Summary
Despite the critical regulatory function of promoter-proximal pausing, the influence of pausing kinetics on transcriptional control remains an active area of investigation. Here, we present Start-TimeLapse-seq (STL-seq), a method that captures the genome-wide kinetics of short, capped RNA turnover and reveals principles of regulation at the pause site. By measuring the rates of release into elongation and premature termination through inhibition of pause release, we determine that pause-release rates are highly variable and most promoter-proximal paused RNA Polymerase II molecules prematurely terminate (~80%). The preferred regulatory mechanism upon a hormonal stimulus (20-hydroxyecdysone) is to influence pause-release rather than termination rates. Transcriptional shutdown occurs concurrently with induction of promoter-proximal termination under hyperosmotic stress but paused transcripts from TATA box-containing promoters remain stable, demonstrating an important role for cis-acting DNA elements in pausing. STL-seq dissects the kinetics of pause release and termination, providing an opportunity to identify mechanisms of transcriptional regulation.
Keywords: STL-seq, scRNA, TimeLapse, promoter-proximal pausing, release into elongation, premature termination
In Brief
Zimmer et al. report an RNA sequencing method, STL-seq, which captures steady-state dynamics of promoter-proximal pausing. The authors use STL-seq to dissect the effects of release into elongation and premature termination on Pol II pausing dynamics and to reveal the principles of regulation at the pause site.
Introduction
Promoter-proximal pausing is a dynamic step in transcription that occurs at most RNA polymerase II (Pol II)-transcribed genes in metazoans and is an important point of regulatory input controlling gene expression (Guenther et al., 2007; Core and Adelman, 2019). Promoter-proximal pausing is the process by which Pol II stalls 20–60 bp downstream of the transcription start site (TSS), forming a stable complex engaged on chromatin with a short nascent transcript (Fraser et al., 1978; Gariglio et al., 1981; Gilmour and Lis, 1986; Adelman and Lis, 2012; Vos et al., 2018). To proceed through pausing and synthesize a full-length transcript, Pol II must be released into elongation, a step promoted by the kinase activity of positive transcription elongation factor b (P-TEFb, Marshall and Price, 1995; Wada et al., 1998). Several studies, however, have demonstrated that not all paused Pol II molecules are released into elongation and some prematurely terminate through eviction from the DNA and rapid degradation of the nascent transcript (Brannan et al., 2012; Henriques et al., 2013; Buckley et al., 2014; Nilson et al., 2017; Steurer et al., 2018; Elrod et al., 2019; Tatomer et al., 2019; Beckedorff et al., 2020; Huang et al., 2020). Pause release and termination are in kinetic competition and determine the fate of paused Pol II; changing the rate of either can regulate gene expression.
The pause site represents a major node of regulatory input but how pause release and termination respond to regulatory signals genome-wide remains unclear (Henriques et al., 2013; Williams et al., 2015; Nilson et al., 2017; Bartman et al., 2019). While nascent RNA sequencing can reveal an increase in the number of Pol II molecules released into elongation, it remains ambiguous whether such observations are due to the distinct biochemical activities of increasing the pause-release rate or of decreasing the premature-termination rate. In addition, measuring premature termination is challenging because terminated transcripts are rapidly degraded and thus difficult to directly observe. These obstacles limit our understanding of pausing regulation and prompt a more systematic analysis of pausing dynamics that includes determining the rates of release into elongation and premature termination and the regulation of each.
Conventional RNA-seq experiments do not robustly capture the short transcripts associated with paused Pol II and therefore are poorly suited to study pausing. However, an RNA sequencing-based method, Start-seq (Nechaev et al., 2010), specifically enriches short, capped RNA transcripts (scRNA) associated with paused Pol II such that each read represents a single engaged Pol II molecule paused at the promoter-proximal site. While Start-seq has provided important insights into steady-state levels of paused RNA Pol II (Williams et al., 2015; Henriques et al., 2018), analyzing the dynamics and turnover of these paused transcripts has proven more challenging. Thus, many questions about pausing kinetics remain unanswered, including the fraction of Pol II molecules that are released into elongation from the pause site.
Previous studies estimated paused Pol II half-lives by blocking initiation of new transcripts using triptolide (Trp), an inhibitor of TFIIH helicase activity (Henriques et al., 2013; Jonkers et al., 2014; Krebs et al., 2017; Shao and Zeitlinger, 2017; Erickson et al., 2018; Tettey et al., 2019). Yet kinetics upon Trp inhibition may not be reflective of kinetics of the uninhibited state, making these estimates unreliable (Nilson et al., 2017; Erickson et al., 2018; Dienemann et al., 2019). Efforts to estimate half-lives of paused Pol II in a Trp-independent manner have been performed by integrating information from multiple nascent RNA-seq methods but require the assumption that premature termination occurs rarely (Gressel et al., 2017; Jaeger et al., 2020). To our knowledge, these are the only two strategies applied to study paused Pol II behavior in a genome-wide and TSS-specific manner.
We sought to develop an approach that focuses on short nascent transcripts with the specificity of Start-seq and also captures the dynamics of RNA transcripts using RNA metabolic labeling and nucleotide recoding (TimeLapse-seq, Schofield et al., 2018). Here we present Start-TimeLapse-seq (STL-seq), which measures steady-state kinetics of paused Pol II genome-wide without blocking transcription initiation. We apply STL-seq to fly and human cells and find very similar half-lives of paused Pol II in both systems. We demonstrate that STL-seq, when combined with P-TEFb inhibition, allows deconstruction of Pol II turnover into components of premature termination and release into elongation. We find that Pol II prematurely terminates at a similar rate at nearly all promoter-proximal pause sites. While release into elongation is infrequent when compared to termination, it is highly variable across the genome and is the primary target of regulation in response to hormonal stimulus by 20-hydroxyecdysone treatment in Drosophila. On the other hand, termination is largely unaffected by the stimulus but is induced upon hyperosmotic stress. Our work provides the first direct, global measurements of pausing dynamics using non-perturbing methods and supports a model in which release into elongation regulates expression levels while premature termination functions as a quality control mechanism to ensure competent elongation.
Results
Short, capped transcripts can be metabolically labeled using s4U
We sought to develop a method that directly measures the steady-state kinetics of Pol II pausing. We were inspired by Start-seq, which enriches for the short, capped RNA (scRNA) associated with the paused complex (Nechaev et al., 2010). While Start-seq does not inherently capture transcript dynamics, we reasoned that if we could combine it with TimeLapse-seq, an enrichment-free method capable of capturing transcriptional dynamics, we could distinguish newly synthesized and preexisting scRNA through 4-thiouridine (s4U) metabolic labeling. The fraction of scRNA synthesized during labeling can be revealed by chemically converting s4U to a cytidine analogue which manifests as an apparent T-to-C mutation in sequencing data (Schofield et al., 2018). Start-TimeLapse-seq (STL-seq) therefore combines the power of metabolic labeling with the specificity of scRNA enrichment to reveal dynamics of promoter-proximal Pol II pausing.
We treated D. melanogaster S2 cells with s4U for 5 mins (Figure 1A), a time well validated for studying transient transcripts using other s4U-based methods (Schwalb et al., 2016; Schofield et al., 2018) and generally in line with previous pause duration estimates (Core and Adelman, 2019). We found that s4U-treated samples, but not controls, were enriched for TimeLapse-dependent T-to-C mutations (Figure 1B). Use of an alignment strategy that does not penalize T-to-C mismatches improved mapping of STL-seq reads, particularly shorter reads with two or more T-to-C mutations, while maintaining low background mutations (Figure S1A-C, Bismark, Krueger and Andrews, 2011).
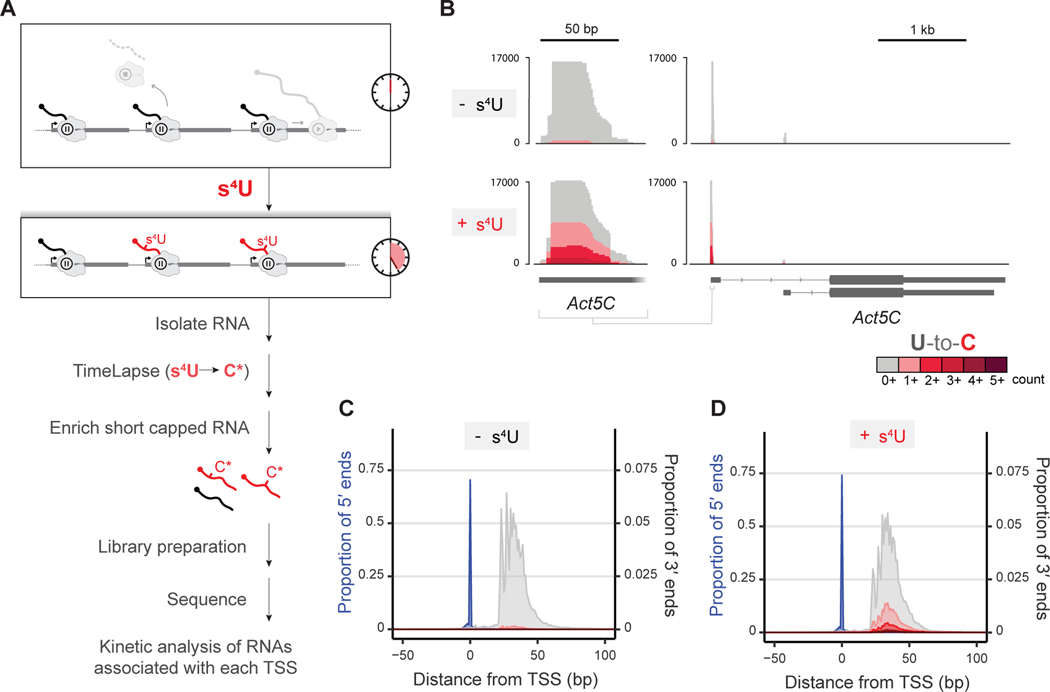
Figure 1. STL-seq captures turnover dynamics of transcripts from promoter-proximal paused polymerase.
(A) Scheme of STL-seq. Native RNA is metabolically labeled with s4U for a short time before isolating RNA. TimeLapse chemistry is performed prior to enriching for short, capped RNA transcripts which are then sequenced.
(B) Example STL-seq tracks demonstrating typical Start-seq coverage with elevated T-to-C TimeLapse mutations only in s4U-labeled samples. The entire Act5C locus is shown (right) with an expanded view of the major TSS (left).
(C and D) Metaplots of STL-seq 5´ and 3´ read ends identify the TSS and promoter-proximal pause site relative to the observed TSS location. The single nucleotide location of the TSS (blue, 5´ end of read) and pausing position (grey and red, 3´ end of read) are depicted separately. The 3´ ends are colored by the read’s mutational content while the 5´ ends are not. Read ends at each distance from the TSS for the unlabeled (C) and labeled (D) samples are shown as a proportion of the total number of reads. The proportion of 5´ ends corresponds to the left y-axis scale and the proportion of 3´ ends corresponds to the right y-axis scale.
We found that STL-seq reads provide similar profiles from s4U-labeled and unlabeled samples, demonstrating that the metabolic labeling and chemical treatment do not interfere with measurements of scRNAs (Figures 1C & 1D). STL-seq signals at each TSS are highly reproducible, both at the level of total reads (Pearson’s r = 0.94, Figure S1D) and T-to-C mutation-containing reads in the labeled samples (Pearson’s r = 0.91, Figure S1E). Correlation between total read counts of labeled and unlabeled samples is also high (Figure S1D). Together, these results demonstrate that s4U can be introduced to label newly synthesized scRNA transcripts without adversely altering the scRNA levels.
To verify that mutated reads are synthesized only by newly initiated Pol II, we inhibited initiation by treating cells with Trp prior to metabolic labeling. We did not observe accumulation of STL-seq reads containing T-to-C mutations, indicating that Trp efficiently blocks new initiation and that scRNAs recently released into elongation are not a significant source of signal (Figures S1F & S1G). Furthermore, previous work has demonstrated scRNA released from chromatin are rare, suggesting that most scRNA are degraded rapidly upon dissociation from chromatin (Henriques et al., 2013). We conclude that the mutations derived from labeled scRNAs in STL-seq are a result of the transcription of new scRNAs and therefore reflective of newly initiated Pol II which have not yet been released into elongation or terminated.
STL-seq data can be used to quantify scRNA turnover accurately and robustly
Data from our single timepoint STL-seq experiment suggested diverse kinetics of scRNA turnover at different TSSs. To further explore scRNA dynamics, we performed an independent STL-seq time series (1.5, 3, 5, 7.5, 10, and 120 minutes of s4U labeling) such that nearly all scRNAs across all TSSs were predicted to turn over within the longest labeling period. We observed the expected time-dependent accumulation of T-to-C mutations and found that the rate of accumulation varied at different TSSs, illustrating the capability of STL-seq to reveal a range of pausing kinetics across the genome (Figures 2A & S2A).
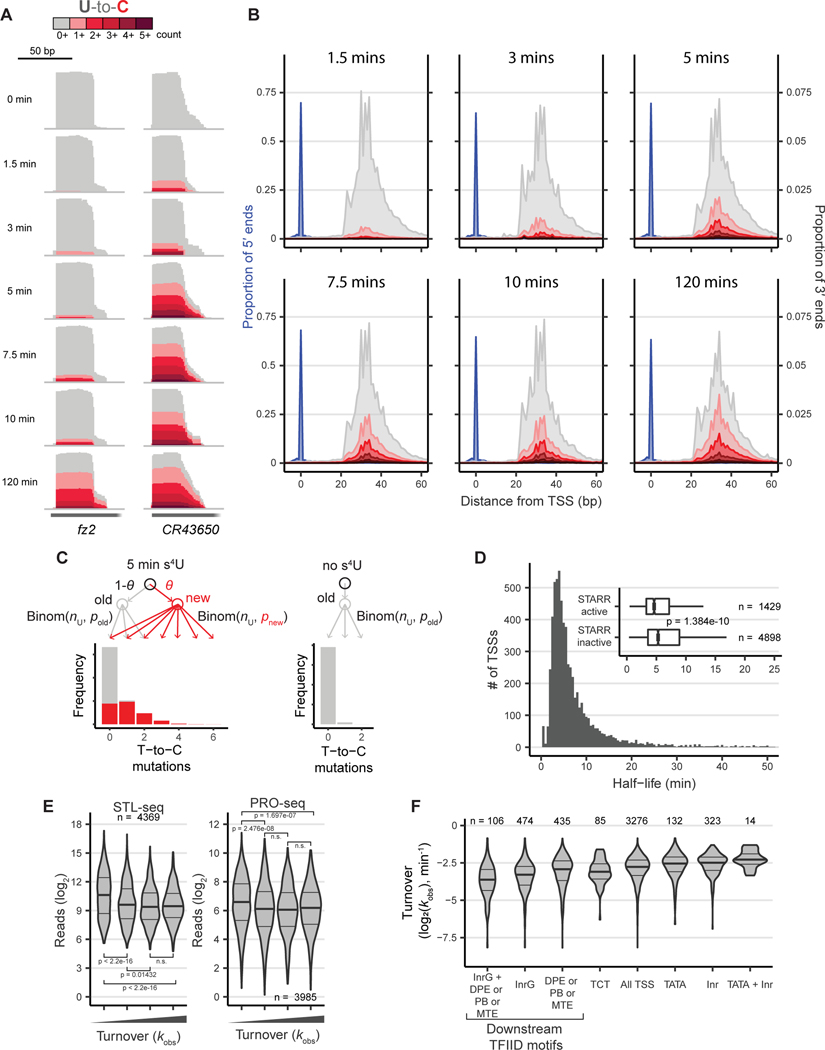
Figure 2. Estimation of scRNA transcript turnover from STL-seq.
(A) STL-seq tracks of the fz2 and CR43650 TSSs labeled with s4U for the indicated times. Tracks are autoscaled to show relative proportion of mutated reads.
(B) Metaplots of STL-seq 5´ (blue) and 3´ (grey and red) read ends relative to the TSS labeled with s4U for the indicated times with similar presentation to Figure 1C&D.
(C) The fraction of new scRNA (θ) is estimated with a mixed binomial model. The model estimates the background mutation rate (pold) with the unlabeled control and uses the number of U’s in each read (nU) and the distribution of T-to-C mutations in the labeled samples to estimate the TimeLapse-dependent mutation rate (pnew). In this simulated example, each read derives from a TSS with a 5 min half-life and average read length of 35 nt with a uridine every 4 nts. Newly synthesized transcripts (red) are synthesized with pnew = 10% and preexisting reads (grey) are synthesized with pold = 0.25%. See STAR methods for more details.
(D) Histogram of scRNA half-life estimates made with STL-seq from S2 cells. The inset boxplot separates scRNA half-lives into those aligned to regions with and without STARR-seq enhancer activity. Significance was assessed by a two-sided Wilcoxon rank sum test.
(E) The distribution of either STL-seq reads (left) or promoter-proximal PRO-seq reads (right, Elrod et al., 2019) grouped into even quartiles by observed scRNA turnover. Significance was assessed by a two-sided Wilcoxon rank sum test.
(F) Distribution of the total observed turnover rate constant at promoters grouped by motif content. All motifs are known TFIID binding elements except the polypyrimidine initiator (TCT) and the degenerate initiator (Inr). The pause button (PB), downstream promoter element (DPE), and motif ten element (MTE) were grouped together such that promoters may have one or a combination of these within 50 bp downstream.
To use the mutational content of STL-seq reads to study scRNA dynamics, we developed statistical methods to robustly quantify turnover. Determining the fraction of newly made transcripts (θ) allows estimation of turnover using a first-order observed rate constant (, min−1) for transcripts initiated from each TSS. It is important to use a statistical model to infer the fraction of scRNA that are newly made because some newly made reads will not contain any mutations (Figure S2B). The lack of mutations in some newly synthesized reads could lead to a global underestimation of scRNA turnover rates if only mutation-containing reads were considered newly made. Instead we use a binomial mixture model. We define θ in relation to with an exponential model such that
where t is the labeling time. Similar to our previous analyses (Schofield et al., 2018), we estimated the fraction of newly made transcripts using a binomial model of the number of mutations observed (tc) and uridines (nu) present in each read (Figure 2C), thereby accounting for variable uridine content across TSSs. The model also depends on the TSS-specific background (po) mutation rate, which is determined from the unlabeled controls, and the TSS-specific TimeLapse mutation rate (pn). The probability mass function is
which is parameterized on the logit scale to avoid hard upper and lower bounds.
We used a Bayesian hierarchical modeling approach to estimate these parameters using RStan software (Version 2.19.3, Carpenter et al., 2017) that implements no-U-turn Markov Chain Monte Carlo (MCMC) sampling. We defined hierarchical parameters for the global background () and TimeLapse () mutation rates to account for local variability while allowing for information sharing between TSSs to benefit those with lower coverage. The local mutation rates for the sth TSS were defined with a non-centered parameterization as follows
where σn and σo are the standard deviations of the global TimeLapse and background mutations rates, respectively, and zn and zo are TSS-specific z-scores for the TimeLapse and background mutations rates, respectively. For the complete parameterization and prior definition, see STAR methods. Simulations of scRNA with variable kinetics and uridine content supported the feasibility of using mutational content from STL-seq data to infer scRNA half-lives with this model (Figures S2B & S2C, see STAR methods).
STL-seq reveals high turnover of scRNAs at most TSSs
We applied the binomial mixture model to our genome-wide STL-seq data and found that median estimates of high confidence TSSs (low uncertainty in parameter estimates, see STAR Methods) agree well between replicates (Figure S2D). By combining both replicates to estimate a single for each TSS, we find the median half-life of scRNA to be about five minutes with half-lives spanning from minutes to tens of minutes (inner 90% range spanning 2.1 to 24 min, Figure 2D). In agreement with previous findings (Henriques et al., 2018), scRNA initiated from regions with enhancer activity as measured by STARR-seq turn over with half-lives faster than those initiated from regions without enhancer activity. However, we do not find evidence of scRNA with extremely long average half-lives (one hour or longer) that were observed in previous Trp inhibition experiments (Chen et al., 2015; Krebs et al., 2017; Shao and Zeitlinger, 2017; Henriques et al., 2018). More generally, STL-seq estimates show moderate agreement with estimates made with previously published Trp inhibition data (Figures S2E & S2F); however, the slower estimates made with Trp inhibition data (Shao and Zeitlinger, 2017) buttress previous concerns that Trp may stabilize paused Pol II. These results demonstrate that the overall rate of paused scRNA turnover is fast regardless of the TSS type and led us to investigate what TSS and promoter features associate with variability in scRNA turnover.
We asked if the level of Pol II occupancy at the pause site influences pausing kinetics. As Pol II spends little time loaded on the promoter in the preinitiation complex (PIC), promoter-proximal pausing is a major rate-limiting step during early transcription (Krumm et al., 1995; Core and Adelman, 2019). Accordingly, pause sites should always be close to fully occupied so long as the promoter is in an active state. We used STL-seq read counts from high confidence TSSs (see STAR methods) as an indicator of Pol II occupancy and found that slow turnover is not strongly correlated with higher occupancy (Figures 2E & S2G). To further probe this relationship, we reanalyzed available PRO-seq data (Elrod et al., 2019) and counted reads in the promoter-proximal region. This analysis showed a similar relationship where slow turnover is weakly associated with higher read counts (Figures 2E & S2H). Thus, STL-seq data provide further evidence that pausing is a principal rate-limiting step prior to elongation.
TFIID, a bridge-like PIC component, is sufficient to induce pausing in vitro (Fant et al., 2020). Cis-acting DNA elements, especially those related to TFIID binding, have been shown to influence Pol II pausing (Hendrix et al., 2008; Shao and Zeitlinger, 2017). TFIID contacts the TATA box through its TATA-binding protein (TBP) subunit and makes additional DNA contacts downstream of the promoter at the initiator motif (InrG), downstream promoter element (DPE), motif ten element (MTE), and pause button (PB). Presence of these downstream motifs tends to extend pausing half-lives while the TATA box tends to shorten them (Hendrix et al., 2008; Shao and Zeitlinger, 2017). Our data recapitulate these results at high confidence TSSs and demonstrate the destabilizing effect of the degenerate, G-less initiator motif (Inr, Shao et al., 2019) (Figure 2F). The polypyrimidine initiator (TCT) motif, which is similar to InrG but does not bind TFIID, appears to be associated with similar kinetics as InrG. Our robust and reproducible measurements of support previous observations and provide the foundation to further examine the principles underlying promoter-proximal pausing.
Termination is generally faster but less variable than release into elongation
Next, we sought to determine the proportion of paused Pol II molecules that are prematurely terminated at each TSS prior to entering productive elongation. Previous work established that premature termination is an important fate of the paused complex, but the relative contributions of pause release and premature termination were not determined for TSSs genome-wide (Brannan et al., 2012; Henriques et al., 2013; Buckley et al., 2014; Jonkers et al., 2014; Nilson et al., 2017; Steurer et al., 2018; Elrod et al., 2019; Tatomer et al., 2019). We used flavopiridol (FP) treatment (prior to s4U labeling) to inhibit release into elongation and allow for measurement of premature termination (Figure 3A). FP increases STL-seq reads at the majority of TSSs, except those with the most scRNA reads, perhaps because they are already fully saturated with paused Pol II (Figure S3A). This increase in STL-seq reads indicates a stabilization of the paused complex due to inhibition of release into elongation by FP.
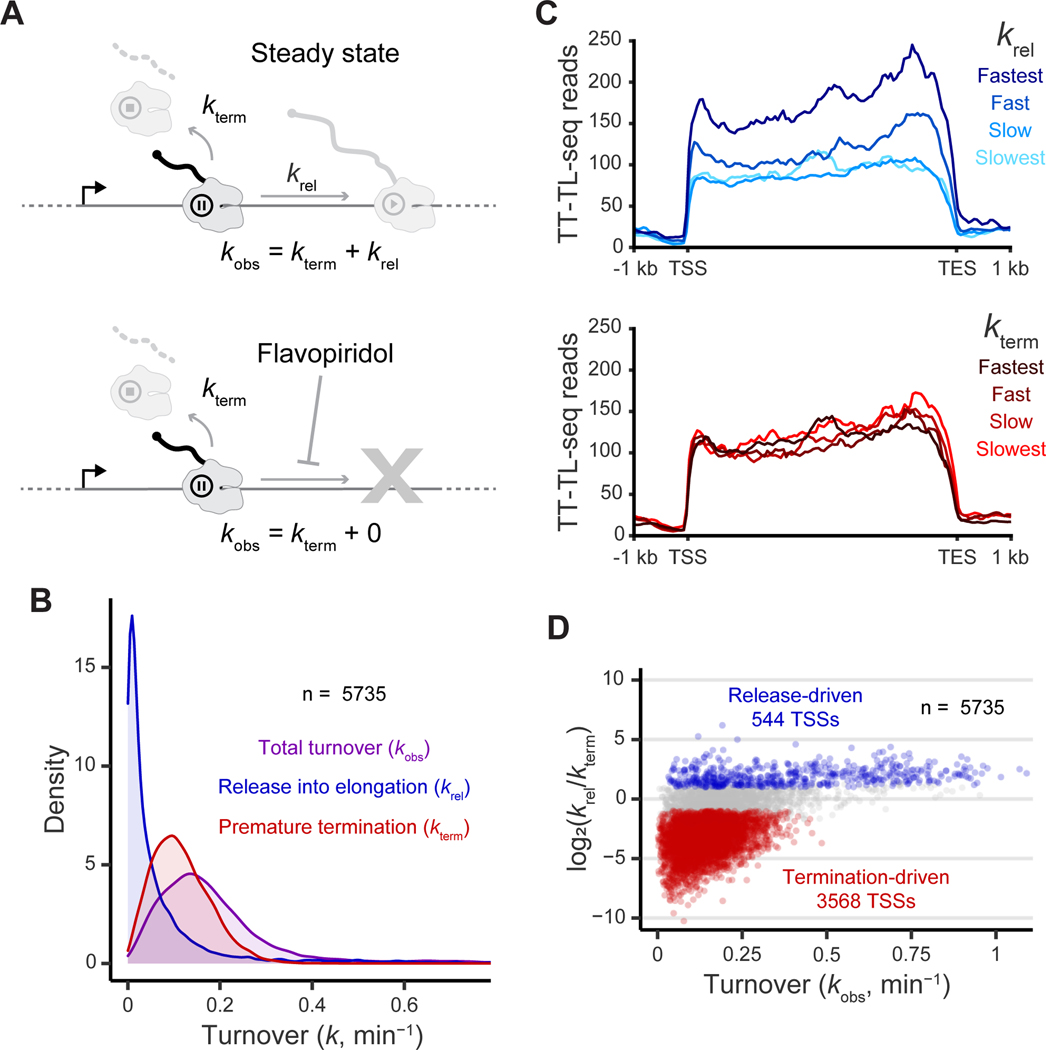
Figure 3. Termination is fast while release into elongation explains variability of Pol II turnover at pause sites.
(A) Representation of pausing kinetics under steady-state and flavopiridol-inhibited conditions. The steady-state observed turnover ()is the sum of the rates of release into elongation () and premature termination (). Upon flavopiridol inhibition, observed turnover is caused only by premature termination.
(B) The distribution of the first-order rate constants for total turnover, release into elongation, and premature termination.
(C) Metaplots of TT-TL-seq signal grouped into even quartiles by release and termination of the respective high confidence TSS (n = 2422 genes). Coverage is determined over 50 nt bins.
(D) Total observed rate constant plotted versus the log2 ratio of the release rate and termination rate. Points are colored if the 80% credible interval of the log2 ratio does not overlap zero and the median value is greater than 1 (blue) or less than −1 (red).
We further developed the model described above to assume that the observed turnover rate constant () at steady state is the sum of termination and pause-release rate constants (Figure 3A, see STAR methods). Previous studies demonstrated that FP does not perturb premature termination (Buckley et al., 2014; Krebs et al., 2017). Therefore, under FP inhibition, the observed turnover is attributed only to premature termination (). We calculate the pause-release rate constant ()as the difference between and . We find that pause-release constants on average are slow but vary widely (median 0.027 min−1; inner 90% range 0.0015–0.31 min−1), while termination constants are fast and more tightly distributed (median 0.11 min−1; inner 90% range 0.027–0.23 min−1) (Figure 3B).
Because polymerases must be released from the pause site to transcribe the rest of the gene body, we expected that transcriptional activity in the gene body should be a function of how quickly scRNA are released into elongation. Transient-Transcriptome-TimeLapse-seq (TT-TL-seq) enriches for nascent RNA and is therefore a good measure of transcriptional activity. We performed TT-TL-seq and compared coverage to STL-seq estimates at TSSs where we could make high confidence estimates of (n = 2865; see STAR methods). As expected, we found that is the best predictor of TT-TL-seq signal when compared to and (Figures 3C & S3B-E). We also assessed this relationship using an orthogonal measure of elongating Pol II activity (gene-body PRO-seq reads; Elrod et al., 2019; Figure S3F), which further supported our conclusion that STL-seq pause-release rates are more tightly linked to gene body transcription.
To provide additional validation of our approach to estimate and we reasoned that genes with significant levels of paused polymerase at the TSS but very low transcriptional activity in their gene body must have low . Therefore, we expect at these TSSs and those rate constants should not be perturbed by FP. We used TT-TL-seq data to identify genes with the lowest 10% of transcriptional activity where we expect . We then identified confident TSSs at these genes and found and using a model designed to estimate differences (see STAR methods). We found that turnover under FP inhibition and in the uninhibited state were not substantially different at these TSSs, further supporting our assumption that FP does not alter (Figure S3G).
To examine the relative amount of Pol II terminated or released into elongation at each TSS, we took the log2-transformed ratio of the rates of pause release versus termination. We found that termination is faster than pause release at most TSSs (62%) while the converse is uncommon (9%) (Figures 3D & S3H). However, TSSs with the fastest total turnover ( > 0.5 min−1) are more likely to release scRNA into productive elongation than terminate the transcript. We compared the change in scRNA read counts upon FP treatment to the ratio of to and found that more frequent pause release is associated with the accumulation of more reads, as would be expected (Figure S3I). Taken together, these results reveal that on average, termination is about four times faster than pause release and therefore ~80% of total turnover, while pause release is typically slower but has a larger dynamic range.
Certain histone tail modifications are associated with less permissive pausing dynamics
The local chromatin environment around promoters is important for the regulation and maintenance of transcriptional activity. To examine how the local chromatin landscape around pause sites is related to scRNA dynamics revealed by STL-seq, we focused our analysis on high confidence promoter TSSs. We found that pause sites with the least stable scRNA are modestly enriched for chromatin marks typically associated with active promoters. For example, low monomethylation and high trimethylation of histone 3 lysine 4 (H3K4me1/me3) indicate high promoter activity (Heintzman et al., 2007). We find that slower turnover of scRNA is related to a larger ratio of mono-to-trimethylation at H3K4 (Figures S4A & S4B). Additionally, trimethylated histone 3 lysine 36 (H3K36me3), whose deposition is signaled by active elongation (reviewed in Gates et al., 2017), is enriched immediately downstream of fast turnover sites (Figures S4A & S4C).
We found distinct H3K4 methylation profiles to be associated with promoters depending on their relative rates of pause release and termination (Figures 4A & S4A). H3K4me3 promotes PIC assembly and transcription initiation (Lauberth et al., 2013). We observe that relative rates of pause release and termination are not significantly related to H3K4me3 levels (Figure S4D), supporting the notion that H3K4me3 only behaves as a signal to activate promoters. On the other hand, H3K4me1 is depleted at promoters that are the most likely to release Pol II into elongation (Figure 4B). By examining the relationship of pause release and termination with H3K4me1 separately, we find that the modification is more strongly related to pause release (Figures 4C & S4E). This negative correlation suggests H3K4me1 could play a role in suppressing the release of Pol II into elongation, an idea consistent with the fact that H3K4me1 is enriched at enhancers where productive elongation is rare (reviewed in Calo and Wysocka, 2013). In further agreement, scRNA initiated from promoters with low H3K4me1/me3 ratios are more likely to be released into elongation and behave the least similarly to scRNA initiated from enhancer TSSs (eTSSs, Figure 4D).
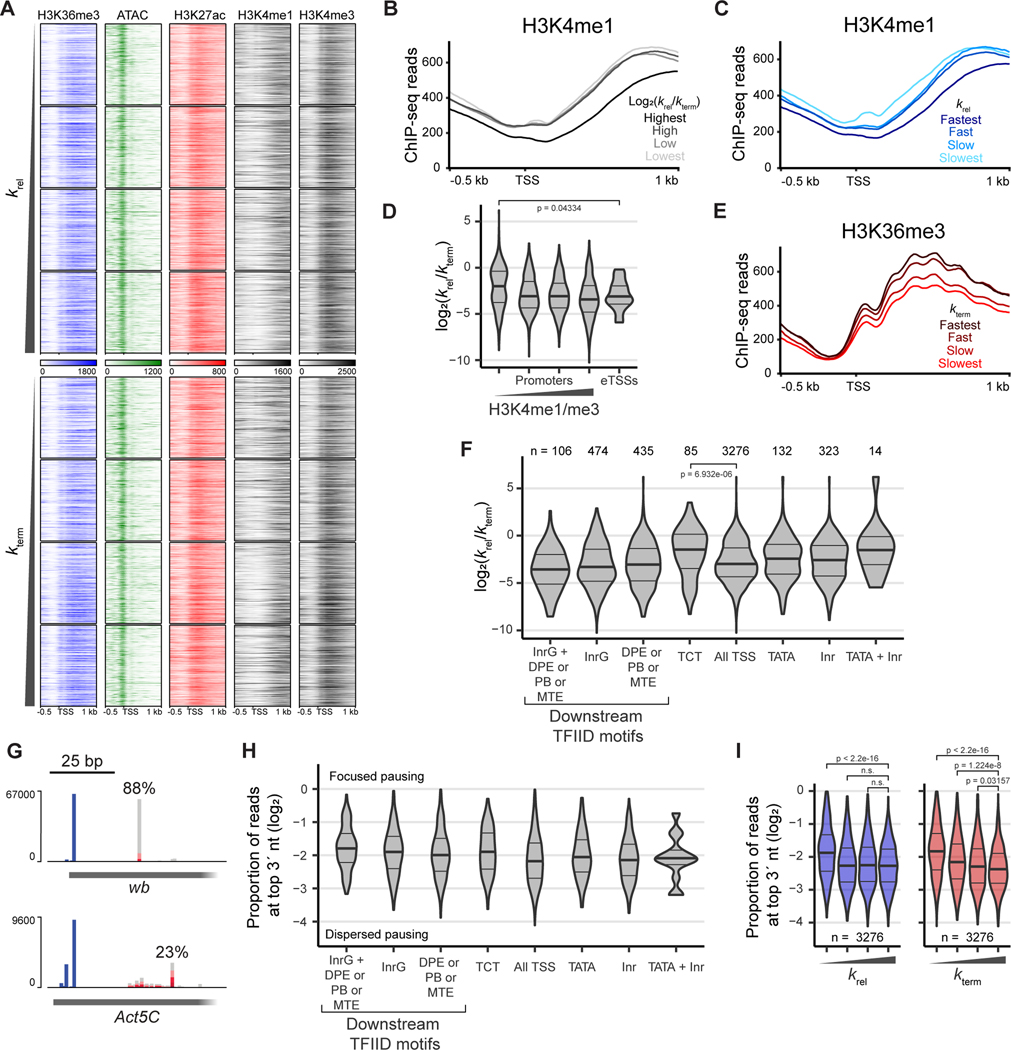
Figure 4. Weak promoter architecture leads to rapid termination of paused Pol II.
(A) Heat maps of ATAC-seq (Elrod et al., 2019) and H3K27ac, H3K4me1, H3K4me3 (Henriques et al., 2018), and H3K36me3 (Chen et al., 2012) ChIP-seq around promoters grouped into even quartiles and order by pause release and termination at the respective TSS. Heatmaps are centered on the STL-seq TSS with a window of 0.5 kb upstream and 1 kb downstream.
(B) Metaplot of H3K4me1 ChIP-seq data around promoters with a window of 0.5 kb upstream and 1 kb downstream of the TSS grouped into even quartiles by the log2 ratio of the pause-release rate to the termination rate (n = 3270 promoters).
(C) Metaplot of H3K4me1 ChIP-seq data around promoters with a window of 0.5 kb upstream and 1 kb downstream of the TSS grouped into even quartiles by pause release (n = 3270 promoters).
(D) Distribution of the log2 ratio of the release rate to the termination rate at promoters grouped by whether TSSs are of high confidence promoters or enhancers (eTSSs, n = 21). Promoters are further grouped into even quartiles by H3K4me1 enrichment determined by ChIP-seq. Significance was assessed by a two-sided Wilcoxon rank sum test.
(E) Metaplot of H3K36me3 ChIP-seq data around promoters with a window of 0.5 kb upstream and 1 kb downstream of the TSS grouped into even quartiles by termination (n = 3270 promoters).
(F) Distribution of the log2 ratio of the release rate to the termination rate at promoters grouped by motif content. The pause button (PB), downstream promoter element (DPE), and motif ten element (MTE) were grouped together such that promoters may have one or a combination of these in the downstream region. Significance was assessed by a two-sided Wilcoxon rank sum test.
(G) Example STL-seq tracks where the single nucleotide location of the TSS (blue, 5´ end of read) and pausing position (grey and red, 3´ end of read) are depicted separately. The 3´ ends are colored by the read’s mutational content while the 5´ ends are not. The maximum percent of reads with the same 3´ end is shown above the read position.
(H) Distribution of the proportion of reads with 3´ ends located at the most frequent pause position. At each promoter, the most common position of the 3´ read end was identified, and the proportion of reads at this position was determined. Promoters are separated by promoter motif as in F.
(I) Distribution of the proportion of reads with 3´ ends located at the most frequent pause position as in H but promoters are grouped into even quartiles by pause release (left) or termination (right). Significance was assessed by a two-sided Wilcoxon rank sum test.
We examined the relationship between scRNA dynamics and H3K36me3 levels and found the mark to exhibit a significant positive relationship with premature termination (Figure 4E). We also find that H3K36me3 is depleted from promoter-proximal regions at the slowest releasing sites but has similar levels at the remaining promoters (Figure S4F). As H3K36me3 suppresses cryptic initiation from weak downstream promoters (Xie et al., 2011), our data raise the intriguing possibility that this suppression is supported by promoting premature termination. Fast termination is also found at less accessible promoters as measured by ATAC-seq, bolstering the association between premature termination and weak promoters (Figure S4G).
In summary, H3K4me1 and H3K36me3 are enriched at pause sites that are less likely to release Pol II into elongation. It is possible that histone tail modifications locally repress gene expression by influencing dynamics at promoter-proximal pause sites. Our data support a model in which H3K4me1 blocks release into elongation, while H3K36me3 recruits factors that promote premature termination.
Promoter and pause-site architecture are associated with stability of the paused complex
Together with previous work (Hendrix et al., 2008; Shao and Zeitlinger, 2017; Fant et al., 2020), our findings demonstrate the importance of TFIID binding elements in pausing and led us to examine how promoter motif content relates to both pause-release and termination kinetics. We find no evidence that downstream TFIID binding motifs substantially alter the proportion of Pol II which is terminated or released into elongation (Figure 4F). As would therefore be expected from our estimates, downstream TFIID motifs are associated with slow rates for both and (Figures S4H & S4I). The TCT motif marks TSS with a high proportion of Pol II that is released into elongation. The TCT motif is primarily found at promoters of ribosomal proteins which are typically among the most highly expressed genes (Parry et al., 2010). Therefore, it is unsurprising to observe elevated pause-release rates at TSSs with the TCT motif (Figure S4H). Further investigation of these TSSs will likely provide a deeper understanding of how cis-acting DNA can promote release into elongation.
Strong downstream TFIID binding stabilizes the paused Pol II complex and coordinates the focused pausing of Pol II molecules (Kwak et al., 2013; Shao and Zeitlinger, 2017). We reasoned that if the total scRNA turnover and pause site dispersion is influenced by the organization of the PIC, relative rates of pause release and termination could also be differentially affected. As a measure of focused versus dispersed pausing, we determined the proportion of scRNA with the identical and most common 3´ position at each TSS (Figure 4G). Consistent with previous findings (Kwak et al., 2013), focused pause sites are associated with downstream TFIID binding motifs (Figure 4H). When comparing pause site dispersion with scRNA kinetics, the fastest terminating pause sites are associated with less focused pausing profiles while dispersion had little bearing on the pause-release rate (Figure 4I). These data demonstrate that cis-regulatory DNA elements in the promoter-proximal region mark TSSs with distinct kinetic and physical pausing profiles. In addition, premature termination is more likely to occur at TSSs that do not reproducibly pause Pol II in the same position.
Enhanced release into elongation is the major response to hormone stimulus
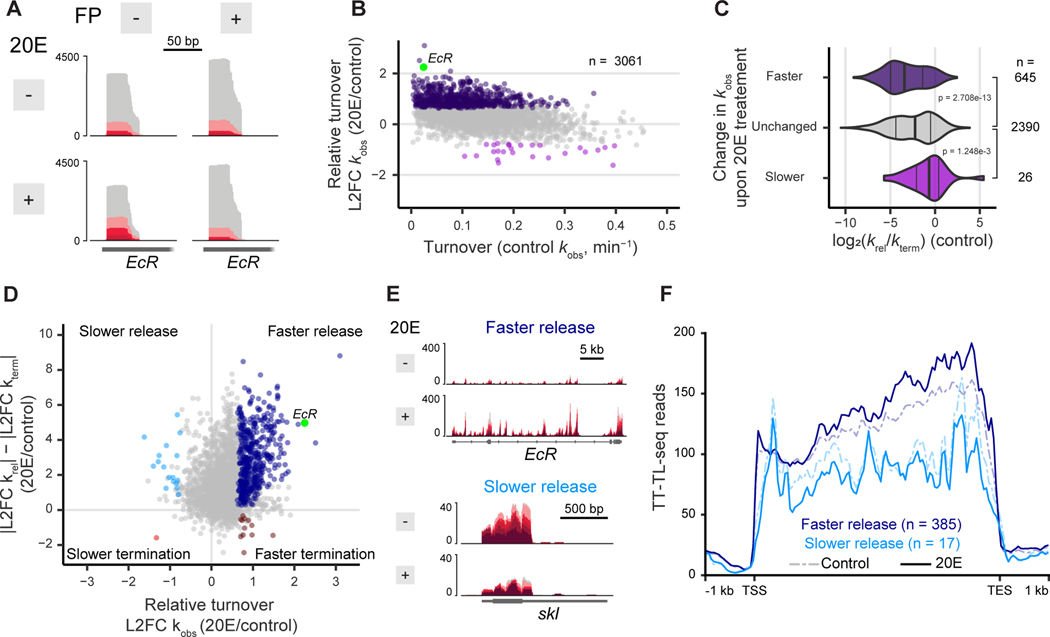
A major outstanding question which has not been broadly addressed is whether release into elongation, premature termination, or both are targets for the regulation of gene expression. STL-seq presents an opportunity to quantify changes in pause release and termination in response to a regulatory stimulus. By treating cells with 20-hydroxyecdysone (20E), a hormone known to both induce and repress expression of target genes (Yamanaka et al., 2013; Uyehara and McKay, 2019), we can determine the preferred mechanism of regulation at the pause site. If altered initiation rates were solely responsible for the transcriptional response, we would expect to see correlation between changes in STL-seq reads and TT-TL-seq reads, but this was not the case (Figure S5A). To dissect the relative changes in pause-release and termination rates, we pretreated 20E-stimulated cells with FP and performed STL-seq. In uninhibited samples, 20E stimulus markedly increased the proportion of mutation-containing scRNA from TSSs of genes well-characterized as 20E targets (e.g., Figure 5A). To quantify these changes genome-wide, we used the same model as described above to estimate termination and pause-release rates. At high confidence TSSs, we find that 20E stimulus generally increases the total observed turnover of scRNA at many TSSs (Figure 5B).
Figure 5. Hormonal stimulus by 20E preferably regulates release into elongation.
(A) STL-seq tracks of the EcR gene TSS when treated with or without 20E and with or without flavopiridol inhibition.
(B) Total observed rate constants of high confidence promoters plotted versus the log2 fold change when S2 cells are stimulated with 20E for thirty minutes. Points are colored if the 80% credible interval is greater than log2(1.5) (dark purple) or less than -log2(1.5) (light purple). The EcR TSS shown in A is highlighted in green.
(C) Distribution of the log2 ratio of the pause-release rate versus termination rate at promoters grouped by the change in the total turnover as determined in B. Significance was assessed by a two-sided Wilcoxon rank sum test.
(D) The log2 fold change of promoters plotted versus the difference of the magnitudes of the log2 fold change of the release and termination constants upon 20E stimulus. Points are colored if the 80% credible interval of L2FC is entirely greater than log2(1.5) or less than -log2(1.5) and if the 80% credible interval of the difference in magnitudes does not overlap zero. The EcR TSS shown in A is highlighted in green.
(E) TT-TL-seq tracks of EcR and skl +/− 20E stimulus as examples of genes where 20E-induced changes in scRNA turnover are driven by faster or slower pause release, respectively.
(F) Metaplots of TT-TL-seq signal without (dashed) and with (solid) 20E stimulus separated by whether release from the TSS was faster (dark blue) or slower (light blue) upon 20E stimulus as determined in D. Coverage is determined over 50 nt bins.
Because 20E-stimulated TSSs tend to release Pol II into elongation slowly under normal conditions, we expected upregulation of to be the more likely response (Figure 5C). Indeed, the inflation of pause-release rates is more dramatic than the diminution of termination both in effect size and in the number of TSSs (Figures S5B & S5C). At each TSS, we examined the difference in magnitude of the log2 fold change of and in the context of the change in (Figures 5D & S5D). The four possible regulatory options of induction or suppression of either pause release or termination are separated into the four quadrants. Independent of whether a TSS is repressed or induced, our data reveal that most are regulated primarily at the level of release into elongation. Even at the fastest terminating TSSs where suppression of premature termination has the most potential to lead to increased expression, we see that pause release is preferably regulated (Figures S5E & S5F). To confirm these findings, we performed TT-TL-seq with the same RNA as collected for STL-seq (Figure 5E). We binned genes by the quadrant in which their TSSs are found in Figure 5D. In support of the predicted transcriptional changes detected by STL-seq, we observed that 20E treatment leads to increased transcriptional activity over genes with induced pause release, as well as loss of transcription over genes with repressed pause release (Figure 5F).
In summary, we observed that hormone treatment broadly elevates scRNA turnover at many TSSs. STL-seq demonstrates that 20E-induced changes in the total turnover rate constants in most cases are driven by regulation of release into elongation while premature termination contributes only minor regulatory effects.
Hyperosmotic stress induces premature termination
While hormone treatment primarily regulates , we wondered if other stimuli might influence . Hyperosmotic stress alters the transcriptional landscape of human cells by inducing readthrough transcription as well as widespread transcriptional repression (Vilborg et al., 2015; Rosa-Mercado et al., 2021). Previous Pol II ChIP-seq experiments under the same conditions revealed loss of Pol II over the body of repressed genes. This observation suggests that salt stress-induced transcriptional repression is at least partially accomplished at or prior to promoter-proximal pausing. Therefore, STL-seq is uniquely suited to provide insight into the mechanism accounting for this transcriptional repression.
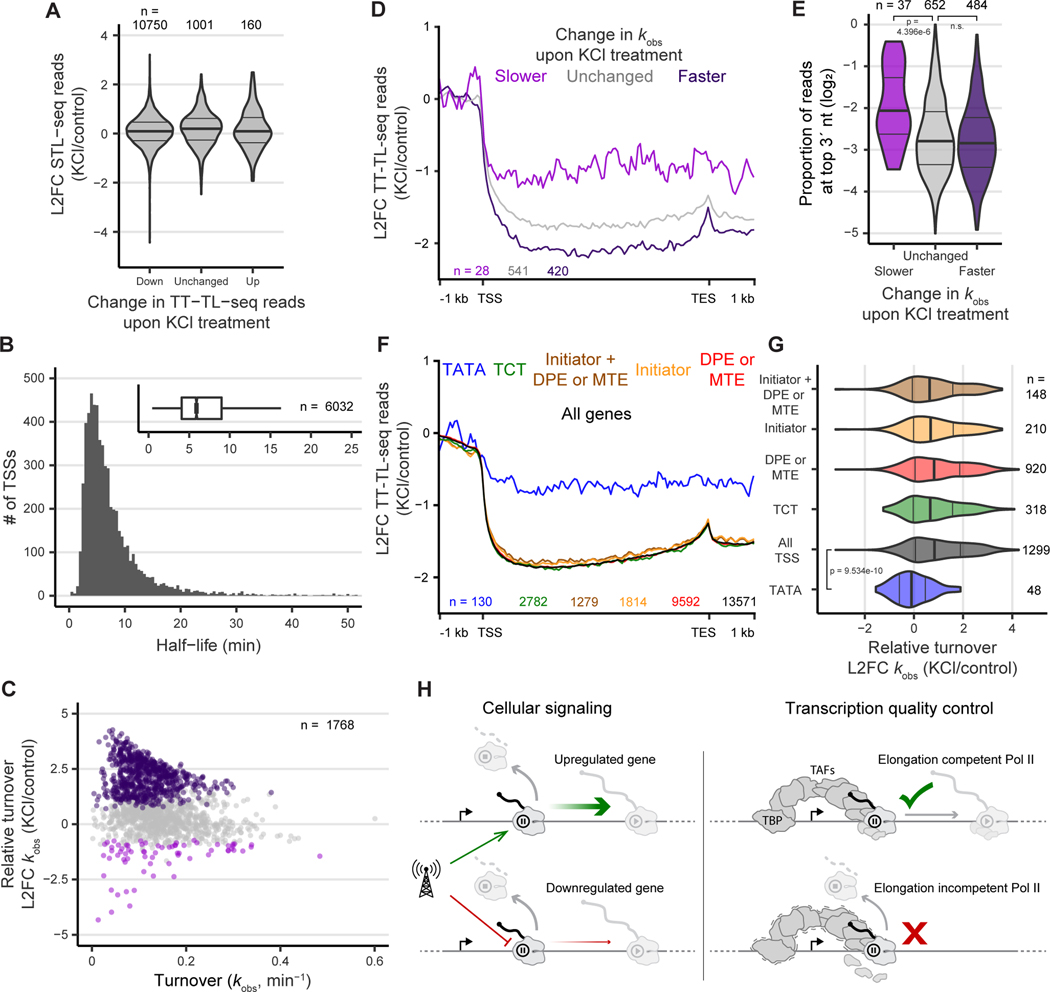
We treated h0uman 293T cells with 80 mM KCl for one hour and performed STL-seq to assess changes in initiation, termination, and pause release. We found that STL-seq signals were highly reproducible for both total and mutation-containing reads (Figures S6A & S6B). If reduced initiation were solely responsible for transcriptional downregulation, we would expect to see substantial loss of STL-seq reads at the promoters of downregulated genes, but we did not (Figure 6A). Thus, hyperosmotic stress must induce transcriptional repression via a reduction in the pause-release rate or an increase in the termination rate.
Figure 6. Hyperosmotic stress induces premature termination at TATA-less promoters.
(A) Change STL-seq reads at promoter TSSs grouped by the change in TT-TL-seq signal over the gene body.
(B) Histogram of scRNA half-life estimates made with STL-seq from human 293T cells. The inset boxplot depicts the distribution of all TSSs.
(C) Total observed rate constants of TSSs plotted versus the log2 fold change when cells are exposed to hyperosmotic stress for one hour. Points are colored if the 80% credible interval is entirely greater than log2(1.5) (dark purple) or less than -log2(1.5) (light purple).
(D) Metaplots of the log2 fold change of TT-TL-seq signal upon hyperosmotic stress grouped by the change in turnover of scRNA at the gene’s TSS as determined by STL-seq. Coverage was determined over 50 nt bins before calculating the fold change of each bin.
(E) Distribution of the proportion of reads at promoters with 3´ ends located at the most frequent pause position grouped by the change in scRNA turnover and colored as in D. At each TSS the most common position of the 3´ read end was identified, and the proportion of reads at this position was determined.
(F) Metaplots of the log2 fold change of TT-TL-seq signal upon hyperosmotic stress grouped by the motif content of the associated STL-seq TSS. Coverage was determined over 50 nt bins before calculating the fold change of each bin.
(G) The distribution of the log2 fold change of scRNA turnover rate constants at promoters upon hyperosmotic stress. TSSs are grouped by motif content and colored as in F. Significance was assessed by a two-sided Wilcoxon rank sum test.
(H) Proposed model for the distinct roles of release into elongation and premature termination at the promoter-proximal pause site. To alter gene expression, cells signal for either an increase or decrease in release into elongation (left). Premature termination does not contribute greatly to the response to cellular signaling but acts to evict paused Pol II whose elongation factors do not assemble properly (right). Coordinated binding of TFIID subunits, TBP and TAFs, is important for maturation of an elongation-competent Pol II and significantly stabilizes the mature complex.
We applied the same model as described above to estimate of scRNA genome wide. Notably, the distribution of steady-state scRNA half-life estimates in human 293T cells is very similar to that of fly S2 cells (Figure 6B). When comparing stressed and unstressed cells, we found scRNA transcripts from many high confidence TSSs in untreated conditions to be much less stable upon hyperosmotic stress (Figure 6C). This observation and the loss of gene body transcription suggest that induction of Pol II premature termination at the pause site is a major response to hyperosmotic stress.
We then compared changes in turnover to previously published TT-TL-seq data (Rosa-Mercado et al., 2021). Supporting our model that hyperosmotic stress induces premature termination, active transcription is more repressed over genes with destabilized scRNA (Figures 6D & S6C). Promoters with decreased turnover produce more focused pause sites than those with unchanged or induced turnover (Figure 6E). Upon hyperosmotic stress, the induced TSSs become even less focused (Figure S6D). Together, these results suggest that the promoters of genes downregulated upon KCl treatment are prone to stress-induced termination at the pause site.
We hypothesized that TSSs prone to stress-induced termination may lack cis-acting DNA elements that recruit components of the PIC or other pausing factors. We again binned promoters by their motif content (using the consensus TATA box described by Juven-Gershon et al. (2008), see STAR methods). Strikingly, genes with promoters containing a TATA box were protected from transcriptional repression (Figures 6F & S6E). The TSSs of these genes were also protected from stress-induced termination at the pause site (Figure 6G). However, none of the downstream TFIID motifs generally protected genes from transcriptional repression or premature termination despite their ability to stabilize the paused Pol II complex. Taken together, these results indicate that termination at the pause site is an important regulatory process that is associated with cis-acting DNA elements at the promoter.
Discussion
STL-seq provides genome-wide insight into the dynamics of promoter-proximal pausing by combining the time resolution of metabolic labeling (Schofield et al., 2018) with the TSS specificity of Start-seq (Nechaev et al., 2010). Our results demonstrate that STL-seq reliably captures kinetic information of scRNA, allowing inference of the kinetic behavior of the Pol II paused complex.
We found total observed turnover are similar between human and fly (Figures 2D & 6B), suggesting that pausing dynamics and regulation may be conserved across metazoans. To better understand the complex behavior of Pol II at the pause site, we dissected total observed turnover into rates of release into elongation and premature termination. This revealed that while only a small fraction of paused Pol II enters productive elongation, pause release is highly dynamic and responds to 20E stimulus in Drosophila. On the other hand, premature termination does not determine gene expression and is insensitive to the same hormonal stimulus. These findings provide detailed kinetic support for the concept that active regulation at the pause site occurs by altering the rate of release into elongation (Figure 6H).
We also sought to identify a function for premature termination. Similar to Beckedorff et al. (2020), we favor a model in which termination at the promoter-proximal pause site occurs as a quality check mechanism to ensure that members of a mature elongation complex (EC) correctly and completely assemble on Pol II (Figure 6H). In support of this model, we found relatively fast termination rates at TSSs with features that we view as hallmarks of inefficient EC assembly. These features include high H3K36me3, the lack of downstream TFIID binding motifs, and less focused pause sites. H3K36me3 functions to repress cryptic initiation and leads to the erasure of other activating histone tail modifications (Vermeulen et al., 2007; Xie et al., 2011; Lauberth et al., 2013). The absence of downstream TFIID binding motifs leads to less focused pausing, which we suspect is a symptom of poorly assembled ECs.
Previous work (Kwak et al., 2013) demonstrated that weaker contacts between the PIC and the paused complex lead to less focused pause sites. Our data supports an extension of this model in which weaker interactions between the PIC and the paused complex lead to faster premature termination. We hypothesized that stressing cells in a manner that disrupts the transcriptional machinery may lead to increased premature termination at the pause site. Hyperosmotic stress alters the Pol II interactome and leads to transcriptional silencing genome-wide (Rosa-Mercado et al., 2021). We showed that induction of premature termination is at least partly responsible for the response to hyperosmotic stress that results in genome-wide transcriptional repression. Therefore, we speculate that hyperosmotic stress disrupts elongation factor assembly and results in a larger proportion of incompetent ECs reaching the pause site. These incompetent ECs are then signaled for premature termination before they can enter productive elongation. However, leaky pause release of ECs which lack critical processing machinery may lead to production of downstream-of-gene containing transcripts (DoGs), which are a product of readthrough transcription (Vilborg et al., 2015), and/or misspliced transcripts (Reimer et al., 2021). In this manner, premature termination at the pause site may provide a form of kinetic proofreading.
Recently, the Integrator complex has been the focus of studies examining premature termination at the pause site (Elrod et al., 2019; Tatomer et al., 2019; Beckedorff et al., 2020; Huang et al., 2020), and one study proposes that Integrator acts in early elongation (Lykke-Andersen et al., 2021). Interestingly, hyperosmotic stress causes dissociation of Integrator from Pol II (Rosa-Mercado et al., 2021), suggesting that Integrator is not responsible for the induction of premature termination observed under hyperosmotic stress. More generally, future STL-seq experiments may help clarify the roles of Integrator at the promoter-proximal pause site.
Here, TFIID has emerged as a critical factor that acts beyond initiation to establish and maintain proper kinetics of promoter-proximal pausing. Strikingly, we found that the presence of TATA box prevents stress-induced premature termination, highlighting the vital role of TFIID through initiation and pause release. Our findings illustrate the unique capabilities of STL-seq to reveal the dynamics and regulation of promoter-proximal pausing as well as to identify essential pausing factors. This method will enable future studies investigating the mechanism of promoter-proximal pausing which were not previously possible.
Limitations of the Study
STL-seq is a powerful tool to study pausing kinetics and provide mechanistic insight into promoter-proximal pausing at most TSSs. Accurate estimation of kinetic parameters using STL-seq is limited by the read depth and mutational content of reads mapping to each TSS. In practice, we have found that rate estimates at a TSS are less reliable when the mutational content at the TSS is low. In this case, there may not be enough information to use our Bayesian models to confidently determine rate constants, leading to large credible intervals for our parameter estimates. Low numbers of observed mutations could result from low read coverage, few uridines in a scRNA, or from a low s4U incorporation rate. We developed criteria for identification and removal of unreliable TSSs from our analyses (see STAR Methods), allowing us to restrict analyses to the thousands of TSSs where kinetic parameters can be confidently estimated.
To dissect the steady-state observed turnover of scRNA at the pause site, we took advantage of the rapid inhibition of P-TEFb activity by flavopiridol (FP) to block pause release. We provide evidence that FP does not perturb premature-termination rate constants, but the accuracy of our and estimates would be reduced at some TSSs if P-TEFb inhibition directly or indirectly influences premature-termination rates constants. Nonetheless, our hyperosmotic stress experiments demonstrate that additional information about transcription over the gene body (e.g., from TT-TimeLapse-seq data) is sufficient to identify changes in and without the need for P-TEFb inhibition.
STAR Methods
Resource Availability
Lead Contact
Communication with authors of this manuscript should be directed to Matthew D. Simon (matthew.simon@yale.edu).
Material Availability
Requests for materials generated in this study should be directed to the lead contact. The authors will readily share reagents upon request.
Data and Code Availability
Raw data of TT-TL-seq and STL-seq libraries generated in this study are publicly available and accessible on Gene Expression Omnibus (GEO) under GSE166202.
Mutation calling was performed with a custom analysis pipeline freely available at https://bitbucket.org/mattsimon9/timelapse_pipeline/src/master/. Kinetic parameter estimates were made with custom RStan models freely available in the same repository. Custom scripts written for downstream analyses are available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
HEK293T cells were grown at 37°C in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. D. melanogaster S2 cells were grown at 27°C in Schneider’s Drosophila Medium supplemented with 10% FBS and 1% penicillin and streptomycin.
Method details
Cell lines and s4U metabolic labeling
Metabolic labeling of cells was performed as described previously (Schofield et al., 2018). For STL-seq in Drosophila, S2 cells were grown to approximately 3–4 million cells/mL and spiked with s4U (1 mM). Cells were incubated at 27°C for the appropriate labeling time, fully resuspended and transferred to 4–5 volumes of ice-cold PBS in an ice bath. Cells were pelleted by centrifuging at 500Xg for 5 mins. PBS was removed, cells were resuspended in 1 mL TRIzol, and frozen at −80°C. HEK293T cells were grown to approximately 60% confluency when the media was spiked with s4U (1 mM). Immediately after 5 mins of labeling, plates were placed on ice and washed with ice-cold PBS. Cells were scraped from plates and pelleted by centrifuging at 500Xg for 5 mins. PBS was removed and cells were resuspended in 1 mL TRIzol and frozen at −80°C. HEK293T cells were spiked into S2 TRIzol samples at 5% by cell count. Total Drosophila RNA was spiked into total human RNA at 5% by mass.
Drug and KCl treatments
For STL-seq and TT-TL-seq, D. melanogaster S2 cells were treated with 42 μM 20-hydroxyecdysone or DMSO for 30 minutes. S2 were treated with either 10 μM Triptolide for 10 minutes, 500 nM Flavopiridol for 40 minutes, or DMSO for the same time as a control. For combined Flavopiridol and 20-hydroxyecdysone treatments, S2 cells were pretreated with 500 nM Flavopiridol for 10 minutes before adding 20-hydroxyecdysone directly to cell media. Labeling times were always the last 5 minutes of any treatment.
To induce hyperosmotic stress, HEK293T cells were treated with 80 mM KCl for a total of 1 hour as described in Vilborg et al., 2015. Metabolic labeling was performed during the last 5 minutes of stress.
STL-Seq
Total RNA from S2 and 293T cells suspended in TRIzol was purified as described previously with minor changes (Schofield et al., 2018). Following TRIzol extraction, RNA was precipitated with one volume of isopropanol supplemented with 1 mM DTT. Extracted RNA was immediately subjected to TimeLapse chemistry as previously described with minor modifications. The oxidant used was meta-chloroperoxybenzoic acid (mCPBA) to avoid modifying the 3´ ends of RNA and interfering with downstream ligations. All purifications with Agencourt RNAClean XP beads were performed with 2 volumes of beads and supplemented with isopropanol to improve recovery of short RNA. Start-seq was performed on total RNA essentially as previously described with minor modifications (Nechaev et al., 2010). Briefly, total RNA was electrophoresed on a 15% denaturing Urea-PAGE gel for 1 hr at 200 V. RNA between the sizes of ~20 and ~80 nt was excised, extracted from the gel with a crush-soak method, and ethanol precipitated. The short RNA was then treated successively with RNA 5´ polyphosphatase (VWR), Terminator 5´-phosphate-dependent exonuclease (Lucigen), and ligated to a custom, pre-adenylated DNA adapter with T4 RNA ligase 2 truncated (NEB). Short, capped RNA was then electrophoresed on a 15% denaturing Urea-PAGE gel for 1 hr at 200 V. RNA between the sizes ~40 and ~100 nt was excised, extracted, and ethanol precipitated. Ligated RNA was treated successively with calf intestinal alkaline phosphatase (NEB), RNA 5´ Pyrophosphohydrolase with ThermoPol buffer (NEB), and T4 RNA ligase 1 (NEB) to ligate a custom RNA oligo. RNA was reverse transcribed with SuperScript RT III and finally amplified with Phusion polymerase. Amplified libraries were purified by electrophoresis on a 6% native TBE PAGE gel, extraction, and ethanol precipitation. Libraries were sequenced either on a NovaSeq 6000 2X100bp or HiSeq 4000 2X150bp.
STL-seq alignment, mutational analysis, and TSS calling
For STL-seq, filtering and alignment to the human GRCh38 genome version 26 (Ensembl 88) or the Drosophila dm6 genome were performed as described previously with some modifications (Schofield et al., 2018). Paired-end sequencing formats with 100 bp reads or longer caused low quality score for most second reads in each pair. Consequently, data was treated as single-end data by using only the first read in each pair. Reads were trimmed of adaptor sequences with Cutadapt v1.16 (Martin, 2011) and aligned to GRCh38 or dm6 using the Bowtie 2 option of Bismark v0.22.2 (Krueger and Andrews, 2011) with default parameters except --local. Bismark was used in concert with Bowtie2 v2.2.9 (Langmead and Salzberg, 2012). Bismark alignment was a critical step as standard alignment software does not efficiently align short reads with one or more T-to-C mutations. Reads aligning to transcripts were quantified with HTSeq (Anders et al., 2015) htseq-count. SAMtools v1.5 (Li et al., 2009) was used to collect only read pairs with a mapping quality greater than 2 and concordant alignment (sam FLAG = 0/16). Mutation calling was performed essentially as described previously (Schofield et al., 2018). Briefly, T-to-C mutations were only considered if they met several conditions. Mutations must have a base quality score greater than 40 and be more than 3 nucleotides from the read’s end. Sites of likely single-nucleotide polymorphisms (SNPs) and alignment artefacts were identified with bcftools or from sites of high mutation levels in the nons4U treated controls (binomial likelihood of observation p < 0.05). These sites were not considered in mutation calling. Browser tracks were made using STAR v2.5.3a (Dobin et al., 2013). Reads which did not align in the initial alignment step were aligned to either the dm6 or GRCh38 genome (according to the spike-in species) in the same manner as above. Normalization scale factors were calculated with edgeR (Robinson et al., 2010) using read counts from the spike-in species (calcNormFactors using method = ‘upperquartile’).
TSS calling was performed with TSScall to identify annotated (obsTSS) and unannotated (uTSS) transcription start sites (Nechaev et al., 2010). Aligned sequencing reads from all samples of one species were pooled and analyzed with the TSScall pipeline. For Drosophila data, default settings were used except --annotation_search_window 500, --annotation_join_distance 100, and --call_method global. For human data, default settings were used except --annotation_search_window 1000, --annotation_join_distance 200, and -call_method big_winner. BEDTools (Quinlan and Hall, 2010) was used to assign reads to the nearest TSS within a 200bp window upstream and downstream of the read’s ends. TSSs are considered to be promoters if they are classified as an obsTSS. See STARR-seq methods below for eTSS calling.
TT-TL-seq
RNA previously collected for STL-seq was used for TT-TL-seq. Genomic DNA was depleted by treating with TURBO DNase and total RNA was extracted with one equivalent volume of Agencourt RNAClean XP beads according to manufacturer’s instructions. 50 μg of total RNA was subjected to MTS chemistry and biotinylation followed by streptavidin enrichment essentially as previously described (Schofield et al., 2018). TimeLapse chemistry was performed as described above. For each sample, 10 ng of RNA input was used to prepare sequencing libraries from the Clontech SMARTer Stranded Total RNA-Seq kit (Pico Input) with ribosomal cDNA depletion. Libraries were sequenced on a NovaSeq 6000 2X100bp. Filtering and alignment to the Drosophila dm6 genome were performed as described previously (Schofield et al., 2018).
Alignment of previously published sequencing data
For TT-TL-seq from 293T cells, filtering and alignment to the human hg38 genome were performed as described previously (Schofield et al., 2018). PRO-seq sequencing data was filtered and aligned in the same manner as TT-TL-seq data except during HISAT2 alignment of PRO-seq data, the default mismatch penalties were used, and mutation calling was not performed.
All ChIP-seq, ATAC-seq, and STARR-seq data were treated identically. Reads were filtered to remove duplicate sequences with FastUniq (Xu et al., 2012), trimmed of adaptor sequences with Cutadapt v1.16 and aligned to the Drosophila dm6 genome using the Bowtie2 v2.2.9 (Langmead and Salzberg, 2012). SAMtools v1.5 was used to collect reads with a mapping quality greater than 2 and concordant alignment (sam FLAG = 0/16 for single-end data and 147/99 or 83/163 for paired-end data).
Previously published Start-seq data was processed identically to STL-seq data to align reads and normalize counts.
Estimation of Pol II turnover with previous data under triptolide inhibition
To estimate scRNA half-lives from previously published Start-seq data under Trp inhibition, TSSs with low read counts in the uninhibited control samples were removed and all samples were normalized to the control. The data were transformed to the log scale and each TSS was fit with a linear model. We found that normalized Start-seq signal increases at many TSSs upon Trp treatment, suggesting that Trp affects the kinetics of Pol II at the promoter-proximal pause site and making it an unreliable approach. This artifact produced negative estimates which are biologically impossible. TSSs demonstrating this behavior and were calculated to have a negative rate constant were removed from our analysis.
For previously published ChIP-nexus data under Trp inhibition, pausing half-lives were published with the associated transcript isoform.
Estimation of the new fraction of scRNA and kinetic parameters of scRNA
All samples treated with 5-minute s4U feeds were modeled with the same Binomial model. For each treatment, the number of uridines (nu) and T-to-C mutations (TCi) in each read is determined and reads are grouped by the TSS to which they map. The s4U-untreated samples were used as unlabeled controls (c) to determine the background mutation rate attributed to reverse transcription mistakes, sequencing error, or other sources. The new fraction of scRNA (θ) and mutation rate were modeled as a mixture of two binomial distributions of either true TimeLapse or background mutations parametrized on the logistic scale. The probability mass function of the model is:
where pn is the TimeLapse mutation rate in new transcripts and po is the background mutation rate. Under normal steady-state conditions, we assume an exponential model relating the new fraction of transcripts at the sth TSS and the observed turnover rate constant for scRNA () such that
where t is the s4U labeling time of the experiment. To estimate termination and release, the termination rate constant at each TSS ()was defined with an upper boundary of the total observed rate constant such that
where a is a real value with lower limit of 0. The new fraction of transcripts under FP inhibition was related to in the same manner as .
The TSS specific pause release rate constant () was calculated as the difference between and . This parameterization of and avoided cases where release is very slow, and the tail of the posterior distribution may extend into the negative range due to an unrestricted model.
To estimate these parameters, we used a Bayesian hierarchical modeling approach using RStan software (Version 2.19.3, Carpenter et al., 2017) that implements no-U-turn Markov Chain Monte Carlo (MCMC) sampling. We designed non-centered hierarchical models to estimate global TimeLapse mutation rate () for the jth treatment condition while also allowing for variability by estimating TSS-specific mutation probabilities (). For the background mutation rate, we estimated a single global parameter () while allowing for local variation among TSSs by estimating TSS-specific mutation probabilities (). We used weakly informative priors for global mutation rates on the logistic scale which covered the range of previously observed mutation rates that could be reasonably expected. The TSS-specific mutation rates were found by estimating a standard deviation (σ) for each global parameter and a TSS-specific z-score (z). Finally, s4U-labeled and unlabeled control samples are indicated by I where if sample c is labeled with s U I = 1 and zero if the sample is unlabeled.
Global parameter priors:
Local parameter priors:
For reads :
The definition of θ in terms of and is included in the model which allows retrieval of posterior distributions of all parameters. Similarly, is calculated within the model as a generated quantity thereby generating a posterior distribution of estimates. Fits from these models converged well at all TSSs when run on the complete dataset with a minimum average read cutoff in the s4U-untreated controls (50 reads from fly cells and 100 reads from human cells). We limited our analysis to TSSs with an 80% CI size for was smaller than 1 on the natural log scale to avoid TSSs where we could not make a precise estimate. To identify the high confidence TSSs, we further limited our analysis to TSSs with an 80% CI that was smaller than 0.5 on the natural log scale. We only consider estimates to be high confidence if both and for the TSS qualified as high confidence. In all cases, we report estimates of the parameters using the median value of the posterior distribution.
Estimation of the global effect of flavopiridol on premature termination
To assess if flavopiridol influences , we developed a model designed to test for flavopiridol-induced changes in turnover. This model is similar to the model described above. We defined a TSS-specific effect parameter (f[s]) such that the turnover at a TSS under FP inhibition depends on and f[s] as defined below
where f is unrestricted and the scaled value of is guaranteed to be greater than zero. Therefore, the definition of θFP is
While the definition of θSS is unchanged in this model. In addition, a hierarchical parameter for the effect of FP (fg) was defined so that the prior for the local effect at each TSS depends on the global effect of FP and the standard deviation of the global effect (σf).
Global parameter priors:
Local parameter prior:
Parameterizations and definitions for the other parameters described above remain unchanged. Estimates for all kinetic parameters were made within the same model and f[s] was transformed into the log2 fold change within the generated quantities of the model. This model converged well when run on the complete dataset with a minimum average read cutoff of 50 reads in the s4U-untreated controls. We performed the same filtering as described above to determine high confidence TSSs under uninhibited and inhibited conditions. We identified TSSs where should be very close to zero by those whose gene-body coverage in TT-TL-seq was in the bottom 10% of all genes. As previously, the median value for the FP-induced log2 fold change at each TSS was used as a point estimate for the true value.
Simulation of STL-seq data
Local TSS-specific mutation rates were randomly chosen from a normal distribution centered on 0.1 and 0.0025 on the logistic scale for true TimeLapse (pn) and background (po) mutations, respectively. The mean values were chosen based on observed mutation rates observed in previous TimeLapse data. For nreads as defined in the text, we simulated scRNA reads from a TSS with half-life hl using the following model:
where the ith read contains nu[i] uridines which are evenly spaced along the read every nt nucleotides. The uridine frequency was chosen this way because scRNA initiated from the same TSS will contain identical sequences which only vary by the distance transcribed (l[i]). The mean length and standard deviation were selected to closely reflect the true distribution of read lengths across all scRNA reads. The new fraction of reads (θ) depends on the half-life (hl) of scRNA and the s4U labeling time (t) of the experiment. Whether a read is new was randomly assigned with a Bernoulli distribution with probability θ. If a read is new, the mutation rate and the number of mutations observed in a read (TC[i]) is determined according to a binomial distribution with nu[i] trials and probability pn. If a read is old, the number of mutations is determined similarly but with probability po. Five TSSs with half-lives 1, 2.5, 5, 7.5, and 10 mins were simulated together with varying degrees of coverage (25 to 1000 reads) and treated as data from a STL-seq experiment. These data were modeled with the same binomial model described above to estimate the scRNA half-life. The estimated turnover rates were compared to the true values used as input to the simulation.
TT-TL-seq data analysis
RPKM was calculated with the total length of each transcript isoform. TSScall identified transcript isoforms associated with each called TSS. If a single isoform cannot be unambiguously assigned to a TSS, the longest isoform was chosen. Transcripts were grouped into equal quartiles by kinetic parameters of their TSS or subsets as defined in the text. Metaplots and heatmaps were produced with deepTools2 (Ramirez et al., 2016).
PRO-seq data analysis
As a measure of promoter-proximal pausing, PRO-seq reads were counted within the first 250bp downstream of every TSS called from STL-seq. To determine transcriptional activity over the gene body, PRO-seq reads were counted in the range of 251–1250bp downstream of every TSS called from STL-seq.
ChIP-seq and ATAC-seq data analysis
Aligned ChIP-seq reads for all datasets were counted within the first 500bp downstream of all TSSs identified in STL-seq data. Aligned ATAC-seq reads within the window of −200 to +100 around each TSS were counted. TSSs were grouped into equal quartiles by kinetic parameters and deepTools2 was used to generate metaplots and heatmaps.
STARR-seq data analysis and eTSS identification
To identify STARR-seq peaks from previously published data, aligned bam files of STARR-seq biological replicates from either a developmental core promoter (dCP) or housekeeping core promoter (hkCP) were merged and analyzed with the STARRpeaker tool using default parameters (except --mincov 1). Resulting peak calls for dCP and hkCP were merged and TSSs were assigned as a STARR-seq active TSS if they were within 500 bp of a peak. TSSs were considered to be enhancer TSSs (eTSS) if they are classified as a uTSS by TSScall and have STARR-seq enhancer activity.
Identification of Promoter motifs
PWMTools was used to search for consensus sequences of each motif within the specified window around annotated promoter TSSs identified in STL-seq data. Matches to the consensus sequence were not allowed to contain any mismatches.
| Drosophila Motif | Consensus sequence | TSS Search window |
|---|---|---|
| TATA box | STATAWAWR (Ohler et al., 2002) | −110 to +1 |
| Initiator +G (InrG) | TCAGTY (Ohler et al., 2002; Hendrix et al., 2008) | −5 to −1 |
| Initiator -G (Inr) | TCAHTY (Ohler et al., 2002; Hendrix et al., 2008; Shao et al., 2019) | −5 to −1 |
| TCT motif | YYCTTTYY (Parry et al., 2010) | −5 to −1 |
| Downstream promoter element (DPE) | KCGGTTSK (Ohler et al., 2002) | +1 to +50 |
| Motif ten element (MTE) | CSARCSSA (Lim et al., 2004) | +1 to +50 |
| Pause Button (PB) | KCGRWCG (Hendrix et al., 2008) | +1 to +50 |
| Human Motif | Consensus sequence | TSS Search window |
|
| ||
| TATA box | TATAWAAR (Carninci et al., 2006; Ponjavic et al., 2006; Juven-Gershon et al., 2008) | −110 to +1 |
| Initiator | YYANWYY (Javahery et al., 1994) | −5 to −1 |
| TCT motif | YCTYTYY (Parry et al., 2010) | −5 to −1 |
| Downstream promoter element (DPE) | RGWYV (Burke and Kadonaga, 1996) | +1 to +50 |
| Motif ten element (MTE) | CSARCSSA (Lim et al., 2004) | +1 to +50 |
Quantification and Statistical Analysis
Information for the quantification and statistical analyses of data presented here can be found in the STAR methods subsections described above or in the relevant figure legends.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-thiouridine (s4U) | Alfa Aesar | AAJ60679-MC |
| Methane thiosulfonate biotin-XX (MTSEA-biotin-XX) | Biotium | 91022 |
| Dynabeads MyOne Streptavidin C1 beads | Thermo Fisher Scientific | 65002 |
| meta-chloroperoxybenzoic acid (mCPBA) | Alfa Aesar | AAAL00286-14 |
| 2,2,2-trifluoroethylamine (TFEA) | Thermo Fisher Scientific | AC303500010 |
| Flavopiridol | Sigma Aldrich | F3055-1MG |
| Triptolide | Invivogen | Ant-tpl |
| Agencourt RNAClean XP beads | Beckman Coulter | A63987 |
| SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input | Takara Bio USA | 634413 |
| Deposited Data | ||
| S2 Start-seq | Krebs et al., 2017; Elrod et al., 2019 | GEO: GSE77369 |
| Kc167 ChIP-nexus pausing half-lives | Shao and Zeitlinger, 2017 | https://github.com/zeitlingerlab/Shao_NG_2017 |
| S2 PRO-seq | Elrod et al., 2019 | GEO: GSE114467 |
| S2 H3K4me3, H3K4me1, H3K27ac ChIP-seq | Henriques et al., 2018 | GEO: GSE85191 |
| S2 H3K36me3 | Chen et al., 2012 | GEO: GSE27679 |
| S2 ATAC-seq | Elrod et al., 2019 | GEO: GSE114467 |
| S2 STARR-seq | Arnold et al., 2013; Zabidi et al., 2015 | GEO: GSE57876 |
| HEK293T TT-TL-seq | Rosa-Mercado et al., 2021 | GEO: GSE152063 |
| Raw and analyzed S2 STL-seq data | This paper | GEO: GSE166202 |
| Raw and analyzed 293T STL-seq data | This paper | GEO: GSE166202 |
| Raw and analyzed S2 TT-TL-seq data | This paper | GEO: GSE166202 |
| Experimental Models: Cell Lines | ||
| HEK293T cells | Steitz Lab | NA |
| S2 cells | Simon Lab | NA |
| Oligonucleotides | ||
| STL-seq custom 3´ DNA adapter | Table S1 | NA |
| STL-seq custom 5´ RNA adapter | Table S1 | NA |
| STL-seq custom RT primer | Table S1 | NA |
| STL-seq custom indexing primers | Table S1 | NA |
| Software and Algorithms | ||
| SAMtools | Li et al., 2009 | NA |
| BEDTools | Quinlan and Hall, 2010 | NA |
| FastUniq | Xu et al., 2012 | NA |
| Cutadapt | Martin, 2011 | NA |
| Bismark | Krueger and Andrews, 2011 | NA |
| Bowtie2 | Langmead and Salzberg, 2012 | NA |
| HISAT2 | Kim et al., 2019 | NA |
| HTSeq | Anders et al., 2015 | NA |
| edgeR | Robinson et al., 2010 | NA |
| deepTools | Ramirez et al., 2016 | NA |
| Stan | Carpenter et al., 2017 | NA |
| STAR | Dobin et al., 2013 | NA |
| PWMTools | Ambrosini, PWMTools | NA |
| TSScall | Nechaev et al., 2010 | NA |
| STARRpeaker | Lee et al., 2020 | NA |
| STL-seq mutation calling pipeline | This paper | https://bitbucket.org/mattsimon9/timelapse_pipeline/src/master/ |
| RStan models to estimate kinetic parameters | This paper | https://bitbucket.org/mattsimon9/timelapse_pipeline/src/master/ |
Highlights.
STL-seq measures the kinetics of promoter-proximal paused Pol II transcripts
Four out of five initiated transcripts are terminated at the pause site on average
Cell signaling regulates release into elongation to alter gene expression
Hyperosmotic stress induces termination at pause sites of TATA-less promoters
Acknowledgments
We thank N. Dimitrova, K. Neugebauer, and all members of the Simon Lab for valuable discussion and feedback, M. Machyna for improvements to computational analysis of sequencing data, and L. Kiefer, M. Sullivan, and I. Vock for assistance with Bayesian modeling and simulation. This work was supported by the NIH NIGMS T32GM007223 (J.T.Z.), CA200147 (J.A.S. & N.A.R.M.), T32AI055403 (N.A.R.M.), NIH New Innovator Award DP2 HD083992-01 (M.D.S.), and NIH R01 GM137117 (M.D.S.). N.A.R.M. is a Ford Foundation Predoctoral Fellow and J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests
J.A.S. is a member of the Molecular Cell Advisory Board. M.D.S. is the inventor on a patent application related to nucleotide recoding. The authors declare no further competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K. and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet, 13(10), 720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, PWMTools, http://ccg.epfl.ch/pwmtools
- Anders S, Pyl PT, and Huber W. (2015). HTSeq--a Python framework to work with highthroughput sequencing data. Bioinformatics, 31(2), 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń Ł M, Rath M, and Stark A. (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science, 339(6123), 1074–7. [DOI] [PubMed] [Google Scholar]
- Bartman CR, Hamagami N, Keller CA, Giardine B, Hardison RC, Blobel GA, and Raj A. (2019). Transcriptional Burst Initiation and Polymerase Pause Release Are Key Control Points of Transcriptional Regulation. Mol Cell, 73(3), 519–532 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckedorff F, Blumenthal E, Dasilva LF, Aoi Y, Cingaram PR, Yue J, Zhang A, Dokaneheifard S, Valencia MG, Gaidosh G, et al. (2020). The Human Integrator Complex Facilitates Transcriptional Elongation by Endonucleolytic Cleavage of Nascent Transcripts. Cell Rep, 32(3), 107917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. (2012). mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell, 46(3), 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley MS, Kwak H, Zipfel WR, and Lis JT (2014). Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev, 28(1), 14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW and Kadonaga JT (1996). Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters Genes Dev, 10, 711–724. [DOI] [PubMed] [Google Scholar]
- Calo E. and Wysocka J. (2013). Modification of Enhancer Chromatin: What, How, and Why? Mol Cell, 49(5), 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet, 38(6), 626–35. [DOI] [PubMed] [Google Scholar]
- Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, Brubaker M, Guo J, Li P, and Riddell A. (2017). Stan: A Probabilistic Programming Language. J Stat Softw, 76(1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gao X, and Shilatifard A. (2015). Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev, 29(1), 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim T-K, He HH, Zieba J, et al. (2012). Systematic evaluation of factors influencing ChIP-seq fidelity. Nature Methods, 9(6), 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. and Adelman K. (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienemann C, Schwalb B, Schilbach S, and Cramer P. (2019). Promoter Distortion and Opening in the RNAPolymerase II Cleft. Mol Cell, 73, 97–106. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, and Adelman K. (2019). The Integrator Complex Attenuates Promoter-Proximal Transcription at ProteinCoding Genes. Mol Cell, 76(5), 738–752 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson B, Sheridan RM, Cortazar M, and Bentley DL (2018). Dynamic turnover of paused Pol II complexes at human promoters. Genes Dev, 32(17–18), 1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant CB, Levandowski CB, Gupta K, Maas ZL, Moir J, Rubin JD, Sawyer A, Esbin MN, Rimel JK, Luyties O, et al. (2020). TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser NW, Sehgal PB, and Darnell JE (1978). DRB-induced premature termination of late adenovirus transcription. Nature, 272, 590–593. [DOI] [PubMed] [Google Scholar]
- Gariglio P, Bellard M, and Chambon P. (1981). Clustering of RNA polymerse B molecules in the 5’ moiety of the adult β-globin gene of hen erythocytes. Nucleic Acids Res, 9(11), 2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates LA, Foulds CE, and O’malley BW (2017). Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem Sci, 42(12), 977989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS and Lis JT (1986). RNA Polymerase II Interacts with the Promoter Region of the Non-induced hsp70 Gene in Drosophila melanogaster Cells. Mol Cell Biol, 6(11), 39843989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, Decker TM, Qin W, Leonhardt H, Eick D, and Cramer P. (2017). CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, and Young RA (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell, 130(1), 7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet, 39(3), 311–8. [DOI] [PubMed] [Google Scholar]
- Hendrix DA, Hong J-W, Zeitlinger J, Rokhsar DS, and Levine MS (2008). Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. PNAS, 105(22), 7762–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, and Adelman K. (2013). Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell, 52(4), 517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, and Adelman K. (2018). Widespread transcriptional pausing and elongation control at enhancers. Genes Dev, 32(1), 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-L, Jee D, Stein CB, Elrod ND, Henriques T, Mascibroda LG, Baillat D, Russell WK, Adelman K, and Wagner EJ (2020). Integrator Recruits Protein Phosphatase 2A to Prevent Pause Release and Facilitate Transcription Termination. Mol Cell, 80(2), 345358.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger MG, Schwalb B, Mackowiak SD, Velychko T, Hanzl A, Imrichova H, Brand M, Agerer B, Chorn S, Nabet B, et al. (2020). Selective Mediator dependence of cell-typespecifying transcription. Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, and Smale ST (1994). DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biochem, 14(1), 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, and Lis JT (2014). Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife, 3, e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, and Kadonaga JT (2008). The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol, 20(3), 253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology, 37(8), 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, and Schubeler D. (2017). Genome-wide Single-Molecule Footprinting Reveals High RNA Polymerase II Turnover at Paused Promoters. Mol Cell, 67(3), 411–422 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. and Andrews SR (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27(11), 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, and Groudine M. (1995). Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation Genes Dev, 9, 559–572. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, and Lis JT (2013). Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods, 9(4), 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, and Roeder RG (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell, 152(5), 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Shi M, Moran J, Wall M, Zhang J, Liu J, Fitzgerald D, Kyono Y, Ma L, White KP, et al. (2020). STARRPeaker: uniform processing and accurate identification of STARRseq active regions. Genome Biology, 21(1), 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Santoso B, Boulay T, Dong E, Ohler U, and Kadonaga JT (2004). The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev, 18(13), 1606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Žumer K, Molska EŠ, Rouvière JO, Wu G, Demel C, Schwalb B, Schmid M, Cramer P, and Jensen TH (2021). Integrator is a genome-wide attenuator of non-productive transcription. Molecular Cell, 81(3), 514–529.e6. [DOI] [PubMed] [Google Scholar]
- Marshall NF and Price DH (1995). Purification of P-TEFb, a Transcription Factor Required for the Transition into Productive Elongation. Journal of Biological Chemistry, 270(21), 12335–12338. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet journal, 17, 10–12. [Google Scholar]
- Nechaev S, Fargo DC, Santos GD, Liu L, Gao Y, and Adelman K. (2010). Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science, 327, 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson KA, Lawson CK, Mullen NJ, Ball CB, Spector BM, Meier JL, and Price DH (2017). Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res, 45(19), 11088–11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao G-C, Niemann H, and Rubin GM (2002). Computational analysis of core promoters in the Drosophila genome Genome Biol, 3(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, and Kadonaga JT (2010). The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev, 24(18), 2013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, and Sandelin A. (2006). Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol, 7(8), R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res, 44(W1), W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer KA, Mimoso CA, Adelman K, and Neugebauer KM (2021). Co-transcriptional splicing regulates 3′ end cleavage during mammalian erythropoiesis. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Mercado NA, Zimmer JT, Apostolidi M, Rinehart J, Simon MD, and Steitz JA (2021). Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, and Simon MD (2018). TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods, 15(3), 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, and Cramer P. (2016). TT-seq maps the human transient transcriptome. Science, 352(6290), 1225–1228. [DOI] [PubMed] [Google Scholar]
- Shao W, Alcantara SGM, and Zeitlinger J. (2019). Reporter-ChIP-nexus reveals strong contribution of the Drosophila initiator sequence to RNA polymerase pausing. eLife, 8, e41461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W. and Zeitlinger J. (2017). Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet, 49, 1045–1051. [DOI] [PubMed] [Google Scholar]
- Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, Van Cappellen WA, et al. (2018). Live-cell analysis of endogenous GFPRPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. PNAS, 115(19), E4368–E4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer DC, Elrod ND, Liang D, Xiao MS, Jiang JZ, Jonathan M, Huang KL, Wagner EJ, Cherry S, and Wilusz JE (2019). The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev, 33(21–22), 1525–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettey TT, Gao X, Shao W, Li H, Story BA, Chitsazan AD, Glaser RL, Goode ZH, Seidel CW, Conaway RC, et al. (2019). A Role for FACT in RNA Polymerase II Promoter-Proximal Pausing. Cell Rep, 27(13), 3770–3779 e7. [DOI] [PubMed] [Google Scholar]
- Uyehara CM and Mckay DJ (2019). Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. PNAS, 116(20), 9893–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, Van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, and Timmers HTM (2007). Selective Anchoring of TFIID to Nucleosomes by Trimethylation of Histone H3 Lysine 4. Cell, 131(1), 58–69. [DOI] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, and Steitz JA (2015). Widespread Inducible Transcription Downstream of Human Genes. Mol Cell, 59(3), 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, and Cramer P. (2018). Structure of paused transcription complex Pol II-DSIF-NELF. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, and Handa H. (1998). Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. The EMBO journal, 17(24), 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, and Adelman K. (2015). Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell, 58(2), 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, and Impey S. (2011). KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J, 30(8), 1473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, and Chen S. (2012). FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One, 7(12), e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, and O’connor MB (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol, 58, 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, and Stark A. (2015). Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature, 518(7540), 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of TT-TL-seq and STL-seq libraries generated in this study are publicly available and accessible on Gene Expression Omnibus (GEO) under GSE166202.
Mutation calling was performed with a custom analysis pipeline freely available at https://bitbucket.org/mattsimon9/timelapse_pipeline/src/master/. Kinetic parameter estimates were made with custom RStan models freely available in the same repository. Custom scripts written for downstream analyses are available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.