LETTER
Understanding the mechanism of the rapid spread of the SARS-CoV-2 B.1.1.529 (Omicron) variant is of great importance to devising public health interventions. Increased viral load has been reported as one cause of increased infectiousness in prior emergent variants and has been associated with symptomatic versus asymptomatic infections (1). While initial reports have attributed Omicron’s exponential growth and increased contagiousness to evasion of humoral memory due to altered spike protein antigens (2, 3), it is unclear to what extent viral load has contributed to its dominance (4, 5). Here, we examine whether symptomatic individuals and asymptomatic carriers infected with Omicron demonstrate differences in viral load by examining the reverse transcriptase PCR (RT-PCR) cycle thresholds (CT) in sequence-confirmed cases, compared with prior infections with the Alpha and Delta variants of SARS-CoV-2.
Since March 2020, the University of Washington Virology Laboratory has performed approximately one-third of the testing for the state of Washington, with a majority of samples collected at open-access community testing sites throughout the state. Samples were collected using anterior nasal swabs observed by health care professionals and tested on four assays (Roche Cobas 6800, Abbott Alinity m, Panther Hologic Fusion, and a Centers for Disease Control and Prevention (CDC) assay-based lab-developed test [6, 7]). The lowest CT value output from each platform was chosen as the single representative value. The symptomatic status of the individuals presenting to the testing sites was determined from free-text reasons for seeking testing. Using keywords present in such text responses, we categorized individuals as either symptomatic, exposed, or asymptomatic (see Table S1 in the supplemental material). To control for variations in community-wide viral RNA loads, we restricted our analysis to time periods when the Alpha, Delta, and Omicron variants were increasing in prevalence.
We examined 2001 Alpha samples collected between 1 March and 8 May 2021, 792 Delta samples collected between 1 June and 15 July 2021, and 1,935 Omicron samples collected between 1 December 2021 and 2 January 2022. The median CT values were 22.0 ± 4.8 (Alpha), 19.7 ± 4.8 (Delta), and 20.8 ± 4.5 (Omicron). Overall, Omicron demonstrated a significantly different CT distribution compared to both Alpha and Delta (Wilcoxon rank sum test, P < 2e-16).
The Omicron infections did not have higher viral loads than those with Delta when stratified by the major PCR platforms used and by symptomatic versus asymptomatic status (Fig. 1). Consistent with prior reports (4, 5), the symptomatic individuals across each variant had higher viral loads than did the asymptomatic carriers. Within each clinical category examined, the individuals with Omicron did not have higher viral loads than did those infected with Delta (Wilcoxon rank sum test: symptomatic, P = 2.7e-6; asymptomatic, P = 2.8e-4; exposure, P = 0.01) (Fig. 1). The viral loads from symptomatic individuals measured on the Abbott platform were significantly lower for Omicron versus Delta infections (P < 0.0001) but were not significantly different on the Roche platform (P = 0.29). The viral loads from asymptomatic individuals trended lower for Omicron infections compared to Delta but were not statistically significant on either the Roche (P = 0.06) or Abbott platforms (P = 0.14).
FIG 1.
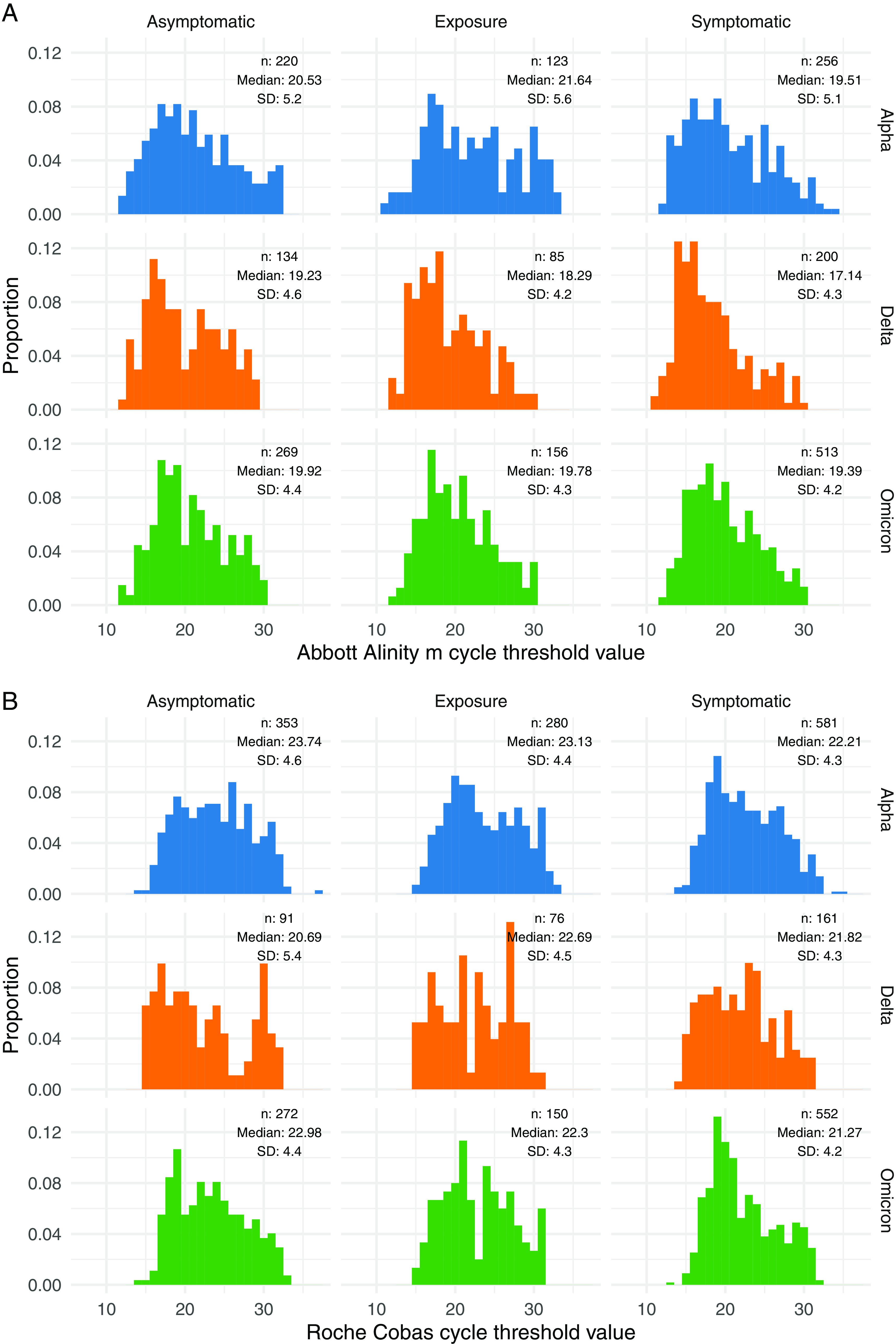
Histogram of RT-PCR cycle threshold distributions among variants, stratified by clinical status and RT-PCR platform. The samples and clinical status are shown for the Abbott Alinity m (A) and Roche cobas (B) platforms.
Our data suggest that the spread of Omicron is unlikely to be attributed to higher nasal viral loads compared to prior variants. Limitations to this analysis include the lack of stratification by the specific day of symptom onset, lack of longitudinal data to measure the peak viral load, use of CT as a surrogate for viral load, and restriction of the analysis to only anterior nasal swab viral loads. While recent data suggest that SARS-CoV-2 may be detectable earlier in saliva (8), our data capturing almost 5,000 sequence-confirmed nasal collections across multiple clinical categories and stratified by PCR platform demonstrate that Omicron is not associated with a higher nasal viral load compared to previous variants.
ACKNOWLEDGMENTS
A.L.G. reports central testing contracts with Abbott and research support from Gilead and Merck outside the submitted work.
Footnotes
Supplemental material is available online only.
Contributor Information
Alexander L. Greninger, Email: agrening@uw.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Teyssou E, Delagrèverie H, Visseaux B, Lambert-Niclot S, Brichler S, Ferre V, Marot S, Jary A, Todesco E, Schnuriger A, Ghidaoui E, Abdi B, Akhavan S, Houhou-Fidouh N, Charpentier C, Morand-Joubert L, Boutolleau D, Descamps D, Calvez V, Marcelin AG, Soulie C. 2021. The Delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19. J Infect 83:e1–e3. 10.1016/j.jinf.2021.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carreño JM, Alshammary H, Tcheou J, Singh G, Raskin AJ, Kawabata H, Sominsky LA, Clark JJ, Adelsberg DC, Bielak DA, Gonzalez-Reiche AS, Dambrauskas N, Vigdorovich V, Srivastava K, Sather DN, Sordillo EM, Bajic G, van Bakel H, Simon V, Krammer F, PSP-PARIS Study Group. 2022. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602:682–688. 10.1038/d41586-021-03846-z. [DOI] [PubMed] [Google Scholar]
- 3.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, et al. 2022. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185:467–484.e15. 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 25:2001483. 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvatore PP, Dawson P, Wadhwa A, Rabold EM, Buono S, Dietrich EA, Reses HE, Vuong J, Pawloski L, Dasu T, Bhattacharyya S, Pevzner E, Hall AJ, Tate JE, Kirking HL. 2021. Epidemiological correlates of PCR cycle threshold values in the detection of SARS-CoV-2. Clin Infect Dis 72:e761–e767. 10.1093/cid/ciaa1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman JA, Pepper G, Naccache SN, Huang M-L, Jerome KR, Greninger AL. 2020. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol 58:e00821-20. 10.1128/JCM.00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perchetti GA, Pepper G, Shrestha L, LaTurner K, Yae Kim D, Huang M-L, Jerome KR, Greninger AL. 2021. Performance characteristics of the Abbott Alinity m SARS-CoV-2 assay. J Clin Virol 140:104869. 10.1016/j.jcv.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamson B, Sikka R, Wyllie AL, Premsrirut P. 2022. Discordant SARS-CoV-2 PCR and rapid antigen test results when infectious: a December 2021 occupational case series. medRxiv. 10.1101/2022.01.04.22268770. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1. Download jcm.00139-22-s0001.xlsx, XLSX file, 0.2 MB (213KB, xlsx)


