Abstract
Mutant prevention concentration (MPC) has been proposed as a new measure of antibiotic potency by which the ability to restrict selection of resistant mutants is evaluated. To determine whether MPC provides potency information unavailable from the more customary measurement of the MIC, 18 fluoroquinolones were examined for their ability to block the growth of Mycobacterium smegmatis and to select resistant mutants from wild-type populations. Both MPC and MIC were affected by changes in the moiety at the fluoroquinolone C-8 position and in alkyl groups attached to the C-7 piperazinyl ring. When eight resistant mutants, altered in the gyrase A protein, were tested with fluoroquinolones having either a methoxy or a hydrogen at the C-8 position, the MIC for the most resistant mutant correlated better with the MPC than did the MIC for wild-type cells. For C-8-fluorine derivatives, which were generally less active than the C-8-methoxy compounds but which were more active than C-8-hydrogen derivatives, the MICs for both the mutant and the wild type correlated well with the MPCs. Thus, measurement of the MICs for wild-type cells can reflect the ability of a quinolone to restrict the selection of resistance, but often it does not. With the present series of compounds, the most potent contained a C-8-methoxy and a small group attached to the C-7 ring.
To develop more effective antituberculosis agents, we have been studying the fluoroquinolones. These compounds, which act by trapping gyrase on DNA, are generally used only as second-line therapeutics because resistance mutations in Mycobacterium tuberculosis often render them ineffective. For example, in the early 1990s a strain of M. tuberculosis resistant to isoniazid, rifampin, streptomycin, and ethambutol spread among immunocompromised persons in New York City (1). Patients were treated with the fluoroquinolone ciprofloxacin, and within a few months some were found harboring strains of M. tuberculosis that were resistant to fluoroquinolones as well as to the four other agents (11). When we examined these strains, we noticed that some fluoroquinolones (e.g., sparfloxacin) were more effective than others (e.g., ciprofloxacin) at blocking the growth of resistant mutants after the data were normalized to the results obtained with wild-type cells (5, 12). This observation led to the idea that some quinolones might be better than others at trapping resistant gyrase in mycobacteria. Subsequent work showed that compounds with a methoxy group attached to the C-8 position (C-8-OMe) were particularly effective against resistant mutants (5, 13).
While examining the effects of C-8-OMe groups we observed a complex relationship between the recovery of resistant mutants and the fluoroquinolone concentration (6). Increases in fluoroquinolone concentration cause the fraction of cells that form colonies on quinolone-containing agar to drop sharply to a plateau and then drop sharply a second time. We proposed that the plateau is due to the presence of first-step, resistant mutants in the population. If this were true, the second drop in mutant recovery should occur when the MIC for these mutants is approached: once the growth of first-step mutants is blocked, no mutant should be recovered at the cell amounts used because a rare, double mutation would be required to overcome the quinolone effect. The minimal quinolone concentration that allows no mutant recovery when more than 1010 cells are applied to drug-containing agar was defined as the mutant prevention concentration (MPC). If relevant concentrations of an antibiotic in tissue can be maintained above the MPC, selection of resistant mutants should be severely restricted.
MPC has been measured for only a few fluoroquinolones; consequently, structure-activity relationships are still poorly defined. In the present work we determined the MICs and MPCs for Mycobacterium smegmatis of fluoroquinolones that differed in moieties attached to the C-8 position and to the C-7 piperazinyl ring. C-8 substituents influenced the effect that C-7-ring alkyl groups have on fluoroquinolone MICs and MPCs. When resistant gyrA (gyrase) mutants were examined, fluoroquinolone susceptibility varied with drug structure in a way that allowed inferences about quinolone binding to gyrase. Data obtained with the mutants also revealed that the MPC often correlates better with the MIC for the most resistant first-step mutant than with the MIC for wild-type cells, consistent with the two-mutation explanation for MPC. Collectively, these data show that the MPCs for and the susceptibilities of resistant mutants can be used to discriminate among closely related fluoroquinolones.
MATERIALS AND METHODS
Bacterial strains and growth.
The relevant features of M. smegmatis mc2155 and its gyrA mutants are listed in Table 1. The cells were grown as liquid cultures in Middlebrook 7H9 medium containing 10% albumin dextrose complex and 0.05% Tween 80 (8). Colonies were grown on Middlebrook 7H10 agar plates containing a fluoroquinolone. The incubation temperature for growth was 37°C.
TABLE 1.
Bacterial strains used in the study
| M. smegmatis strain | Relevant genotypea | Source or reference |
|---|---|---|
| KD1163 | mc2155 (wild type) | S. Cole (4) |
| KD1987 | KD1163 gyrA (A91V) | 14 |
| KD1991 | KD1163 gyrA (D95Y) | 14 |
| KD1995 | KD1163 gyrA (D95H) | 14 |
| KD1998 | KD1163 gyrA (D95N) | 14 |
| KD1999 | KD1163 gyrA (D95A) | 14 |
| KD2003 | KD1163 gyrA (D95G) | 14 |
| KD2008 | KD1163 gyrA (G89C) | 14 |
| KD2012 | KD1163 gyrA (G89A) | 14 |
Amino acid abbreviations: A, alanine; D, aspartic acid; C, cysteine; H, histidine; G, glycine; N, asparagine; V, valine; and Y, tyrosine. The first letter represents the wild-type amino acid at the position indicated by the number. The second letter indicates the identity of the mutant amino acid at that position.
The MIC was determined graphically from log-log plots of the fluoroquinolone concentration on agar plates versus the fraction of CFU recovered, and it was defined as the concentration required to inhibit colony formation by 99% (MIC99) after incubation for 3 days. Cultures contained about 5 × 108 CFU per ml prior to plating. For these determinations, fluoroquinolone concentration increments differed by 0.01 μg/ml for wild-type cells and by 0.1 or 0.5 μg/ml for resistant mutants, depending on the compound and the mutant. To assay the selection of fluoroquinolone-resistant mutants, M. smegmatis was grown to the stationary phase (350 Klett units), harvested by centrifugation (6,000 × g for 15 min), and resuspended in the same volume of fresh growth medium. Incubation was continued until the turbidity of the culture reached 400 to 450 Klett units, and the culture was again concentrated by centrifugation. The cells were resuspended to about 109 CFU per ml in growth medium and were then applied at various volumes and concentrations to fluoroquinolone-containing 7H10 agar at a maximum of 2 × 109 CFU per 100-mm-diameter agar plate. The plates were allowed to dry in a biosafety cabinet for about 30 min at room temperature, sealed with electrical tape to minimize evaporation, and inverted for incubation at 37°C for 3 to 7 days. After the colonies were counted, they were confirmed to be composed of resistant mutants by regrowth on fluoroquinolone-containing agar.
Fluoroquinolones.
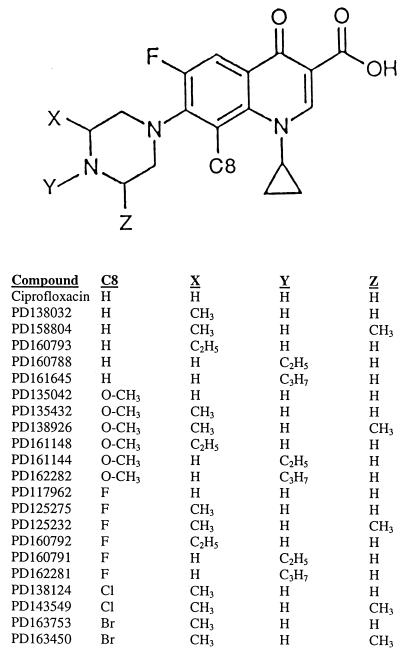
Several sets of fluoroquinolones were provided by Parke-Davis Pharmaceutical Company, Ann Arbor, Mich. (for structures, see Fig. 1). PD135432 has the same structure as AM1155 (gatifloxacin). Ciprofloxacin was obtained from Miles Laboratories (West Haven, Conn.). The fluoroquinolones (10 mg) were dissolved in 0.1 ml of 1 N NaOH (1/10 of the final volume), and then sterile water was added to give a final concentration of 10 mg/ml. This stock solution was divided into 50-μl aliquots and was stored at −80°C. Dilution series were prepared with autoclaved water. Solutions were occasionally stored at −20°C for several weeks.
FIG. 1.
Fluoroquinolone structures. A structure common to the compounds tested is shown, with the variable groups marked X, Y, Z, and C8. Identification numbers for the compounds are listed below the structures. Due to rotation at the C-7 position, substitutions at the 3′ (X) and 5′ (Z) positions are identical.
RESULTS AND DISCUSSION
Effects of C-8- and C-7-ring substituents on inhibition of wild-type growth by fluoroquinolones.
The bacteriostatic effects of substituents attached to the C-8 position and to the C-7 piperazinyl ring of N-1-cyclopropyl fluoroquinolones were examined by measuring the MIC99 for wild-type M. smegmatis. Three sets of compounds differing at the C-8 position (C-8-H, C-8-OMe, and C-8-F) were compared; within each set, the compounds differed in substituents attached to the C-7 piperazinyl ring (Fig. 1). With the C-8-H series, optimal activity was observed when two methyl groups were attached to the C-7 ring (Table 2, strain KD1163), as reported previously for Mycobacterium avium (9). For the C-8-OMe compounds, several C-7-ring substituents (dimethyl, monomethyl, N-ethyl, and C-3′-ethyl) improved the bacteriostatic activity by three- to fourfold, while the N-isopropyl substituent had no effect (Table 2, strain KD1163). Each member of the C-8-OMe set had a lower MIC99 than its counterpart in the C-8-H set. The activity of the most potent C-8-F compound, in terms of absolute MIC99, was in the same range as that of the most active C-8-OMe compounds. However, the C-8-F series exhibited a pattern different from that seen with the two other sets in that the compound lacking a C-7-ring alkyl was quite active. As the size of the alkyl groups increased beyond the monomethyl level, the activities of the C-8-F derivatives decreased. Collectively, these data establish that the nature of the C-8 moiety influences the bacteriostatic effect of C-7-ring alkyl groups. Additional experiments are now required to attribute specific structures to effects on drug uptake, efflux, and trapping of gyrase on DNA.
TABLE 2.
Effects of fluoroquinolone C-8- and C-7-ring substituents on MICs for M. smegmatis
| Straina and C-7-ring alkyl | MIC99 (μg/ml)
|
||||
|---|---|---|---|---|---|
| C-8-H | C-8-OMe | C-8-F | C-8-Cl | C-8-Br | |
| KD1163 (GryA+) | |||||
| None | 0.18 | 0.14 | 0.044 | ||
| Methyl | 0.10 | 0.030 | 0.036 | 0.029 | 0.032 |
| Dimethyl | 0.043 | 0.028 | 0.049 | 0.058 | 0.11 |
| C-3′-Ethyl | 0.12 | 0.029 | 0.068 | ||
| N-Ethyl | 0.11 | 0.033 | 0.10 | ||
| N-Isopropyl | 0.16 | 0.14 | 0.31 | ||
| KD2012 (GyrA G89A) | |||||
| None | 0.86 | 0.18 | 0.42 | ||
| Methyl | 0.92 | 0.18 | 0.24 | ||
| Dimethyl | 0.63 | 0.18 | 0.24 | ||
| C-3′-Ethyl | 1.0 | 0.28 | 0.37 | ||
| N-Ethyl | 1.1 | 0.37 | 0.43 | ||
| N-Isopropyl | 1.6 | 0.85 | 1.6 | ||
| KD2008 (GyrA G89C) | |||||
| None | 4.7 | 0.94 | 0.94 | ||
| Methyl | 2.6 | 0.73 | 1.0 | ||
| Dimethyl | 3.8 | 0.91 | 1.2 | ||
| C-3′-Ethyl | 3.2 | 0.65 | 0.73 | ||
| N-Ethyl | 4.4 | 2.4 | 3.2 | ||
| N-Isopropyl | 7.4 | 7.5 | 9.6 | ||
| KD1987 (GyrA A91V) | |||||
| None | 1.7 | 0.41 | 0.54 | ||
| Methyl | 1.2 | 0.23 | 0.46 | ||
| Dimethyl | 0.98 | 0.41 | 0.47 | ||
| C-3′-Ethyl | 1.2 | 0.38 | 0.53 | ||
| N-Ethyl | 1.7 | 0.51 | 0.97 | ||
| N-Isopropyl | 1.5 | 1.4 | 2.3 | ||
| KD1990 (GyrA D95A) | |||||
| None | 1.8 | 0.31 | 0.65 | ||
| Methyl | 1.2 | 0.22 | 0.47 | ||
| Dimethyl | 0.96 | 0.28 | 0.43 | ||
| C-3′-Ethyl | 2.6 | 0.3 | 0.67 | ||
| N-Ethyl | 1.7 | 0.41 | 1.5 | ||
| N-Isopropyl | 1.5 | 1.2 | 2.4 | ||
| KD2003 (GyrA D95G) | |||||
| None | 4.5 | 0.83 | 1.3 | ||
| Methyl | 2.8 | 0.49 | 1.2 | ||
| Dimethyl | 3.0 | 0.71 | 1.2 | ||
| C-3′-Ethyl | 3.8 | 0.65 | 1.4 | ||
| N-Ethyl | 4.3 | 1.0 | 2.6 | ||
| N-Isopropyl | 4.3 | 3.0 | 5.3 | ||
| KD1995 (GyrA D95H) | |||||
| None | 3.5 | 0.57 | 1.1 | ||
| Methyl | 2.7 | 0.46 | 1.0 | ||
| Dimethyl | 3.6 | 0.59 | 0.92 | ||
| C-3′-Ethyl | 3.8 | 0.54 | 1.2 | ||
| N-Ethyl | 3.2 | 0.76 | 2.0 | ||
| N-Isopropyl | 3.6 | 2.3 | 3.8 | ||
| KD1998 (GyrA D95N) | |||||
| None | 4.6 | 0.81 | 1.3 | ||
| Methyl | 3.3 | 0.66 | 1.3 | ||
| Dimethyl | 3.4 | 0.93 | 1.3 | ||
| C-3′-Ethyl | 3.6 | 0.63 | 1.4 | ||
| N-Ethyl | 4.9 | 1.2 | 2.6 | ||
| N-Isopropyl | 4.3 | 4.1 | 6.8 | ||
| KD1991 (GyrA D95Y) | |||||
| None | 4.0 | 0.61 | 1.6 | ||
| Methyl | 2.6 | 0.47 | 1.1 | ||
| Dimethyl | 2.3 | 0.51 | 0.91 | ||
| C-3′-Ethyl | 3.8 | 0.47 | 1.3 | ||
| N-Ethyl | 3.6 | 0.81 | 2.3 | ||
| N-Isopropyl | 4.6 | 2.4 | 5.8 | ||
| Avg for GyrA variants | |||||
| None | 3.2 | 0.58 | 0.98 | ||
| Methyl | 2.2 | 0.43 | 0.86 | ||
| Dimethyl | 2.3 | 0.56 | 0.84 | ||
| C-3′-Ethyl | 2.9 | 0.49 | 0.96 | ||
| N-Ethyl | 3.1 | 0.93 | 2.0 | ||
| N-Isopropyl | 3.6 | 2.8 | 4.7 | ||
See footnote a of Table 1 for amino acid abbreviations.
Since the optimal C-7-ring substituents were one and two methyl groups, we added C-8-Cl and C-8-Br fluoroquinolones to the series to assess the effect of altering the C-8 moiety. For the monomethyl series, compounds containing halogens or a C-8-OMe group exhibited similar effectiveness, which was about threefold better than that of the compound lacking a C-8 substituent (Table 2, strain KD1163). In the case of the dimethyl series, the C-8-OMe derivative was the most active; compounds with C-8 halogens varied considerably in activity, and the compound lacking a C-8 group was as effective as some halogenated compounds. Collectively, these measurements indicate that fluoroquinolones with a C-8-OMe group are generally more effective against wild-type M. smegmatis than compounds with fluorine or hydrogen at position C-8 and that they are most potent when a small alkyl group is attached to the C-7 ring.
Effect of C-8- and C-7-ring substituents on inhibition of growth of first-step fluoroquinolone-resistant gyrase mutants.
The relationship between fluoroquinolone structure and activity against resistant gyrA mutants was examined by determining the MIC99s of the C-8-OMe, C-8-H, and C-8-F fluoroquinolones for eight mutants of M. smegmatis (Table 2, strains labeled as GyrA amino acid changes). The C-8-OMe compounds were the most effective, followed by the C-8-F derivatives (see averages for GyrA variants at the bottom of Table 2). Within the C-8-OMe series, the C-7-ring monomethyl derivative was generally the most effective; for both the C-8-F and C-8-H series, the monomethyl compound was optimal about as often as the dimethyl derivative was. Compounds with large alkyl groups on the C-7 ring were generally least effective.
The GyrA G89C variant and several substitutions at position 95 were associated with greater resistance than that for the other variants examined (summarized in Table 3). A comparison of two C-7-ring ethyl compounds showed that the position of the C-7-ring alkyl affects the identity and susceptibility of the most resistant variant, particularly when there is a methoxy or fluorine attached to the C-8 position: for the N-ethyl compounds, the G89C amino acid change was associated with the most resistance; for the C-3′-ethyl derivatives, the D95 substitutions were associated with resistance similar to or greater than that for the G89C alteration (Table 3). When the mutants were compared for the effects of the position of the piperazinyl ring ethyl by determination of the MIC ratio (Table 4), the G89C variant was distinctly less susceptible than the A91V and D95 mutants to the N-linked ethyl compounds, at least when a methoxy or fluorine substituent was attached to the C-8 position. In other work we found that GyrA G89C variants are preferentially selected by high concentrations of the C-7-ring N-ethyl compound, while D95G variants are selected by the C-3′-ethyl derivative (14), consistent with the data in Tables 3 and 4. The N-isopropyl group also preferentially lowered the activity against the G89C GyrA variant (Table 4). If we assume that comparisons among gyrase mutants reflect differences in intracellular quinolone binding to gyrase, these data indicate that N-linked alkyl groups interfere with fluoroquinolone binding to a GyrA protein that contains a Cys-89 mutation, particularly when a methoxy or halogen is attached to the C-8 position.
TABLE 3.
Effect of fluoroquinolone C-8- and C-7-ring substituents on the identities and susceptibilities of the most resistant gyrA mutant of M. smegmatis
| C-7-ring group | Most resistant mutanta with C-8 substituent of:
|
||
|---|---|---|---|
| Methoxy | Fluorine | Hydrogen | |
| None | D95G,N; G89C (0.94) | D95Y (1.6) | D95G,N,Y; G89C (4.7) |
| Methyl | D95N; G89C (0.73) | D95G,N (1.3) | D95G,N (3.3) |
| Dimethyl | D95N; G89C (0.93) | D95G,N; G89C (1.3) | D95H,N; G89C (3.8) |
| C-3′-Ethyl | D95G,N; G89C (0.65) | D95G,H,N,Y (1.4) | D95G,H,N,Y (3.8) |
| N-Ethyl | G89C (2.4) | G89C (3.2) | D95G,N; G89C (4.9) |
| N-Isopropyl | G89C (7.5) | G89C (9.6) | G89C (7.4) |
Effects of mutations were considered indistinguishable when the difference among MICs was less than 15%. Numbers in parentheses are MIC99s (in micrograms per milliliter) for the most resistant mutant obtained by use of the data listed in Table 2.
TABLE 4.
MIC ratios for fluoroquinolones differing in the C-7-ring ethyl and N-isopropyl groups
| GyrA amino acid substitution | MIC ratio
|
|||||
|---|---|---|---|---|---|---|
|
N-Ethyl/C-3′-ethyla with C-8 substituent of:
|
N-Isopropyl/no alkylb with C-8 substituent of:
|
|||||
| Methoxy | Fluorine | Hydrogen | Methoxy | Fluorine | Hydrogen | |
| G89C | 3.7 | 4.3 | 1.4 | 8.0 | 10 | 1.6 |
| A91V | 1.3 | 1.8 | 1.3 | 3.4 | 4.3 | 0.88 |
| D95avec | 1.6 | 1.9 | 1.4 | 4.1 | 4.1 | 1.0 |
Ratio of MICs of fluoroquinolones differing in the position of the C-7-ring ethyl group, using the data listed in Table 2.
Ratio of MICs of fluoroquinolones having a C-7-ring N-isopropyl group relative to the MIC of the fluoroquinolone lacking a C-7-ring alkyl group, using the data listed in Table 2.
The average MIC99 (in micrograms per milliliter) was determined with D95G, D95H, D95N, and D95Y GyrA variants, as listed in Table 2.
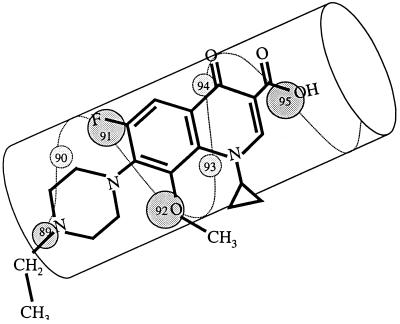
Interference between Cys-89 and the N-alkylated C-7 piperazinyl ring can be explained by the fluoroquinolone orientation on α-helix 4 of the GyrA breakage-rejoining fragment. From X-ray crystallography of the Escherichia coli GyrA protein it has been argued that α-helix 4 is located on the surface of the protein, where it interacts with DNA (10). Amino acid sequence similarities between the GyrA proteins of E. coli and M. smegmatis indicate that amino acids 89, 91, and 95 of the M. smegmatis GyrA protein reside in α-helix 4. A negative interaction between Cys-89 and N-linked C-7-ring alkyls suggests that the C-7-ring binds to gyrase near amino acid 89, while the remainder of the quinolone interacts with amino acids 91 and 95, the other two positions where strong resistance mutations map. Such an alignment, which is shown schematically in Fig. 2, explains why a variant of E. coli with a GyrA G81D variation, which is equivalent to a position 89 mutation in M. smegmatis, exhibits resistance to fluoroquinolones but not to nalidixic acid (3): nalidixic acid lacks the C-7 ring required for interference.
FIG. 2.
Orientations of fluoroquinolones and GyrA α-helix 4. α-helix 4, adapted from the crystal structure of the breakage-reunion domain of the GyrA protein of E. coli (10), is drawn parallel to the long axis of PD161144. Amino acid numbers represent positions in the M. smegmatis GyrA protein.
Effects of C-8- and C-7-ring substituents on selection of resistant mutants.
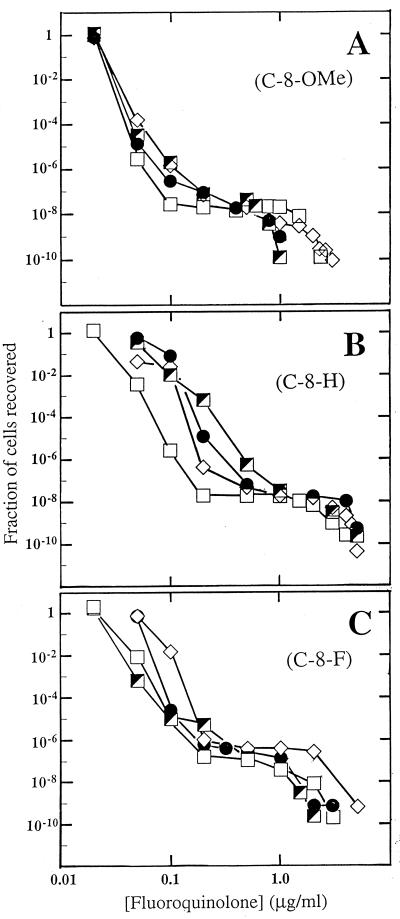
When large numbers of cells, on the order of 108 to 1010, were incubated on agar plates containing various fluoroquinolone concentrations, colony numbers dropped sharply in the presence of concentrations slightly above the MIC99. At higher concentrations a plateau was seen for many compounds, followed by a second sharp drop at very high concentrations (Fig. 3). To assess the effect of C-7-ring substituents on the ability of a C-8-OMe group to suppress selection of resistant mutants, four C-8-OMe compounds that had similar MIC99s for wild-type cells were compared (Fig. 3A). Differences among the four compounds were observed with respect to the concentration at which the high-concentration drop in recovery occurred: compounds with a C-3′-ethyl or monomethyl substituent on the C-7 ring were the most effective, followed by the dimethyl and N-ethyl compounds (Fig. 3A). The compound with the lowest MIC99 for wild-type cells, the C-7-ring dimethyl quinolone, ranked third when compared for the concentration at which the second drop occurred. Thus, the MIC for wild-type cells does not tightly correspond to the ability to suppress selection of resistant mutants. The N-ethyl compound ranked fourth, as expected, because the MIC99 of this compound for the most resistant mutants was generally higher than those for the other mutants examined (Table 3).
FIG. 3.
Effect of small C-7-ring alkyls on selection of resistant mutants by fluoroquinolones. M. smegmatis strain KD1163 was applied to agar plates at the indicated fluoroquinolone concentrations. Fluoroquinolones contained a C-8-OMe group (A), a C-8-H group (B), and a C-8-F group (C). Symbols for panel A: squares, PD138926, C-8-OMe C-7-ring dimethyl; diamonds, PD161144, C-8-OMe C-7-ring N-ethyl; filled circles, PD161148, C-8-OMe C-7-ring 3′-ethyl; half-filled squares, PD135432, C-8-OMe C-7-ring methyl. Symbols for panel B: squares, PD158804, C-8-H C-7-ring dimethyl; diamonds, PD160788, C-8-H C-7-ring N-ethyl; filled circles, PD160793, C-8-H C-7-ring 3′-ethyl; half-filled squares, PD138032, C-8-H C-7-ring methyl. Symbols for panel C, squares, PD125232, C-8-F C-7-ring dimethyl; half-filled squares, PD125275, C-8-F C-7-ring 3′-methyl; diamonds, PD160791, C-8-F C-7-ring N-ethyl; filled circles, PD160792, C-8-F C-7-ring 3′-ethyl. A replicate experiment focusing on the second sharp drop in colony recovery gave results similar to those shown.
To determine whether the effects of the C-7-ring substituents for which data are shown in Fig. 3A are influenced by the presence of the C-8-OMe group, we examined a set of compounds that differed from the first series in having either a C-8-H or a C-8-F moiety rather than a C-8-OMe group. In the C-8-H series, the variation was wider for MIC99 but narrower for the concentration at which the high-concentration drop occurred (Fig. 3B). With the C-8-F series, the MIC99 paralleled the concentration at the second sharp drop in mutant recovery (Fig. 3C). Thus, the group at the C-8 position appears to influence the relationship between wild-type MIC99 and the concentration needed to cause the second drop in mutant recovery.
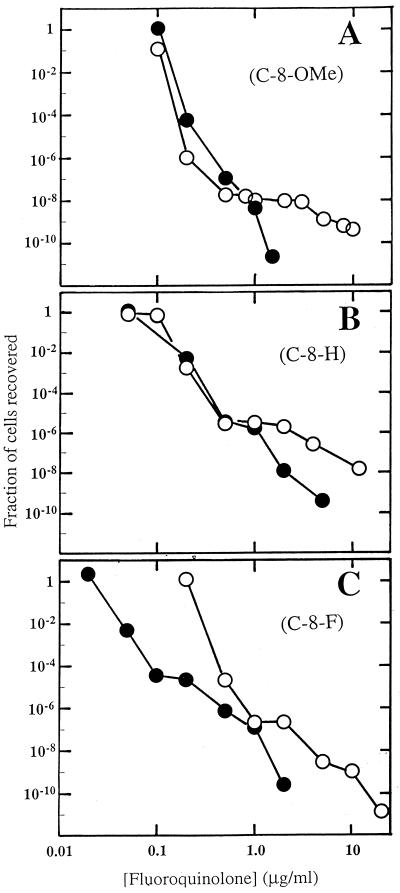
Two other C-8-OMe compounds, one with a C-7-ring N-isopropyl group (PD162282) and one lacking a C-7-ring substituent (PD135042), had MICs that were fivefold higher than those discussed above (Table 2, strain KD1163). For this pair, the compound lacking a C-7-ring alkyl substituent was remarkable for the absence of a plateau (Fig. 4A). The C-8-H and C-8-F derivatives of this compound also exhibited little plateau regions (Fig. 4B and C). The addition of an N-isopropyl group to the C-7 ring lengthened the plateau for the C-8-OMe and C-8-H compounds (Fig. 4A and B), consistent with the poor activity of the N-isopropyl compounds against resistant mutants (Table 2). For the C-8-F compounds, an isopropyl group attached to the C-7 ring simply shifted the mutant recovery curve to higher concentrations (Fig. 4C). This behavior is currently unexplained. Collectively, the data in Fig. 4 emphasize that the C-8 moiety influences the relationship between MIC99 and the concentration associated with the second drop in mutant recovery.
FIG. 4.
Effect of C-7-ring isopropyl group on selection of resistant mutants by fluoroquinolones. M. smegmatis was applied to agar plates containing the indicated fluoroquinolone concentrations. Fluoroquinolones contained a C-8-OMe group (A), a C-8-H group (B), and a C-8-F group (C). Symbols for panel A: filled circles, PD135042, C-8-OMe with no addition to C-7-ring; open circles, PD162282, C-8-OMe C-7-ring N-isopropyl. Symbols for panel B: filled circles, ciprofloxacin, C-8-H with no addition to the C-7 ring; open circles, PD161645, C-8-H C-7-ring N-isopropyl. Symbols for panel C: filled circles, PD117962, C-8-F with no addition to C-7 ring; open circles, PD162281, C-8-F C-7-ring N-isopropyl. A replicate experiment focusing on the second sharp drop in colony recovery gave results similar to those shown.
MPC.
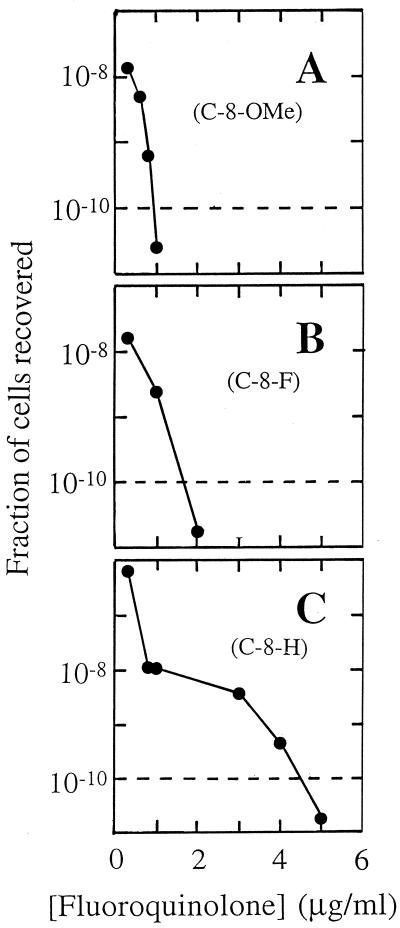
For comparative purposes, we define MPC as the concentration at which no colony is recovered when more than 1010 cells are applied to agar plates. MPC can be determined by agar dilution, as is customary for measurement of MIC, using concentration increments as fine as necessary. We prefer to estimate MPC by interpolation, as shown in Fig. 5. The dashed line in Fig. 5 represents a threshold below which more than 1010 cells must be applied to agar plates to recover a single colony. The intersection of the dashed line with the experimental data is used to estimate the MPC. Table 5 shows the MPCs of the three sets of fluoroquinolones by using the type of analysis used to obtain the results presented in Fig. 5. The most potent compounds by this assay were C-8-OMe derivatives with a C-7-ring methyl or ethyl group, provided that the latter is attached to a ring carbon.
FIG. 5.
Determination of MPC. Resistant M. smegmatis mutants were recovered from agar plates containing the indicated concentrations of PD161148 (C-8-OMe, C-7-ring 3′-ethyl) (A), PD160792 (C-8-F, C-7-ring 3′-ethyl) (B), and PD160793 (C-8-H, C-7-ring 3′-ethyl) (C). MPC1010 is defined as the concentration at which less than one colony is recovered when 1010 cells are applied to agar plates. MPC1010 is approximated by the intersection of the data curve and the dashed line in the figure.
TABLE 5.
Effect of fluoroquinolone C-8 and C-7-ring substituents on MPC of M. smegmatis
| C-7-ring group | MPC (μg/ml)
|
||
|---|---|---|---|
| C-8-H | C-8-OMe | C-8-F | |
| None | 6.0 | 1.4 | 2.0 |
| Methyl | 5.2 | 1.0 | 2.2 |
| Dimethyl | 4.2 | 2.0 | 3.1 |
| C-3′-Ethyl | 4.5 | 0.9 | 1.7 |
| N-Ethyl | 4.9 | 3.1 | 5.3 |
| N-Isopropyl | 19 | 11 | 13 |
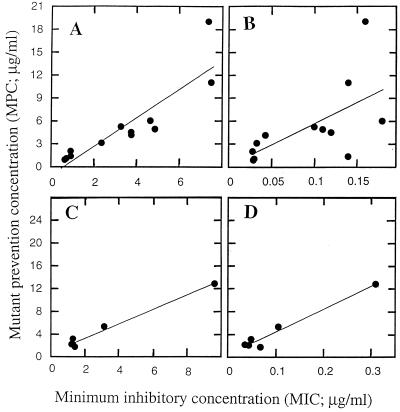
A plot of the MPCs of the C-8-H and C-8-OMe compounds against the MICs of each compound for the most resistant of the eight gyrA mutants is shown in Fig. 6A. When we assumed a linear relationship, the value of r (2) was 0.92. Much more scatter was evident when the MPC was plotted against the MIC99 for the wild-type cells (Fig. 6B). In this case r was 0.39. For the C-8-F compounds, the MPCs correlated well with the MICs for both mutant and wild-type cells (Fig. 6C and D). Thus, the MIC for the most resistant mutant correlates well with the MPC; whether the MICs for wild-type cells also correlate with the MPCs depends on the nature of the moiety at the C-8 position of the fluoroquinolone. A correlation of the MPC with the MIC for the mutant is consistent with the second drop in colony recovery on quinolone-containing agar occurring at the point where the quinolone concentration reaches the MIC for the most resistant mutant. At that point resistance in a wild-type population would require two concurrent mutations, which for M. smegmatis would occur at a frequency of 10−14 to 10−16 (single-mutation frequencies were about 10−7 to 10−8; see the plateau region in Fig. 3 and see reference 14). Such a double-mutation frequency is orders of magnitude too low to be detected by the methods used in the present work.
FIG. 6.
Relationship between MPC and MIC. The MPC of each fluoroquinolone, determined as described in the legend to Fig. 5, was plotted against the MICs of C-8-OMe and C-8-H fluoroquinolones with the C-7-ring moieties indicated in Fig. 2 for the most resistant gyrA mutant (A) or wild-type cells (B). The relationships between the MPC and the MIC for a gyrA mutant or the MIC for the wild type are shown in panels C and D, respectively, for the C-8-F compounds.
Concluding remarks.
The present work expands the initial description of MPC (6) by showing that differences can be detected among a set of closely related fluoroquinolones and by showing that the MPC correlates well with the MIC for the most resistant, first-step gyrA mutant. Measurement of MIC and MPC also revealed an unexplained interaction between C-8 substituents and C-7-ring alkyl groups. To be clinically useful, the MPC must be below the concentration in serum or tissue at the site of infection. Thus, use of a combination of MPC and pharmacokinetic parameters provides a way to compare antibacterial agents for their potential ability to restrict the selection of resistant mutants. Of the first-line antituberculosis agents tested, none has an MPC below the maximum concentration in serum attained during therapy (7). However, the MPCs of both PD135432 (gatifloxacin) and moxifloxacin, another C-8-OMe fluoroquinolone, are below maximum concentration in serum attained with doses recommended by the manufacturers (7). Thus, administration of fluoroquinolones such that concentrations in serum are above the MPC is feasible with existing compounds. Animal and clinical studies are now needed to determine whether that will severely restrict the selective enrichment of resistant mutants.
ACKNOWLEDGMENTS
We thank Marila Gennaro and Samuel Kayman for critical comments on the manuscript.
This work was supported by NIH grant AI35257.
Footnotes
This is publication 74 from the Public Health Research Institute Tuberculosis Center.
REFERENCES
- 1.Bifani P, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lusfty M, Moghazeh S, Eisner W, Daniel T, Kaplan M, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug resistant Mycobacterium tuberculosis clone family: adverse implications for tuberculosis control in the 21st century. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 2.Bishop O N. Statistics for biology. New York, N.Y: Houghton-Mifflin Co.; 1966. p. 65. [Google Scholar]
- 3.Cambau E, Borden F, Collatz E, Gutmann L. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob Agents Chemother. 1993;37:1247–1252. doi: 10.1128/aac.37.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S, Eiglmeier K, Gas S, Barry C E, Tekaia F, Babcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on bacterial growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Y, Zhao X, Kreiswirth B, Drlica K. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000;44:2581–2584. doi: 10.1128/aac.44.9.2581-2584.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs W R, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems in mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 9.Klopman G, Fercu D, Renau T, Jacobs M. N-1-tert-Butyl-substituted quinolones: in vitro anti-Mycobacterium avium activities and structure-activity relationship studies. Antimicrob Agents Chemother. 1996;40:2637–2643. doi: 10.1128/aac.40.11.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morais-Cabral J H, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan E A, Kreiswirth B N, Palumbo L, Kapur V, Musser J M, Ebrahimzadeh A, Frieden T R. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet. 1995;345:1148–1150. doi: 10.1016/s0140-6736(95)90980-x. [DOI] [PubMed] [Google Scholar]
- 12.Xu C, Kreiswirth B N, Sreevatsan S, Musser J M, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174:1127–1130. doi: 10.1093/infdis/174.5.1127. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B-Y, Pine R, Domagala J, Drlica K. Fluoroquinolone action against clinical isolates of Mycobacterium tuberculosis: effects of a C-8-methoxyl group on survival in liquid media and in human macrophages. Antimicrob Agents Chemother. 1999;43:661–666. doi: 10.1128/aac.43.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J-F, Dong Y, Zhao X, Lee S, Amin A, Ramaswamy S, Domagala J, Musser J M, Drlica K. Selection of antibiotic resistance: allelic diversity among fluoroquinolone-resistant mutations. J Infect Dis. 2000;182:517–525. doi: 10.1086/315708. [DOI] [PubMed] [Google Scholar]