT | Millipore | Cat# 70922-4 |
| PI(3,4,5)P3 diC8 (PIP3) | Echelon Biosciences | Cat# P-3908-3 |
| Malachite Green Solution | Echelon Biosciences | Cat# K-1501 |
| eBioscience IC Fixation Buffer | ThermoFisher Scientific | Cat# 00-8222-49 |
| eBioscience Permeabilization Buffer (10X) | ThermoFisher Scientific | Cat# 00-8333-56 |
| Zombie Aqua Fixable Viability Kit | Biolegend | Cat #423102 |
| Molecular Probes DAPI (4',6 Diamidino 2 Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat#D1306 |
| Propidium iodide (PI) | Sigma | Cat#P4170 |
| Mowiol (Polyvinyl Alcohol) | Sigma Merck | Cat#81381 |
| Critical commercial assays | ||
| Plasmid Midi Kit (25) | Qiagen | Cat#12143 |
| 5’PtdIns(3,4,5)P3 Phophatase Activity Fluorescence Polarization Assay | Echelon Biosciences | Cat# K-1400 |
| SuperSignal™ West Femto Maximum Sensitivity Substrate | Thermo Scientific™ | Cat#34095 |
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | Thermo Scientific™ | Cat#34080 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (100X) | Thermo Scientific™ | Cat#78440 |
| Syn PER Synaptic Protein Extraction Reagent | Thermo Fisher Scientific | Cat#87793 |
| 4–15% Mini-PROTEAN® TGX™ Precast Protein Gels | Bio-Rad | Cat#4561084 |
| ELISA MAX™ Standard Set Mouse IL-6 | BioLegend | Cat#431301 |
| ELISA MAX™ Standard Set Mouse TNF-α | BioLegend | Cat#430901 |
| TMB Substrate Set | BioLegend | Cat#421101 |
| Cell Counting KIT-8 CCK-8 | Dojindo Molecular Technologies Inc | Cat#CK04-20 |
| Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent | Thermo Fisher Scientific | Cat#CMAX00008 |
| Truecut™ Cas9 Protein v2 | Thermo Fisher Scientific | Cat#A36498 |
| TrueGuide™ Synthetic sgRNA – Inpp5d mouse | Thermo Fisher Scientific | Cat#CRISPR494808_SGM |
| TrueGuide™ Synthetic sgRNA – Negative control | Thermo Fisher Scientific | Cat#A35526 |
| Syn PER Synaptic Protein Extraction Reagent | Thermo Fisher Scientific | Cat#87793 |
| V-PLEX Aβ Peptide Panel 1 (6E10) Kit | Meso Scale Discovery | Cat#K15200E-2 |
| In Vitro Toxicology Assay Kit, Resazurin based | Sigma-Aldrich | Cat#TOX8-1KT |
| Phrodo™ iFL Green STP Ester (amine-reactive) | Thermo Fisher Scientific | Cat# P36012 |
| Deposited data | ||
| SHIP2 structure | (Mills et al., 2012) | PDBID: 4A9C |
| SHIP2 structure | (Le Coq et al., 2017) | PDBID: 5OKM |
| INPP5B structure | (Tresaugues et al., 2014) | PDBID: 3MTC |
| Mcule library | (Hsieh et al., 2019) | N/A |
| Experimental models: Cell lines | ||
| BV2 | ATCC | Cat# CRL-2467 |
| Mouse BV2 scramble clone | Generated by Paolicelli’s lab | NA |
| Mouse BV2 Inpp5d clone 1 | Generated by Paolicelli’s lab | NA |
| Mouse BV2 Inpp5d clone 2 | Generated by Paolicelli’s lab | NA |
| Mouse BV2 cell-line | Kind gift of Marc Suter (UNIL) | CVCL_0182 |
| Human Stably expressing-APPsw Hela cells | Kind gift from Lawrence Rajendran (UZH) | N/A |
| L929 mouse fibroblast cells | Kind gift of Steven Taffet (UPSTATE) | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J | Jackson Laboratory | Cat#000664 |
| B6.129S6-Inpp5dtm1Wgk/J | Jackson Laboratory | Cat#028255 |
| B6.129P2-Lyz2tm1(cre)Ifo/J | Jackson Laboratory | Cat#004781 |
| Mouse: B6.Cg-Tg(Camk2a-cre)T29-1Stl/J | Jackson Laboratory | Cat#005359 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG tdTomato)Hze/J | Jackson Laboratory | Cat#007914 |
| Oligonucleotides | ||
| See Table S2 for: SHIP1 cloning primers (pS97splitRBP_S1-Enz) | This Paper | N/A |
| See Table S2 for: SHIP1deltaC2 cloning primers (pS97splitRBP_ S1ΔC2) | This Paper | N/A |
| See Table S2 for: SHIP2 cloning primers (pS97splitRBP_ S2-Enz) | This Paper | N/A |
| Recombinant DNA | ||
| pET24TEV-tSHIP1 | (Brooks et al., 2015) | Addgene ID 183770 |
| pET24TEV-tSHIP2 | (Russo et al., 2015) | Addgene ID 183771 |
| pS97splitRBP | (Ha et al., 2019) | N/A |
| pS97splitRBP_S1ΔC2 | This paper | Addgene ID 183769 |
| pS97splitRBP_S1-Enz | This paper | Addgene ID 183767 |
| pS97splitRBP_S2-Enz | This paper | Addgene ID 183768 |
| Software and algorithms | ||
| AtomNet® AI technology | This paper | N/A |
| FlowJo v10.8.1 | BD Biosciences | https://www.flowjo.com |
| SpectroFlo Version 2.2.0.2 | Cytek | https://cytekbio.com/pages/aurora |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| Imaris | Bitplane | http://www.bitplane.com/imaris/imaris |
| Prism 9.2 | GraphPad | https://www.graphpad.com/ |
| BioTek Gen2 Data analysis software | Agilent | https://www.biotek.com/ |
| Image Lab Software | Bio-rad | Cat# 12012931 |
| Thunder Imaging System | Leica Microsystems | https://www.leica-microsystems.com |
| Other | ||
| HisTrap™ High Performance 5mL column | Cytiva Life Sciences | Cat# GE17-5248-01 |
| Pierce Protein Concentrators 30KMWCO PES | Thermo Fisher Scientific | Cat#88529 |
| DMEM/Ham's F-12 50/50 Mix [+] L-glutamine | Corning® | Cat#10-090-CV |
| Cellstripper™, Liquid | Corning® | Cat# 25-056-CI |
| Gibco™ DMEM, low glucose, pyruvate | ThermoFisher Scientific | Cat# 31885049 |
| Trypsin-EDTA (0.05%), phenol red | ThermoFisher Scientific | Cat# 25300062 |
| Poly-D-lysine Hydrobromide | Sigma Merck | Cat# P7886 |
| μ-Slide 18 Well | IBI | Cat# 81817 |
| Geneticin disulphate (G418) | Carl Roth | Cat# 108321-42-2 |
| Zeocin™ Selection Reagent | ThermoFisher Scientific | Cat# R25001 |
| Microvette® 100 Serum, 300 μL | Sarstedt | Cat#20.1308.100 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, William G. Kerr (wgkerr@mac.com).
Materials availability
The following plasmids generated and used in this study have recently been deposited to Addgene (Deposit Number: 80865).
They are:
pS97splitRBP_S1-Enz
pS97splitRBP_S2-Enz
pS97splitRBP_S1dC2
pET24TEV-tSHIP1
pET24TEV-tSHIP2
The SHIP1flox mice used in this study have been deposited at Jackson Laboratories.
Experimental model and subject details
For the LPS-induced endotoxemia study, C57BL6/J (Male) mice (8-10 weeks old) were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were housed at the SUNY Upstate Medical University vivarium under conventional, nonspecific-pathogen-free conditions.CamKIIa-CRE mice crossed to Rosa26-fl-STOP-fl-TdTomato adult male mice were housed at the UNIL animal facility in nonspecific-pathogen-free conditions and used to produce synaptosomes for uptake and degradation assay.
Method details
Computational method
VHTS screen: At the onset of the virtual high throughput screen for SHIP1 modulators, no experimentally determined structure of the protein was available and hence homology models were built using known SHIP2 structures (PDBID: 4A9C & 5OKM). Virtual screening was performed using the AtomNet® AI technology, for structure-based drug design trained to predict protein-ligand binding affinity. The screening site was centered around the SHIP1 residues N520, K521, H541, S544, Y644, R645 and N667, determined as the potential binding site for inositol phosphate, based on alignment to the INPP5B complex (PDBID: 3MTC). The Mcule library of approximately 4M commercially available organic small molecule compounds was prepared and screened, as described previously(Hsieh et al., 2019), using an ensemble of protein-ligand conformations. Each of the 4M molecules was scored and ranked by the AtomNet® technology, following which a top set of chemically diverse compounds was further inspected for undesirable substructures and molecular properties before 76 compounds were purchased for experimental testing. Following initial testing and identification, K306 was synthetized in house with the scheme described in the Materials.
Cloning and expression of SHIP1-Enz, SHIP1ΔC2 and SHIP2-Enz
Cloning and expression of tSHIP1 from human whole blood mRNA have been previously described (Brooks et al., 2015). Truncated human SHIP1 protein containing phosphatase and C2 domain (SHIP1-Enz, residues 397-864) and the truncated SHIP1 protein without the C2 domain (SHIP1ΔC2 residues 397-730) were amplified from pET24TEV-tSHIP1 (Brooks et al., 2015). Truncated human SHIP2 protein containing phosphatase and C2 domain (SHIP2-Enz, residues 414-872) was amplified from pET24TEV-tSHIP2 (Russo et al., 2015). All primers used for cloning are listed in Table S2. These three constructs were then individually cloned into pS97splitRBP(Ha et al., 2019) between NotI and BamHI sites, in E.Coli DH5 alpha (New England Biolabs). Plasmids were extracted with Midi Prep (Qiagen) and sequences were verified by sequencing of the entire expression regions. The plasmids were then transformed into E. coli BL21 (New England Biolabs). Fresh colonies were amplified in 1L LB at 37°C, shaking at 250rpm until OD600nm reached 0.6. Protein expression was induced with 200μM IPTG and incubation was continued at 16°C, shaking at 250rpm for 16h. Following centrifugation, bacteria pellets were stored overnight at −20°C. Protein was extracted form bacterial cell pellet using BugBuster HT (Millipore), according to manufacturer’s recommendations. His-tagged proteins were purified by FPLC using HisTrapHP 5mL column (Cytiva Life Sciences) at a 5 mL/min flow rate and 10mM-1M imidazole gradient in Buffer A (20mM Tris pH 8.0, 300mM NaCl, 10mM imidazole, 5mM β-Mercaptoethanol). Fractions containing active protein by Malachite Green Phosphatase Release Assay (Echelon, see below) were pooled, concentrated and buffer exchanged to reduce imidazole concentration by centrifugation using Pierce Protein Concentrators 30KMWCO PES, (ThermoFisher Scientific). Protein was aliquoted and stored at −80°C in Buffer A without β-ME and containing 50% glycerol.
Malachite green phosphatase release assays
Malachite Green Phosphatase Release Assays (Echelon Biosciences) were performed with recombinant human truncated SHIP1 (tSHIP1) (Brooks et al., 2015) and SHIP2 (Echelon Biosciences). Briefly, serial dilutions of the compounds dissolved in appropriate solvent (DMSO for newly discovered agonists or ETOH for AQX-MN100) were added to the recombinant enzymes diluted in reaction buffer Rx (50mM Hepes pH 7.4, 150mM NaCl, 1mM MgCl2, 0.25mM EDTA) in triplicate reactions in 96-well plates. Reactions were incubated for 2 min at room temperature. 2.5 μL of 1mM Phosphatidylinositol 3,4,5-trisphosphate diC8 (PI(3,4,5)P3diC8) (Echelon Biosciences) was added to each reaction to a final concentration of 100μM in a final volume of 25 μL/well. Following 20 min incubation at 37°C, 100μL of Malachite Green Solution (Echelon Biosciences) was added to each well and plates were incubated at room temperature in the dark for 15 min. Plates were then read at 620nm on a plate reader (Synergy 2, BioTek).
5′PtdIns(3,4,5)P3 phosphatase activity fluorescence polarization (FP) assay
FP Assay for detection of PI(3,4)P2 (Echelon Biosciences) was performed using tSHIP1 (Brooks et al., 2015), SHIP1-Enz or SHIP2-Enz with serial dilutions of K306 (in DMSO) or AQX-MN100 (in ETOH) according to manufacturer’s recommendation as previously described (Fuhler et al., 2012). Briefly, serial dilution of the compounds and a control with 2.5%solvent only (0μM) is added to the enzyme in a volume of 40μL of enzyme specific reaction buffer (Rx) (S1 Reaction buffer for tSHIP1 and SHIP1-Enz: 20mM Tris pH7.5, 150mM NaCl, 0.05% Tween 20, 10mM MgCl2), (S2 reaction buffer for tSHIP2 or SHIP2-Enz: 50mM Hepes pH 7.4, 150mM NaCl, 1mM MgCl2, 0.25mM EDTA). 40μL of 4μM PIP3 (PI(3,4,5)P3 diC8) in Rx buffer is added to each reaction and incubated at 37°C for 20min, followed by heat inactivation for 3 min at 95°C. Reactions were spun down briefly and 10μL of each reaction was added in 6 replicate wells to a black 384-well plate provided with the FP kit. 10μL of 1X reconstituted Detector in PBS is then added to each well, followed by 5μL of 1X freshly diluted Probe (in PBS) protected from light. Assay plates are gently tapped to mix, the plate is sealed and spun down briefly and reaction in allowed to equilibrate for 60 min at room temperature protected from light. Control wells are included with each assay (No Enzyme control (NE):5μL Rx buffer with 2.5% solvent+ 5μL PIP3 (4μM), 10μL Detector, 5 μL Probe, Probe Alone (PA) control (10μL enzyme buffer, 10μL PBS, 5 μL Probe). Fluorescence polarization is then read on a Bio-Tek Synergy 2 (Bio-Tek) plate reader and is expressed in milli Polarization (mP) units.
Cell culture
For synaptosomes uptake and degradation studies and the beta amyloid clearance assay BV2 cells were cultured in low glucose DMEM (ThermoFisher Scientific) supplemented with 10% fetal bovine serum and 1% Penicillin Streptomycin, at 37 °C and 5% CO2 in a humidified incubator. Cells were split with Trypsin-EDTA 0.05% (ThermoFisher Scientific) and seeded in Poly-D-Lysine (0.1 mg/mL, Sigma Merck) coated 96-well plates (VWR) or μ-Slide 18 Well Glass Bottom (Vitaris) according to the experiment performed. For inflammatory cytokines production studies, iNOS detection studies, beta amyloid and dead neuron uptake and degradation studies, BV2 were cultured in DMEM/F12 (Corning) 10% HI FBS at 37 °C and 5% CO2 in a humidified incubator and split 1:6 every 3 days with Cellstripper™ (Corning). BMDM were prepared as described previously (Saz-Leal et al., 2018) from LysMCre + SHIP1 Flox/Flox mice, LysMCre negative SHIP1 Flox/Flox mice and C57BL6/JL WT mice with L929 supernatant. For amyloid production in the Clearance Assay, HeLa cells expressing the APP Swedish mutation (HeLa swAPP) were cultured in DMEM (Invitrogen), supplemented with 10% FCS and 100 U mL-1 penicillin/streptomycin, 0.1% G418 antibiotic (Carl Roth), and 0.1% Zeocin (Invitrogen). Cells were kept at 37 °C and 5% CO2 in a humidified incubator.
Generation of Inpp5d single clones through CRISPR/Cas9 genome editing
24 h after seeding (2000 cells/well) in coated 96-well plates, Inpp5d genetic editing was performed using 0.3 μL/100μL Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent (ThermoFisher Scientific, CMAX00008), 15nM Inpp5d Mouse TrueGuide Synthetic gRNA (ThermoFisher Scientific, A35533), and 250 ng Truecut™ Cas9 Protein v2 (ThermoFisher Scientific, A36499). After 72 h, cells were split, and single cells were seeded by means of serial dilution. SHIP1 reduction in single clones was assessed by means of Western Blot (WB), Intracellular Flow Cytometry (ICFC) and Immuno-Fluorescence (IF) as reported in Figure S1.
Immunofluorescence for SHIP1 on BV2 clones
Cells were fixed with 4% PFA for 20 min at RT. After 15 min of permeabilization (PBS, Triton 0.25%), samples were incubated with Blocking Buffer (BF = PBS, Triton 0.25%, BSA 2%) for 1h at RT. Cells were incubated with primary antibody ON at 4 °C (SHIP1 P1C1, 1:200 in BF, Santa Cruz Biotechnology; PI(3,4)P2 Purified Anti-PtdIns(3,4)P2 IgG, Echelons Biosciences, #Cat Z-P034, final concentration 5 μg/mL). After three washes with PBS, Alexa Fluor Plus 555 Goat anti-Mouse IgG (H + L) (1:1000 in BF, Thermofisher Scientific) was added on the samples for 1h at RT. Other three washes with PBS were performed, followed by DAPI incubation (1 μg/ml in PBS, Thermo Fisher Scientific) for 10 min at RT. Cells were rinsed once with PBS and coverslips were mounted on glass slide with Mowiol. Images were acquired with a fluorescence microscope (Thunder Imaging System, Leica Microsystems) or with a Leica Stellaris 5 Confocal microscope (Leica Microsystems), with APO 63x/1.40 oil objective. Two biological replicates per condition were acquired and analyzed. Image analysis was performed using ImageJ software. Fixed thresholds for signal intensity were applied across experiments, and area covered was measured per field of view, then rationalized for the number of cells, given by DAPI counting.
WB of SHIP1 edited clones and parental BV2
Cells were harvested from an 80% confluent P2 culture. After two washes with cold PBS, cells were spun down at 10,000xg for 1 min and lysed on ice for 20 min with Lysis buffer (20mM Tris, 150mM NaCl, 1mM EGDA, 1mM EGTA, 1%Triton X-100) containing 1x protease and phosphatases inhibitor Halt (ThermoFisher Scientific). The cell lysate was spun down for 20 min at 10,000xg to eliminate cell debris. Western blot was then performed as described previously (Pedicone et al., 2019). Briefly, 25μg of cleared lysate in loading buffer containing DTT was loaded on 4-15% Precast gels (Bio-Rad). The gel was run 30 min at 180V followed by transfer to a nitrocellulose membrane with Trans-Blot Turbo (Bio-Rad). The membrane was blocked in 5% Nonfat milk (Cell Signaling) in TBST (Tris-Buffer Saline, 0.1% Tween 20) O/N and incubated 2h with 1:500 SHIP1 P1C1 (Santa Cruz Biotechnology), 1:1000 β-actin C-4 (Santa Cruz Biotechnology), or 1:1000 HSP90 α−β F-8 (Santa Cruz Biotechnology). After three-10-min washes in TBST the membrane was incubated with1:1000 m-IgGkBP-HRP (Santa Cruz Biotechnology) in 5% Nonfat milk for 45 min. To detect the chemiluminescence signal, we used a Bio-Rad Chemidoc with ECL Pico(ThermoFisher Scientific) for housekeeping proteins and ECL Femto (ThermoFisher Scientific) for SHIP1.
ICFC of SHIP1 edited clones and parental BV2
Parental BV2 cells, Clone 1 and Clone2 were seeded at 5X105 cells/mL, 0.5 mL/well in 24 wells plates. 16h after seeding cells were washed with PBS, harvested with PBS 1%EDTA, and spun down 5 min 350xg. Cells were then incubated with 0.25 μL of Zombie Aqua (Biolegend) in 100 μL of PBS for 20 min on ice. Cells were washed with FACS buffer (FB: PBS, 3%FBS HI,0.5%HEPES) and spun down 5 min at 350xg. Cells were fixed with 200μL eBioscience IC Fixation Buffer (ThermoFisher Scientific) for 20min on ice and washed/permeabilized with 2mL 1X eBioscience Permeabilization Buffer (1XPB, ThermoFisher Scientific) and spun down 5 min at 400xg. Cells were incubated with 1 μL of Fc Block (TruStain FcX, Biolegend) in 50 μL 1XPB followed by in 2 μL of SHIP1-PE P1C1 (Biolegend) in 50 μL of 1XPB. Cells were incubated for 15 min on ice, washed with 2 mL of 1XPB, spun down 5 min at 4000xg and acquired with LSRII (BD). A biological duplicate was performed, 20.000 live cells were acquired in each sample and analyze with FlowJo10.
Flow cytometric assays for phagocytosis measurements
BV2 cells were seeded at 5X105 cells/mL, 0.5 mL/well in 24 wells plates. K306 or Vehicle control (0.5%DMSO) was added to each well and incubated for 1h before adding the phagocytic cargo. 0.5 μg/mL of beta amyloid 1-42 Hylite-488 labeled (Anaspec) or 5x105/mL Propidium Iodide (PI)-labeled dead neurons (DN) were then added to each well and incubated for 1h at 37°C. After the incubation the cells were washed with 1 mL of cold PBS and incubated for 0,2 or 6h with fresh media at 37°C containing K306 or solvent control. At the end of the incubation cells were washed with cold PBS, detached from the plate with PBS 1%EDTA, and incubated with antibody for surface staining in FB. 1 μL of Fc Block (TruStain FcX, Biolegend) in 50 μL FB was added to each tube followed 0.5 μL of CD45-APCCy7 (Biolegend) in 50 μL in FB/tube incubated for 15 min on ice. Cells were washed twice and resuspended in 200 μL of cold FB containing 0.1 μg/mL PI for beta amyloid assay or 0.1 μg/mL DAPI for DN assay as live/dead staining incubated on ice for 5 min, as previously described (Pedicone et al., 2019). Samples were kept on ice, acquired with BD LSR II and analyzed with FlowJo10.
Synaptosome isolation
Synaptosomes from adult mouse brains were isolated using Syn-PER™ Synaptic Protein Extraction Reagent (ThermoFisher Scientific, 87793). Briefly, brains from CamKIIcre/+; Rosa26-TdTomatoFlox/+ mice were collected and Dounce homogenization was performed on ice. The solution was centrifuged at 1200 × g for 10 min at 4 °C. The pellet was discarded, and the supernatant was further centrifuged at 15,000 × g for 20 min at 4 °C. The pellet containing synaptosomes was resuspended to obtain a final concentration of 6.4 μg/μL (5% v/v DMSO in Syn-PER™ reagent) and the suspension was aliquoted and stored at −80°C.
Synaptosomal uptake and degradation assay
48h after seeding (2000 cells/well) in coated μ-Slide 18 Well Glass Bottom, BV2 cells were treated with DMSO (1:1000, 1 h) and K306 (10 μM, 1 h). At the end of the treatment, cells were incubated with TdTomato-synaptosomes at a final concentration of 65 μg/ml. After three PBS washes, cells were fixed with 4% PFA to evaluate the amount of the cargo that was taken up (T0) or incubated with fresh medium for 6 h (T6H), prior to fixation, to assess synaptosome degradation. Four independent biological replicates were performed.
pHrodo-green synaptosome conjugation
TdTomato labelled-synaptosomes were diluted to a concentration of 1 mg/mL and mixed with 12 μg of Phrodo™ iFL Green STP Ester (amine-reactive) (Thermo Fisher Scientific, #Cat P36012) in 100 mM sodium bicarbonate pH 8.3, for 1h at RT rotating in the dark. After pelleting down, one wash with PBS was performed to remove unbound pHrodo. Synaptosomes were centrifuged at 15,000 × g for 20 min at 4 °C and then the pellet was resuspended in Syn-PER™ Synaptic Protein Extraction Reagent with 5% (v/v) DMSO and kept at −80°C for long storage.
Microscopy and live imaging of synaptosome uptake and degradation
Images were acquired as z-stack with a fluorescence microscope (Thunder Imaging System, Leica Microsystems), with APO 40x/0.95 dry and APO 63x/1.40 oil objectives. Two technical replicates per condition were acquired and analyzed within each biological replicate. Image analysis was performed on max-projections using ImageJ Software; fixed thresholds for signal intensity were applied across experiments, and mean intensity and area covered per cell were measured.
Live imaging degradation of Synaptosomes by BV2 was acquired by light sheet fluorescent microscopy (InVi-SPIM, Luxendo) and reported as Time-lapse micrographs, after initial 1h uptake, at 0, 120, 345 min in Figure S3 and or as full Video.S1
Three-dimensional reconstruction of BV2 cells engulfing synaptosomes was produced using Imaris Software (Bitplane, Switzerland) from z-stack acquisition using a confocal microscope (Stellaris 5, Leica). Briefly, after 1 hour incubation with TdTomato-labelled synaptosomes, cells were fixed with 4% PFA for 20 minutes at RT. After one wash with PBS, samples were incubated with 647 conjugated-Phalloidin CruzFluor (Santa Cruz Biotechnology, 1:1000 in PBS) for 30 minutes at RT. Cells were rinsed three times with PBS and coverslips were mounted on glass slide with Mowiol. 3D reconstruction was generated by applying surface rendering of confocal stacks for phalloidin-647 and TdTomato in their respective channels.
Aβ clearance assay
BV2 microglia cells were incubated with HeLa swAPP-conditioned medium, and kept at 37 °C, 5% CO2. Medium was collected after 6 h of incubation with cells and used for measurement of residual Aβ. Cell viability was subsequently analyzed with Resazurin assay (Sigma-Aldrich) using a fluorescent plate reader (Tecan) for cell viability normalization. 96-well MULTI-ARRAY Multiplex Kit (V-PLEX Aβ Peptide, Meso Scale Discovery) were used to measure the level of Aβ38, Aβ40, Aβ42 by electrochemiluminescence (ECL). Aβ species peptides were detected with a monoclonal antibody and quantified by using a Meso QuickPlex SQ120 reader (Meso Scale Discovery). Values were normalized to DMSO control and to cell viability.
Intracellular flow for INOS detection
BV2 cells were plated at 5x105 cells/mL, 0.5 mL/well in 24 well plates and allowed to adhere 2-4h at 37°C, 5% CO2. Cells were treated with K306 (10μM) or Vehicle (Veh: 0.25%DMSO) for 1h. LPS (100 ng/ml) or PBS were added to appropriate wells and incubation was continued for 1h. The second round of K306 (10μM) or Veh was added to each well such that the final concentration of DMSO was 0.5% in each well and the concentration of K306 was 10μM or 20μM as indicated. Cells were washed with PBS (1mL/well) and were harvested with PBS 1% EDTA. Cells were collected in FACS tubes (BD Biosciences) and washed with 2mL cold PBS and spun down 5 min at 350xg. Cell pellets were stained with Zombie Aqua (0.4μL of stock diluted 1:100 in 50μL of PBS/test) for 20min on ice, washed with 2mL cold PBS and spun down 5 min at 350xg. Pellets were resuspended in 15μL Fc Block (TruStain FcX, Biolegend), and stained with surface antibody cocktail (CD11b-APC_Cyanine7 (Clone M1/70) and CD45-PerCP (Clone 30-F11) in 35μL FB/sample for 20min on ice. Cells were washed with 2mL cold PBS and spun down 5 min at 350xg. Cells were fixed with 200μL eBioscience IC Fixation Buffer (ThermoFisher Scientific) for 20min on ice and washed with 1XPB and spun down 5 min at 400xg. Pellets were resuspended in 25μL Fc Block in 1XPB and stained with iNOS-PE_eFlour610 (CloneCxNFT). Cells were washed twice with 2mL 1XPB and spun down 5 min at 400xg. FMO control was stained with the cocktail minus the iNOS antibody. Final pellets were resuspended in 150μL 1XPB and samples were acquired on the Cytek Aurora, after acquiring single-stained colors and unstained control cells. Samples were unmixed using SpectroFlo Version 2.2.0.2. FSC files were exported and further analyzed with FlowJo Version 10.8.1. An example of gating strategy is found in Figure S2.
ELISA measurements
Parental (scrambled guide control) BV2 cells or SHIP1 edited clones were seeded at 5x105/mL, 250 μL/well in 48 well plate. Cell were treated with 10μM K306 or vehicle control (0.5% DMSO) and incubated for 1h before addiction of stimuli. 100 ng/mL of beta amyloid 1-42 (Abcam) or 10 ng/mL of LPS (E. coli O 111:B4 sigma) were added to the cells and incubated for 2 h to detect TNF-α or 6 h to detect IL-6 production. Supernatants were spun down at 400xg for 5 min to remove cell debris and froze at −20°C for ELISAs. TNF-α and IL-6 levels were normalized to percentage of live cells as compared to control (vehicle) determined by the Cell Counting KIT-8 (CCK-8, Dojindo Molecular Technologies Inc), that was diluted 1:10 in media, incubated with the cells for 2h and detected at 460nm with a plate reader (Synergy 2, BioTek). following manufacturer’s recommendations. Fully differentiated BMDM were seeded at 2x105/mL, 250 μL/well in 48 well plates and treated with the potential agonists and LPS stimuli as described for BV2. TNF-α and IL-6 levels were quantified from cellular supernatants or serum with ELISA MAX™ Standard Set Mouse kit for TNF-α (Biolegend) and IL-6 (Biolegend), following manufacturer’s recommendations. We detected absorbance at 450nm and removed 570nm background absorbance, with a plate reader (Synergy 2, BioTek). All experiments were repeated three times and each value was reported as mean ± SEM from biological replicates in the same assay.
Preparation of SHIP agonists for in vitro studies
All the agonists discovered in this study were resuspended from dry powder at a final concentration 20mM in DMSO (Sigma) for in vitro studies. AQXMN100 was resuspended in ethanol (ETOH) at 20mM. The agonists were diluted in solvent to 200X of desired final concentration and diluted 1:10 in media. The 20X solution was prepared fresh before each use and added to the seeded cells for a final solvent concentration of 0.5%(v/v).
Murine LPS-induced endotoxemia model
C57BL6/J mice (8-10 weeks old) were purchased from Jackson Laboratories (Bar Harbor, ME). Prior to and during LPS-induced endotoxemia, mice were housed at the SUNY Upstate Medical University vivarium under conventional, nonspecific-pathogen-free conditions. K306 was dissolved in water with 5% DMSO and administered to the mice with Intraperitoneal injection (IP). Two doses of K306 (20 mg/kg) or solvent control (5% DMSO) were administered: 30 min before and 30 min after LPS challenge. LPS (E. Coli O111:B4, Sigma) was delivered with IP injection at 2 mg/kg in PBS. Mice were euthanized 1h after LPS challenge by CO2 inhalation and blood was immediately collected by cardiac puncture and placed in clotting activator coated tubes (Sarstedt). After 20 min at room temperature, serum was separated from the blood cellular fraction by centrifugation and stored in aliquots at −80°C for ELISAs. The experiment was repeated with two independent cohort of mice with at least 5 mice in each group (N = 5). TNF-α ELISA (Biolegend) was performed on duplicate wells/mouse under manufacturer’s recommendation as reported above. Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at SUNY Upstate Medical University.
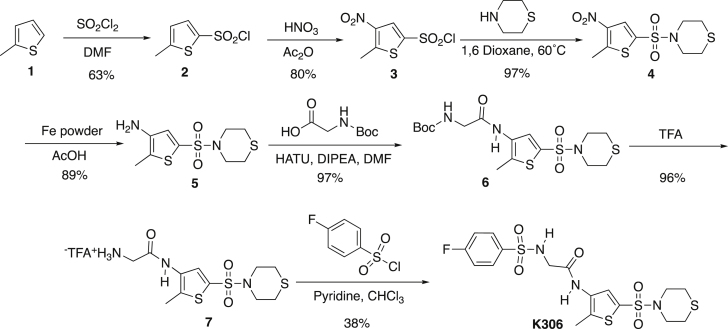
Synthesis of K306
The synthesis of K306 proceeded by the synthetic route shown below. Chlorosulfonylation and nitration of 2-methylthiophene 1 gave the sulfonyl chloride 3, which was then used to form the sulfonamide 4. Reduction of the nitro group and coupling of the resulting amine to Boc-glycine gace the amide 6. Removal of the Boc protecting group with trifluoroacetic acid (TFA) followed by formation of a second sulfonamide with 4-fluorobenzenesulfonyl chloride gave the K306 structure.
5-Methylthiophene-2-sulfonyl chloride (2)
Lit Ref
Dry DMF (1.02 mL, 13.23 mmol) was added to a flame-dried round bottom flask under argon and cooled to 0°C. Sulfuryl Chloride (1.07 mL, 13.23 mmol) was added dropwise to the solution and stirred for 40 min at 0°C forming a white crystal complex. 2-methylthiophene (1) (1.00 mL, 10.19 mmol) was then added dropwise to the complex and heated to 100°C and stirred for 1 h. The mixture was poured into ice-water and extracted with CHCl3 (3x25 mL). The organics were washed with a 5% NaHCO3 solution followed by water, dried over Na2SO4, and concentrated yielding the pure black oil (1.24g, 63%). TLC Rf = 0.655 (hexane: ethyl acetate, 80%:20%); 1H NMR (400 MHz, CDCl3) δ 7.70 (d, J = 4.0 Hz, 1H), 6.85 (dd, J = 3.9, 0.8 Hz, 1H), 2.61 (s, 3H); 13C(Baran et al. 2003) NMR (100 MHz, CDCl3) δ 152.6, 140.7, 135.5, 126.3, 12.9 (Sone et al., 1985)
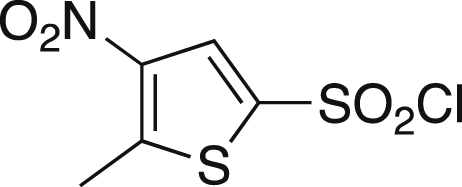
5-Methyl-4-nitrothiophene-2-sulfonyl chloride (3)
Lit Ref
Cooled fuming nitric acid (1.53 mL, 42.58) was added dropwise to a round bottom flask of acetic anhydride (1.66 mL, 17.57 mmol) and stirred at 0°C. A solution of sulfonyl Chloride 2 (0.58 g, 2.95 mmol) in acetic anhydride (1.66 mL, 17.57 mmol) was added dropwise over a 30 min period. The temperature of the mixture was raised to 15°C and stirred for 3 h. After stirring the mixture the was poured over crushed-ice, extracted with diethyl ether (3x15 mL), and washed with saturated NaHCO3 until a neutral pH was achieved. The organics were collected and dried over Na2SO4, filtered and concentrated yielding a pure orange powder (0.50 g, 70.4%). mp = 77.8-83.1°C; TLC Rf = 0.491 (30% EA/70% hexanes); IR (ATR) 3657, 3103, 2980, 1543, 1369, 1316, 1160, 1008, 541 cm−1; 1H NMR (CDCl3, 400 MHz) δ 8.19 (s, 1H), 2.86 (s, 3H); 13C[1H] NMR (CDCl3, 100 MHz) δ 152.3, 143.2, 138.8, 130.8, 16.2. Anal. Calcd for C5H4ClNO4S2: C, 55.49; H, 6.81; N, 4.98. Found: C, 55.11; H, 6.44; N, 4.90 (Gronowitz and Ander, 1975).
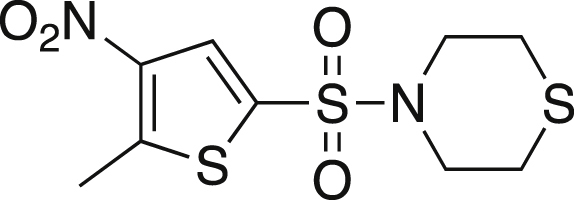
4-[(5-Methyl-4-nitrothiophene-2-yl)sulfonyl]thiomorpholine (4)
Thiomorpholine (0.46 mL, 4.55 mmol) and nitrothiophene 3 (0.50 g, 2.07 mmol) was dissolved in 2 mL of dry 1,4-dioxane and heated to 60°C. The mixture was stirred for 1 h at 60°C, after which the reaction was cooled to rt and 20 mL of water was added. The mixture was extracted with DCM (3x20 mL), and the organics were dried over MgSO4, filtered and concentrated. The crude mixture was purified via silica gel chromatography in 70% hexanes:30% ethyl acetate yielding a maroon solid (0.490g, 78%). mp = 142.7-147.0°C; TLC Rf = 0.531 (30% EA/70% hexanes); IR (ATR) 3655, 3117, 2980, 2913, 1545, 1507, 1368, 1163, 1153, 569 cm−1; 1H NMR (CDCl3, 400 MHz) δ 7.94 (s, 1H), 3.43 (t, J = 4.8 Hz, 4H), 2.86 (s, 3H), 2.76 (t, J = 4.8, 4H); 13C[1H] NMR (CDCl3, 100 MHz) δ 149.3, 143.7, 133.1, 127.9, 48.0, 27.3, 16.0. Anal. Calcd for C9H12N2O4S3: C, 35.05; H, 3.92; N, 9.08. Found: C, 35.13; H, 4.06; N, 8.91.
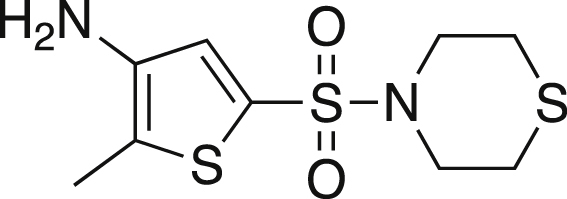
2-Methyl-5-thiomorpholine-4-sulfonyl)thiophen-3-amine (5)
Nitrothiophene 4 (0.49 g, 1.59 mmol) was dissolved in acetic acid (5.3 mL, 92.67 mmol) and iron powder (0.44 g, 7.95 mmol) was added. The mixture was heated to 60°C and stirred for 1 h, after which the acetic acid was removed via vacuo. The residue was dissolved in ethyl acetate and washed with saturated NaHCO3 until a pH of eight was reached. The organics were washed with brine, dried over MgSO4, filtered and concentrated. The crude mixture was purified via silica gel chromatography in a gradient of 70% hexanes:30% ethyl acetate to 50% hexanes:50% ethyl acetate yielding a dark solid (0.230g, 52%). mp = 139.7-146.7°C; TLC Rf = 0.121 (30% EA/70% hexanes); IR (ATR) 3361, 2913, 2852, 1565, 1348, 1327, 1144, 898, 699 cm−1; 1H NMR (CDCl3, 400 MHz) δ 7.54 (s, 1H), 3.72 (br s, 2H), 3.64 (t, J = 4.6 Hz, 4H), 3.01 (t, J = 4.8, 4H), 2.54 (s, 3H); 13C[1H] NMR (CDCl3, 100 MHz) δ 140.0, 130.5, 125.6, 122.0, 48.0, 27.3, 11.8. Anal. Calcd for C9H14N2O2S3: C, 38.83; H, 5.09; N, 10.06. Found: C, 39.17; H, 4.73; N, 9.84.
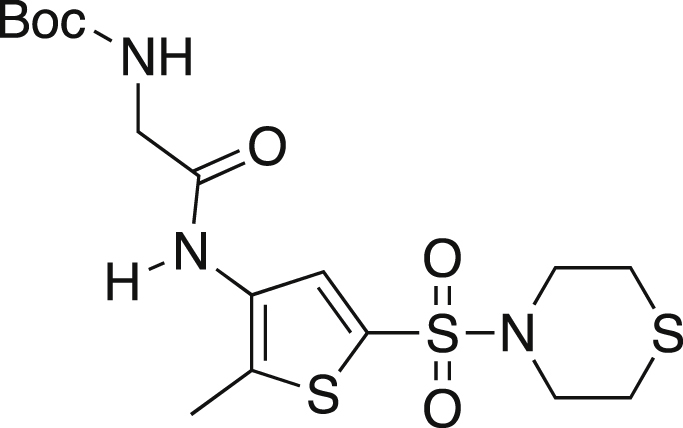
tert-butyl N-(([2-methyl-5-(thiomorpholine-4-sulfonyl)thiophen-3-yl]carbamoyl)methyl) carbamate (6)
Boc-glycine (0.159g, 0.905 mmol), amine 5 (0.210 g, 0.754 mmol), HATU (0.574 g, 1.509 mmol) and DIPEA (0.262 mL, 1.509) were dissolved in 10.5 mL of dry DMF under argon. The mixture was stirred for 24 h at rt. The reaction was diluted with ethyl acetate, washed with NH4Cl (3x15 mL) and brine. The organics were dried over MgSO4, filtered and concentrated. The crude mixture was purified via silica gel chromatography using 50% hexanes:50% ethyl acetate yielding a yellow oil (0.250g, 76%). TLC Rf = 0.308 (50% EA/50% hexanes); IR (ATR) 3656, 3254, 2980, 2971, 1665, 1350, 1154, 1141 cm−1; 1H NMR (CDCl3, 400 MHz) δ 8.55 (br s, 1H), 7.78 (s, 1H), 5.60 (t, J = 5.7 Hz, 1H), 3.89 (d, J = 5.6 Hz, 2H), 3.32 (t, J = 4.0 Hz, 4H), 2.68 (t, J = 5.2, 4H), 2.32 (s, 3H), 1.43 (s, 9H); 13C[1H] NMR (CDCl3, 100 MHz) δ 168.0, 156.9, 132.8, 132.2, 130.9, 128.6, 80.9, 47.9, 45.1, 28.3, 27.2, 12.4.
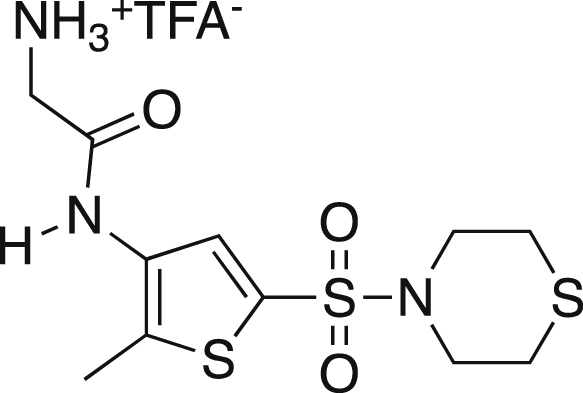
2-Aminotrifluoroacetate-N-[2-methyl-5-(thiomorpholine-4-sulfonyl)thiophene-3-yl]acetamide (7)
Boc protected amine 6 was dissolved in TFA and stirred at rt for 0.5 h. The solvent was removed under vacuo and a pure off-white solid was recovered (1.57g, 96.3%). mp = 190°C dc; IR (ATR) 3255, 2980, 2915, 1665, 1350, 1329, 1153 cm−1; 1H NMR (MeOD, 400 MHz) δ 7.79 (s, 1H), 3.93 (s, 2H), 3.37 (t, J = 5.0 Hz, 4H), 2.74 (t, J = 5.2, 4H), 2.45 (s, 3H); 13C[1H] NMR (MeOD, 100 MHz) δ 164.5, 138.9, 134.7, 131.7, 131.4, 128.9, 40.4, 26.7, 11.1. Anal. Calcd for C13H18F3N3O5S3: C, 34.74; H, 4.04; N, 9.35. Found: C, 34.97; H, 4.16; N, 8.96.
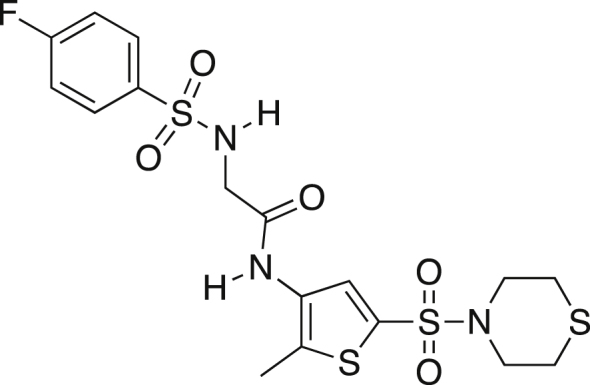
K306 (8)
Amine salt 7 (0.600g, 1.384 mmol) was dissolved in dry dichloromethane (2.15 mL) and dry TEA (0.425 mL, 3.045 mmol) and stirred at rt. 4-fluorobenzenesulfonyl chloride (0.296g, 1.523 mmol) was added and the mixture was stirred at rt for 24 h. The mixture was washed with water (3x 5 mL) followed by a 5% HCl solution. The organics were collected, dried over MgSO4, filtered and concentrated. The crude mixture was purified by precipitation from dichloromethane yielding a white powder (0.213g, 31.2%). mp = 177.2-180.3°C; TLC Rf = 0.371 (1% MeOH/99% DCM); IR (ATR) 3301, 3259, 2959, 2915, 1665, 1352, 1324, 1166, 1141 cm−1; 1H NMR (DMSO, 400 MHz) δ 7.82 (dd, J = 5.0, 8.5 Hz, 2H), 7.60 (s, 1H), 7.36 (t, J = 8.7 Hz, 2H), 3.63 (s, 2H), 3.19 (t, J = 4.4, 4H), 2.68 (t, J = 4.4 Hz, 4H), 2.31 (s, 3H); 13C[1H] NMR (DMSO, 100 MHz) δ.167.7, 138.5, 133.4, 133.2, 130.1, 130.0, 129.9, 129.3, 116.5, 116.3, 48.3, 26.8, 12.8; Anal. Calcd for C17H20FN3O5S4: C, 41.37; H, 4.08; N, 8.51. Found: C, 41.66; H, 3.80; N, 8.87.
K306 water solubility
Lipophilicity (CLogP) and Solubility (CLogS) were calculated for K306 and AQXMN100 using respectively XLOGP version 3.2.2 for CLogP (Wang et al., 1997) and topological method for CLogS (Delaney, 2004). Results are reported in Table S1.
Quantification and statistical analysis
All statistical analysis were performed using GraphPad Prism (version 9.2). two-way ANOVA was used to analyze all data with two independent variables as time and treatment or concentration and treatment. One-way ANOVA was used to analyze data with only one variable but comparing different treatments or concentrations. The Students T-test was used to analyze data comparing only two treatments. The statistical analysis used is reported in the figure legend for each assay, Data are reported as Mean ± SEM.
Acknowledgments
This work was supported by NIH grant RO1 AG059717(to WGK and JDC), an ERC Starting Grant (REMIND 804949) (to RCP) and the Synapsis Foundation - Alzheimer Research Switzerland ARS (to RCP).
Author contributions
W.G.K, C.P, A.M., and R.C.P. interpreted the data and wrote the manuscript with inputs from S.F.. D.B. ran the computational screening. J.C. and S.M. designed the K306 synthesis, calculated K306 solubility, and provided K306 along with Atomwise for the studies. S.L. and J.H.H. provided the new cloning vector and contributed with molecular cloning skills with S.F. and C.P. for vector design. S.F. was responsible for the protein production. S.F. and C.P tested the agonists with in silico screening. C.P. performed in vitro IL-6 and TNF-α studies for BMDM and BV2 cells. A.M. and C.P. performed respectively confocal and flow cytometry assays for cargo uptake and degradation, PI(3,4)P2 studies and SHIP expression studies on gene-edited cells. Gene-edited cells were generated by A.M. and R.C.P. S.F. ran the iNOS study with a spectral analyzer. C.P. and S.F. performed in vivo studies for LPS endotoxemia. All the author edited and approved the final version of the paper
Declaration of interests
D.B. is an Atomwise employee.
The other authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104170.
Contributor Information
Rosa Chiara Paolicelli, Email: rosachiara.paolicelli@unil.ch.
William G. Kerr, Email: wgkerr@mac.com.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baran C.P., Tridandapani S., Helgason C.D., Humphries R.K., Krystal G., Marsh C.B. The inositol 5'-phosphatase SHIP-1 and the Src kinase Lyn negatively regulate macrophage colony-stimulating factor-induced Akt activity. J. Biol. Chem. 2003;278:38628–38636. doi: 10.1074/jbc.M305021200. [DOI] [PubMed] [Google Scholar]
- Bradshaw W.J., Williams E.P., Fernandez-Cid A., Burgess-Brown N., von Delft F., Arrowsmith C.H., Edwards A., Bountra C., Gileadi O. 2019. (Released: 2019-01-16). PDB ID: 6IBD The phosphatase and C2 domains of human SHIP1. [DOI] [Google Scholar]
- Brooks R., Fuhler G.M., Iyer S., Smith M.J., Park M.Y., Paraiso K.H., Engelman R.W., Kerr W.G. SHIP1 inhibition increases immunoregulatory capacity and triggers apoptosis of hematopoietic cancer cells. J. Immunol. 2010;184:3582–3589. doi: 10.4049/jimmunol.0902844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks R., Iyer S., Akada H., Neelam S., Russo C.M., Chisholm J.D., Kerr W.G. Coordinate expansion of murine hematopoietic and mesenchymal stem cell compartments by SHIPi. Stem Cells. 2015;33:848–858. doi: 10.1002/stem.1902. [DOI] [PubMed] [Google Scholar]
- Brown G.C., Vilalta A. How microglia kill neurons. Brain Res. 2015;1628:288–297. doi: 10.1016/j.brainres.2015.08.031. [DOI] [PubMed] [Google Scholar]
- Chamberlain T.C., Cheung S.T., Yoon J.S.J., Ming-Lum A., Gardill B.R., Shakibakho S., Dzananovic E., Ban F., Samiea A., Jawanda K., et al. Interleukin-10 and small molecule SHIP1 allosteric regulators trigger anti-inflammatory effects through SHIP1/STAT3 Complexes. iScience. 2020;23:101433. doi: 10.1016/j.isci.2020.101433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Shojaee S., Buchner M., Geng H., Lee J.W., Klemm L., Titz B., Graeber T.G., Park E., Tan Y.X., et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015;521:357–361. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement S., Krause U., Desmedt F., Tanti J.F., Behrends J., Pesesse X., Sasaki T., Penninger J., Doherty M., Malaisse W., et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409:92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- Colonna M., Butovsky O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 2017;35:441–468. doi: 10.1146/annurev-immunol-051116-052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J.S. ESOL: estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004;44:1000–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- Ecker V., Stumpf M., Brandmeier L., Neumayer T., Pfeuffer L., Engleitner T., Ringshausen I., Nelson N., Jucker M., Wanninger S., et al. Targeted PI3K/AKT-hyperactivation induces cell death in chronic lymphocytic leukemia. Nat. Commun. 2021;12:3526. doi: 10.1038/s41467-021-23752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S., Brooks R., Gumbleton M., Park M.Y., Russo C.M., Howard K.T., Chisholm J.D., Kerr W.G. SHIPi enhances autologous and allogeneic hematopoietic stem cell transplantation. EBioMedicine. 2015;2:205–213. doi: 10.1016/j.ebiom.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S., Srivastava N., Sudan R., Middleton F.A., Shergill A.K., Ryan J.C., Kerr W.G. SHIP1 deficiency in inflammatory bowel disease is associated with severe crohn’s disease and peripheral T cell reduction. Front. Immunol. 2018;9:1100. doi: 10.3389/fimmu.2018.01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhler G.M., Brooks R., Toms B., Iyer S., Gengo E.A., Park M.Y., Gumbleton M., Viernes D.R., Chisholm J.D., Kerr W.G. Therapeutic potential of SH2 domain-containing inositol-5'-phosphatase 1 (SHIP1) and SHIP2 inhibition in cancer. Mol. Med. 2012;18:65–75. doi: 10.2119/molmed.2011.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Watanabe R.M., Valle T.T., Hauser E.R., Magnuson V.L., Langefeld C.D., Ally D.S., Mohlke K.L., Silander K., Kohtamaki K., et al. The Finland-united states investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. an autosomal genome scan for genes that predispose to type 2 diabetes. Am. J. Hum. Genet. 2000;67:1174–1185. [PMC free article] [PubMed] [Google Scholar]
- Gronowitz S., Ander I. On the base-catalysed reaction of some methyl nitrothiophenes with aldehydes, an unexpected cyclobutane formation. Acta Chem. Scand. Ser. B-Organic Chem. Biochem. 1975;29:513–523. [Google Scholar]
- Gumbleton M., Sudan R., Fernandes S., Kerr W.G., Engelman R.W., Russo C.M., Chisholm J.D., Kerr W.G. Dual enhancement of T and NK cell function by pulsatile inhibition of SHIP1 improves antitumor immunity and survival. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aam5353. [DOI] [PubMed] [Google Scholar]
- Ha J.H., Presti M.F., Loh S.N. A single protein disruption site results in efficient reassembly by multiple engineering methods. Biophys. J. 2019;117:56–65. doi: 10.1016/j.bpj.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.V., Hanson J.E., Sheng M. Microglia in alzheimer's disease. J. Cell Biol. 2018;217:459–472. doi: 10.1083/jcb.201709069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason C.D., Damen J.E., Rosten P., Grewal R., Sorensen P., Chappel S.M., Borowski A., Jirik F., Krystal G., Humphries R.K. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Develop. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.H., Li L., Vanhauwaert R., Nguyen K.T., Davis M.D., Bu G., Wszolek Z.K., Wang X. Miro1 marks Parkinson's disease subset and miro1 reducer rescues neuron loss in Parkinson's models. Cell Metab. 2019;30:1131–1140.e7. doi: 10.1016/j.cmet.2019.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C., Choi M.L., Yi J.H., Kim S.C., Drews A., George-Hyslop P.S., Bryant C., Gandhi S., Cho K., Klenerman D. Beta amyloid aggregates induce sensitised TLR4 signalling causing long-term potentiation deficit and rat neuronal cell death. Commun. Biol. 2020;3:79. doi: 10.1038/s42003-020-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A., Perry V.H., Gordon S. Immunohistochemical localization of a macrophage-specific antigen in developing mouse retina: phagocytosis of dying neurons and differentiation of microglial cells to form a regular array in the plexiform layers. J. Cell Biol. 1983;97:253–257. doi: 10.1083/jcb.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa S., Sasaoka T., Yaguchi S., Ishihara H., Tsuneki H., Murakami S., Fukui K., Wada T., Kobayashi S., Kimura I., et al. Impact of SRC homology 2-containing inositol 5'-phosphatase 2 gene polymorphisms detected in a Japanese population on insulin signaling. J. Clin. Endocrinol. Metab. 2005;90:2911–2919. doi: 10.1210/jc.2004-1724. [DOI] [PubMed] [Google Scholar]
- Kaisaki P.J., Delepine M., Woon P.Y., Sebag-Montefiore L., Wilder S.P., Menzel S., Vionnet N., Marion E., Riveline J.P., Charpentier G., et al. Polymorphisms in type II SH2 domain-containing inositol 5-phosphatase (INPPL1, SHIP2) are associated with physiological abnormalities of the metabolic syndrome. Diabetes. 2004;53:1900–1904. doi: 10.2337/diabetes.53.7.1900. [DOI] [PubMed] [Google Scholar]
- Kerr W.G. Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann. N. Y Acad. Sci. 2011;1217:1–17. doi: 10.1111/j.1749-6632.2010.05869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr W.G., Park M.Y., Maubert M., Engelman R.W. SHIP deficiency causes Crohn's disease-like ileitis. Gut. 2011;60:177–188. doi: 10.1136/gut.2009.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr W.G., Pedicone C., Dormann S., Pacherille A., Chisholm J.D. Small molecule targeting of SHIP1 and SHIP2. Biochem. Soc. Trans. 2020;48:291–300. doi: 10.1042/BST20190775. [DOI] [PubMed] [Google Scholar]
- Le Coq J., Camacho-Artacho M., Velazquez J.V., Santiveri C.M., Gallego L.H., Campos-Olivas R., Dolker N., Lietha D. Structural basis for interdomain communication in SHIP2 providing high phosphatase activity. eLife. 2017;6 doi: 10.7554/eLife.26640. e26640/26641-e26640/26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Coq J., Lopez Navajas P., Rodrigo Martin B., Alfonso C., Lietha D. A new layer of phosphoinositide-mediated allosteric regulation uncovered for SHIP2. FASEB J. 2021;35 doi: 10.1096/fj.202100561R. [DOI] [PubMed] [Google Scholar]
- Lemm E.A., Valle-Argos B., Smith L.D., Richter J., Gebreselassie Y., Carter M.J., Karolova J., Svaton M., Helman K., Weston-Bell N.J., et al. Preclinical evaluation of a novel SHIP1 phosphatase activator for inhibition of PI3K signaling in malignant B cells. Clin. Cancer Res. 2020;26:1700–1711. doi: 10.1158/1078-0432.CCR-19-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J., Iram T., Zardeneta M., Lee S.E., Lehallier B., Haney M.S., Pluvinage J.V., Mathur V., Hahn O., Morgens D.W., et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren J.E., Michael D.R., Ashlin T.G., Ramji D.P. Cytokines, macrophage lipid metabolism and foam cells: implications for cardiovascular disease therapy. Prog. Lipid Res. 2011;50:331–347. doi: 10.1016/j.plipres.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Mills S.J., Persson C., Cozier G., Thomas M.P., Tresaugues L., Erneux C., Riley A.M., Nordlund P., Potter B.V.L. A synthetic polyphosphoinositide headgroup surrogate in complex with SHIP2 provides a rationale for drug discovery. ACS Chem. Biol. 2012;7:822–828. doi: 10.1021/cb200494d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J.C., Moldwin R., Hanno P., Dmochowski R., Peters K.M., Payne C., Wein A. Targeting the SHIP1 pathway fails to show treatment benefit in interstitial cystitis/bladder pain syndrome: lessons learned from evaluating potentially effective therapies in this enigmatic syndrome. J. Urol. 2019;202:301–308. doi: 10.1097/JU.0000000000000192. [DOI] [PubMed] [Google Scholar]
- Ong C.J., Ming-Lum A., Nodwell M., Ghanipour A., Yang L., Williams D.E., Kim J., Demirjian L., Qasimi P., Ruschmann J., et al. Small-molecule agonists of SHIP1 inhibit the phosphoinositide 3-kinase pathway in hematopoietic cells. Blood. 2007;110:1942–1949. doi: 10.1182/blood-2007-03-079699. [DOI] [PubMed] [Google Scholar]
- Panhuysen C.I., Cupples L.A., Wilson P.W., Herbert A.G., Myers R.H., Meigs J.B. A genome scan for loci linked to quantitative insulin traits in persons without diabetes: the Framingham offspring study. Diabetologia. 2003;46:579–587. doi: 10.1007/s00125-003-1066-z. [DOI] [PubMed] [Google Scholar]
- Paolicelli R.C., Jawaid A., Henstridge C.M., Valeri A., Merlini M., Robinson J.L., Lee E.B., Rose J., Appel S., Lee V.M., et al. TDP-43 depletion in microglia promotes amyloid clearance but also induces synapse loss. Neuron. 2017;95:297–308.e6. doi: 10.1016/j.neuron.2017.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso K.H., Ghansah T., Costello A., Engelman R.W., Kerr W.G. Induced SHIP deficiency expands myeloid regulatory cells and abrogates graft-versus-host disease. J. Immunol. 2007;178:2893–2900. doi: 10.4049/jimmunol.178.5.2893. [DOI] [PubMed] [Google Scholar]
- Park M.Y., Srivastava N., Sudan R., Viernes D.R., Chisholm J.D., Engelman R.W., Kerr W.G. Impaired T-cell survival promotes mucosal inflammatory disease in SHIP1-deficient mice. Mucosal. Immunol. 2014;7:1429–1439. doi: 10.1038/mi.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedicone C., Fernandes S., Dungan O.M., Dormann S.M., Viernes D.R., Adhikari A.A., Choi L.B., De Jong E.P., Chisholm J.D., Kerr W.G. Pan-SHIP1/2 inhibitors promote microglia effector functions essential for CNS homeostasis. J. Cell Sci. jcs. 2019 doi: 10.1242/jcs.238030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Malhotra S., Torchia J.A., Kerr W.G., Coggeshall K.M., Humphrey M.B. TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci. Signal. 2010;3:ra38. doi: 10.1126/scisignal.2000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Heilmann S., Becker T., Hernandez I., Wagner H., Thelen M., Mauleon A., Rosende-Roca M., Bellenguez C., Bis J.C., et al. Follow-up of loci from the international genomics of alzheimer's disease project identifies TRIP4 as a novel susceptibility gene. Transl. Psychiatry. 2014;4:e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo C.M., Adhikari A.A., Wallach D.R., Fernandes S., Balch A.N., Kerr W.G., Chisholm J.D. Synthesis and initial evaluation of quinoline-based inhibitors of the SH2-containing inositol 5'-phosphatase (SHIP) Bioorg. Med. Chem. Lett. 2015;25:5344–5348. doi: 10.1016/j.bmcl.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C., Wolfs L., Fattorelli N., Thrupp N., Voytyuk I., Schmidt I., Mancuso R., Chen W.T., Woodbury M.E., Srivastava G., et al. The major risk factors for alzheimer's disease: age, sex, and genes modulate the microglia response to abeta plaques. Cell Rep. 2019;27:1293–1306.e6. doi: 10.1016/j.celrep.2019.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saz-Leal P., Del Fresno C., Brandi P., Martinez-Cano S., Dungan O.M., Chisholm J.D., Kerr W.G., Sancho D. Targeting SHIP-1 in myeloid cells enhances trained immunity and boosts response to infection. Cell Rep. 2018;25:1118–1126. doi: 10.1016/j.celrep.2018.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Rasband W., Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman M.W., Wortley K.E., Lai K.M., Gowen L.C., Kintner J., Kline W.O., Garcia K., Stitt T.N., Yancopoulos G.D., Wiegand S.J., et al. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- Somasundaram R., Fernandes S., Deuring J.J., de Haar C., Kuipers E.J., Vogelaar L., Middleton F.A., van der Woude C.J., Peppelenbosch M.P., Kerr W.G., et al. Analysis of SHIP1 expression and activity in crohn's disease patients. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T., Abe Y., Sato N., Ebina M. The Use of N,N-dimethylformamide–sulfonyl chloride complex for the preparation of thiophenesulfonyl chlorides. Bull. Chem. Soc. Jpn. 1985;58:1063–1064. [Google Scholar]
- Srivastava N., Iyer S., Sudan R., Youngs C., Engelman R.W., Howard K.T., Russo C.M., Chisholm J.D., Kerr W.G. A small-molecule inhibitor of SHIP1 reverses age- and diet-associated obesity and metabolic syndrome. JCI Insight. 2016;1 doi: 10.1172/jci.insight.88544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre M.-K., Simoncicova E., Bögi E., Tremblay M.-È. Shedding light on the dark side of the microglia. ASN Neuro. 2020;12 doi: 10.1177/1759091420925335. 175909142092533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresaugues L., Silvander C., Flodin S., Welin M., Nyman T., Graeslund S., Hammarstroem M., Berglund H., Nordlund P. Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases. Structure (Oxford, United Kingdom) 2014;22:744–755. doi: 10.1016/j.str.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Ulland T.K., Wang Y., Colonna M. Regulation of microglial survival and proliferation in health and diseases. Semin. Immunol. 2015;27:410–415. doi: 10.1016/j.smim.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A., Benjaminsen J., Moritz C., Henke K., Hartmann J., Norlin N., Richter K., Schieber N.L., Franke T., Schwab Y., et al. Clearance by microglia depends on packaging of phagosomes into a unique cellular compartment. Dev. Cell. 2019;49:77–88.e7. doi: 10.1016/j.devcel.2019.02.014. [DOI] [PubMed] [Google Scholar]
- Wahle J.A., Paraiso K.H.T., Kendig R.D., Lawrence H.R., Chen L., Wu J., Kerr W.G. Inappropriate recruitment and activity by the src homology region 2 domain-containing phosphatase 1 (SHP1) is responsible for receptor dominance in the SHIP-deficient NK cell. J. Immunol. 2007;179:8009. doi: 10.4049/jimmunol.179.12.8009. [DOI] [PubMed] [Google Scholar]
- Wallach I., Dzamba M., Heifets A. AtomNet: A deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv. 2015 doi: 10.48550/arXiv.1510.02855. Preprint at. [DOI] [Google Scholar]
- Wang R., Fu Y., Lai L. A new atom-additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 1997;37:615–621. [Google Scholar]
- Wang Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H., et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Rogus J.J., Terwedow H.A., Yang J., Wang Z., Chen C., Niu T., Wang B., Xu H., Weiss S., et al. An extreme-sib-pair genome scan for genes regulating blood pressure. Am. J. Hum. Genet. 1999;64:1694–1701. doi: 10.1086/302405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanuck S.F. Microglial phagocytosis of neurons: diminishing neuronal loss in traumatic, infectious, inflammatory, and autoimmune CNS disorders. Front. Psychiatry. 2019;10:712. doi: 10.3389/fpsyt.2019.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.H., Fu Y.C., Zhang D.W., Yin K., Tang C.K. Foam cells in atherosclerosis. Clin. Chim. Acta. 2013;424:245–252. doi: 10.1016/j.cca.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Zhan Y., Paolicelli R.C., Sforazzini F., Weinhard L., Bolasco G., Pagani F., Vyssotski A.L., Bifone A., Gozzi A., Ragozzino D., et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang F.W., Yao L.L., Hao A.J. Microglia--friend or foe. Front. Biosci. (Schol Ed) 2011;3:869–883. doi: 10.2741/193. [DOI] [PubMed] [Google Scholar]
- Zhang M., Schmitt-Ulms G., Sato C., Xi Z., Zhang Y., Zhou Y., St George-Hyslop P., Rogaeva E. Drug repositioning for alzheimer's disease based on Systematic 'omics' data mining. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.