ABSTRACT
Emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with enhanced transmissibility, pathogenicity, and immune escape ability have ravaged many countries and regions, which has brought substantial challenges to pandemic prevention and control. Real-time reverse transcriptase PCR (rRT-PCR) is widely used for SARS-CoV-2 detection but may be limited by the continuous evolution of the virus. However, the sensitivity of Chinese commercial rRT-PCR kits to critical SARS-CoV-2 variants remains unknown. In this study, contrived MS2 virus-like particles were used as reference materials to evaluate the analytical sensitivity of Daan, BioGerm, EasyDiagnosis, Liferiver, and Sansure kits when detecting six important variants (Alpha, Beta, Gamma, Delta, Omicron, and Fin-796H). The Beta and Delta variants adversely affected the analytical sensitivity of the BioGerm ORF1ab gene assay (9.52% versus 42.96%, P = 0.014, and 14.29% versus 42.96%, P = 0.040, respectively), whereas the N gene assay completely failed in terms of the Fin-796H variant. The Gamma and Fin-796H variants impeded the PCR amplification efficiency for the Sansure ORF1ab gene assay (33.33% versus 66.67%, P = 0.031, and 66.67% versus 95.24%, P = 0.040, respectively), and the Delta variant compromised the E gene assay (52.38% versus 85.71%, P = 0.019). The Alpha and Omicron variants had no significant effect on the kits. This study highlights the necessity of identifying the potential effect of viral mutations on the efficacy and sensitivity of clinical detection assays. It can also provide helpful insights regarding the development and optimization of diagnostic assays and aid the strategic management of the ongoing pandemic.
KEYWORDS: SARS-CoV-2 variants, mutation, rRT-PCR, analytical sensitivity
INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has severely endangered national security and human health on a global scale. From late December 2019 to March 1, 2022, there have been more than 435.62 million confirmed cases, including 5.95 million deaths (1). As the global pandemic continues to flare, the viral genome has accumulated a considerable number of mutations, and circulating variants with enhanced transmissibility, pathogenicity, and immune escape ability have ravaged many countries and regions. The large-scale, sustained emergence of novel SARS-CoV-2 variants has brought substantial challenges to pandemic prevention and control.
For real-time investigation and tracking of SARS-CoV-2 evolution, three nomenclature systems have been established by the Global Initiative of Sharing All Influenza Data (GISAID) (2), Nextstrain (3), and Pango (4). The World Health Organization (WHO) recommends the use of the Greek alphabet to distinguish important variants (5), which will facilitate public discussion and understanding of variants. Variants of concern (VOCs), including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529), and variants of interest (VOIs), including Lambda (C.37) and Mu (B.1.621), have been classified based on the increased risk posed to global public health (6). Some genetic variations have been documented to cause significant changes in transmissibility, pathogenicity, immune escape ability, available diagnostics, vaccines, and therapeutics of the virus, making them more detrimental to human beings. For example, the N501Y mutation contained in the Alpha, Beta, Gamma, and Omicron variants can enhance the binding affinity of the receptor-binding domain (RBD) for human angiotensin-converting enzyme 2 (ACE2) (7), leading to the enhanced cellular infectivity of the virus. The E484K mutation presented in the Beta and Gamma variants can facilitate evasion from the host immune system (8), thereby reducing the neutralization activity of the vaccine against the variants (9). Furthermore, the L452R mutation of the Delta variant can escape the cellular immune response, stabilize the spike protein, and boost the infectivity and replication capability of the virus (10). The Fin-796H (B.1.1.318) variant, identified by researchers from Finland, was found to present mutations similar to those in the Alpha and Beta variants and an additional mutation in the area targeted by the N gene assay recommended by China’s Center for Disease Control and Prevention (CDC).
Real-time reverse transcriptase PCR (rRT-PCR) is recognized as the “gold standard” for COVID-19 diagnosis and is widely used in clinics, but may be challenged and limited by the continuous evolution of the virus. The variation in SARS-CoV-2 is of high frequency and wide range (11). However, most nucleic acid detection reagents were established in the early stages of the pandemic, and primer/probe oligonucleotides were designed based on the initial viral sequence. As SARS-CoV-2 continues to mutate, an increasing number of mismatched sites have appeared in the primer/probe binding regions, which may affect the analytical performance of the detection reagents and even lead to false-negative results. Previous studies have compared the performance characteristics of commercial rRT-PCR kits for the detection of nonmutated SARS-CoV-2. However, the sensitivity of Chinese commercially available rRT-PCR kits to critical SARS-CoV-2 variants remains unknown. According to the external quality assessment of SARS-CoV-2 conducted by China’s National Center for Clinical Laboratories, of all the SARS-CoV-2 RNA detection kits approved by China’s National Medical Products Administration (NMPA), Daan Gene Co., Ltd. of Sun Yat-sen University, Shanghai BioGerm Medical Technology Co., Ltd., Wuhan EasyDiagnosis Biomedicine Co., Ltd., Shanghai Liferiver Bio-Tech Co., Ltd., and Sansure Biotech Co., Ltd. are the top five tests most commonly used in Chinese laboratories, accounting for more than 75% of the total market share. Therefore, this study aimed to evaluate the analytical sensitivity of these five kits when detecting six important variants, namely, Alpha, Beta, Gamma, Delta, Omicron, and Fin-796H, and to investigate the potential effects of virus mutations on these tests.
MATERIALS AND METHODS
Preparation of samples.
The genome sequence of the SARS-CoV-2 Wuhan-Hu-1 strain (accession number MN908947.3) was obtained from the National Center for Biotechnology Information (NCBI) GenBank and used as a reference sequence for viral genetic variations. The whole genome sequences of the SARS-CoV-2 variants, including Alpha, Beta, Gamma, Delta, Omicron, and Fin-796H, were downloaded from the GISAID (Table 1). Genome sequence alignments between individual variants and the Wuhan-Hu-1 strain were performed using Nucleotide BLAST (12). Only genome sequences with no more than three degenerate bases were selected for further studies. A preliminary investigation was conducted on the target detection segments of the NMPA-approved assays and laboratory-developed tests. All of these assays targeted sequences in the ORF1ab, N, or E genes. The length of the RNA sequence enveloped was 12795 bp, consisting of a partial region of the ORF1ab, full region of the S, N, and E genes. Ultimately, target detection segments of nucleotide sequences from the SARS-CoV-2 reference genome and six different mutated genomes were outsourced to Sangon Biotech Co., Ltd. (Shanghai, China) for synthesis.
TABLE 1.
Information on the six variants in the study
| WHO label | Pango lineage | Virus name | Accession ID | Earliest documented samples | Date of designation |
|---|---|---|---|---|---|
| Alpha | B.1.1.7 | BetaCoV/England/QEUH-F56F0F/2021 | EPI_ISL_852526 | United Kingdom, Sep-2020 | 18-Dec-2020 |
| Beta | B.1.351 | BetaCoV/South_Africa/KRISP-K007869/2020 | EPI_ISL_860630 | South Africa, May-2020 | 18-Dec-2020 |
| Gamma | P.1 | BetaCoV/Brazil/AM-994/2020 | EPI_ISL_833174 | Brazil, Nov-2020 | 11-Jan-2021 |
| Delta | B.1.617.2 | BetaCoV/India/ILSGS00922/2021 | EPI_ISL_1663498 | India, Oct-2020 | 11-May-2021b |
| Omicron | B.1.1.529 | BetaCoV/South Africa/CERI-KRISP-K032233 | EPI_ISL_6699757 | Multiple countries, Nov-2021 | 26-Nov-2021c |
| Fin-796Ha | B.1.1.318 | BetaCoV/Finland/FinD796H/2021 | EPI_ISL_1061414 | Finland, Feb-2021 | /d |
Unlike other variants, the “Fin-796H” label is not recommended by WHO.
The Delta variant was designated as VOI on April 4, 2021, and as VOC on May 11, 2021.
The Omicron variant was designated as VUM on November 24, 2021, and as VOC on November 26, 2021.
The Fin-796H variant is not a VOC designated by WHO.
The synthesized sequences were double-enzyme excised, gel-purified, and subcloned into Kpn I/Pac I or AatII/Pac I sites of the dual expression vector pACYC-MS2 that was able to stably express MS2 coat protein and maturase. The recombinant plasmids successfully verified by Sanger sequencing were transformed into Escherichia coli strain BL21(DE3) to express armored RNAs, and the MS2 virus-like particles (VLPs) were purified according to previous studies in our laboratory (13). The obtained MS2-VLPs were then verified by DNase I and RNase A digestion and real-time quantitative PCR (RT-qPCR), after which each sample was quantitated using droplet digital PCR (ddPCR, Bio-Rad). First, the RNAs of diluted VLPs were isolated using the QIAamp Viral RNA minikit (Qiagen, Hilden, Germany), followed by being reverse transcribed into cDNA using the PrimeScript RT reagent kit (Perfect Real Time; TaKaRa, Japan). Then, ddPCR was performed on the Bio-Rad QX-200 system with ddPCR Supermix for Probes (Bio-Rad, Hercules, USA). The QuantaSoft software version 1.7.4 (Bio-Rad) was used to analyze the copy number of cDNA in the ddPCR reaction system, which can be used to deduce the initial concentration of VLPs. All primers and probes used for RT-qPCR and ddPCR are listed in Table S1 in the supplemental material.
Preparation of commercial rRT-PCR kits.
Five commercial rRT-PCR detection kits most commonly used in Chinese laboratories were selected for evaluation: Daan, BioGerm, EasyDiagnosis, Liferiver, and Sansure. The characteristics of the five kits are summarized in Table 2. To eliminate the potential influence of nucleic acid extraction on detection, a commercial NP968-C automatic nucleic acid extraction system (Xi’an Tianlong Science and Technology Co., Ltd.) was used to separate and purify nucleic acids. A total sample of 200 μL of the SARS-CoV-2 variant-specific MS2-VLPs was used for extraction, and RNA was eluted with 80 μL elution buffer. All the procedures were performed according to the manufacturer’s instructions.
TABLE 2.
Characteristics of five SARS-CoV-2 rRT-PCR detection kitsa
| Detection kit | Specimens | Target genes | Claimed LOD (copies/mL) | RNA input (μL) | Total reaction vol (μL) | Total PCR cycle no. |
|---|---|---|---|---|---|---|
| Daan | NP, OP, sputum | ORF1ab, N | 500 | 5 | 25 | 45 |
| BioGerm | OP, sputum | ORF1ab, N | 500 | 5 | 25 | 45 |
| EasyDiagnosis | NP, OP, sputum | ORF1ab, N | 500 | 5 | 25 | 40 |
| Liferiver | NP, sputum | ORF1ab, N, E | 200 | 5 | 25 | 45 |
| Sansure | NP, OP, BALF | ORF1ab, N | 200 | 20 | 50 | 45 |
NP, nasal pharyngeal; OP, oral pharyngeal; BALF, bronchoalveolar lavage fluid; ORF, open reading frame; N, nucleocapsid protein gene; E, envelope protein gene; LOD, the limit of detection.
Limits of detection (LODs) of different kits for different strains.
The six types of quantified VLPs were, respectively, serially diluted 3-fold to 9000, 3000, 1000, 333.33, 111.11, 37.04, 12.35, and 4.12 copies/mL using Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) and dispensed in 0.5-mL aliquots for detection. For the reference strain and six variants, a total of 168 positive samples were tested using the five commercial kits on the Applied Biosystems 7500 Fast real-time PCR system (Thermo Fisher Scientific, USA), with 21 samples per concentration. In addition, 21 negative samples were detected in parallel. Retesting of the samples and interpretation of results were carried out according to the manufacturer’s specifications (Table S2). Ultimately, the test results were uniformly classified into two categories: positive and negative.
Statistical analysis.
MedCalc Statistical Software version 19.6.1 (MedCalc Software Ltd., Ostend, Belgium) was used for probit regression analysis, and the LODs of five detection assays were estimated. Other statistical analyses were performed using SPSS Statistics for Windows (version 19.0; IBM Corp., Armonk, NY, USA). The variance among different groups was compared using Pearson’s chi-square test, and a P value <0.05 was regarded as statistically significant.
RESULTS
Evaluation of samples.
MS2-VLPs of the reference strain and six variants of SARS-CoV-2 were constructed and expressed successfully, which were verified using sequencing and double enzymatic digestion. After quantification using RT-qPCR and ddPCR with specific primers and probes, the VLPs containing different target genes were blended in identical proportions at a concentration of 1 × 108 copies/mL. The samples were then diluted in a 3-fold series and tested from 9,000 copies/mL to 4.12 copies/mL.
Retest rates of different samples.
Based on the retest rules for instructions, samples from suspected positive cases in the first test were retested. The retest rates of five detection kits for different samples are presented in Table S3. In general, the retest rates of samples at 111.11 and 37.04 copies/mL were higher than those of samples at lower or higher concentrations. However, this was not the case when the BioGerm kit was used to detect serially diluted Fin-796H variants. The percentage of Fin-796H samples with concentrations ranging from 9,000 to 333.33 copies/mL that required retesting were all 100% using the BioGerm kit. Besides, when detecting Beta, Delta, and Omicron variants at 111.11 copies/mL using the BioGerm kit, and Beta, Gamma, and Delta variants at 111.11 copies/mL using the EasyDiagnosis kit, the retest rates all exceeded 70%. However, for the Sansure kit, retesting of single-target positive samples was not required, the retest rates of which were 0%.
Evaluation of LODs of rRT-PCR kits.
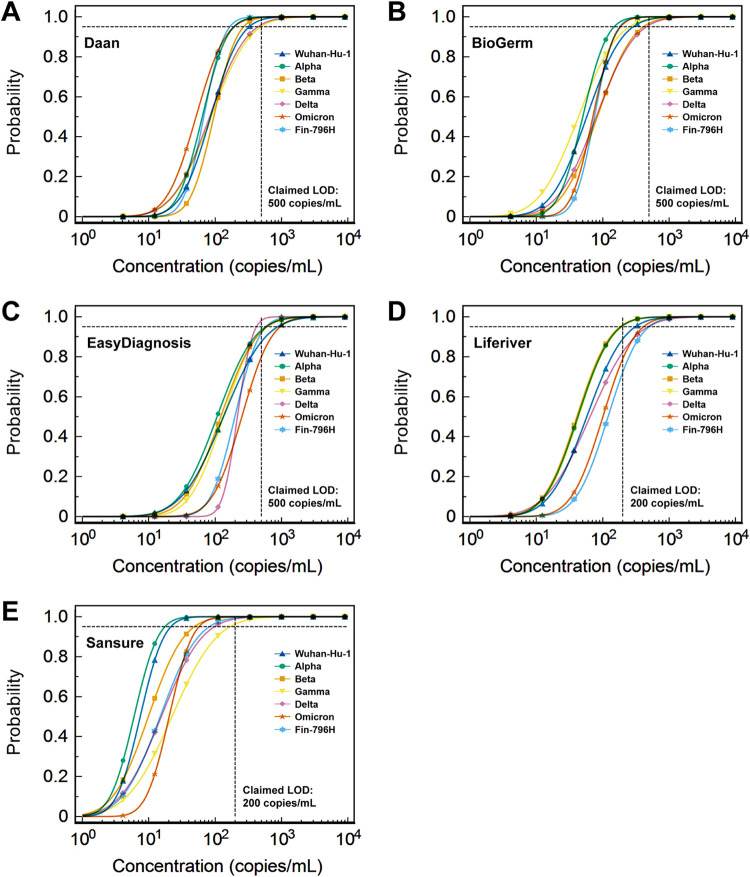
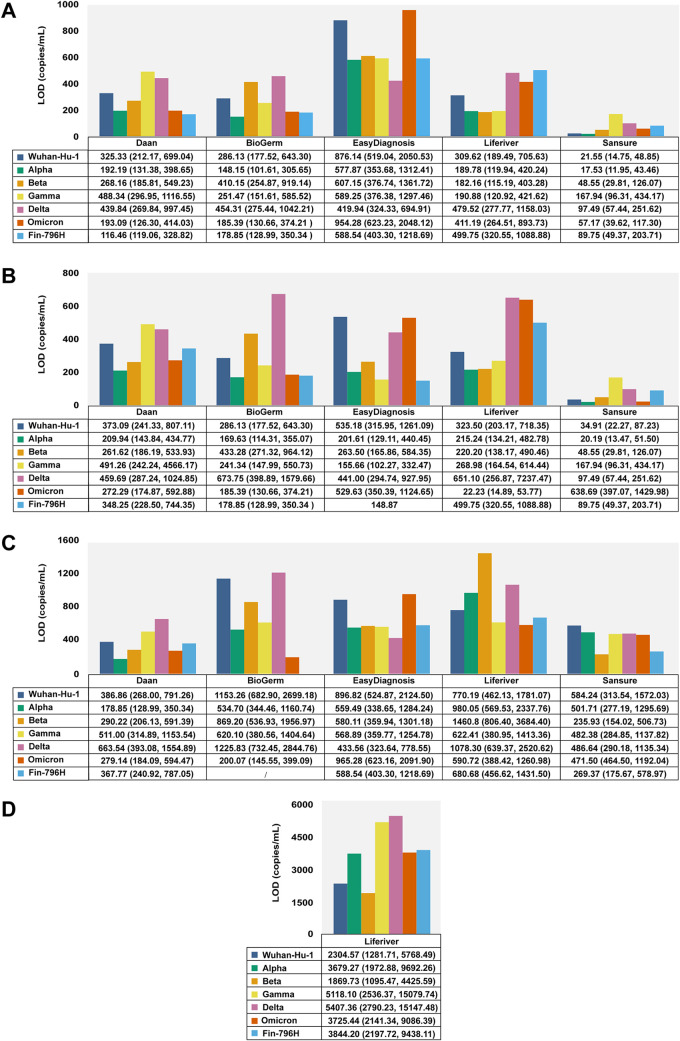
The results of the probit regression analysis performed to calculate the LODs of the six detection assays for different strains with a 95% repeatable probability are shown in Table 3, Fig. 1, and Fig. 2A. The LODs of Gamma and Delta variants detected using the Daan kit were 488.34 copies/mL (95% confidence interval [CI]: 296.95–1116.55 copies/mL) and 439.84 copies/mL (95% CI: 269.84–997.45 copies/mL), which were higher than that of the Wuhan-Hu-1 strain (325.33 copies/mL, 95% CI: 212.17–699.04 copies/mL) (Fig. 1A, 2A). The BioGerm kit showed higher LODs for the Beta (410.15 copies/mL, 95% CI: 254.87–919.14 copies/mL) and Delta (454.31 copies/mL, 95% CI: 275.44–1042.21 copies/mL) variants than the Wuhan-Hu-1 strain (286.13 copies/mL, 95% CI: 177.52–643.30 copies/mL) (Fig. 1B, 2A). The LOD of the EasyDiagnosis for the Omicron variant (954.28 copies/mL, 95% CI: 623.23–2,048.12 copies/mL) was higher than the Wuhan-Hu-1 strain (876.14 copies/mL, 95% CI: 519.04–2,050.53 copies/mL) (Fig. 1C, 2A). The Liferiver kit showed higher LODs for the Delta (479.52 copies/mL, 95% CI: 277.77–1,158.03 copies/mL), Omicron (411.19 copies/mL, 95% CI: 264.51–893.73 copies/mL), and Fin-796H (499.75 copies/mL, 95% CI: 320.55–1,088.88 copies/mL) variants than the Wuhan-Hu-1 strain (309.62 copies/mL, 95% CI: 189.49–705.63 copies/mL) (Fig. 1D, 2A). The LODs of the Beta (48.55 copies/mL, 95% CI: 29.81–126.07 copies/mL), Gamma (167.94 copies/mL, 95% CI: 96.31–434.17 copies/mL), Delta (97.49 copies/mL, 95% CI: 96.31–434.17 copies/mL), Omicron (57.17 copies/mL, 95% CI: 39.62–117.33 copies/mL), and Fin-796H (81.74 copies/mL, 95% CI: 49.37–203.71 copies/mL) variants detected using the Sansure kit were all higher than the Wuhan-Hu-1 strain (21.55 copies/mL, 95% CI: 14.75–48.85 copies/mL) (Fig. 1E, 2A).
TABLE 3.
Summary of results for positive rates (21 samples tested in total)
| Concentration (copies/mL) | Daan (retest rate, %) |
BioGerm (retest rate, %) |
EasyDiagnosis (retest rate, %) |
Liferiver (retest rate, %) |
Sansure (retest rate, %) |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wuhan-Hu-1 | Alpha | Beta | Gamma | Delta | Omicron | Fin-796H | Wuhan-Hu-1 | Alpha | Beta | Gamma | Delta | Omicron | Fin-796H | Wuhan-Hu-1 | Alpha | Beta | Gamma | Delta | Omicron | Fin-796H | Wuhan-Hu-1 | Alpha | Beta | Gamma | Delta | Omicron | Fin-796H | Wuhan-Hu-1 | Alpha | Beta | Gamma | Delta | Omicron | Fin-796H | |
| 9000 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 3000 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 1000 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 95.24 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 333.33 | 100 | 100 | 100 | 100 | 90.48 | 100 | 100 | 100 | 100 | 90.48 | 100 | 85.71 | 100 | 100 | 80.95 | 85.71 | 90.48 | 80.95 | 85.71 | 90.48 | 57.14 | 100 | 100 | 95.24 | 100 | 100 | 95.24 | 95.24 | 100 | 100 | 100 | 100 | 95.24 | 100 | 100 |
| 111.11 | 47.62 | 76.19 | 52.38 | 38.10 | 61.90 | 80.95 | 80.95 | 61.90 | 85.71 | 66.67 | 80.95 | 71.43 | 76.19 | 76.19 | 23.81 | 42.86 | 23.81 | 42.86 | 4.76 | 14.29 | 9.52 | 57.14 | 80.95 | 95.24 | 76.19 | 47.62 | 33.33 | 42.86 | 100 | 100 | 95.24 | 100 | 100 | 100 | 100 |
| 37.04 | 23.81 | 23.81 | 9.52 | 19.05 | 19.05 | 14.29 | 33.33 | 42.86 | 38.10 | 14.29 | 28.57 | 19.05 | 9.52 | 14.29 | 19.05 | 23.81 | 23.81 | 9.52 | 0 | 0 | 4.76 | 47.62 | 47.62 | 42.86 | 57.14 | 28.57 | 4.76 | 19.05 | 100 | 100 | 95.24 | 42.86 | 76.19 | 66.67 | 100 |
| 12.35 | 0 | 0 | 0 | 9.52 | 4.76 | 0 | 4.76 | 4.76 | 0 | 4.76 | 23.81 | 4.76 | 0 | 0 | 4.76 | 0 | 0 | 0 | 0 | 0 | 0 | 4.76 | 9.52 | 9.52 | 4.76 | 19.05 | 4.76 | 0 | 76.19 | 85.71 | 71.43 | 33.33 | 47.62 | 66.67 | 80.95 |
| 4.12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.76 | 19.05 | 9.52 | 14.29 | 9.52 | 4.76 | 23.81 |
FIG 1.
Results of probit regression analysis performed to calculate the LODs of five rRT-PCR detection kits for different strains. (A–E) Daan, BioGerm, EasyDiagnosis, Liferiver, and Sansure, respectively. LOD, the limit of detection.
FIG 2.
Estimated LODs of five rRT-PCR detection kits for different strains. (A–D) The overall LODs, LODs of ORF1ab gene, N gene, and E gene, respectively. LOD, the limit of detection.
Evaluation of LODs of target genes.
The effects of SARS-CoV-2 variants on the LODs of target genes assayed by the five commercial detection kits were analyzed. The positive rates of the ORF1ab gene of Beta and Delta variants detected using the BioGerm kit at 37.04 copies/mL were 9.52% (2/21) and 14.29% (3/21), respectively, which were much lower than that of the Wuhan-Hu-1 strain (42.96%, 9/21), indicating the potential effect of genetic mutations carried by the two variants on the sensitivity of the BioGerm ORF1ab gene (P = 0.014 and 0.040, respectively) (Fig. 2B). Most remarkably, BioGerm failed to detect the N gene in the Fin-796H variant (Fig. 2C). Accordingly, contrasted with the Wuhan-Hu-1 strain, the lower positive rates of the ORF1ab gene for the Gamma variant at 37.04 copies/mL (42.86%, 9/21, versus 95.24%, 20/21, P = 0.000) and 12.35 copies/mL (33.33%, 7/21, versus 66.67%, 14/21, P = 0.031), as well as the Fin-796H variant at 37.04 copies/mL (66.67%, 14/21, versus 95.24%, 20/21, P = 0.040), showed that the Gamma and Fin-796H variants might affect the performance of Sansure (Fig. 2B). Furthermore, similar performance compromising effects on the Liferiver were also observed during the detection of the E gene of the Delta variant at 1,000 copies/mL (52.38%, 11/21, versus 85.71%, 18/21, P = 0.019) (Fig. 2D). The Alpha and Omicron variants had no significant effect on the analytical sensitivity of these five kits.
DISCUSSION
SARS-CoV-2 replicates rapidly and is prone to replication errors upon entry into the host cells. Moreover, its wide range of hosts and structural characteristics predispose it to mutations or genetic recombination in the process of evolution, leading to extensive genetic diversity. Some variations have been documented to cause considerable alterations in transmissibility, pathogenicity, immune escape ability, available diagnostics, vaccines, and therapeutics of the virus. Variations in the primer/probe binding regions may potentially affect the PCR amplification efficiency and even lead to erroneous results. The 28881–28883GGG>AAC trinucleotide mutation, located at the 5′ end of the forward primer in the N gene assay recommended by China’s CDC, has a reported mutation rate of 36.11% in the 84,305 available high-quality genomes submitted to the NCBI and GISAID databases (14). Substantial mismatches and polymorphisms have also been identified in the reverse primer from the RdRp gene assay of Charité hospital and the reverse primer from the N gene assay of Japan’s National Institute of Infectious Diseases (NIID) (15). In addition, a study from Belgium observed that the C-to-U transition at position 26340 in the SARS-CoV-2 genome restrained the Roche Cobas SARS-CoV-2 E gene assay in eight clinically positive samples (16). The B.1.177.75 lineage containing 28948C>T coupled with 28932C>T was speculated to be associated with the failure of the Thermo Fisher TaqPath N gene assay (17). Isolates containing the 29200C>T single nucleotide polymorphism have also been reported to suppress the N gene detection in the Cepheid Xpert Xpress assay (18). In response to the emergence of new variants, reagent manufacturers in China have performed sequence alignment with publicly available variants and optimized their tests by changing target sequences, thus ensuring the accurate detection of variants. As a result, the five detection kits, all of which are commonly used large-scale population screening reagents, can effectively detect positive infections in the 2021 Delta and Omicron epidemics (Delta: in Guangzhou in May, Nanjing, and Yangzhou in July, Putian and Xiamen in September, and Ejina Banner in October; Omicron: in Tianjin and Anyang in December).
In this study, by comparing the detection results from six different SARS-CoV-2 variants with those from the nonmutated Wuhan-Hu-1 strain, we analyzed the underlying effects of different variants on the sensitivity of commercially available rRT-PCR detection kits. According to the results, the Beta and Delta variants affected the analytical sensitivity of the BioGerm ORF1ab gene assay, whereas the N gene assay completely failed in terms of the Fin-796H variant. Furthermore, the Gamma and Fin-796H variants impeded the PCR amplification efficiency of the Sansure ORF1ab gene assay, and the Delta variant compromised the Liferiver E gene assay. The Alpha and Omicron variants had no significant effect on the analytical sensitivity of these five kits. The sensitivity of different target genes will affect the retest rate, thus delaying detection results reported to clinics, leading to more consumption of reagents, and aggravating the workload of medical staff.
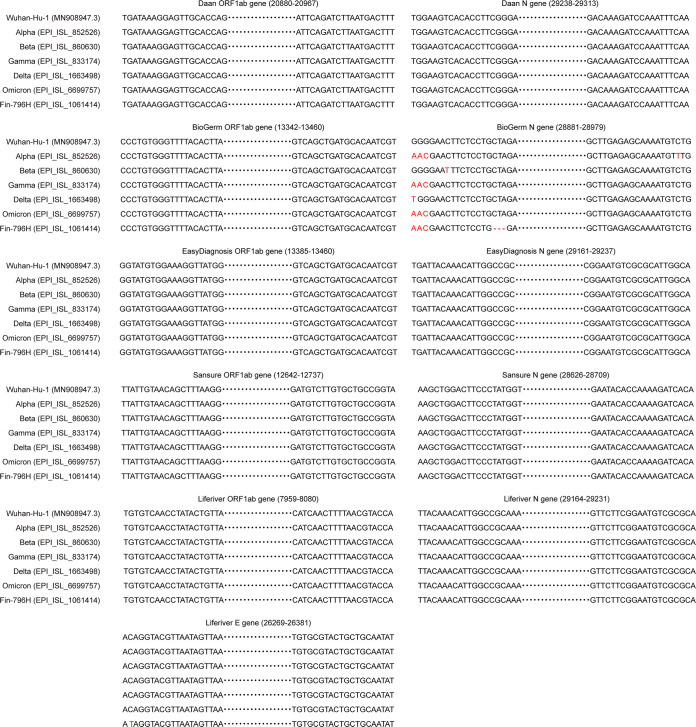
The severity of the influence of mismatches on PCR amplification efficiency is dependent on numerous factors, among which the position and number of mismatches are the most important (19, 20). The presence of four mismatches in a single primer is known to block amplification almost entirely, and the degree of inhibition is negatively related to the distance of the mismatches from the 3′ end of the primers (21). Mismatches situated at the probe binding sites have the greatest influence on the detection efficiency (22). The rRT-PCR products of these kits were sequenced, and the target regions are shown in Fig. 3. Based on the sequencing results, the target regions of the BioGerm kit were consistent with China’s CDC-developed SARS-CoV-2 assay (23). The 28881–28883GGG>AAC combined with 28896_28898delCTA in the N gene assay suggest that they may be responsible for the failure of BioGerm to detect the Fin-796H variant. Apart from the BioGerm kit, the combined mutations may also affect the performance of other commercial or laboratory-developed tests that are based on the same set of primers and probe as China’s CDC-developed N gene assay. The adverse influence of pyrimidine-pyrimidine and purine-pyrimidine mismatches is more substantial than purine-purine mismatches, with G:G and C:C as exceptions because of the hydrogen-bonded structure and stacking force (24). Additionally, the DNA polymerase binds to the terminus to be extended by recognizing its hydrogen bonding, base stacking, and overall steric structure, which may be altered by mismatches between primers and templates (20). The incorporation of mismatched base pairs has been shown to destabilize the primer–template duplexes to a certain extent (25), the overall stability of which may determine the binding of the DNA polymerase to the target sites. In addition, the polymerase may be much more easy to dissociate from mismatched duplexes, contributing to incompletely amplified DNA products (26).
FIG 3.
Sequencing results of rRT-PCR products of detection kits. The bases marked in red are mismatched compared to the Wuhan-Hu-1 strain.
However, the increased or decreased analytical sensitivity in the absence of mutations in the amplified region was observed, indicating that other factors might affect the assay efficiency in a particular sequence context (22). Templates with mutations might facilitate the formation of secondary structures that are conducive or adverse to opening during reverse transcription and amplification processes (27). Moreover, the primers might hybridize with the stem structure or ring structure of the mutated template, which is less stable and more susceptible to hydrolysis by RNase A (28–30). There is another possibility that mismatches might neutralize or aggravate the stem-loop interference in the primer binding site previously free of mutation (31). Therefore, there are many factors that might affect the PCR amplification efficiency, including the number, position, and type of mismatches, the inherent flexibility of DNA with many conformational states (32), and the adjacent sequence context (varying steric hindrance and/or duplex stability) (33). Notably, the effects of spatial structures are minor compared to those of the mismatches on the primer/probe binding sites.
Furthermore, our results revealed that the affected detection of one of the target genes did not necessarily change the overall LODs of the detection kits, because the undesired effect of one target gene could be countered by another impervious gene after retesting. This phenomenon can be explained by the interpretation criteria of different stringencies for these detection kits. For example, a single positive result for the target genes required to be retested for the Liferiver kit, and two positive retested results, could be interpreted as positive. Therefore, although the LODs of the Alpha variant for the Liferiver N gene and E gene assays were higher compared to those of the Wuhan-Hu-1 strain, the overall LOD was unaffected due to the ORF1ab gene. In contrast, although the influence of each target gene was small, the superposition effects of the combined target genes possibly resulted in more pronounced differences in the overall LODs.
To minimize the effect of mutations on the detection performance of commercial kits in the context of emerging and new variants, improvements can be made from multiple perspectives. First, in the design of the primers and probes used in a diagnostic assay for SARS-CoV-2, research personnel should not only focus on the expected sensitivity and specificity of nonmutated sequences, but should also take circulating variants into consideration. In the design of primers/probe sets, the selection from conserved regions of the genome has been suggested to minimize the number of mismatches. A single T or double T at the 3′ terminal end has been shown to aid in alleviating amplification bias derived from mismatches (20). Given the multiple rRT-PCR assays employed by almost all reagent manufacturers, in silico alignment of the primer and probe sequences with sufficient SARS-CoV-2 genomic sequences should be emphasized. The NCBI Primer-BLAST (34) can be used to confirm primer specificity in the case of different variants. And the secondary structures of variant sequences can be predicted using ProbKnot (35) or Mfold (36) to avoid ring structures recognized by primers. In addition, lowering the annealing temperature (37), increasing the extension cycles in the low template-concentration reaction (21), replacement of modified bases such as 5-meC (38), and introducing degenerate primers (39) can also be exploited to improve the mismatch tolerance. Moreover, after completing the optimization of the reaction system design, different variant samples are recommended to be evaluated for performance validation. Second, laboratories should comprehensively interpret the detection results in consideration of the feasible genetic mutations. Contradictory results must be cautiously combined with clinical observations, medical history, and epidemiological information, which can be verified using other detection kits to prevent incorrect reports to clinics. Besides, several vital variants can be added to regular quality control materials in routine work. Third, genomic surveillance based on ongoing sequencing is of tremendous importance for monitoring strains of concern and localized epidemic outbreaks.
In conclusion, this study revealed the potential influence of six significant SARS-CoV-2 variants on the analytical sensitivities of five commercial rRT-PCR detection kits and analyzed the underlying mechanisms. The implications of this study remind us that the effect of mutations on detection should not be ignored. To ensure the validity of commercial rRT-PCR kits in the context of genomic diversity, it is necessary to strengthen genomic surveillance and sequencing capacities of SARS-CoV-2 samples, as well as improve the analytical performance of the kits.
ACKNOWLEDGMENTS
The work was supported by the National Key Research and Development Program of China grant 2021YFC0863300 (J.L.).
We thank all the reagent manufacturers that provided the automatic nucleic acid extraction system and rRT-PCR kits for SARS-CoV-2 detection in this study.
Footnotes
Supplemental material is available online only.
Contributor Information
Jinming Li, Email: jmli@nccl.org.cn.
Rui Zhang, Email: ruizhang@nccl.org.cn.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.World Health Organization . 2022. Coronavirus (COVID-19) dashboard with vaccination data. https://covid19.who.int/. Accessed 1 March 2022.
- 2.Global Initiative of Sharing All Influenza Data. 2021. Clade and lineage nomenclature aids in genomic epidemiology studies of active hCoV-19 viruses. https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/. Accessed 1 March 2022.
- 3.Bedford T, Hodcroft EB, Neher RA. 2021. Updated Nextstrain SARS-CoV-2 clade naming strategy. Nextstrain. https://nextstrain.org/blog/2021-01-06-updated-SARS-CoV-2-clade-naming. Accessed 1 March 2022.
- 4.Rambaut A, Holmes EC, O'Toole A, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2021. WHO announces simple, easy-to-say labels for SARS-CoV-2 variants of interest and concern 2021. https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern. Accessed 1 March 2022.
- 6.World Health Organization. 2021. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 1 March 2022.
- 7.Shahhosseini N, Babuadze GG, Wong G, Kobinger GP. 2021. Mutation signatures and in silico docking of novel SARS-CoV-2 variants of concern. Microorganisms 9:926. 10.3390/microorganisms9050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise J. 2021. Covid-19: the E484K mutation and the risks it poses. BMJ 372:n359. 10.1136/bmj.n359. [DOI] [PubMed] [Google Scholar]
- 9.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29:463–476.e6. 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, Ngare I, Kimura I, Uriu K, Kosugi Y, Yue Y, Shimizu R, Ito J, Torii S, Yonekawa A, Shimono N, Nagasaki Y, Minami R, Toya T, Sekiya N, Fukuhara T, Matsuura Y, Schreiber G, Genotype to Phenotype Japan (G2P-Japan) Consortium, Ikeda T, Nakagawa S, Ueno T, Sato K. 2021. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 29:1124–1136.e11. 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan T. 2020. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 81:104260. 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madden T. 2013. The BLAST sequence analysis tool. The NCBI Handbook 2:425–436. [Google Scholar]
- 13.Lin G, Zhang K, Zhang D, Han Y, Xie J, Li J. 2017. Fast preparation of a long chimeric armored RNA as controls for external quality assessment for molecular detection of Zika virus. Clin Chim Acta 466:138–144. 10.1016/j.cca.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Gand M, Vanneste K, Thomas I, Van Gucht S, Capron A, Herman P, Roosens NHC, De Keersmaecker SCJ. 2021. Deepening of in silico evaluation of SARS-CoV-2 detection RT-qPCR assays in the context of new variants. Genes 12:565. 10.3390/genes12040565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchinski KS, Jassem AN, Prystajecky NA. 2020. Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline. J Clin Virol 131:104581. 10.1016/j.jcv.2020.104581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, Meex C, Melin P, Hayette M-P, Bours V, Durkin K. 2020. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol 58:e01598-20. 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amato L, Jurisic L, Puglia I, Di Lollo V, Curini V, Torzi G, Di Girolamo A, Mangone I, Mancinelli A, Decaro N, Calistri P, Di Giallonardo F, Lorusso A, D'Alterio N. 2021. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerg Microbes Infect 10:1148–1155. 10.1080/22221751.2021.1933609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. 2020. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill 25:2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christopherson C, Sninsky J, Kwok S. 1997. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res 25:654–658. 10.1093/nar/25.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, Sninsky JJ. 1990. Effects of primer–template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res 18:999–1005. 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefever S, Pattyn F, Hellemans J, Vandesompele J. 2013. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem 59:1470–1480. 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- 22.Smith S, Vigilant L, Morin PA. 2002. The effects of sequence length and oligonucleotide mismatches on 5' exonuclease assay efficiency. Nucleic Acids Res 30:e111. 10.1093/nar/gnf110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Institute for Viral Disease Control and Prevention. 2020. Specific primers and probes for detection 2019 novel coronavirus. http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html. Accessed 1 March 2022.
- 24.Aboul-Ela F, Koh D, Tinoco I, Jr, Martin FH. 1985. Base–base mismatches: thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res 13:4811–4824. 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikuta S, Takagi K, Wallace RB, Itakura K. 1987. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatched base pairs. Nucleic Acids Res 15:797–811. 10.1093/nar/15.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petruska J, Goodman MF, Boosalis MS, Sowers LC, Cheong C, Tinoco I. 1988. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci USA 85:6252–6256. 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranauskas A, Paliksa S, Alzbutas G, Vaitkevicius M, Lubiene J, Letukiene V, Burinskas S, Sasnauskas G, Skirgaila R. 2012. Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng Des Sel 25:657–668. 10.1093/protein/gzs034. [DOI] [PubMed] [Google Scholar]
- 28.Prats-Ejarque G, Blanco JA, Salazar VA, Nogues VM, Moussaoui M, Boix E. 2019. Characterization of an RNase with two catalytic centers: human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates. Biochim Biophys Acta Gen Subj 1863:105–117. 10.1016/j.bbagen.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Smith RM, Walton CM, Wu CH, Wu GY. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3'-terminal sequences. J Virol 76:9563–9574. 10.1128/jvi.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villegas J, Burzio V, Villota C, Landerer E, Martinez R, Santander M, Martinez R, Pinto R, Vera MI, Boccardo E, Villa LL, Burzio LO. 2007. Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res 35:7336–7347. 10.1093/nar/gkm863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Brownley A, Stockwell TB, Beeson K, McIntosh TC, Busam D, Ferriera S, Murphy S, Levy S. 2008. Novel computational methods for increasing PCR primer design effectiveness in directed sequencing. BMC Bioinformatics 9:191. 10.1186/1471-2105-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson SJ, Beese LS. 2004. Structures of mismatch replication errors observed in a DNA polymerase. Cell 116:803–816. 10.1016/S0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 33.Stadhouders R, Pas SD, Anber J, Voermans J, Mes THM, Schutten M. 2010. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Molecular Diagnostics 12:109–117. 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellaousov S, Mathews DH. 2010. ProbKnot: fast prediction of RNA secondary structure including pseudoknots. RNA 16:1870–1880. 10.1261/rna.2125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishii K, Fukui M. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol 67:3753–3755. 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutyavin IV. 2008. Use of base-modified duplex-stabilizing deoxynucleoside 5′-triphosphates to enhance the hybridization properties of primers and probes in detection polymerase chain reaction. Biochemistry 47:13666–13673. 10.1021/bi8017784. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Zhang J, Li J. 2020. Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2. Theranostics 10:7150–7162. 10.7150/thno.47649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download jcm.02374-21-s0001.pdf, PDF file, 0.2 MB (191.2KB, pdf)