LETTER
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has proceeded in multiple waves since late 2019, partly driven by the emergence of successive variants. Worldwide sequencing of the viral genome has yielded unprecedented real-time tracking of the evolution of SARS-CoV-2 (1). Alpha emerged in late 2020 (2) and was succeeded by Delta, which predominated in the latter portion of 2021. In late 2021, Omicron displaced Delta (3; https://covid.cdc.gov/covid-data-tracker/#variant-proportions).
Detection of SARS-CoV-2 infection using a PCR-based method is the gold standard for molecular diagnosis. At our institution, the TaqPath COVID-19 multiplex PCR assay (Thermo Fisher, USA) was deployed in October 2020 and became the primary clinical testing modality. It includes primers for three viral targets: N, ORF1AB, and S genes (4). Deletion of codons 69 and 70 in the S gene, common to Alpha (5) and Omicron (6), results in S gene target failure (SGTF). We used SGTF with N gene cycle threshold (Ct) < 30 as a proxy for lineage to monitor the early growth rate of Alpha and Omicron by calculating case doubling time. Lineage designation was later supported by viral genome sequencing via NextSeq (Illumina, USA), also performed on samples with N gene Ct < 30. For comparison, Delta case doubling time was calculated using sequencing-confirmed samples.
To estimate the case doubling time of the variants based on frequency of cases rather than count, a model was constructed to show growth relative to existing variants. Following the approach outlined by Volz et al. (7), in a model where strains are competing in the population and growth of existing strains is assumed to be constant at some level, R, prior to the introduction of the new strain, the variant can be assumed to have a reproduction number of R plus some percentage (1+s). In this model, the log odds of the variant share will be proportional to (R/g)st, where g is the generation time, s is a selection coefficient, and t is time. If a logistic model is fit with the shares of variant and time, the estimated coefficient on time is thus equal to (R/g)s. If R is assumed to be 1, reflecting stable growth conditions, and g is assumed to be 6.5 days, then b = (1/6.5)*s and s = b*6.5. This estimate of the growth rate per generation can be converted to doubling days using quotient of natural logs whereby the case doubling rate (in days) is equal to natural log (2)/natural log(s).
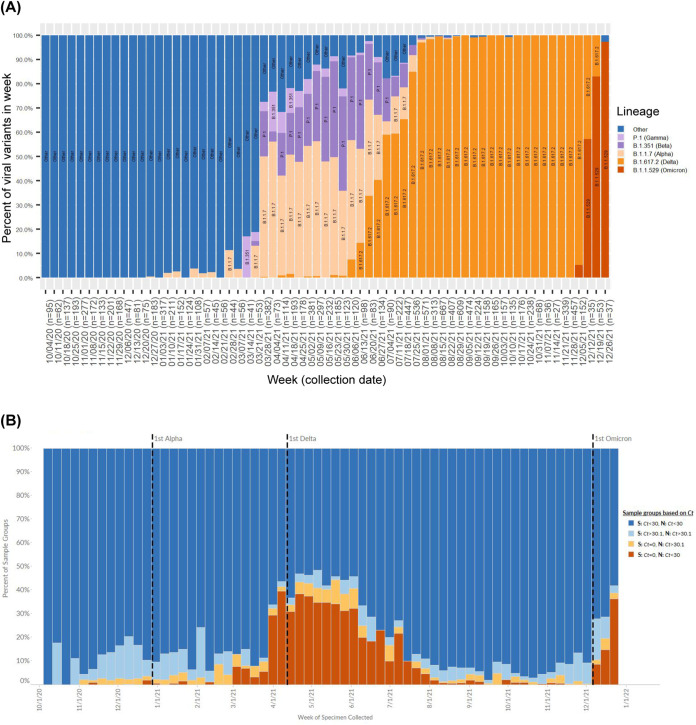
At our institution, the first sequencing-confirmed Alpha was collected on December 29, 2020, the first Delta on April 15, 2021, and the first Omicron on December 8, 2021 (Fig. 1A), with SGTF appearing shortly after Alpha and Omicron (Fig. 1B). Alpha had a case doubling time of 9.54 days based on 6,931 samples with SGTF. Delta’s case doubling time was 19.30 days based on 447 sequenced samples. Omicron’s case doubling time is 4.28 days, nearly half that of Alpha, based on 1,078 samples with SGTF since December 8, 2021.
FIG 1.
(A) At OHSU, 12,474 viruses were sequenced with collection dates between October 4, 2020 and December 29, 2021. A total of 1,415 sequences were identified as Alpha, collected between December 29, 2020 and August 30, 2021. There were 6,399 sequences identified as Delta, collected between April 15, 2021 and December 22, 2021. There were 108 sequences identified as Omicron, collected between December 8 and 29, 2021. (B) SGTF appeared concurrently with sequencing-confirmed samples of the Alpha and Omicron variants, with very low levels of SGTF reported in the period of Delta predominance (August through mid-December, 2021). SGTF is defined by samples with S Ct = 0 with N Ct < 30 (dark orange). Samples with S Ct = 0 and N Ct > 30.1 (light orange) or S Ct > 30.1 and N Ct > 30.1 (light blue) were considered nonspecific. Samples with S Ct < 30 and N Ct < 30 (dark blue) were negative for SGTF.
This shortened case doubling time is additional evidence of the increased transmissibility of Omicron. Using SGTF provides a real-time estimate of viral epidemiology for Omicron; however, there are limitations with this analysis. Rarely, SGTF can be detected in sublineages of the Delta variant that are simultaneously in circulation (8). Also, the time it took for Delta to come to predominance was complicated by several factors including remote schooling, increased outdoor activities during summer, the protection from vaccination in the population, and the simultaneous presence of Alpha, Beta, and Gamma variants in circulation. All of these may have contributed to Delta’s prolonged case doubling time compared with that of Alpha and Omicron. Nonetheless, clinical laboratory data are essential for monitoring the SARS-CoV-2 pandemic. The case doubling time in our community reflects the nationwide trend toward Omicron predominance and immune evasion, and can inform local pandemic mitigation efforts. In conjunction with early reports of the clinical severity of and immunologic responses to a given variant, case doubling time can be used to inform whether to implement a variety of public health measures, e.g., masking policies, hospital visitor policies, vaccination booster recommendations, scale-up of laboratory testing, and widespread distribution of at-home testing.
ACKNOWLEDGMENTS
SARS-CoV-2 genome sequencing was funded in part by the Oregon Public Health Authority (OHA Agreement Number 170015). We thank OHSU clinical laboratory scientists for their high-quality performance and documentation of SARS-CoV-2 test results. We thank Brian O’Roak’s lab for their sustained contributions to OHSU’s sequencing operations. Doris Y. Yang was a freshman attending Harvard College while working as a student intern on Weekly Oregon State Genome Dashboard for OHSU.
Contributor Information
Xuan Qin, Email: qinxu@ohsu.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Belman S, Saha S, Beale MA. 2022. SARS–CoV-2 genomics as a springboard for future disease mitigation in LMICs. Nat Rev Microbiol 20:3. 10.1038/s41579-021-00664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. 2021. Emergence of SARS-CoV-2 B.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep 70:95–99. 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2021. Science brief: Omicron (B.1.1.529) variant, CDC COVID-19 science briefs. CDC, Atlanta, GA. [PubMed] [Google Scholar]
- 4.Thermo Fisher Scientific. 2021. TaqPath COVID-19 Combo Kit and TaqPath COVID-19 Combo Kit advanced instructions for use. Thermo Fisher Scientific, Waltham, MA. [Google Scholar]
- 5.Vogels CBF, Breban MI, Ott IM, Alpert T, Petrone ME, Watkins AE, Kalinich CC, Earnest R, Rothman JE, Goes de Jesus J, Morales Claro I, Magalhães Ferreira G, Crispim MAE, Singh L, Tegally H, Anyaneji UJ, Hodcroft EB, Mason CE, Khullar G, Metti J, Dudley JT, MacKay MJ, Nash M, Wang J, Liu C, Hui P, Murphy S, Neal C, Laszlo E, Landry ML, Muyombwe A, Downing R, Razeq J, de Oliveira T, Faria NR, Sabino EC, Neher RA, Fauver JR, Grubaugh ND, Network for Genomic Surveillance in South Africa. 2021. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol 19:e3001236. 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. 2021. SARS-CoV-2 B.1.1.529 (Omicron) variant—United States, December 1–8, 2021. MMWR Morb Mortal Wkly Rep 70:1731–1734. 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. 2021. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 8.Lythgoe KA, Golubchik T, Hall M, House T, MacIntyre-Cockett G, Fryer H, Thomson L, Nurtay A, Buck D, Green A, Trebes A, Piazza P, Lonie LJ, Studley R, Rourke E, Cook D, Smith D, Bashton M, Nelson A, Crown M, McCann C, Young GR, dos Santos RAN, Richards Z, Tariq A, Team Wsic S, Group C-IS, Consortium T-GU, Fraser C, Diamond I, Barrett J, Walker S, Bonsall D. 2022. Lineage replacement and evolution captured by the United Kingdom Covid Infection Survey. medRxiv. 10.1101/2022.01.05.21268323. [DOI] [PMC free article] [PubMed] [Google Scholar]