ABSTRACT
Genome sequencing is a powerful tool for identifying SARS-CoV-2 variant lineages; however, there can be limitations due to sequence dropout when used to identify specific key mutations. Recently, ThermoFisher Scientific has developed genotyping assays to help bridge the gap between testing capacity and sequencing capability to generate real-time genotyping results based on specific variants. Over a 6-week period during the months of April and May 2021, we set out to assess the ThermoFisher TaqMan mutation panel genotyping assay, initially for three mutations of concern and then for an additional two mutations of concern, against SARS-CoV-2-positive clinical samples and the corresponding COVID-19 Genomics UK Consortium (COG-UK) sequencing data. We demonstrate that genotyping is a powerful in-depth technique for identifying specific mutations, is an excellent complement to genome sequencing, and has real clinical health value potential, allowing laboratories to report and take action on variants of concern much more quickly.
KEYWORDS: SARS-CoV-2, variants of concern, genotyping, genome sequencing, real time, SNPs, PCR
INTRODUCTION
Viruses mutate, and SARS-CoV-2 is no exception. As the COVID-19 pandemic continues around the world, mutations are naturally occurring, resulting in the emergence of divergent clusters/variants containing sets of mutations. These clusters/variants have differing prevalence in different geographical regions, likely in response to changing immune profiles of the human population (1). Movement of people enabled by global air travel allows these variants to spread and mutate further under differing selection pressures. In the United Kingdom, the Alpha variant (B.1.1.7), first identified in Kent, rapidly swept to dominance by December 2020 (2). In April 2021, the Delta variant (B.1.617.2), first identified in India, rapidly outcompeted the Alpha variant to become dominant in a matter of weeks (3). The geographical location where these variants emerged is not relevant (4); rather, it is the specific mutations present which greatly impact virus characteristics including transmissibility and antigenicity, where mutations of significance have been identified in the SARS-CoV-2 Spike protein of these variants of concern (VOC) that contribute to enhanced transmission and/or immune invasion (5). Other VOC have been identified and characterized, including both Beta (B.1.351, first identified in South Africa) and Gamma (P.1, first identified in Brazil) (5).
Previously at the University of Birmingham, we set up a SARS-CoV-2 modular testing facility at the request of the United Kingdom Department of Health and Social Care (6). Our PCR assay of choice was the 3-target design (ORF1ab and S and N genes) TaqPath COVID-19 CE-IVD (European CE marking for In Vitro Diagnostic (IVD) medical devices) real-time reverse-transcriptase polymerase chain reaction (RT-PCR) kit, where target areas are unique to SARS-CoV-2 to reduce detection of other coronaviruses and compensate for virus mutations. Initially, we detected all three genes in SARS-CoV-2 samples; however, during November 2020, we along with other laboratories using the same PCR assay started to notice a drop-off in the detection of the S gene and then a rapid rise in this S-gene target failure (SGTF) in early December 2020 (7). From discussions with the University of Birmingham genome sequencing laboratory (as part of the COVID-19 Genomics UK Consortium [COG-UK]) and as confirmed by other laboratories, it was demonstrated that the S gene did amplify, therefore confirming that this gene was still present but was not being detected using the TaqPath COVID-19 assay. This was attributed to a 6-bp deletion (Δ69/70) in the middle of the S gene where the fluorescent specific probe binds, thus negating the probe’s ability to fluoresce (8). Simultaneously, these findings of a new VOC were reported by our laboratory and multiple testing facilities across the United Kingdom, where this finding identified the VOC B.1.1.7 (Alpha variant). Although SGTF identification with the ThermoFisher TaqPath real-time RT-PCR assay was not intentional, this observation with this PCR assay allowed us and other COVD-19 testing facilities to use this assay as an accurate epidemiological tool to track the rise, spread, and dominance of this VOC in the United Kingdom. By April 2021, COVID-19 TaqPath PCR testing facilities, including our laboratory, noted an increasing number of samples without SGTF, where the 3 target genes were amplifying, and upon genome sequencing analysis, these samples were identified as the Delta (B.1.617.2) variant (9).
To better understand viral transmission and evolution and to inform public health responses and vaccine development, genomic sequencing is essential. In March 2020, the COVID-19 Genomics UK Consortium (COG-UK) was created for this purpose (10). To date, COG-UK has sequenced over 1,100,000 SARS-CoV-2 samples, providing a vast amount of data to the global COVID-19 response. This yields crucial information about the number of variants circulating in the population and possible lines of transmission; however, sequencing can be time-consuming and costly, and in some cases full coverage of the virus is not possible.
Recently, ThermoFisher has developed genotyping assays to help bridge the gap between testing capacity and sequencing capability to receive results in real time, so in addition to our modular system for COVID-19 testing, we decided to build the ThermoFisher TaqMan SARS-CoV-2 mutation panel into our workflow (Fig. 1). Here, we present our data showing that the TaqMan genotyping assays identify variants in all samples tested with a zero failure rate and that often the assay confirms mutations in a viral isolate that cannot be definitively identified from genome sequence data alone. We conclude that the genotyping assay is an excellent complement to genome sequencing efforts and allows rapid, point-of-testing determination of the presence of any genetic variant for SARS-CoV-2 for which an assay can be designed.
FIG 1.
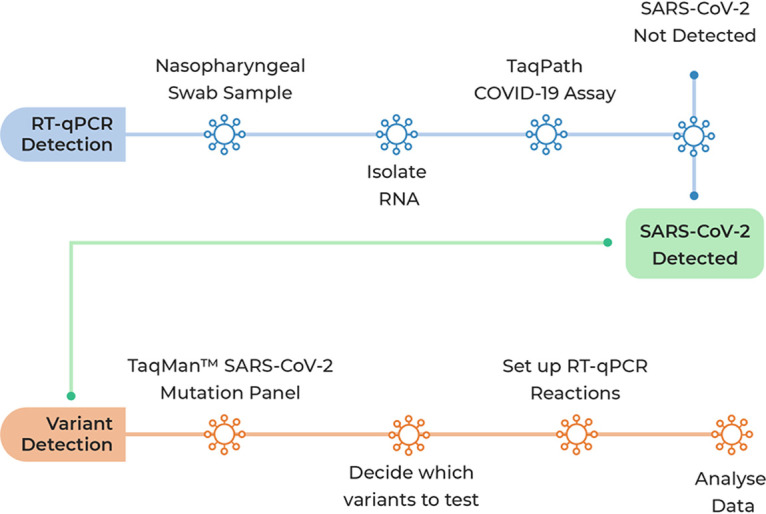
Workflow for ThermoFisher TaqMan SARS-CoV-2 mutation panel assay. Blue workflow, protocol used to detect SARS-CoV-2. Orange workflow, protocol using SNP assays to confirm mutations associated with SARS CoV-2 emerging variants.
MATERIALS AND METHODS
Patient samples.
Five hundred nasopharyngeal Pillar 1 swab samples (testing for those with a clinical need, and health and care workers) in virus transfer medium (VTM) were sent to the University of Birmingham during April and May 2021 from Birmingham Health Trust Hospital. SARS-CoV-2-positive samples from the University of Birmingham lateral flow testing site were also sent and archived. Five Pillar 2 SARS-CoV-2 samples (swab testing for the wider population) previously identified in February 2021 at the University of Birmingham Turnkey laboratory were also utilized. The use of anonymized samples in this study was allowed under ethics gained to aid assay development (NRES Committee West Midlands - South Birmingham 2002/201 amendment number 4, 24 April 2013).
RNA extraction.
RNA was extracted from 200 μl of patient sample using the ThermoFisher MagMAX Viral/Pathogen II nucleic acid isolation kits with MagMAX magnetic beads and MS2 phage internal control, using the automated ThermoFisher Kingfisher flex magnetic particle processor (11).
TaqPath COVID-19 assay.
Reaction mixtures were prepared using the ThermoFisher TaqPath COVID-19 CE-IVD RT-PCR kit protocol (12). RT-PCR of reaction mixtures was performed using the Applied Biosystems QuantStudio 5 real-time PCR instrument. Subsequent EDT files were transferred to a computer with QuantStudio Design and Analysis desktop software v2.5.1 for analysis of exponential curves. The TaqPath COVID-19 assay coamplifies three target genes: ORF1ab, N gene, and S gene. Results were classified as positive with respect to at least 2 single-target genes (ORF1ab, N, and S) provided the raw CT (cycle threshold) values were below 37 for single gene target signals. Bacteriophage MS2 RNA was added to each sample as an internal positive control for each sample and to monitor potential sample inhibition. A negative control (distilled water [dH2O]) is included on every plate. The SGTF of the TaqPath COVID-19 CE-IVD RT-PCR kit was considered a proxy for the presence of Δ69/70 in the S gene of SARS-CoV-2.
TaqMan SARS-CoV-2 mutation panel workflow.
Sample inclusion for the mutation assay required RNA extracts from positive samples (CT of ≤30, as defined in the manufacturer’s protocol, available from ThermoFisher) for the TaqMan SARS-CoV-2 mutation panel workflow. From our pool of positive samples, 185 were randomly selected for the mutation panel assay following the assay workflow (Fig. 1). Samples containing S-gene single-target failure (SGTF) were included in the assay as long as Orf1a and N-gene CT values were within range as there was no compromise of assay accuracy.
Designing a genotyping panel for the mutation assay.
TaqMan probes specific to single nucleotide polymorphisms (SNPs) found in variants known to be circulating widely within the United Kingdom around March to May 2021 were used in this study. S-gene mutations chosen were N501Y, E484K, K417N, P618R, and L452R (Fig. 2). Probes detect both the reference and mutation sequences of SARS-CoV-2. Reporter dye information for the TaqMan SARS-CoV-2 mutation panel is represented in the assay context sequence, which is the nucleotide sequence surrounding the mutation site in the SARS-CoV-2 reference genome (hCoV-19/Wuhan/2019; GISAID EPI_ISL_402124). The variant allele is detected by 6-carboxyfluorescein (FAM) dye, and the reference allele is detected by VIC dye.
FIG 2.
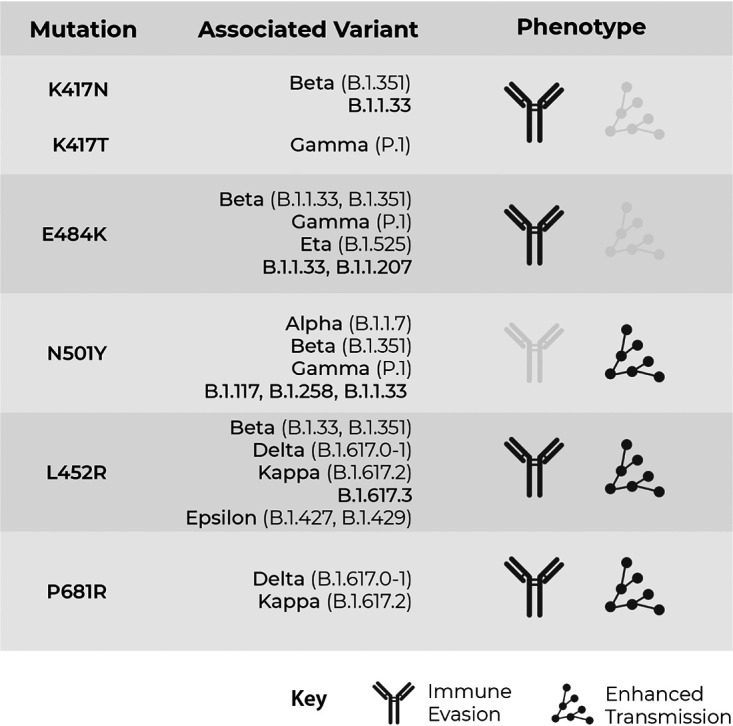
ThermoFisher mutation panel assay targets with associated SARS-CoV-2 variants and phenotype.
The presence of SGTF and N501Y was indicative of the Alpha variant; the Beta variant was defined by the presence of K417N, E484K, and N501Y; and Gamma was defined as E484K and N501Y without the presence of K417N, while the presence of L452R and P681R was indicative of the Delta variant.
Positive controls for both the original SARS-CoV-2 sequence and chosen SNP mutations were used in the assay. The AcroMetrix coronavirus 2019 (COVID-19) RNA control (low positive control), prepared using full-length genomic RNA from SARS-CoV-2, was used as a positive control for SARS-CoV-2. A plasmid control containing mutation sequences for N501Y, E484K, and K417N was used as a positive control for SNP mutations. However, at the time of running these experiments a plasmid control for mutations P681R and L452R was unavailable.
RT-PCR mix was prepared per the mutation assay protocol (13). For samples with CT values of <30, 5 μL of RNA was added to the reaction mixture, and for samples with CT values of <16, 2.5 μL of RNA was added to the reaction mixture. Reactions and the real-time PCR program were set up according to the mutation panel assay protocol (13).
Library preparation and sequencing.
Library preparation of positive SARS-CoV-2 samples (cycle threshold, <30) was performed using the nCoV-2019 LoCost sequencing protocol version 3 (14), using normalized primers (New England Biolabs) for the V3 ARTIC primer scheme (ARTIC network) (15). Sequencing was performed on a MinION flow cell (R9.4.1) run on a GridION sequencing device (Oxford Nanopore Technologies).
The ARTIC network nCoV-2019 novel coronavirus bioinformatics protocol was used to process the raw sequencing data including genome assembly and variant calling using nanopolish 0.11.3 (15). The genotyping was then performed using a nextflow pipeline (https://github.com/BioWilko/genotyping-pipeline). Briefly, genotypes were called using aln2type (https://github.com/connor-lab/aln2type) utilizing custom variant definition files for each mutation (included in repository), and lineages were called using Pangolin (16).
Data analysis.
Results were plotted as allelic discrimination plots using the QuantStudio Design and Analysis v2.5 with the genotyping analysis module.
RESULTS
Allelic discrimination plots show clear discrimination between wild-type samples and mutation samples using QuantStudio Design and Analysis.
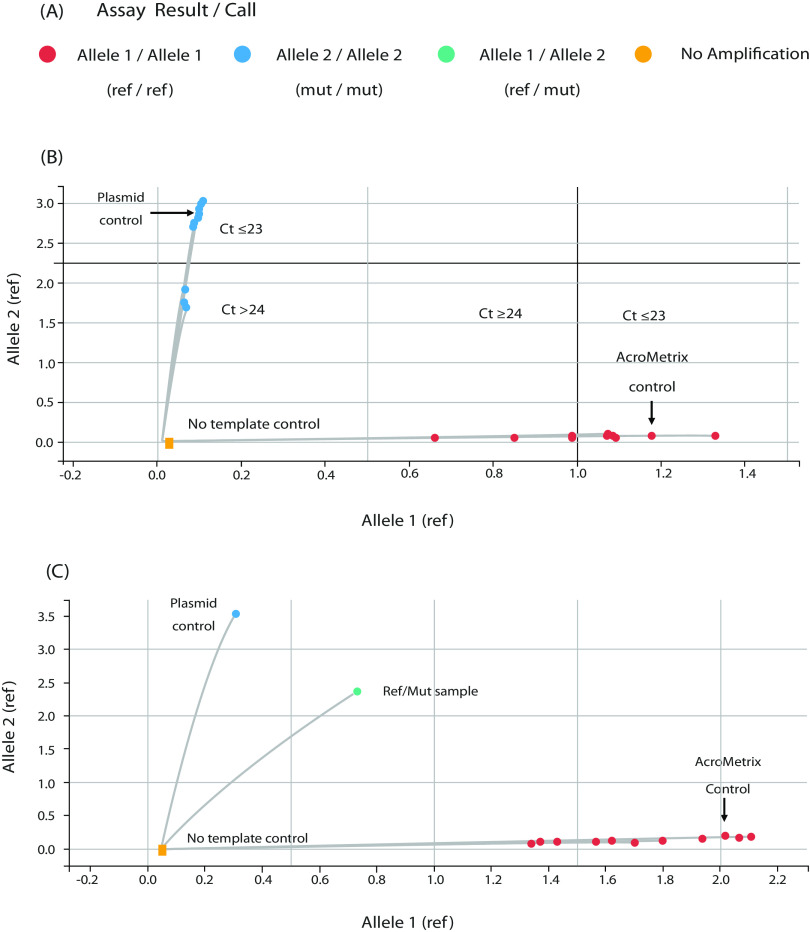
The Design and Analysis software genotype-calling algorithm was initially designed for diploid organism genotype calling. This is leveraged for the TaqPath assay by amplification and detection of both wild-type (WT) and variant alleles (labeled alleles 1 and 2, respectively). This allows the software to identify samples with a clear amplification curve matching either allele 1 (reference/wild type) or allele 2 (variant) for each specific mutation, respectively (Fig. 3). In some instances, the software will identify heterozygosity (i.e., presence of both wild-type and variant alleles), which indicates the need for further inspection of that sample. Heterozygosity may indicate a mutation at the underlying assay binding site or a truly heterologous sample (e.g., multiple strains present in the sample).
FIG 3.
Mutation assay results viewed using QuantStudio Design and Analysis software with the genotyping analysis module. (A) Assay results are “called” in 4 colors according to their outcome. Red indicates allele 1/allele 1 (ref/ref) for WT, blue indicates allele 2/allele 2 (mut/mut) for mutation present, green refers to allele 1/allele 2 (ref/mut), and orange means that results show no amplification. (B) Allelic discrimination plot showing clear discrimination between wild-type (WT) samples (red dots along the x axis) and the mutation samples (blue dots along the y axis) with high and low viral loads. (C) Allelic discrimination plot showing an example of a ref/mut sample (green dots). CT, cycle threshold. Cutoffs are determined by the QuantStudio Design and Analysis software.
Each probe is labeled with VIC dye to detect the reference (WT) sequence and FAM dye to detect the mutation sequence, which has one nucleotide difference. This allows clear discrimination on a cluster plot between WT and mutation samples, as seen with the AcroMetrix control (reference sequence) and plasmid control containing the mutation sequence (Fig. 3B).
Where input samples have similar CT values, they appear as clusters on the discrimination plot, as seen in the mut/mut samples (Fig. 3B), or in the case of a range of CT values, samples are dispersed along/up the axis as seen in ref/ref samples (Fig. 3B and C). We were able to detect samples with CT values varying from 12 to 29, in respect to ORF1ab gene, N gene, and S gene. In one case we could detect mutations with a CT value of 33, albeit with slightly reduced sensitivity. Samples with a high viral load cluster in the upper x/y axis, and those with a low viral load cluster in the lower x/y axis.
Independent mutations that are located next to one another in the SARS-CoV-2 virus genome, such as P681R and P681H, can complicate genotype analysis, as probes of an assay for one mutation will fail to bind to viral sequences that contain other adjacent mutations. Mutations under the probe can appear as ref/mut and slope away from ref/ref or mut/mut samples, clustering along the x axis, near the no-template controls (NTCs), thus exhibiting weak amplification due to the probes’ nonspecific activity (Fig. 3C). Genotyping calls can manually be adjusted to “no amp,” or two separate assays can be run, such as P681R and P681H, to compare and facilitate accurate genotype analysis. If it is not possible to make a call, then further characterization by genome sequencing will be necessary.
The mutation panel assay is extremely effective at identifying mutations in laboratory samples.
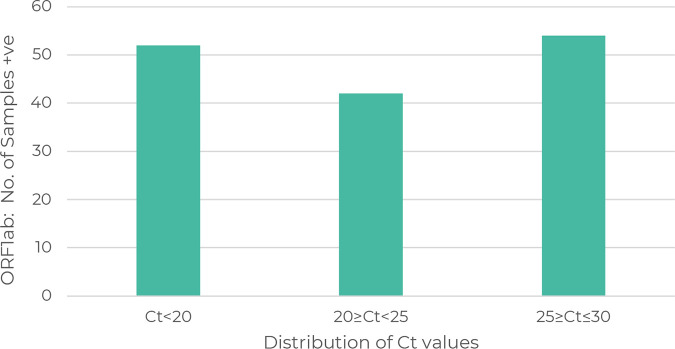
All samples run through the mutation panel assay produced a result, with either mutation present, mutation absent, or reference/mutation, indicating that another mutation within that SNP was present. Input samples had various CT values (CT, 12 to 29) with regard to each of the three single-target genes ORF1ab, N, and S (see Data Set S1 in the supplemental material). Using the ORF1ab gene as a marker for CT distribution across a CT value group, good distribution of different CT values was observed (Fig. 4). RNA quality was not measured, and in some cases, samples had been stored at −80°C for up to 2 months and through no more than 2 freeze/thaw cycles. However, no effect on the performance of the mutation assay was observed. Furthermore, we also noted that for positive samples with a CT of ≤16, RNA was added at 2.5 μL instead of 5 μL into the mutation panel PCR, therefore allowing more RNA to be archived.
FIG 4.
Distribution of TaqPath COVID-19 PCR CT values for the ORF1ab gene. The y axis indicates the number of samples positive for the SARS-CoV-2 ORF1ab gene. The x axis is grouped into ranges for CT values up to and including 30. Abbreviations: CT, cycle threshold; ORF1ab, open reading frame 1ab.
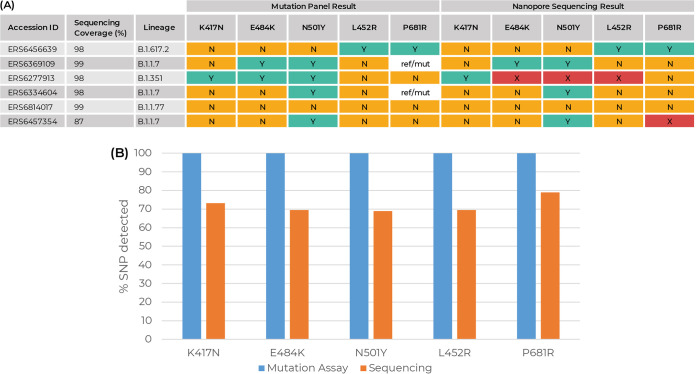
All samples run through this assay were sent for sequencing onsite at the University of Birmingham as part of COG-UK. This provided us with the ability to compare the mutation panel assay results with those of genome sequencing. By cross-referencing the genome sequencing results for each SNP, we could identify whether the mutation assay correctly called each SNP mutation. Our data show that for all samples, where sequencing data were available, the mutation panel is in 100% agreement (Fig. 5A; also refer to Data Set S1 for the complete data set for all samples run). While Nanopore sequencing may miss an SNP mutation (Fig. 5B), the mutation assay can identify this. This can be due to the RNA quality, the sequencing platform used, or issues with primers required for genome sequencing but highlights the importance of the genotyping assay for rapid identification and subsequent action.
FIG 5.
(A) Example of mutation assay results compared with Nanopore sequencing results from a selection of samples run through the assay. All results for all samples run are included in Data Set S1 in the supplemental material. Orange square “N,” mutation not present; green square “Y,” mutation present; white “ref/mut,” mutation present on one allele only. The red square “X” indicates that there was not sufficient coverage of that SNP after sequencing. (B) Percent comparison results of all mutation assay results and corresponding Nanopore sequencing data. Blue bars (mutation assay), percent (%) SNP agreement compared to the corresponding sequencing data. Orange bars (sequencing), percent SNPs identified when sequencing data for each sample were assigned a lineage by Pangolin.
The mutation assay cannot distinguish what the substitution is in “ref/mut” results, which highlights the continued importance of sequencing and updating SNP mutations which can be added when designing an assay panel. The mutation assay cannot be used to identify variant lineages; however, it can, due to the detection of specific mutations, give an indication as to which variant the sample may be and can also exclude the presence of a VOC in a sample based on the absence of key characterizing mutations of significance. This is of particular importance as case numbers rise and sequencing capacity and turnaround time may not be matched. Specifically, for VOC Alpha (B.1.1.7) ref/mut further analysis of genome sequencing data revealed that this variant, although negative for P681R, was positive for P681H. Importantly, it was noted that one ref/mut was also positive for E484K, and clarification from the University of Birmingham arm of COG-UK confirmed that this was a small cluster of B1.1.7 variants that was being monitored and acted on in the Birmingham area.
The mutation panel assay is highly adaptable to newly emerging variants and mutations.
The mutation panel designed for this assay was intended to identify samples containing SNPs associated with variants of SARS-CoV-2 widely circulating within the United Kingdom from March 2021 to May 2021; this included mutations found in the table in Fig. 2. Between April and May B.1.617 variant numbers were increasing rapidly and beginning to overtake the B.1.1.7 variant within the UK population. Therefore, as experiments were being conducted in real time the addition of P618R and L452R assays was essential for rapid identification of B.1.617 variants. Samples that had previously been run for the original assays and sent for sequencing meant that RNA availability was limited. However, freshly isolated samples from May provided the opportunity to run all assays at once.
One hundred eighty-six samples were assayed, with 182 samples run for N501Y, 183 for E484K, and 178 for K417N, and a total of 42 samples were assayed for all 5 mutations; 68.7% of samples assayed were positive for N501Y (WT ref/ref = 31.3%), 2% of samples were positive for E484K (WT ref/ref = 97.2%), 2.8% of samples assayed were positive for K417N (WT ref/ref = 97.2%), 57.1% of samples were positive for L425R (WT ref/ref = 42.9%), and 61% were positive for P681R (WT ref/ref = 19%). A ref/mut “call” was also noted for E484K (1 sample, 0.6%) and P681R (8 samples, 19%).
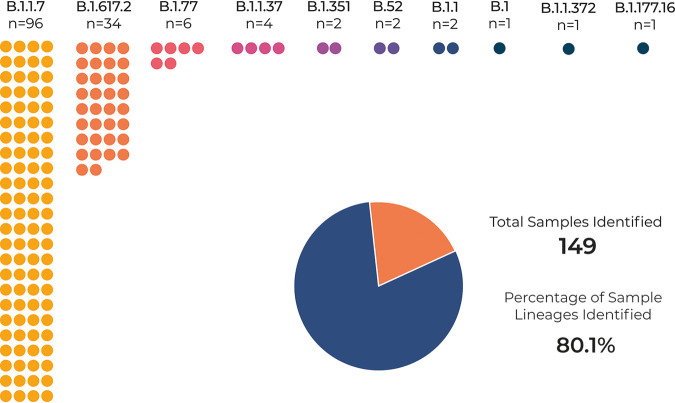
Lineage was identifiable in 80.1% (Fig. 6) of samples sent for sequencing, which as mentioned previously may be due to the sequencing protocol used and/or RNA quality. From our pool of samples, 65.4% of samples were the B.1.1.7 variant, 18.3% were B.1.617.2, 3.2% were B.1.177, 2.2% were B.1.137, and 1.1% were B.1.351 and B.52, with B.1.177.16, B.1.1.372, B.1.1, and B.1 variants making up 4%. Cases of B.1.617.2 were first identified at the end of April 2021.
FIG 6.
Lineage of sequenced samples identified by Pangolin. (Top and left) Number of samples assigned a SARS CoV-2 lineage. (Bottom right) Percentage of samples assigned a lineage compared to the total number of samples assigned a lineage.
DISCUSSION
Genomic epidemiology is a powerful tool for tracking transmission and importation of SARS-CoV-2 as well as assessing the effectiveness of public health measures (1–3). Tracking transmission within the population in real time enables laboratories to report and governments to act. Recently, ThermoFisher has developed genotyping assays to help bridge the gap between testing capacity and sequencing capability to receive results in real time. A previous study from our laboratory demonstrated that B.1.1.7 was associated with significantly higher viral loads (17), and had a genotyping assay been available at the time, this would have helped to identify the Alpha variant much more quickly and to identify speed and spread of infection for quicker action and containment.
In our laboratory, an opportunity arose to genotype RNA extracted in real time from positive SARS-CoV-2 samples from Birmingham Trust Hospital, Pillar 2, and the University of Birmingham lateral flow site. Initially using three verified SNP assays from ThermoFisher’s Applied Biosystems TaqMan SARS-CoV-2 mutation panel and then expanding to five to include SNPs for the Delta variant, we matched genotypes for specific mutations in variants Alpha (B.1.1.7), Beta (B.1.351), and Delta (B.1.617.2).
Here, we demonstrate that these mutation panels provide robust detection of VOC even if RNA is of low quality and after more than one freeze/thaw step. Importantly, a specific mutation can be identified on the same day that a nasopharyngeal swab tests positive by RT-PCR. Where sequencing data were available, the genotyping assay always matched 100% with the correct lineage.
The ref/mut function is a key to the genotyping assay, as when detected, this will imply an amino acid change for the specific mutation at the genomic site where the primers amplify. This is crucial in alerting researchers to the potential rise of a new VOI (variant of interest) so it can be monitored to see its potential to become a VOC.
Our study confirms that same-day rapid real-time detection of variants present in the population is very achievable—from a swab entering the lab and then being processed through the TaqPath COVID-19 RT-PCR and mutation assay, where results were then available in approximately 5 h. Confirmation of mutations present and lineage identification following genomic sequencing onsite were provided in approximately 72 h; however, when onsite sequencing is not available, turnaround time may increase significantly. The mutation panel is also significantly less expensive than sequencing, at ∼£0.45p per reaction, compared to ∼£35 per genome when operating at scale. One limitation of the TaqPath assay is the limit of detection, which may impact laboratories wishing to test extremely low-yield samples. This is also an issue for genome sequencing of samples, which in the United Kingdom is possible only on samples with CT values of <30 on the TaqPath assay.
Rapid identification of VOC enables test-and-trace efforts to identify regional clusters and perform targeted testing to prevent spread of more transmissible variants. Having both PCR setups in our modular testing system and onsite sequencing removes the logistics, costs, and time of moving samples between testing labs and sequencing labs and reporting the results.
Importantly, the genotyping panel can be updated readily as new SNP mutations are identified via genome sequencing data from COG-UK. Delta variants emerged quickly over a few weeks, and during our study we were able to source very quickly two mutation panels for the Delta variant (L452R and P681R), and on their arrival, we were able to act on the same day.
Genome sequencing can be inconclusive for identifying key mutations found by the mutation panels—we observed several instances where the TaqPath assay identified specific alleles that were not detected via genome sequencing (see Data Set S1 in the supplemental material)—but nonetheless, it is a powerful tool for identifying SARS-CoV-2 variant lineages through phylogenetic trees. However, as previous studies have discussed, there are limitations due to sequence dropout when genome sequencing is used to identify specific key mutations (18–21). The presence of SNPs in the forward and/or reverse primer binding sites may lead to complete or partial lack of amplification. These allelic dropouts specifically affect PCR-based (tile amplicon) targeted sequencing, thus resulting in incomplete genome coverage, especially at smaller amounts, resulting in the loss of both 5′ and 3′ regions that fall outside primer binding positions. This can be mitigated through adjustments to the specific primer scheme used, something that happens at regular intervals for the ARTIC protocol employed worldwide for Nanopore-based SARS-CoV-2 sequencing.
For the study presented here, we demonstrated that genotyping has two major functions. (i) Genotyping is a powerful additional, more in-depth, assay for identifying specific mutations and has real clinical health value allowing laboratories to report and act on VOC much more quickly than genome sequencing. (ii) Genotyping is an excellent additional complement to the already powerful tool of genome sequencing already proven for assigning lineages via phylogenetic trees.
Our data confirm that SARS-CoV-2 genotyping is essential for real-time identification of VOC here now and tracking of those that emerge for informing public health strategy.
ACKNOWLEDGMENTS
Reagents and technical assistance for this study were provided free of charge by Jelena Feenstra, Tamra Hill, Manoj Ghandi, Camilla Ulekleiv, and Carl Borufka at ThermoFisher.
The UoB Turnkey COVID-19 diagnostic lab was created wholly from funds directly attributed by DHSC. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research &hx0026; Innovation (UKRI), the National Institute of Health Research (NIHR) (grant code: MC_PC_19027), and Genome Research Limited, operating as the Wellcome Sanger Institute.
ThermoFisher had no influence or role in study design, data analysis, data interpretation, or construction of the manuscript.
All authors except A.M. confirm that they do not have any commercial or other potential conflicts of interest relevant to the publication of this research. A.M. has delivered paid presentations on behalf of ThermoFisher on the use of the original TaqMan assay in the UoB laboratory.
The members of The COVID-19 Genomics UK (COG-UK) Consortium are as follows:Dinesh Aggarwal (University of Cambridge, Department of Medicine, Cambridge, UK), Beth Blane (University of Cambridge, Department of Medicine, Cambridge, UK), Ellena Brooks (University of Cambridge, Department of Medicine, Cambridge, UK), Alessandro M. Carabelli (University of Cambridge, Department of Medicine, Cambridge, UK), Carol M. Churcher (University of Cambridge, Department of Medicine, Cambridge, UK), Katerina Galai (University of Cambridge, Department of Medicine, Cambridge, UK), Sophia T. Girgis (University of Cambridge, Department of Medicine, Cambridge, UK), Ravi K. Gupta (University of Cambridge, Department of Medicine, Cambridge, UK), Nazreen F. Hadjirin (University of Cambridge, Department of Medicine, Cambridge, UK), Danielle Leek (University of Cambridge, Department of Medicine, Cambridge, UK), Catherine Ludden (University of Cambridge, Department of Medicine, Cambridge, UK; Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Georgina M. McManus (University of Cambridge, Department of Medicine, Cambridge, UK), Sophie Palmer (University of Cambridge, Department of Medicine, Cambridge, UK), Sharon J. Peacock (University of Cambridge, Department of Medicine, Cambridge, UK; Public Health England, 61 Colindale Ave, London NW9 5EQ, UK; Cambridge University Hospital NHS Foundation Trust, Cambridge, UK; Wellcome Sanger Institute, Hinxton, Cambridge, UK), Kim S. Smith (University of Cambridge, Department of Medicine, Cambridge, UK), Elias Allara (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), David Bibby (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Chloe Bishop (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Andrew Bosworth (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Daniel Bradshaw (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Vicki Chalker (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Meera Chand (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Gavin Dabrera (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Nicholas Ellaby (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Eileen Gallagher (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Natalie Groves (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Ian Harrison (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Hassan Hartman (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Richard Hopes (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Jonathan Hubb (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Stephanie Hutchings (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Angie Lackenby (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Juan Ledesma (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), David Lee (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Nikos Manesis (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Carmen Manso (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Tamyo Mbisa (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Shahjahan Miah (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Peter Muir (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Richard Myers (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Husam Osman (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Vineet Patel (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Clare Pearson (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Steven Platt (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Hannah M. Pymont (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Mary Ramsay (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Esther Robinson (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Ulf Schaefer (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Alicia Thornton (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Katherine A. Twohig (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), Ian B. Vipond (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), David Williams (Public Health England, 61 Colindale Ave, London NW9 5EQ, UK), William L. Hamilton (University of Cambridge, Department of Medicine, Cambridge, UK; Cambridge University Hospital NHS Foundation Trust, Cambridge, UK; Wellcome Sanger Institute, Hinxton, Cambridge, UK), Ben Warne (University of Cambridge, Department of Medicine, Cambridge, UK; Cambridge University Hospital NHS Foundation Trust, Cambridge, UK; Cambridge Institute for Therapeutic Immunology and Infectious Disease, University of Cambridge, Cambridge, UK; West of Scotland Specialist Virology Centre, NHS Greater Glasgow and Clyde, Glasgow, UK), Louise Aigrain (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Alex Alderton (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Roberto Amato (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Cristina V. Ariani (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Jeff Barrett (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Andrew R. Bassett (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Mathew A. Beale (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Charlotte Beaver (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Katherine L. Bellis (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Emma Betteridge (Wellcome Sanger Institute, Hinxton, Cambridge, UK), James Bonfield (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Iraad F. Bronner (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Michael H. S. Chapman (Wellcome Sanger Institute, Hinxton, Cambridge, UK), John Danesh (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Robert Davies (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Matthew J. Dorman (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Eleanor Drury (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Jillian Durham (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Ben W. Farr (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Luke Foulser (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Sonia Goncalves (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Scott Goodwin (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Marina Gourtovaia (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Ewan M. Harrison (University of Cambridge, Department of Medicine, Cambridge, UK; Public Health England, 61 Colindale Ave, London NW9 5EQ, UK; Wellcome Sanger Institute, Hinxton, Cambridge, UK; Cardiff and Vale University Health Board, Cardiff, UK), David K. Jackson (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Keith James (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Dorota Jamrozy (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Ian Johnston (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Leanne Kane (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Sally Kay (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Jon-Paul Keatley (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Dominic Kwiatkowski (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Cordelia F. Langford (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Mara Lawniczak (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Stefanie V. Lensing (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Steven Leonard (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Laura Letchford (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Kevin Lewis (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Jennifier Liddle (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Rich Livett (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Stephanie Lo (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Alex Makunin (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Inigo Martincorena (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Shane McCarthy (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Samantha McGuigan (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Robin J. Moll (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Rachel Nelson (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Karen Oliver (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Steve Palmer (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Naomi R. Park (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Minal Patel (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Liam Prestwood (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Christoph Puethe (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Michael A. Quail (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Diana Rajan (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Shavanthi Rajatileka (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Nicholas M. Redshaw (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Carol Scott (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Lesley Shirley (Wellcome Sanger Institute, Hinxton, Cambridge, UK), John Sillitoe (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Scott A. J. Thurston (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Gerry Tonkin-Hill (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Jaime M. Tovar-Corona (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Danni Weldon (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Andrew Whitwham (Wellcome Sanger Institute, Hinxton, Cambridge, UK), Yasmin Chaudhry (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Iliana Georgana (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Ian G. Goodfellow (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Grant Hall (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Myra Hosmillo (Cambridge University Hospital NHS Foundation Trust, Cambridge, UK; University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Aminu S. Jahun (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Malte L. Pinckert (University of Cambridge, Department of Pathology, Division of Virology, Cambridge, UK), Stephen W. Attwood (Department of Zoology, University of Oxford, Oxford, UK), Louis du Plessis (Department of Zoology, University of Oxford, Oxford, UK), Marina Escalera Zamudio (Department of Zoology, University of Oxford, Oxford, UK), Sarah Francois (Department of Zoology, University of Oxford, Oxford, UK), Bernardo Gutierrez (Department of Zoology, University of Oxford, Oxford, UK), Moritz U. G. Kraemer (Department of Zoology, University of Oxford, Oxford, UK), Oliver G. Pybus (Department of Zoology, University of Oxford, Oxford, UK), Jayna Raghwani (Department of Zoology, University of Oxford, Oxford, UK), Tetyana I. Vasylyeva (Department of Zoology, University of Oxford, Oxford, UK), Alex E. Zarebski (Department of Zoology, University of Oxford, Oxford, UK), Verity Hill (Institute of Evolutionary Virology, University of Edinburgh, Edinburgh, UK), Nabil-Fareed Alikhan (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Alp Aydin (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), David J. Baker (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Leonardo de Oliveira Martins (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Gemma L. Kay (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Thanh Le-Viet (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Alison E. Mather (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Lizzie Meadows (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Justin O'Grady (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Andrew J. Page (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Steven Rudder (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Alexander J. Trotter (Quadram Institute Bioscience, Norwich Research Park, Norwich NR4 7UQ, UK), Chris J. Illingworth (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Chris Jackson (MRC Biostatistics Unit, University of Cambridge, East Forvie Building, Forvie Site, Robinson Way, Cambridge CB2 0SR, UK), Elihu Aranday-Cortes (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Patawee Asamaphan (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Alice Broos (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Stephen N. Carmichael (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Ana da Silva Filipe (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Joseph Hughes (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Natasha G. Jesudason (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Natasha Johnson (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Kathy K. Li (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Daniel Mair (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Jenna Nichols (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Seema Nickbakhsh (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Marc O. Niebel (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Kyriaki Nomikou (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Richard J. Orton (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), David L. Robertson (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Rajiv N. Shah (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), James G. Shepherd (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Joshua B. Singer (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Igor Starinskij (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Emma C. Thomson (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Lily Tong (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Sreenu Vattipally (MRC-University of Glasgow Centre for Virus Research, Glasgow, UK), Claire Cormie (University of Cambridge, Cambridge, UK), Joana Dias (University of Cambridge, Cambridge, UK), Sally Forrest (University of Cambridge, Cambridge, UK), Harmeet K. Gill (University of Cambridge, Cambridge, UK), Ellen E. Higginson (University of Cambridge, Cambridge, UK), Leanne M. Kermack (University of Cambridge, Cambridge, UK), Mailis Maes (University of Cambridge, Cambridge, UK), Chris Ruis (University of Cambridge, Cambridge, UK), Sushmita Sridhar (University of Cambridge, Cambridge, UK), Jamie Young (University of Cambridge, Cambridge, UK), Amy Ash (Barking, Havering and Redbridge University Hospitals NHS Trust, Romford, UK), Cherian Koshy (Guy's and St. Thomas' BRC, London, UK), Nick Cortes (Basingstoke Hospital, Basingstoke, UK), Stephen Kidd (Basingstoke Hospital, Basingstoke, UK), Jessica Lynch (Basingstoke Hospital, Basingstoke, UK), Nathan Moore (Basingstoke Hospital, Basingstoke, UK), Matilde Mori (Basingstoke Hospital, Basingstoke, UK), Emma Wise (Basingstoke Hospital, Basingstoke, UK), Tanya Curran (Belfast Health & Social Care Trust, Belfast, UK), Derek J. Fairley (Belfast Health & Social Care Trust, Belfast, UK), James P. McKenna (Belfast Health & Social Care Trust, Belfast, UK), Helen Adams (Betsi Cadwaladr University Health Board, Bangor, UK), David Bonsall (Big Data Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Christophe Fraser (Big Data Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Tanya Golubchik (Big Data Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Benjamin J. Cogger (Brighton and Sussex University Hospitals NHS Trust, Brighton, UK), Mohammed O. Hassan-Ibrahim (Brighton and Sussex University Hospitals NHS Trust, Brighton, UK), Cassandra S. Malone (Brighton and Sussex University Hospitals NHS Trust, Brighton, UK), Nicola Reynolds (Cambridge Stem Cell Institute, University of Cambridge, Cambridge, UK), Michelle Wantoch (Cambridge Stem Cell Institute, University of Cambridge, Cambridge, UK), Safiah Afifi (Cardiff and Vale University Health Board, Cardiff, UK), Robert Beer (Cardiff and Vale University Health Board, Cardiff, UK), Michaela John (Cardiff and Vale University Health Board, Cardiff, UK), Joshua Maksimovic (Cardiff and Vale University Health Board, Cardiff, UK), Kathryn McCluggage (Cardiff and Vale University Health Board, Cardiff, UK), Sian Morgan (Cardiff and Vale University Health Board, Cardiff, UK), Karla Spellman (Cardiff and Vale University Health Board, Cardiff, UK), Catherine Bresner (Cardiff University, Cardiff, UK), Thomas R. Connor (Cardiff University, Cardiff, UK), William Fuller (Cardiff University, Cardiff, UK), Martyn Guest (Cardiff University, Cardiff, UK), Huw Gulliver (Cardiff University, Cardiff, UK), Christine Kitchen (Cardiff University, Cardiff, UK), Angela Marchbank (Cardiff University, Cardiff, UK), Ian Merrick (Cardiff University, Cardiff, UK), Robert Munn (Cardiff University, Cardiff, UK), Anna Price (Cardiff University, Cardiff, UK), Joel Southgate (Cardiff University, Cardiff, UK), Trudy Workman (Cardiff University, Cardiff, UK), Amita Patel (Centre for Clinical Infection & Diagnostics Research, St. Thomas' Hospital and Kings College London, London, UK), Luke B. Snell (Centre for Clinical Infection & Diagnostics Research, St. Thomas' Hospital and Kings College London, London, UK), Rahul Batra (Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, Guy's and St Thomas' NHS Foundation Trust, London, UK), Themoula Charalampous (Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, Guy's and St Thomas' NHS Foundation Trust, London, UK), Jonathan Edgeworth (Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, Guy's and St Thomas' NHS Foundation Trust, London, UK), Gaia Nebbia (Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, Guy's and St Thomas' NHS Foundation Trust, London, UK), Angela H. Beckett (Centre for Enzyme Innovation, University of Portsmouth (PORT), Portsmouth, UK), Samuel C. Robson (Centre for Enzyme Innovation, University of Portsmouth (PORT), Portsmouth, UK), David M. Aanensen (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Khalil Abudahab (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Mirko Menegazzo (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Ben E. W. Taylor (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Anthony P. Underwood (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Corin A. Yeats (Centre for Genomic Pathogen Surveillance, University of Oxford, Oxford, UK), Louise Berry (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Tim Boswell (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Gemma Clark (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Vicki M. Fleming (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Hannah C. Howson-Wells (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Carl Jones (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Amelia Joseph (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Manjinder Khakh (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Michelle M. Lister (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Wendy Smith (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Iona Willingham (Clinical Microbiology Department, Queens Medical Centre, Nottingham, UK), Paul Bird (Clinical Microbiology, University Hospitals of Leicester NHS Trust, Leicester, UK), Karlie Fallon (Clinical Microbiology, University Hospitals of Leicester NHS Trust, Leicester, UK), Thomas Helmer (Clinical Microbiology, University Hospitals of Leicester NHS Trust, Leicester, UK), Christopher Holmes (Clinical Microbiology, University Hospitals of Leicester NHS Trust, Leicester, UK), Julian Tang (Clinical Microbiology, University Hospitals of Leicester NHS Trust, Leicester, UK), Victoria Blakey (County Durham and Darlington NHS Foundation Trust, Darlington, UK), Sharon Campbell (County Durham and Darlington NHS Foundation Trust, Darlington, UK), Veena Raviprakash (County Durham and Darlington NHS Foundation Trust, Darlington, UK), Nicola Sheriff (County Durham and Darlington NHS Foundation Trust, Darlington, UK), Lesley-Anne Williams (County Durham and Darlington NHS Foundation Trust, Darlington, UK), Matthew Carlile (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Johnny Debebe (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Nadine Holmes (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Matthew W. Loose (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Christopher Moore (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Fei Sang (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Victoria Wright (Deep Seq, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Francesc Coll (Department of Infection Biology, Faculty of Infectious & Tropical Diseases, London School of Hygiene & Tropical Medicine, London, UK), Gilberto Betancor (Department of Infectious Diseases, King's College London, London, UK), Adrian W. Signell (Department of Infectious Diseases, King's College London, London, UK), Harry D. Wilson (Department of Infectious Diseases, King's College London, London, UK), Thomas Davis (Department of Microbiology, Kettering General Hospital, Kettering, UK), Sahar Eldirdiri (Department of Microbiology, Kettering General Hospital, Kettering, UK), Anita Kenyon (Department of Microbiology, Kettering General Hospital, Kettering, UK), M. Estee Torok (Departments of Infectious Diseases and Microbiology, Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK), Hannah Lowe (East Kent Hospitals University NHS Foundation Trust, Canterbury, UK), Samuel Moses (East Kent Hospitals University NHS Foundation Trust, Canterbury, UK), Luke Bedford (East Suffolk and North Essex NHS Foundation Trust, Colchester, UK), Jonathan Moore (Gateshead Health NHS Foundation Trust, Gateshead, UK), Susanne Stonehouse (Gateshead Health NHS Foundation Trust, Gateshead, UK), Ali R. Awan (Gateshead Health NHS Foundation Trust, Gateshead, UK), Chloe L. Fisher (Genomics Innovation Unit, Guy's and St. Thomas' NHS Foundation Trust, London, UK), John BoYes (Gloucestershire Hospitals NHS Foundation Trust, Cheltenham, UK), Laura Atkinson (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Judith Breuer (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Julianne R. Brown (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Kathryn A. Harris (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Jack C. D. Lee (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Divya Shah (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Nathaniel Storey (Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK), Flavia Flaviani (Guy's and St. Thomas' BRC, London, UK), Adela Alcolea-Medina (Guy's and St. Thomas' Hospitals, London, UK), Gabrielle Vernet (Hampshire Hospitals NHS Foundation Trust, Basingstoke, UK), Rebecca Williams (Hampshire Hospitals NHS Foundation Trust, Basingstoke, UK), Michael R. Chapman (Health Data Research UK Cambridge, Cambridge, UK), Wendy Chatterton (Health Services Laboratories, London, UK), Judith Heaney (Health Services Laboratories, London, UK), Lisa J. Levett (Health Services Laboratories, London, UK), Monika Pusok (Health Services Laboratories, London, UK), Li Xu-McCrae (Heartlands Hospital, Birmingham, UK), Matthew Bashton (Hub for Biotechnology in the Built Environment, Northumbria University, Newcastle, UK), Darren L. Smith (Hub for Biotechnology in the Built Environment, Northumbria University, Newcastle, UK), Gregory R. Young (Hub for Biotechnology in the Built Environment, Northumbria University, Newcastle, UK), Frances Bolt (Imperial College Hospitals NHS Trust, London, UK), Alison Cox (Imperial College Hospitals NHS Trust, London, UK), Alison Holmes (Imperial College Hospitals NHS Trust, London, UK), Pinglawathee Madona (Imperial College Hospitals NHS Trust, London, UK), Siddharth Mookerjee (Imperial College Hospitals NHS Trust, London, UK), James Price (Imperial College Hospitals NHS Trust, London, UK), Paul A. Randell (Imperial College Hospitals NHS Trust, London, UK), Olivia Boyd (Imperial College London, London, UK), Fabricia F. Nascimento (Imperial College London, London, UK), Lily Geidelberg (Imperial College London, London, UK), Rob Johnson (Imperial College London, London, UK), David Jorgensen (Imperial College London, London, UK), Manon Ragonnet-Cronin (Imperial College London, London, UK), Aileen Rowan (Imperial College London, London, UK), Igor Siveroni (Imperial College London, London, UK), Graham P. Taylor (Imperial College London, London, UK), Erik M. Volz (Imperial College London, London, UK), Katherine L. Smollett (Institute of Biodiversity, Animal Health & Comparative Medicine, Glasgow, UK), Nicholas J. Loman (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Claire McMurray (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Alan McNally (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Sam Nicholls (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Radoslaw Poplawski (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Joshua Quick (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Will Rowe (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Joanne Stockton (Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK), Rocio T. Martinez Nunez (King's College London, London, UK), Cassie Breen (Liverpool Clinical Laboratories, Liverpool, UK), Angela Cowell (Liverpool Clinical Laboratories, Liverpool, UK), Jenifer Mason (Liverpool Clinical Laboratories, Liverpool, UK), Elaine O'Toole (Liverpool Clinical Laboratories, Liverpool, UK), Trevor I. Robinson (Liverpool Clinical Laboratories, Liverpool, UK), Joanne Watts (Liverpool Clinical Laboratories, Liverpool, UK), Graciela Sluga (Maidstone and Tunbridge Wells NHS Trust, Tunbridge Wells, UK), Shazaad S. Y. Ahmad (Manchester University NHS Foundation Trust, Manchester, UK), Ryan P. George (Manchester University NHS Foundation Trust, Manchester, UK), Nicholas W. Machin (Manchester University NHS Foundation Trust, Manchester, UK), Fenella Halstead (Microbiology Department, Wye Valley NHS Trust, Hereford, UK), Wendy Hogsden (Microbiology Department, Wye Valley NHS Trust, Hereford, UK), Venkat Sivaprakasam (Microbiology Department, Wye Valley NHS Trust, Hereford, UK), Holli Carden (National Infection Service, PHE and Leeds Teaching Hospitals Trust, Leeds, UK), Antony D. Hale (National Infection Service, PHE and Leeds Teaching Hospitals Trust, Leeds, UK), Katherine L. Harper (National Infection Service, PHE and Leeds Teaching Hospitals Trust, Leeds, UK), Louissa R. Macfarlane-Smith (National Infection Service, PHE and Leeds Teaching Hospitals Trust, Leeds, UK), Shirelle Burton-Fanning (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Jennifer Collins (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Gary Eltringham (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Brendan AI. Payne (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Yusri Taha (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Sheila Waugh (Newcastle Hospitals NHS Foundation Trust, Newcastle, UK), Sarah O'Brien (Newcastle University, Newcastle, UK), Steven Rushton (Newcastle University, Newcastle, UK), Rachel Blacow (NHS Greater Glasgow and Clyde, Glasgow, UK), Amanda Bradley (NHS Greater Glasgow and Clyde, Glasgow, UK), Alasdair Maclean (NHS Greater Glasgow and Clyde, Glasgow, UK), Guy Mollett (NHS Greater Glasgow and Clyde, Glasgow, UK), Rebecca Dewar (NHS Lothian, Edinburgh, UK), Martin P. McHugh (NHS Lothian, Edinburgh, UK), Kate E. Templeton (NHS Lothian, Edinburgh, UK), Elizabeth Wastenge (NHS Lothian, Edinburgh, UK), Lindsay Coupland (Norfolk and Norwich University Hospital, Norwich, UK), Samir Dervisevic (Norfolk and Norwich University Hospital, Norwich, UK), Emma J. Meader (Norfolk and Norwich University Hospital, Norwich, UK), Rachael Stanley (Norfolk and Norwich University Hospital, Norwich, UK), Louise Smith (Norfolk County Council, Norwich, UK), Edward Barton (North Cumbria Integrated Care NHS Foundation Trust, Carlisle, UK), Clive Graham (North Cumbria Integrated Care NHS Foundation Trust, Carlisle, UK), Debra Padgett (North Cumbria Integrated Care NHS Foundation Trust, Carlisle, UK), Garren Scott (North Cumbria Integrated Care NHS Foundation Trust, Carlisle, UK), Jane Greenaway (North Tees and Hartlepool NHS Foundation Trust, Stockton on Tees, UK), Emma Swindells (North Tees and Hartlepool NHS Foundation Trust, Stockton on Tees, UK), Clare M. McCann (Northumbria University, Newcastle, UK), Andrew Nelson (Northumbria University, Newcastle, UK), Wen C. Yew (Northumbria University, Newcastle, UK), Monique Andersson (Oxford University Hospitals NHS Foundation Trust, Oxford, UK), Derrick Crook (Oxford University Hospitals NHS Foundation Trust, Oxford, UK), David Eyre (Oxford University Hospitals NHS Foundation Trust, Oxford, UK), Anita Justice (Oxford University Hospitals NHS Foundation Trust, Oxford, UK), Timothy Peto (Oxford University Hospitals NHS Foundation Trust, Oxford, UK), Nichola Duckworth (PathLinks, Northern Lincolnshire & Goole NHS Foundation Trust, Grimsby, UK), Tim J. Sloan (PathLinks, Northern Lincolnshire & Goole NHS Foundation Trust, Grimsby, UK), Sarah Walsh (PathLinks, Northern Lincolnshire & Goole NHS Foundation Trust, Grimsby, UK), Kelly Bicknell (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Anoop J. Chauhan (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Scott Elliott (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Sharon Glaysher (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Robert Impey (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Allyson Lloyd (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Sarah Wyllie (Portsmouth Hospitals University NHS Trust, Portsmouth, UK), Nick Levene (Princess Alexandra Hospital Microbiology Dept, Harlow, UK), Lynn Monaghan (Princess Alexandra Hospital Microbiology Dept, Harlow, UK), Declan T. Bradley (Public Health Agency, Belfast, UK), Tim Wyatt (Public Health Agency, Belfast, UK), Martin D. Curran (Public Health England, Clinical Microbiology and Public Health Laboratory, Cambridge, UK), Surendra Parmar (Public Health England, Clinical Microbiology and Public Health Laboratory, Cambridge, UK), Matthew T. G. Holden (Public Health Scotland, Edinburgh, UK), Sharif Shaaban (Public Health Scotland, Edinburgh, UK), Alexander Adams (Public Health Wales NHS Trust, Cardiff, UK), Hibo Asad (Public Health Wales NHS Trust, Cardiff, UK), Alec Birchley (Public Health Wales NHS Trust, Cardiff, UK), Matthew Bull (Public Health Wales NHS Trust, Cardiff, UK), Jason Coombes (Public Health Wales NHS Trust, Cardiff, UK), Sally Corden (Public Health Wales NHS Trust, Cardiff, UK), Simon Cottrell (Public Health Wales NHS Trust, Cardiff, UK), Noel Craine (Public Health Wales NHS Trust, Cardiff, UK), Michelle Cronin (Public Health Wales NHS Trust, Cardiff, UK), Alisha Davies (Public Health Wales NHS Trust, Cardiff, UK), Elen De Lacy (Public Health Wales NHS Trust, Cardiff, UK), Fatima Downing (Public Health Wales NHS Trust, Cardiff, UK), Sue Edwards (Public Health Wales NHS Trust, Cardiff, UK), Johnathan M. Evans (Public Health Wales NHS Trust, Cardiff, UK), Laia Fina (Public Health Wales NHS Trust, Cardiff, UK), Amy Gaskin (Public Health Wales NHS Trust, Cardiff, UK), Bree Gatica-Wilcox (Public Health Wales NHS Trust, Cardiff, UK), Laura Gifford (Public Health Wales NHS Trust, Cardiff, UK), Lauren Gilbert (Public Health Wales NHS Trust, Cardiff, UK), Lee Graham (Public Health Wales NHS Trust, Cardiff, UK), David Heyburn (Public Health Wales NHS Trust, Cardiff, UK), Ember Hilvers (Public Health Wales NHS Trust, Cardiff, UK), Robin Howe (Public Health Wales NHS Trust, Cardiff, UK), Hannah Jones (Public Health Wales NHS Trust, Cardiff, UK), Rachel Jones (Public Health Wales NHS Trust, Cardiff, UK), Sophie Jones (Public Health Wales NHS Trust, Cardiff, UK), Sara Kumziene-SummerhaYes (Public Health Wales NHS Trust, Cardiff, UK), Caoimhe McKerr (Public Health Wales NHS Trust, Cardiff, UK), Catherine Moore (Public Health Wales NHS Trust, Cardiff, UK), Mari Morgan (Public Health Wales NHS Trust, Cardiff, UK), Nicole Pacchiarini (Public Health Wales NHS Trust, Cardiff, UK), Malorie Perry (Public Health Wales NHS Trust, Cardiff, UK), Amy Plimmer (Public Health Wales NHS Trust, Cardiff, UK), Sara Rey (Public Health Wales NHS Trust, Cardiff, UK), Giri Shankar (Public Health Wales NHS Trust, Cardiff, UK), Sarah Taylor (Public Health Wales NHS Trust, Cardiff, UK), Joanne Watkins (Public Health Wales NHS Trust, Cardiff, UK), Chris Williams (Public Health Wales NHS Trust, Cardiff, UK), Anna Casey (Queen Elizabeth Hospital, London, UK), Liz Ratcliffe (Queen Elizabeth Hospital, London, UK), Erwan Acheson (Queen's University Belfast, Belfast, UK), Zoltan Molnar (Queen's University Belfast, Belfast, UK), David A. Simpson (Queen's University Belfast, Belfast, UK), Thomas Thompson (Queen's University Belfast, Belfast, UK), Cressida Auckland (Royal Devon and Exeter NHS Foundation Trust, Exeter, UK), Sian Ellard (Royal Devon and Exeter NHS Foundation Trust, Exeter, UK), Christopher R. Jones (Royal Devon and Exeter NHS Foundation Trust, Exeter, UK), Bridget A. Knight (Royal Devon and Exeter NHS Foundation Trust, Exeter, UK), Jane A. H. Masoli (Royal Devon and Exeter NHS Foundation Trust, Exeter, UK), Tanzina Haque (Royal Free NHS Trust, London, UK), Jennifer Hart (Royal Free NHS Trust, London, UK), Dianne Irish-Tavares (Royal Free NHS Trust, London, UK), Tabitha W. Mahungu (Royal Free NHS Trust, London, UK), Eric Witele (Royal Free NHS Trust, London, UK), Ashok Dadrah (Sandwell and West Birmingham NHS Trust, Birmingham, UK), Melisa L. Fenton (Sandwell and West Birmingham NHS Trust, Birmingham, UK), Tranprit Saluja (Sandwell and West Birmingham NHS Trust, Birmingham, UK), Amanda Symmonds (Sandwell and West Birmingham NHS Trust, Birmingham, UK), Yann Bourgeois (School of Biological Sciences, University of Portsmouth (PORT), Portsmouth, UK), Garry P. Scarlett (School of Biological Sciences, University of Portsmouth (PORT), Portsmouth, UK), Kate Cook (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Hannah Dent (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Christopher Fearn (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Salman Goudarzi (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Katie F. Loveson (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Hannah Paul (School of Pharmacy and Biomedical Sciences, University of Portsmouth (PORT), Portsmouth, UK), Cariad Evans (Sheffield Teaching Hospitals, Sheffield, UK), Kate Johnson (Sheffield Teaching Hospitals, Sheffield, UK), David G. Partridge (Sheffield Teaching Hospitals, Sheffield, UK), Mohammad Raza (Sheffield Teaching Hospitals, Sheffield, UK), Paul Baker (South Tees Hospitals NHS Foundation Trust, Middlesbrough, UK), Stephen Bonner (South Tees Hospitals NHS Foundation Trust, Middlesbrough, UK), Sarah Essex (South Tees Hospitals NHS Foundation Trust, Middlesbrough, UK), Steven Liggett (South Tees Hospitals NHS Foundation Trust, Middlesbrough, UK), Ronan A. Lyons (Swansea University, Swansea, UK), Adhyana I. K. Mahanama (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), Kordo Saeed (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), Buddhini Samaraweera (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), Siona Silveira (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), Eleri Wilson-Davies (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), P. Emanuela (University Hospitals Southampton NHS Foundation Trust, Southampton, UK), Nadua Bayzid (University College London, London, UK), Marius Cotic (University College London, London, UK), Leah Ensell (University College London, London, UK), John A. Hartley (University College London, London, UK), Riaz Jannoo (University College London, London, UK), Angeliki Karamani (University College London, London, UK), Mark Kristiansen (University College London, London, UK), Helen L. Lowe (University College London, London, UK), Sunando Roy (University College London, London, UK), Adam P. Westhorpe (University College London, London, UK), Rachel J. Williams (University College London, London, UK), Charlotte A. Williams (University College London, London, UK), Sarah Jeremiah (University Hospital Southampton NHS Foundation Trust, Southampton, UK), Jacqui A. Prieto (University Hospital Southampton NHS Foundation Trust, Southampton, UK), Lisa Berry (University Hospitals Coventry and Warwickshire, Coventry, UK), Dimitris Grammatopoulos (University Hospitals Coventry and Warwickshire, Coventry, UK), Katie Jones (University Hospitals Coventry and Warwickshire, Coventry, UK), Sarojini Pandey (University Hospitals Coventry and Warwickshire, Coventry, UK), Andrew Beggs (University of Birmingham, Birmingham, UK), Alex Richter (University of Birmingham, Birmingham, UK), Fiona Ashford (University of Birmingham Turnkey Laboratory, Birmingham, UK), Angus Best (University of Birmingham Turnkey Laboratory, Birmingham, UK), Liam Crawford (University of Birmingham Turnkey Laboratory, Birmingham, UK), Nicola Cumley (University of Birmingham Turnkey Laboratory, Birmingham, UK), Megan Mayhew (University of Birmingham Turnkey Laboratory, Birmingham, UK), Oliver Megram (University of Birmingham Turnkey Laboratory, Birmingham, UK), Jeremy Mirza (University of Birmingham Turnkey Laboratory, Birmingham, UK), Emma Moles-Garcia (University of Birmingham Turnkey Laboratory, Birmingham, UK), Benita Percival (University of Birmingham Turnkey Laboratory, Birmingham, UK), Giselda Bucca (University of Brighton, Brighton, UK), Andrew R. Hesketh (University of Brighton, Brighton, UK), Colin P. Smith (University of Brighton, Brighton, UK), Rose K. Davidson (University of East Anglia, Norwich, UK), Carlos E. Balcazar (University of Edinburgh, Edinburgh, UK), Michael D. Gallagher (University of Edinburgh, Edinburgh, UK), Áine O'Toole (University of Edinburgh, Edinburgh, UK), Andrew Rambaut (University of Edinburgh, Edinburgh, UK), Stefan Rooke (University of Edinburgh, Edinburgh, UK), Thomas D. Stanton (University of Edinburgh, Edinburgh, UK), Thomas Williams (University of Edinburgh, Edinburgh, UK), Kathleen A. Williamson (University of Edinburgh, Edinburgh, UK), Claire M. Bewshea (University of Exeter, Exeter, UK), Audrey Farbos (University of Exeter, Exeter, UK), James W. Harrison (University of Exeter, Exeter, UK), Aaron R. Jeffries (University of Exeter, Exeter, UK), Robin Manley (University of Exeter, Exeter, UK), Stephen L. Michell (University of Exeter, Exeter, UK), Michelle L. Michelsen (University of Exeter, Exeter, UK), Christine M. Sambles (University of Exeter, Exeter, UK), David J. Studholme (University of Exeter, Exeter, UK), Ben Temperton (University of Exeter, Exeter, UK), Joanna Warwick-Dugdale (University of Exeter, Exeter, UK), Alistair C. Darby (University of Liverpool, Liverpool, UK), Richard Eccles (University of Liverpool, Liverpool, UK), Matthew Gemmell (University of Liverpool, Liverpool, UK), Richard Gregory (University of Liverpool, Liverpool, UK), Sam T. Haldenby (University of Liverpool, Liverpool, UK), Julian A. Hiscox (University of Liverpool, Liverpool, UK), Margaret Hughes (University of Liverpool, Liverpool, UK), Miren Iturriza-Gomara (University of Liverpool, Liverpool, UK), Kathryn A. Jackson (University of Liverpool, Liverpool, UK), Anita O. Lucaci (University of Liverpool, Liverpool, UK), Charlotte Nelson (University of Liverpool, Liverpool, UK), Steve Paterson (University of Liverpool, Liverpool, UK), Lucille Rainbow (University of Liverpool, Liverpool, UK), Lance Turtle (University of Liverpool, Liverpool, UK), Edith E. Vamos (University of Liverpool, Liverpool, UK), Hermione J. Webster (University of Liverpool, Liverpool, UK), Mark Whitehead (University of Liverpool, Liverpool, UK), Claudia Wierzbicki (University of Liverpool, Liverpool, UK), Adrienn Angyal (University of Sheffield, Sheffield, UK), Rebecca Brown (University of Sheffield, Sheffield, UK), Thushan I. de Silva (University of Sheffield, Sheffield, UK), Timothy M. Freeman (University of Sheffield, Sheffield, UK), Marta Gallis (University of Sheffield, Sheffield, UK), Luke R. Green (University of Sheffield, Sheffield, UK), Danielle C. Groves (University of Sheffield, Sheffield, UK), Alexander J. Keeley (University of Sheffield, Sheffield, UK), Benjamin B. Lindsey (University of Sheffield, Sheffield, UK), Stavroula F. Louka (University of Sheffield, Sheffield, UK), Matthew D. Parker (University of Sheffield, Sheffield, UK), Paul J. Parsons (University of Sheffield, Sheffield, UK), Nikki Smith (University of Sheffield, Sheffield, UK), Rachel M. Tucker (University of Sheffield, Sheffield, UK), Dennis Wang (University of Sheffield, Sheffield, UK), Max Whiteley (University of Sheffield, Sheffield, UK), Matthew Wyles (University of Sheffield, Sheffield, UK), Peijun Zhang (University of Sheffield, Sheffield, UK), Mohammad T. Alam (University of Warwick, Warwick, UK), Laura Baxter (University of Warwick, Warwick, UK), Hannah E. Bridgewater (University of Warwick, Warwick, UK), Paul E. Brown (University of Warwick, Warwick, UK), Jeffrey K. J. Cheng (University of Warwick, Warwick, UK), Chrystala Constantinidou (University of Warwick, Warwick, UK), Lucy R. Frost (University of Warwick, Warwick, UK), Sascha Ott (University of Warwick, Warwick, UK), Richard Stark (University of Warwick, Warwick, UK), Grace Taylor-Joyce (University of Warwick, Warwick, UK), Meera Unnikrishnan (University of Warwick, Warwick, UK), Alberto C. Cerda (Viapath, Guy's and St Thomas' NHS Foundation Trust, and King's College Hospital NHS Foundation Trust, London, UK), Tammy V. Merrill (Viapath, Guy's and St Thomas' NHS Foundation Trust, and King's College Hospital NHS Foundation Trust, London, UK), Rebekah E. Wilson (Viapath, Guy's and St Thomas' NHS Foundation Trust, and King's College Hospital NHS Foundation Trust, London, UK), Jonathan Ball (Virology, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Joseph G. Chappell (Virology, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Patrick C. McClure (Virology, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), Theocharis Tsoleridis (Virology, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK), David Buck (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Mariateresa de Cesare (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Angie Green (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), George MacIntyre-Cockett (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), John A. Todd (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Amy Trebes (Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK), Rory N. Gunson (West of Scotland Specialist Virology Centre, NHS Greater Glasgow and Clyde, Glasgow, UK).
Footnotes
Supplemental material is available online only.
Contributor Information
Alan McNally, Email: A.McNally.1@bham.ac.uk.
Randall Hayden, St. Jude Children's Research Hospital.
The COVID-19 Genomics UK (COG-UK) Consortium,:
Dinesh Aggarwal, Beth Blane, Ellena Brooks, Alessandro M. Carabelli, Carol M. Churcher, Katerina Galai, Sophia T. Girgis, Ravi K. Gupta, Nazreen F. Hadjirin, Danielle Leek, Catherine Ludden, Georgina M. McManus, Sophie Palmer, Sharon J. Peacock, Kim S. Smith, Elias Allara, David Bibby, Chloe Bishop, Andrew Bosworth, Daniel Bradshaw, Vicki Chalker, Meera Chand, Gavin Dabrera, Nicholas Ellaby, Eileen Gallagher, Natalie Groves, Ian Harrison, Hassan Hartman, Richard Hopes, Jonathan Hubb, Stephanie Hutchings, Angie Lackenby, Juan Ledesma, David Lee, Nikos Manesis, Carmen Manso, Tamyo Mbisa, Shahjahan Miah, Peter Muir, Richard Myers, Husam Osman, Vineet Patel, Clare Pearson, Steven Platt, Hannah M. Pymont, Mary Ramsay, Esther Robinson, Ulf Schaefer, Alicia Thornton, Katherine A. Twohig, Ian B. Vipond, David Williams, William L. Hamilton, Ben Warne, Louise Aigrain, Alex Alderton, Roberto Amato, Cristina V. Ariani, Jeff Barrett, Andrew R. Bassett, Mathew A. Beale, Charlotte Beaver, Katherine L. Bellis, Emma Betteridge, James Bonfield, Iraad F. Bronner, Michael H. S. Chapman, John Danesh, Robert Davies, Matthew J. Dorman, Eleanor Drury, Jillian Durham, Ben W. Farr, Luke Foulser, Sonia Goncalves, Scott Goodwin, Marina Gourtovaia, Ewan M. Harrison, David K. Jackson, Keith James, Dorota Jamrozy, Ian Johnston, Leanne Kane, Sally Kay, Jon-Paul Keatley, Dominic Kwiatkowski, Cordelia F. Langford, Mara Lawniczak, Stefanie V. Lensing, Steven Leonard, Laura Letchford, Kevin Lewis, Jennifier Liddle, Rich Livett, Stephanie Lo, Alex Makunin, Inigo Martincorena, Shane McCarthy, Samantha McGuigan, Robin J. Moll, Rachel Nelson, Karen Oliver, Steve Palmer, Naomi R. Park, Minal Patel, Liam Prestwood, Christoph Puethe, Michael A. Quail, Diana Rajan, Shavanthi Rajatileka, Nicholas M. Redshaw, Carol Scott, Lesley Shirley, John Sillitoe, Scott A. J. Thurston, Gerry Tonkin-Hill, Jaime M. Tovar-Corona, Danni Weldon, Andrew Whitwham, Yasmin Chaudhry, Iliana Georgana, Ian G. Goodfellow, Grant Hall, Myra Hosmillo, Aminu S. Jahun, Malte L. Pinckert, Stephen W. Attwood, Louis du Plessis, Marina Escalera Zamudio, Sarah Francois, Bernardo Gutierrez, Moritz U. G. Kraemer, Oliver G. Pybus, Jayna Raghwani, Tetyana I. Vasylyeva, Alex E. Zarebski, Verity Hill, Nabil-Fareed Alikhan, Alp Aydin, David J. Baker, Leonardo de Oliveira Martins, Gemma L. Kay, Thanh Le-Viet, Alison E. Mather, Lizzie Meadows, Justin O’Grady, Andrew J. Page, Steven Rudder, Alexander J. Trotter, Chris J. Illingworth, Chris Jackson, Elihu Aranday-Cortes, Patawee Asamaphan, Alice Broos, Stephen N. Carmichael, Ana da Silva Filipe, Joseph Hughes, Natasha G. Jesudason, Natasha Johnson, Kathy K. Li, Daniel Mair, Jenna Nichols, Seema Nickbakhsh, Marc O. Niebel, Kyriaki Nomikou, Richard J. Orton, David L. Robertson, Rajiv N. Shah, James G. Shepherd, Joshua B. Singer, Igor Starinskij, Emma C. Thomson, Lily Tong, Sreenu Vattipally, Claire Cormie, Joana Dias, Sally Forrest, Harmeet K. Gill, Ellen E. Higginson, Leanne M. Kermack, Mailis Maes, Chris Ruis, Sushmita Sridhar, Jamie Young, Amy Ash, Cherian Koshy, Nick Cortes, Stephen Kidd, Jessica Lynch, Nathan Moore, Matilde Mori, Emma Wise, Tanya Curran, Derek J. Fairley, James P. McKenna, Helen Adams, David Bonsall, Christophe Fraser, Tanya Golubchik, Benjamin J. Cogger, Mohammed O. Hassan-Ibrahim, Cassandra S. Malone, Nicola Reynolds, Michelle Wantoch, Safiah Afifi, Robert Beer, Michaela John, Joshua Maksimovic, Kathryn McCluggage, Sian Morgan, Karla Spellman, Catherine Bresner, Thomas R. Connor, William Fuller, Martyn Guest, Huw Gulliver, Christine Kitchen, Angela Marchbank, Ian Merrick, Robert Munn, Anna Price, Joel Southgate, Trudy Workman, Amita Patel, Luke B. Snell, Rahul Batra, Themoula Charalampous, Jonathan Edgeworth, Gaia Nebbia, Angela H. Beckett, Khalil Abudahab, Mirko Menegazzo, Ben E. W. Taylor, Anthony P. Underwood, Corin A. Yeats, Louise Berry, Tim Boswell, Gemma Clark, Vicki M. Fleming, Hannah C. Howson-Wells, Carl Jones, Amelia Joseph, Manjinder Khakh, Michelle M. Lister, Wendy Smith, Iona Willingham, Paul Bird, Karlie Fallon, Thomas Helmer, Christopher Holmes, Julian Tang, Victoria Blakey, Sharon Campbell, Veena Raviprakash, Nicola Sheriff, Lesley-Anne Williams, Matthew Carlile, Johnny Debebe, Nadine Holmes, Matthew W. Loose, Christopher Moore, Fei Sang, Victoria Wright, Francesc Coll, Gilberto Betancor, Adrian W. Signell, Harry D. Wilson, Thomas Davis, Sahar Eldirdiri, Anita Kenyon, M. Estee Torok, Hannah Lowe, Samuel Moses, Luke Bedford, Jonathan Moore, Susanne Stonehouse, Ali R. Awan, Chloe L. Fisher, John BoYes, Laura Atkinson, Judith Breuer, Julianne R. Brown, Kathryn A. Harris, Jack C. D. Lee, Divya Shah, Nathaniel Storey, Flavia Flaviani, Adela Alcolea-Medina, Gabrielle Vernet, Rebecca Williams, Michael R. Chapman, Wendy Chatterton, Judith Heaney, Lisa J. Levett, Monika Pusok, Li Xu-McCrae, Matthew Bashton, Darren L. Smith, Gregory R. Young, Frances Bolt, Alison Cox, Alison Holmes, Pinglawathee Madona, Siddharth Mookerjee, James Price, Paul A. Randell, Olivia Boyd, Fabricia F. Nascimento, Lily Geidelberg, Rob Johnson, David Jorgensen, Manon Ragonnet-Cronin, Aileen Rowan, Igor Siveroni, Graham P. Taylor, Erik M. Volz, Katherine L. Smollett, Nicholas J. Loman, Claire McMurray, Alan McNally, Sam Nicholls, Radoslaw Poplawski, Joshua Quick, Will Rowe, Joanne Stockton, Rocio T. Martinez Nunez, Cassie Breen, Angela Cowell, Jenifer Mason, Elaine O’Toole, Trevor I. Robinson, Joanne Watts, Graciela Sluga, Shazaad S. Y. Ahmad, Ryan P. George, Nicholas W. Machin, Fenella Halstead, Wendy Hogsden, Venkat Sivaprakasam, Holli Carden, Antony D. Hale, Katherine L. Harper, Louissa R. Macfarlane-Smith, Shirelle Burton-Fanning, Jennifer Collins, Gary Eltringham, Brendan AI. Payne, Yusri Taha, Sheila Waugh, Sarah O’Brien, Steven Rushton, Rachel Blacow, Amanda Bradley, Alasdair Maclean, Guy Mollett, Rebecca Dewar, Martin P. McHugh, Kate E. Templeton, Elizabeth Wastenge, Lindsay Coupland, Samir Dervisevic, Emma J. Meader, Rachael Stanley, Louise Smith, Edward Barton, Clive Graham, Debra Padgett, Garren Scott, Jane Greenaway, Emma Swindells, Clare M. McCann, Andrew Nelson, Wen C. Yew, Monique Andersson, Derrick Crook, David Eyre, Anita Justice, Timothy Peto, Nichola Duckworth, Tim J. Sloan, Sarah Walsh, Kelly Bicknell, Anoop J. Chauhan, Scott Elliott, Sharon Glaysher, Robert Impey, Allyson Lloyd, Sarah Wyllie, Nick Levene, Lynn Monaghan, Declan T. Bradley, Tim Wyatt, Martin D. Curran, Surendra Parmar, Matthew T. G. Holden, Sharif Shaaban, Alexander Adams, Hibo Asad, Alec Birchley, Matthew Bull, Jason Coombes, Sally Corden, Simon Cottrell, Noel Craine, Michelle Cronin, Alisha Davies, Elen De Lacy, Fatima Downing, Sue Edwards, Johnathan M. Evans, Laia Fina, Amy Gaskin, Bree Gatica-Wilcox, Laura Gifford, Lauren Gilbert, Lee Graham, David Heyburn, Ember Hilvers, Robin Howe, Hannah Jones, Rachel Jones, Sophie Jones, Sara Kumziene-SummerhaYes, Caoimhe McKerr, Catherine Moore, Mari Morgan, Nicole Pacchiarini, Malorie Perry, Amy Plimmer, Sara Rey, Giri Shankar, Sarah Taylor, Joanne Watkins, Chris Williams, Anna Casey, Liz Ratcliffe, Erwan Acheson, Zoltan Molnar, David A. Simpson, Thomas Thompson, Cressida Auckland, Sian Ellard, Christopher R. Jones, Bridget A. Knight, Jane A. H. Masoli, Tanzina Haque, Jennifer Hart, Dianne Irish-Tavares, Tabitha W. Mahungu, Eric Witele, Ashok Dadrah, Melisa L. Fenton, Tranprit Saluja, Amanda Symmonds, Yann Bourgeois, Kate Johnson, David G. Partridge, Mohammad Raza, Paul Baker, Stephen Bonner, Sarah Essex, Steven Liggett, Ronan A. Lyons, Adhyana I. K. Mahanama, Kordo Saeed, Buddhini Samaraweera, Siona Silveira, Eleri Wilson-Davies, P. Emanuela, Nadua Bayzid, Marius Cotic, Leah Ensell, John A. Hartley, Riaz Jannoo, Angeliki Karamani, Mark Kristiansen, Helen L. Lowe, Sunando Roy, Adam P. Westhorpe, Rachel J. Williams, Charlotte A. Williams, Sarah Jeremiah, Jacqui A. Prieto, Lisa Berry, Dimitris Grammatopoulos, Katie Jones, Sarojini Pandey, Andrew Beggs, Alex Richter, Fiona Ashford, Angus Best, Liam Crawford, Nicola Cumley, Megan Mayhew, Oliver Megram, Jeremy Mirza, Emma Moles-Garcia, Benita Percival, Giselda Bucca, Andrew R. Hesketh, Colin P. Smith, Rose K. Davidson, Carlos E. Balcazar, Michael D. Gallagher, Áine O’Toole, Andrew Rambaut, Stefan Rooke, Thomas D. Stanton, Thomas Williams, Kathleen A. Williamson, Claire M. Bewshea, Audrey Farbos, James W. Harrison, Aaron R. Jeffries, Robin Manley, Stephen L. Michell, Michelle L. Michelsen, Christine M. Sambles, David J. Studholme, Ben Temperton, Joanna Warwick-Dugdale, Alistair C. Darby, Richard Eccles, Matthew Gemmell, Richard Gregory, Sam T. Haldenby, Julian A. Hiscox, Margaret Hughes, Miren Iturriza-Gomara, Kathryn A. Jackson, Anita O. Lucaci, Charlotte Nelson, Steve Paterson, Lucille Rainbow, Lance Turtle, Edith E. Vamos, Hermione J. Webster, Mark Whitehead, Claudia Wierzbicki, Adrienn Angyal, Rebecca Brown, Thushan I. de Silva, Timothy M. Freeman, Marta Gallis, Luke R. Green, Danielle C. Groves, Alexander J. Keeley, Benjamin B. Lindsey, Stavroula F. Louka, Matthew D. Parker, Paul J. Parsons, Nikki Smith, Rachel M. Tucker, Dennis Wang, Max Whiteley, Matthew Wyles, Peijun Zhang, Mohammad T. Alam, Laura Baxter, Hannah E. Bridgewater, Paul E. Brown, Jeffrey K. J. Cheng, Chrystala Constantinidou, Lucy R. Frost, Sascha Ott, Richard Stark, Grace Taylor-Joyce, Meera Unnikrishnan, Alberto C. Cerda, Tammy V. Merrill, Rebekah E. Wilson, Jonathan Ball, Joseph G. Chappell, Patrick C. McClure, Theocharis Tsoleridis, David Buck, Mariateresa de Cesare, Angie Green, George MacIntyre-Cockett, John A. Todd, Amy Trebes, and Rory N. Gunson
REFERENCES
- 1.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, COVID-19 Genomics UK (COG-UK) consortium, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. 2021. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593:266–269. 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 3.Mishra S, Mindermann S, Sharma M, Whittaker C, Mellan TA, Wilton T, Klapsa D, Mate R, Fritzsche M, Zambon M, Ahuja J, Howes A, Miscouridou X, Nason GP, Ratmann O, Semenova E, Leech G, Sandkühler JF, Rogers-Smith C, Vollmer M, Unwin HJT, Gal Y, Chand M, Gandy A, Martin J, Volz E, Ferguson NM, Bhatt S, Brauner JM, Flaxman S, COVID-19 Genomics UK (COG-UK) Consortium. 2021. Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. EClinicalMedicine 39:101064. 10.1016/j.eclinm.2021.101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNally A. 2021. What makes new variants of SARS-CoV-2 concerning is not where they come from, but the mutations they contain. BMJ 372:n504. 10.1136/bmj.n504. [DOI] [PubMed] [Google Scholar]
- 5.Public Health England. 2020. Investigation of SARS-CoV-2 variants of concern: technical briefings. Public Health England, London, United Kingdom. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201.
- 6.Richter A, Plant T, Kidd M, Bosworth A, Mayhew M, Megram O, Ashworth F, Crawford L, White T, Moles-Garcia E, Mirza J, Percival B, McNally A. 2020. How to establish an academic SARS-CoV-2 testing laboratory. Nat Microbiol 5:1452–1454. 10.1038/s41564-020-00818-3. [DOI] [PubMed] [Google Scholar]
- 7.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E, on behalf of COVID-19 Genomics Consortium UK (CoG-UK). 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563/1.
- 8.Meng B, Kemp SA, Papa G, Datir R, Ferreira IATM, Marelli S, Harvey WT, Lytras S, Mohamed A, Gallo G, Thakur N, Collier DA, Mlcochova P, Duncan LM, Carabelli AM, Kenyon JC, Lever AM, De Marco A, Saliba C, Culap K, Cameroni E, Matheson NJ, Piccoli L, Corti D, James LC, Robertson DL, Bailey D, Gupta RK, COVID-19 Genomics UK (COG-UK) Consortium. 2021. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep 35:109292. 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Public Health England. 2021. SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 10. Public Health England, London, United Kingdom. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/984274/Variants_of_Concern_VOC_Technical_Briefing_10_England.pdf.
- 10.COVID-19 Genomics UK (COG-UK). 2020. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe 1:e99–e100. 10.1016/S2666-5247(20)30054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ThermoFisher Scientific. 2022. MagMAX Viral/Pathogen II nucleic acid isolation kit. ThermoFisher Scientific, Waltham, MA. https://www.thermofisher.com/order/catalog/product/A48383.
- 12.ThermoFisher Scientific. 2022. ThermoFisher TaqPath COVID-19 RT-PCR testing kit. ThermoFisher Scientific, Waltham, MA. https://www.thermofisher.com/order/catalog/product/A48067.
- 13.ThermoFisher Scientific. 2021. ThermoFisher TaqMan SARS-CoV-2 mutation panel user guide. ThermoFisher Scientific, Waltham, MA. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0024768_TaqManSARS-CoV-2_MutationPanel_UG.pdf.
- 14.Quick J. 2020. nCoV-2019 sequencing protocol v3 (LoCost). Protocols.io. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye.
- 15.ARTIC Network. 2022. ARTIC Nanopore protocol for nCoV2019 novel coronavirus. GitHub. https://github.com/artic-network/artic-ncov2019.
- 16.Loman N, Rowe W, Rambaut A. 2020. nCoV-2019 novel coronavirus bioinformatics protocol. https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html.
- 17.Kidd M, Richter A, Best A, Cumley N, Mirza J, Percival B, Mayhew M, Megram O, Ashford F, White T, Moles-Garcia E, Crawford L, Bosworth A, Atabani SF, Plant T, McNally A. 2021. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J Infect Dis 223:1666–1670. 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, Ouédraogo N, Afrough B, Bah A, Baum JH, Becker-Ziaja B, Boettcher J-P, Cabeza-Cabrerizo M, Camino-Sanchez A, Carter LL, Doerrbecker J, Enkirch T, Dorival IGG, Hetzelt N, Hinzmann J, Holm T, Kafetzopoulou LE, Koropogui M, Kosgey A, Kuisma E, Logue CH, Mazzarelli A, Meisel S, Mertens M, Michel J, Ngabo D, Nitzsche K, Pallash E, Patrono LV, Portmann J, Repits JG, Rickett NY, Sachse A, Singethan K, Vitoriano I, Yemanaberhan RL, Zekeng EG, Trina R, Bello A, Sall AA, Faye O, Faye O, Magassouba N, Williams CV, Amburgey V, Winona L, Davis E, Gerlach J, Washington F, Monteil V, Jourdain M, Bererd M, Camara A, Somlare H, Camara A, Gerard M, Bado G, Baillet B, Delaune D, Nebie KY, Diarra A, Savane Y, Pallawo RB, Gutierrez GJ, Milhano N, Roger I, Williams CJ, Yattara F, Lewandowski K, Taylor J, Rachwal P, Turner D, Pollakis G, Hiscox JA, Matthews DA, O’Shea MK, Johnston AM, Wilson D, Hutley E, Smit E, Di Caro A, Woelfel R, Stoecker K, Fleischmann E, Gabriel M, Weller SA, Koivogui L, Diallo B, Keita S, Rambaut A, Formenty P, Gunther S, Carroll MW. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232. 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan K, Devanga Ragupathi NK, Jacob JJ, Veeraraghavan B. 2020. Highly accurate-single chromosomal complete genomes using IonTorrent and MinION sequencing of clinical pathogens. Genomics 112:545–551. 10.1016/j.ygeno.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Rang FJ, Kloosterman WP, de Ridder J. 2018. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol 19:90. 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 20:8. 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download jcm.02408-21-s0001.xlsx, XLSX file, 0.03 MB (32.5KB, xlsx)