ABSTRACT
Bloodstream infections (BSIs) represent a substantial mortality risk, yet most studies are limited to select pathogens or populations. The aim of this study was to describe the population-wide prevalence of BSIs and examine the associated mortality risk for the responsible microorganisms. We conducted a population-wide retrospective cohort study of BSIs in Ontario in 2017. Blood culture data was collected from almost all microbiology laboratories in Ontario and linked to data sets of patient characteristics. For each organism, we determined the prevalence and crude mortality risk, and using logistic regression models, the adjusted odds of 30-day mortality was calculated relative to patients with negative blood cultures and matched patients without blood culture testing. From 531,065 blood cultures, we identified 22,935 positive BSI episodes in 19,326 patients, for an incidence of 150 per 100,000 population. The most frequently isolated organisms were Escherichia coli, Staphylococcus aureus, coagulase-negative staphylococci, Klebsiella species, and Enterococcus species with 40.2, 22.4, 12.1, 11.1, and 7.1 episodes per 100,000 population respectively. BSI episodes were associated with 17.0% mortality at 30 days. Compared to patients with negative cultures, the adjusted 30-day mortality risk for positive BSIs was 1.47 (95% confidence interval (CI), 1.41 to 1.54) and compared to matched patients without blood culture testing was 2.62 (95% CI, 2.52 to 2.73). Clostridium species were associated with the highest adjusted odds of mortality compared to that of negative cultures (adjusted odds ratio, 5.81; 95% CI, 4.00 to 8.44). Among high incidence pathogens, Staphylococcus aureus had the highest odds ratio of mortality (adjusted odds ratio, 2.14; 95% CI, 1.94 to 2.36). BSIs are associated with increased mortality risk, varying across organisms.
KEYWORDS: bacteremia, bloodstream infection, surveillance, population
INTRODUCTION
Bloodstream infections (BSI) are a growing threat to public health worldwide. The 2 million cases of BSIs that occur annually in North America and Europe are associated with 250,000 deaths, making BSIs the leading cause of mortality from infection (1).
Global surveillance networks have been established to track BSIs (2); however, they are often limited by reliance on voluntary contribution of data from participating institutions. This can introduce selection bias from academic tertiary care centers and underrepresentation of community BSIs. Few studies have been conducted that measure BSIs at a total population level. Fewer among these have also been able to draw associations between patient characteristics or outcomes with individual organisms (3–5).
In Ontario, a universal health care system facilitates linkage of population-wide health care data sets at the patient level, which are now linked to population-wide microbiology data. From this, we constructed a comprehensive view of BSIs in the general population. Our objectives were 3-fold as follows: (i) to quantify the prevalence of BSIs across all health care sectors; (ii) to examine the relative prevalence of BSI organisms across these sectors; and (iii) to examine the odds of mortality associated with these organisms.
MATERIALS AND METHODS
Setting and study population.
This study was conducted in Ontario, Canada, using population-wide information from blood cultures and patient records collected in 2017. The population of Ontario in 2017 was 14.07 million, or 38.5% of the Canadian population (6). Canadian citizens who are residents of the province and those with valid immigration status with a valid health card, totaling 13.3 million people in 2017, are eligible for the universal Ontario Health Insurance Plan (OHIP).
Data sources.
This study was made possible by comprehensive, individual-level, and Ontario-wide administrative data sets, linkable via a unique and confidential identifier held at ICES (formerly called Institute for Clinical Evaluative Sciences). ICES is an independent, nonprofit research institute whose legal status under Ontario’s health information privacy law allows it to collect and analyze health care and demographic data, without consent, for health system evaluation and improvement. Among its many data holdings, ICES integrates hospital data from the Discharge Abstract Database (DAD), emergency department data from the National Ambulatory Care Reporting System (NACRS), and vital statistics and demographic data from the Registered Persons Database (RPDB). ICES recently incorporated data from the Ontario Laboratory Information System (OLIS), which contains microbiology results from 114 hospitals, community laboratories, and public health laboratories.
Definition of a blood culture episode.
Data from blood cultures collected from 1 January 2017 to 31 December 2017 were included. Positive and negative blood cultures were clustered into episodes if they were collected within 7 days of an initial sample. Episodes were excluded for missing demographic data including birth date or sex or if the death date for the patient was >5 days prior to the blood culture date. Cultures reported as “mixed growth” without organism identification were excluded, although this was rare. To avoid the inclusion of contaminated cultures into analysis, many population-based studies of BSIs either exclude typical contaminants entirely (7–9) or apply additional criteria including clinical indicators (2, 10) or multiple positive cultures (3, 11) prior to consideration. As we did not have consistent access to clinical symptoms or signs, which could be used as an indicator of true infection, for organisms which are typically associated with bloodstream culture contamination, we required at least two positive cultures from within the same episode for inclusion as a BSI episode. Similar to other population-wide studies of BSIs (3, 11), the species that were processed with this requirement were coagulase-negative staphylococci (CoNS), Bacillus species, Micrococcus species, Corynebacterium species, Paenibacillus species, Lactobacillus species, and Propionibacterium species (recently reclassified as Cutibacterium species). Cultures in which multiple organisms were identified were counted toward each in the per organism analysis.
Organism groupings.
We aggregated most organisms by genus for ease of displaying data, except for Staphylococcus species and Streptococcus species, which were grouped by commonly isolated species, with aggregate categories of the remaining species in the genus (e.g., “other coagulase-negative staphylococci [CoNS]”).
Patient characteristics.
Demographic data collected included sex, age, location of culture collection (community, long-term care, acute care hospital, and intensive care unit [ICU]), hospitalized days in the 90 days prior to blood culture collection, and Charlson comorbidity index (12). For age, patients were grouped into 0 to 3 months, 3 months to 1 year, 1 to 5 years, 5 to 10 years, 11 to 19 years, and each following decade up to 79 years, and 80 or more years.
Statistical analysis.
BSI episodes for each organism were described by number, proportion, and rate per 100,000 population (using a denominator of 13,278,784 from the RPDB). BSI organism profiles were evaluated across all of the above listed patient characteristics. Crude mortality rates were computed at 7, 30, 60, 90, and 365 days from the date of blood culture collection. We assessed the 30-day odds ratio (OR) of mortality of each organism compared to patients with negative blood cultures, using logistic regression adjusting for age (<18 years, 18 to 65 years, >65 years), sex, location, and hospitalized days in prior 90 days, with random intercepts corresponding to patients. We conducted prespecified subgroup analysis with models stratified by location. As our primary analysis may underestimate associated mortality by comparing to persons unwell enough to warrant blood culture testing, we conducted a separate comparison; we hard-matched patients with BSI with up to 10 other Ontarians without blood culture testing by age (within 2 years), sex, health care location, Deyo-Charlson comorbidity score (12), and days hospitalized in the 90 preceding blood culture (0, 1 to 4, 5 to 9, 10 to 90). This incorporated generalized estimating equations to account for the matched analysis with clusters corresponding to matched groups. Regression results are presented as OR and 95% confidence interval (CI). Analyses were conducted in SAS v9.4 (Cary, NC).
RESULTS
Overall prevalence of blood culture testing and positive blood cultures.
Data collected from microbiology laboratories in 2017 identified 531,065 blood cultures from 191,882 patients, and 252,343 eligible blood culture episodes (Fig. 1). Among these were 22,935 positive BSI episodes occurring in 19,326 patients. This equated to an annual incidence of 150 BSI episodes per 100,000 population.
FIG 1.
Flow diagram of blood culture data processing. Blood culture data collected was first clustered into episodes, followed by exclusion of episodes for which there was incomplete or incorrect data. Remaining episodes were divided into negative or positive episodes. Percentages are expressed relative to the preceding total.
Proportions of BSIs across health sectors and patient groups.
Population characteristics of patients with a blood culture episode, and specifically those with a positive BSI episode, were derived (Table 1). Blood culture collection and BSI episodes increased with age. Between sexes, the distribution of all episodes and positive episodes was similar, with slightly more occurring in men. Although 34.1% of total culture episodes were collected in the community, positive BSI episodes were proportionately less frequent there relative to other locations. The BSI episodes included 3,921 (17.1%) community cases, 12,205 (53.2%) acute care hospital cases, 6,561 (28.6%) intensive care cases, and 248 (1.1%) long-term care cases.
TABLE 1.
Characteristics of patients undergoing blood culture collection and experiencing bloodstream infection episodes
| Demographic characteristic | Total culture episodes (n = 252,343) |
Positive BSI episodes (n = 22,935) |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Age | ||||
| 0–3 mo | 5,810 | 2.3 | 188 | 0.8 |
| 3 mo–1 yr | 1,934 | 0.8 | 65 | 0.3 |
| 1–5 yr | 7,648 | 3.0 | 176 | 0.8 |
| 6–10 yr | 2,998 | 1.2 | 76 | 0.3 |
| 11–19 yr | 5,531 | 2.2 | 194 | 0.8 |
| 20–29 yr | 13,493 | 5.3 | 790 | 3.4 |
| 30–39 yr | 16,997 | 6.7 | 1,217 | 5.3 |
| 40–49 yr | 19,645 | 7.8 | 1,598 | 7.0 |
| 50–59 yr | 32,422 | 12.8 | 3,088 | 13.5 |
| 60–69 yr | 42,187 | 16.7 | 4,417 | 19.3 |
| 70–79 yr | 45,525 | 18.0 | 4,914 | 21.4 |
| 80+ yr | 58,153 | 23.0 | 6,212 | 27.1 |
| Sex | ||||
| Female | 122,520 | 48.6 | 10,320 | 45.0 |
| Male | 129,823 | 51.4 | 12,615 | 55.0 |
| Days in hospitala | ||||
| 0 | 181,857 | 72.1 | 15,272 | 66.6 |
| 1–4 | 19,953 | 7.9 | 1,805 | 7.9 |
| 5–9 | 18,688 | 7.4 | 1,988 | 8.7 |
| 10–90 | 31,845 | 12.6 | 3,870 | 16.9 |
| Location | ||||
| Community | 85,982 | 34.1 | 3,921 | 17.1 |
| Acute care hospital | 117,574 | 46.6 | 12,205 | 53.2 |
| Intensive care unit | 45,502 | 18.0 | 6,561 | 28.6 |
| Long-term care | 3,285 | 1.3 | 248 | 1.1 |
Number of days admitted to hospital in the 90 days prior to blood culture collection date.
Incidence of BSI organisms.
Among the 22,935 BSI episodes, the most common isolate was Escherichia coli, which represented 26.9% of the total. Staphylococcus was the next most common genus, with 25.1% of all isolates. Separating by species, 15.9% of all episodes were Staphylococcus aureus, with the remainder being CoNS (Table 2). This was followed by Streptococcus species, which together accounted for 10.9% of episodes. The most common Streptococcus species was Streptococcus pneumoniae with 3.2% of all BSI episodes. Following this in descending order of prevalence, Klebsiella species, Enterococcus species, Pseudomonas species, and Enterobacter species accounted for 8.2%, 5.8%, 3.4%, and 2.6% of BSI episodes, respectively.
TABLE 2.
Positive BSI episodes by organism isolated
| Organism | Positive BSI episodes (n = 22,935) |
Patients (n = 19,326) |
Annual rate/100,000 population | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Escherichia coli | 5,864 | 26.9 | 5,450 | 28.2 | 40.24 |
| Staphylococci | 5,455 | 25.1 | |||
| Staphylococcus aureus | 3,455 | 15.9 | 3,035 | 15.7 | 22.41 |
| Staphylococcus lugdunensis | 74 | 0.3 | 68 | 0.4 | 0.50 |
| Staphylococcus saprophyticus | 15 | 0.1 | 15 | 0.1 | 0.11 |
| Other CoNS | 1,911 | 8.8 | 1,632 | 8.4 | 12.05 |
| Streptococci | 3,412 | 15.7 | |||
| Streptococcus pneumoniae | 691 | 3.2 | 672 | 3.5 | 4.96 |
| Streptococcus agalactiae | 508 | 2.3 | 487 | 2.5 | 3.60 |
| Viridans group Streptococcus | 469 | 2.2 | 431 | 2.2 | 3.18 |
| Streptococcus pyogenes | 438 | 2.0 | 415 | 2.2 | 3.06 |
| Group G/C Streptococcus | 329 | 1.5 | 312 | 1.6 | 2.30 |
| Streptococcus mitis | 133 | 0.6 | 123 | 0.6 | 0.91 |
| Other streptococci | 844 | 3.9 | 785 | 4.1 | 5.80 |
| Klebsiella species | 1,794 | 8.2 | 1,505 | 7.8 | 11.11 |
| Enterococcus species | 1,267 | 5.8 | 963 | 5.0 | 7.11 |
| Pseudomonas species | 749 | 3.4 | 602 | 3.1 | 4.45 |
| Enterobacter species | 568 | 2.6 | 461 | 2.4 | 3.40 |
| Candida species | 561 | 2.6 | 357 | 1.9 | 2.64 |
| Proteus species | 394 | 1.8 | 369 | 1.9 | 2.72 |
| Bacteroides fragilis | 292 | 1.3 | 268 | 1.4 | 1.98 |
| Serratia species | 226 | 1.0 | 175 | 0.9 | 1.29 |
| Haemophilus influenzae | 195 | 0.9 | 190 | 1.0 | 1.40 |
| Bacillus species | 171 | 0.8 | 136 | 0.7 | 1.00 |
| Clostridium species | 168 | 0.8 | 154 | 0.8 | 1.14 |
| Citrobacter species | 148 | 0.7 | 127 | 0.7 | 0.94 |
| Acinetobacter species | 128 | 0.6 | 106 | 0.6 | 0.78 |
| Salmonella non-Typhi/Paratyphi | 128 | 0.6 | 126 | 0.7 | 0.93 |
| Actinomyces species | 96 | 0.4 | 91 | 0.5 | 0.67 |
| Stenotrophomonas maltophilia | 84 | 0.4 | 55 | 0.3 | 0.41 |
| Aerococcus species | 80 | 0.4 | 77 | 0.4 | 0.57 |
| Fusobacterium species | 78 | 0.4 | 73 | 0.4 | 0.54 |
| Corynebacterium species | 69 | 0.3 | 59 | 0.3 | 0.44 |
| Salmonella Typhi /Paratyphi | 69 | 0.3 | 67 | 0.4 | 0.49 |
| Morganella species | 68 | 0.3 | 65 | 0.3 | 0.48 |
| Bacteroides species | 66 | 0.3 | 60 | 0.3 | 0.44 |
| Others | 813 | 3.7 | 737 | 3.8 | 5.44 |
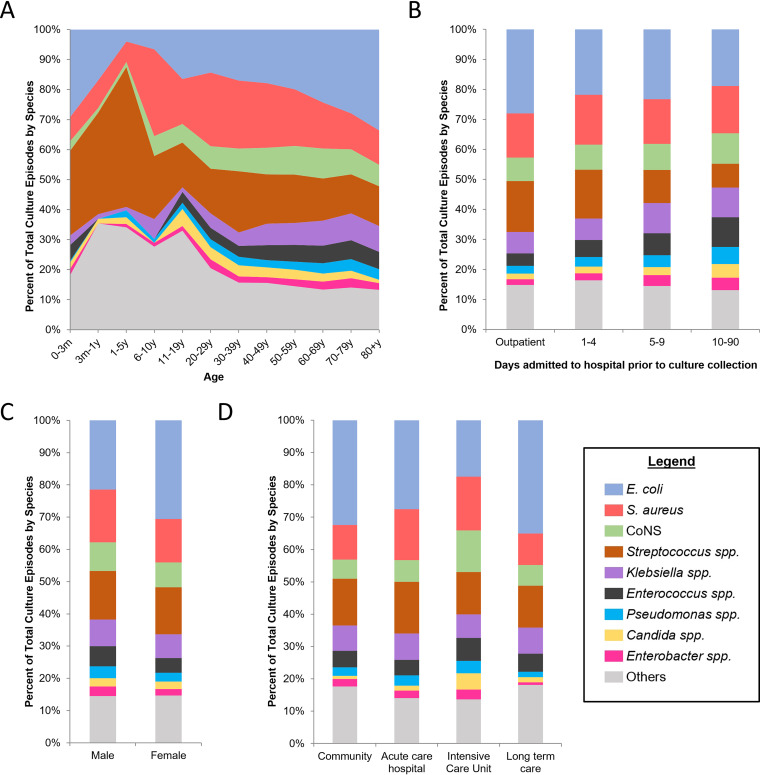
BSI episodes were stratified by patient demographic characteristics and microorganism (Fig. 2). Ranking proportions of species by age group, the overall trends were similar following the second decade of life with an increase in E. coli and decrease in S. aureus (Fig. 2A). Notably, Streptococci species were a prominent cause of BSI episodes from 3 months to 10 years. For inpatients, there was a progressive relative decline in E. coli and increase in Enterococcus species and Candida species (Fig. 2B). Separating by sex, the proportions of culture episodes by organism were generally similar, with more E. coli episodes in women and more Staphylococcus aureus episodes in men (Fig. 2C). Comparing locations, in ICUs there were proportionally fewer E. coli BSI episodes and more Candida species BSIs (Fig. 2D).
FIG 2.
Distribution of microorganisms by demographic characteristics. (A) Plot of microorganisms against age. (B) Bar graphs of microorganisms for outpatient (community and long-term care) and patients admitted to acute care and ICU over the course of hospitalization (1 to 90+ days). (C) Bar graphs comparing microorganisms between men and women. (D) Bar graphs comparing microorganisms by location of culture collection. For panels A to D, data in each column is expressed as a proportion of total positive culture episodes, with legend in bottom right identifying corresponding microorganisms. All microorganisms which each comprised less than 2.5% of total culture episodes are clustered together as “Others.”
Mortality associated with BSI organisms.
We calculated the crude mortality associated with all blood culture episodes and by microorganism (Table 3). In total, 3,509 deaths occurred within 30 days of a positive culture episode. This equated to a mortality rate of 17.0% for positive culture episodes, while the mortality rate associated with a negative culture episode was 9.5%.
TABLE 3.
30-day mortality and adjusted mortality odds ratios by microorganism
| Organism | 30-day mortality |
Mortality OR compared to matched patients without blood culture testinga |
Mortality OR compared to patients with negative culturesb |
|||
|---|---|---|---|---|---|---|
| Deaths | % of episodes | Adjusted OR (95% CI) | P value | Adjusted OR (95% CI) | P value | |
| All positive episodes | 3509 | 17.0 | 2.62 (2.52, 2.73) | <0.0001 | 1.47 (1.41, 1.54) | <0.0001 |
| Staphylococci | ||||||
| Staphylococcus aureus | 764 | 22.8 | 3.53 (3.23, 3.86) | <0.0001 | 2.14 (1.94, 2.36) | <0.0001 |
| Staphylococcus lugdunensis | 13 | 17.6 | 3.39 (1.85, 6.19) | <0.0001 | 1.69 (0.83, 3.45) | 0.1469 |
| Other CoNS | 365 | 19.7 | 2.62 (2.31, 2.97) | <0.0001 | 1.36 (1.19, 1.55) | <0.0001 |
| Escherichia coli | 699 | 12.1 | 1.68 (1.54, 1.83) | <0.0001 | 0.96 (0.87, 1.05) | 0.3270 |
| Streptococci | ||||||
| Streptococcus pneumoniae | 105 | 15.4 | 2.49 (1.99, 3.11) | <0.0001 | 1.46 (1.15, 1.86) | 0.0017 |
| Streptococcus agalactiae | 70 | 13.8 | 2.17 (1.66, 2.84) | <0.0001 | 1.35 (1.01, 1.79) | 0.0397 |
| Viridans group Streptococcus | 66 | 14.2 | 2.18 (1.64, 2.89) | <0.0001 | 1.33 (0.99, 1.78) | 0.0612 |
| Streptococcus pyogenes | 66 | 15.8 | 3.19 (2.38, 4.28) | <0.0001 | 1.88 (1.39, 2.54) | <0.0001 |
| Group G/C Streptococcus | 47 | 14.5 | 2.03 (1.45, 2.85) | <0.0001 | 1.14 (0.81, 1.62) | 0.4457 |
| Streptococcus mitis | 17 | 13.0 | 2.06 (1.24, 3.43) | 0.0054 | 1.23 (0.68, 2.22) | 0.4869 |
| Other streptococci species | 139 | 16.7 | 2.80 (2.31, 3.39) | <0.0001 | 1.58 (1.28, 1.96) | <0.0001 |
| Klebsiella species | 307 | 17.6 | 2.20 (1.92, 2.51) | <0.0001 | 1.32 (1.15, 1.52) | 0.0001 |
| Enterococcus species | 290 | 23.6 | 2.86 (2.46, 3.31) | <0.0001 | 1.68 (1.44, 1.96) | <0.0001 |
| Pseudomonas species | 181 | 24.7 | 2.82 (2.36, 3.37) | <0.0001 | 1.83 (1.50, 2.23) | <0.0001 |
| Candida species | 171 | 32.0 | 4.51 (3.66, 5.56) | <0.0001 | 2.40 (1.93, 2.99) | <0.0001 |
| Enterobacter species | 109 | 19.8 | 2.46 (1.97, 3.08) | <0.0001 | 1.31 (1.03, 1.68) | 0.0286 |
| Proteus species | 79 | 20.7 | 2.42 (1.84, 3.18) | <0.0001 | 1.41 (1.07, 1.87) | 0.0148 |
| Bacteroides fragilis | 73 | 25.3 | 4.40 (3.26, 5.95) | <0.0001 | 2.19 (1.59, 3.00) | <0.0001 |
| Clostridium species | 70 | 41.9 | 6.94 (4.87, 9.89) | <0.0001 | 5.81 (4.00, 8.44) | <0.0001 |
| Serratia species | 46 | 20.7 | 2.76 (1.95, 3.90) | <0.0001 | 1.30 (0.88, 1.90) | 0.1864 |
| Haemophilus influenzae | 38 | 19.5 | 3.48 (2.41, 5.02) | <0.0001 | 2.14 (1.40, 3.27) | 0.0005 |
| Citrobacter species | 22 | 15.6 | 2.12 (1.36, 3.30) | 0.0010 | 1.11 (0.67, 1.82) | 0.6842 |
| Bacillus species | 21 | 13.1 | 2.57 (1.53, 4.29) | 0.0003 | 1.20 (0.72, 1.99) | 0.4860 |
| Acinetobacter species | 19 | 15.5 | 2.18 (1.30, 3.67) | 0.0033 | 1.44 (0.82, 2.51) | 0.2061 |
| Actinomyces species | 19 | 20.0 | 3.33 (2.07, 5.36) | <0.0001 | 2.38 (1.30, 4.34) | 0.0049 |
| Stenotrophomonas maltophilia | 19 | 23.2 | 3.37 (1.87, 6.09) | <0.0001 | 2.25 (1.20, 4.18) | 0.0109 |
| Corynebacterium species | 18 | 27.3 | 4.64 (2.48, 8.68) | <0.0001 | 3.00 (1.59, 5.68) | 0.0007 |
| Aerococcus species | 11 | 13.9 | 2.16 (1.09, 4.29) | 0.0273 | 0.94 (0.46, 1.93) | 0.8750 |
| Fusobacterium species | 10 | 13.0 | 3.87 (1.76, 8.51) | 0.0008 | 2.37 (1.11, 5.08) | 0.0264 |
Compared to up to 10 individuals without blood culture testing matched by age, sex, health care location, Charlson comorbidity score, and number of days in hospital in the 90 days prior to blood culture.
Calculated using a generalized linear mixed model (GLMM) adjusting for age, sex, health care location, and number of days hospitalized in prior 90 days.
Separating by microorganism, the largest number of deaths at 30 days followed S. aureus, E. coli, and CoNS BSI episodes (Table 3). Clostridium species, Candida species, and Bacteroides fragilis had the highest crude 30-day mortality rates with 41.7%, 31.9%, and 25.3%, respectively (Table 3). Short- and long-term mortality also differed by organism (see Table S1 in the supplemental material); 1 year after the index culture, Clostridium species, Candida species, and Stenotrophomonas species were associated with the highest crude mortality rates among all microorganisms.
The 30-day mortality OR for a BSI episode was calculated relative to matched individuals without blood culture testing and patients with negative cultures. Compared to patients with negative blood cultures, BSI carried a significantly increased risk of mortality (Table 3). The risk varied between species, with Clostridium species, Corynebacterium species, and Candida species having the largest fold increase with an OR of 5.81 (95% CI, 4.00 to 8.44), 3.00 (95% CI, 1.59 to 5.68), and 2.40 (95% CI, 1.93 to 2.99), respectively. Compared to matched patients who did not undergo blood culture testing, positive culture episodes altogether were associated with an increased mortality OR of 2.62 (95% CI, 2.52 to 2.73).
Whereas Clostridium species and Candida species are generally considered to be true infections when isolated from blood cultures, Corynebacterium species have been shown to have different degrees of pathogenicity between species, which were specifically elevated for Corynebacterium striatum and Corynebacterium jeikeium (13). To account for the possibility that this increase in mortality risk was due to differences between species, we analyzed the 30-day mortality rate for these two species together compared to other Corynebacterium species. From this, we found that 5/24 (21%) cases of C. striatum and C. jeikeium were associated with mortality compared to 13/45 (29%) for the other Corynebacterium species combined, a difference which was not significant (P = 0.53).
Common and lethal organisms.
In simultaneously comparing organisms by their overall incidence, associated mortality, and adjusted mortality risk, we can identify those which pose a relatively disproportionate impact to the population (Fig. 3). Notable among these is Staphylococcus aureus. Although Staphylococcus aureus has a similar crude mortality rate per BSI episode compared to Enterococcus species and is responsible for less positive culture episodes than E. coli, it is associated with higher adjusted mortality risk and more deaths than these other two organisms.
FIG 3.
Incidence and lethality of bloodstream infection (BSI) organisms. In this bubble plot, microorganisms are plotted by total number of BSI episodes (x axis) against crude percent mortality at 30 days (y axis) following culture episodes yielding that microorganism. Bubble areas are scaled to total number of deaths at 30 days associated with the listed microorganism as expressed in the legend. Color scaling represents the adjusted odds ratio (OR) for 30-day mortality relative to patients with a negative culture episode, calculated by generalized linear mixed models (GLMM), with increasing blue hue when close to or less than an OR of 1 and transitioning through white and red for an increasing OR as expressed in the legend.
Additionally, among Streptococcus species, Streptococcus pyogenes was associated with similar numbers of deaths to S. pneumoniae and Streptococcus agalactiae; however, its adjusted risk is notably higher. Clostridium species are another outlier insofar as their adjusted mortality risk is higher than other organisms with a similar rate of BSI episodes.
DISCUSSION
In this population-wide study of 252,343 blood culture episodes, and 22,935 BSIs among 19,326 patients, we assessed the prevalence and mortality of all BSI organisms. E. coli was the most commonly isolated, followed by Staphylococcus species and Streptococcus species. We demonstrated the mortality associated with each organism, finding that E. coli, S. aureus, and CoNS are associated with the largest number of deaths, while Clostridium species and Candida species have the highest associated mortality rate. Comparing patients with BSI episodes to matched patients without cultures, we found that most microorganisms are associated with an increased risk of mortality, and compared to patients with negative cultures, again Clostridium species and Candida species had the highest 30-day mortality risk. Lastly, Staphylococcus aureus stood out as a pathogen associated with a high incidence, high numbers of deaths, and high adjusted mortality risk.
The most common organisms among positive episodes overall were E. coli, Staphylococcus aureus, Klebsiella species, and Enterococcus species. In comparing these results to similarly designed studies (5, 9), we find that the overall proportion, annualized rate per population, and rank order are generally reproduced. This provides strong external validation for our data collection and processing methodology, namely, that BSI surveillance can be conducted efficiently by merging routine microbiology data across a region.
By integrating culture and patient data, we determined that the organisms with the highest crude mortality rates were Clostridium species and Candida species, with 41.7% and 31.9% mortality at 30 days, respectively. Studies from tertiary hospitals have documented the associated mortality for Clostridium bacteremia as 59.2% at 28 days (14), and for Candidemia as 40.5% at 30 days (15). These two organisms were also associated with the highest adjusted odds of mortality, and so the higher mortality is not accounted for by age, overall comorbidity burden, duration of hospitalization, or health care setting.
Compared to matched controls with negative blood culture results, the adjusted odds of mortality for all positive blood culture episodes was found to be 1.41 (95% CI, 1.41 to 1.54). Although cases were matched by age, sex, health care location, duration of hospitalization, and Charlson comorbidity score, it still remains that there are other cofounders for which a patient for whom blood culture testing is ordered may have a higher risk of mortality, from their specific comorbidities or underlying illness, not directly related to the bacteremia that is identified. Although all individual genera or species analyzed were associated with a significant increase in mortality compared to matched patients who had not undergone blood culture testing, not all were significant compared to patients with negative blood cultures. Again, this may support the idea that the associated mortality is in part due to a patient’s underlying condition and not the specific pathogen; blood culture testing is a marker of a very unwell patient.
While Staphylococcus aureus was not the organism with the largest number of associated BSIs, nor the one with the highest adjusted risk of mortality, it was still prominent in all four dimensions of our composite comparison of microorganisms by the number of positive episodes, associated deaths, crude mortality rate per positive culture episode, and adjusted mortality risk. Its adjusted mortality risk is disproportionately higher than would be expected from its crude mortality rate per episode, and it is associated with more deaths than E. coli despite being identified in about half as many BSI episodes. Overall, this makes Staphylococcus aureus a prominent threat to public health, necessitating ongoing vaccine and drug development as well as surveillance and targeted programs to improve treatment and patient outcomes.
The study had a number of limitations given the use of administrative data. First, we did not have access to clinical symptoms and signs, which could have helped exclude cases of contamination. We may have slightly underestimated the population burden of BSI based on missing data from a small number of laboratories. In addition, the practical aspects of collecting blood cultures are not standardized across the entire province with respect to the number of culture bottles used, or the use of anaerobic and aerobic together in a set. We opted to incorporate simple risk adjustment based on widely available patient characteristics, but there is potential for residual confounding by unmeasured characteristics. Adjustment for Charlson comorbidity score may not have fully accounted for differences in individual illnesses, such as malignancy. Adjustment for ICU location may not have fully accounted for differences in predisposition to some device-associated hospital pathogens, such as CoNS, Lactobacillus species, and Stenotrophomonas species; therefore, we may have overestimated the odds of mortality attributable to these organisms. We have not incorporated data on antibiotic susceptibilities to differentiate the burden of susceptible from resistant pathogens. Differences in mortality rates could be driven by the underlying focus of infection or source of bacteremia, which was not included, as reliable diagnostic codes for infectious source are not always available. However, the use of population-wide microbiology and administrative data also represents an important strength of this study in that it provided comprehensive information on all pathogens across all health care sectors, with linkage to patient characteristics and outcomes.
By merging population-wide microbiology data from over 100 laboratories, we identified the most important BSI organisms across community, long-term care, acute care, and intensive care settings. By identifying the BSI organisms that are most common, that are associated with the most deaths, and that have the highest adjusted odds of mortality, these data can shape targeted approaches to addressing the constantly evolving threat of BSIs.
ACKNOWLEDGMENTS
This work was supported by a project grant from CIHR (grant number 159503 to K.A.B. and N.D.). This study was supported by ICES, which is funded by an annual grant from the Ontario Ministry of Health (MOH) and the Ministry of Long-Term Care (MLTC). Parts of this material are based on data and information compiled and provided by: MOH, OLIS and CIHI. The analyses, conclusions, opinions and statements expressed herein are solely those of the authors and do not reflect those of the funding or data sources; no endorsement is intended or should be inferred. Parts of this material are based on data and/or information compiled and provided by CIHI. However, the analyses, conclusions, opinions and statements expressed in the material are those of the author(s), and not necessarily those of CIHI.
All authors report no potential conflicts of interest
Footnotes
Supplemental material is available online only.
Contributor Information
Nick Daneman, Email: nick.daneman@sunnybrook.ca.
Nathan A. Ledeboer, Medical College of Wisconsin
REFERENCES
- 1.Goto M, Al-Hasan MN. 2013. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19:501–509. 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 2.Diekema DJ, Hsueh P-R, Mendes RE, Pfaller MA, Rolston KV, Sader HS, Jones RN. 2019. The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother 63:e00355-19. 10.1128/AAC.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Søgaard M, Nørgaard M, Dethlefsen C, Schønheyder HC. 2011. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: a population-based cohort study. Clin Infect Dis 52:61–69. 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen SL, Lassen AT, Gradel KO, Jensen TG, Kolmos HJ, Hallas J, Pedersen C. 2015. Bacteremia is associated with excess long-term mortality: a 12-year population-based cohort study. J Infect 70:111–126. 10.1016/j.jinf.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Skogberg K, Lyytikäinen O, Ollgren J, Nuorti JP, Ruutu P. 2012. Population-based burden of bloodstream infections in Finland. Clin Microbiol Infect 18:E170–E176. 10.1111/j.1469-0691.2012.03845.x. [DOI] [PubMed] [Google Scholar]
- 6.Statistics Canada. 2018. Canada at a glance 2018. Statistics Canada, Ottawa, Canada. https://www150.statcan.gc.ca/n1/pub/12-581-x/2018000/pop-eng.htm. [Google Scholar]
- 7.Filice GA, Van Etta LL, Darby CP, Fraser DW. 1986. Bacteremia in Charleston County, South Carolina. Am J Epidemiol 123:128–136. 10.1093/oxfordjournals.aje.a114206. [DOI] [PubMed] [Google Scholar]
- 8.Laupland KB, Gregson DB, Flemons WW, Hawkins D, Ross T, Church DL. 2007. Burden of community-onset bloodstream infection: a population-based assessment. Epidemiol Infect 135:1037–1042. 10.1017/S0950268806007631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas NM, Hennessy JN, Currie BJ, Baird RW. 2020. Trends in bacteremia over 2 decades in the top end of the northern territory of Australia. Open Forum Infect Dis 7:ofaa472. 10.1093/ofid/ofaa472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson J, Elgohari S, Livermore DM, Cookson B, Johnson A, Lamagni T, Chronias A, Sheridan E. 2011. Trends among pathogens reported as causing bacteraemia in England, 2004–2008. Clin Microbiol Infect 17:451–458. 10.1111/j.1469-0691.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- 11.Sjöberg L, Fredlund H. 1988. Survey of blood culture isolates in an area of Sweden from 1980 to 1986. Eur J Clin Microbiol Infect Dis 7:501–504. 10.1007/BF01962600. [DOI] [PubMed] [Google Scholar]
- 12.Deyo RA, Cherkin DC, Ciol MA. 1992. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol 45:613–619. 10.1016/0895-4356(92)90133-8. [DOI] [PubMed] [Google Scholar]
- 13.Yamamuro R, Hosokawa N, Otsuka Y, Osawa R. 2021. Clinical characteristics of Corynebacterium bacteremia caused by different species, Japan, 2014–2020. Emerg Infect Dis 27:2981–2987. 10.3201/eid2712.210473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah M, Bishburg E, Baran DA, Chan T. 2009. Epidemiology and outcomes of clostridial bacteremia at a tertiary-care institution. ScientificWorldJournal 9:144–148. 10.1100/tsw.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muderris T, Kaya S, Ormen B, Aksoy Gokmen A, Varer Akpinar C, Yurtsever Gul S. 2020. Mortality and risk factor analysis for Candida blood stream infection: a three-year retrospective study. J Mycol Med 30:101008. 10.1016/j.mycmed.2020.101008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jcm.02429-21-s0001.pdf, PDF file, 0.1 MB (132.1KB, pdf)