Abstract
To fight against the worldwide COVID-19 pandemic, the development of an effective and safe vaccine against SARS-CoV-2 is required. As potential pandemic vaccines, DNA/RNA vaccines, viral vector vaccines and protein-based vaccines have been rapidly developed to prevent pandemic spread worldwide. In this study, we designed plasmid DNA vaccine targeting the SARS-CoV-2 Spike glycoprotein (S protein) as pandemic vaccine, and the humoral, cellular, and functional immune responses were characterized to support proceeding to initial human clinical trials. After intramuscular injection of DNA vaccine encoding S protein with alum adjuvant (three times at 2-week intervals), the humoral immunoreaction, as assessed by anti-S protein or anti-receptor-binding domain (RBD) antibody titers, and the cellular immunoreaction, as assessed by antigen-induced IFNγ expression, were up-regulated. In IgG subclass analysis, IgG2b was induced as the main subclass. Based on these analyses, DNA vaccine with alum adjuvant preferentially induced Th1-type T cell polarization. We confirmed the neutralizing action of DNA vaccine-induced antibodies by a binding assay of RBD recombinant protein with angiotensin-converting enzyme 2 (ACE2), a receptor of SARS-CoV-2, and neutralization assays using pseudo-virus, and live SARS-CoV-2. Further B cell epitope mapping analysis using a peptide array showed that most vaccine-induced antibodies recognized the S2 and RBD subunits. Finally, DNA vaccine protected hamsters from SARS-CoV-2 infection. In conclusion, DNA vaccine targeting the spike glycoprotein of SARS-CoV-2 might be an effective and safe approach to combat the COVID-19 pandemic.
Keyword: COVID-19, DNA vaccine, Adjuvant, Antibody, T cell activation, Neutralization activity
1. Introduction
The pandemic of COVID-19 spread from the reported cluster of pneumonia cases in Wuhan, Hubei Province, in Dec 2019. Patients with COVID-19 present with viral pneumonia caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) (World Health Organization (WHO) “Novel Coronavirus-China” WHO, 2020: www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/). The number of infected people, and death reached over 178 million, and over 3.8 million, respectively as of May 2021 and is still increasing worldwide. The development of an effective and safe vaccine to combat this unprecedented global pandemic is urgently needed. Many pharmaceutical companies and academia are developing vaccines against SARS-CoV-2, including adenovirus-based, DNA or RNA-based vaccines [1], [2], [3], [4], [5], mostly targeting the spike glycoprotein of SARS-CoV-2, which is essential for virus entry into cells [6]. Some of them are already approved for clinical use in many countries [7].
The advantages of DNA vaccines are that they (1) can be simply and quickly produced by PCR or synthetic methods, (2) can be easily produced at a large scale, (3) are safer than other approaches, such as inactivated virus vaccines, and (4) are more thermostable than other types of vaccines [8], according to WHO DNA vaccine guideline. SARS-CoV-2, classified as the species severe acute respiratory syndrome-related coronavirus, is a member of the family of enveloped positive-sense RNA viruses [9]. The mutation rate of RNA viruses is known to be higher than that of DNA viruses [10], suggesting that developed vaccines for SARS-CoV-2 need to be adapted for its mutation. On the other hand, one concern of development of DNA vaccine using plasmid DNA, especially in human clinical trials, is weak immunogenicity [11,12]. To overcome this limitation, many administrative conditions such as administrative routes, adjuvants have been explored to improve the efficacy of DNA vaccine [13]. In this study, we developed DNA-based vaccine targeting the SARS-CoV-2 spike glycoprotein. In the guidance by the FDA on vaccines to prevent COVID-19, preclinical studies of a COVID-19 vaccine candidate require the evaluation of humoral, cellular, and functional immune responses to support proceeding to initial human clinical trials. Accordingly, in this study, antibody production measured by an antigen-specific enzyme-linked immunosorbent assay (ELISA) was considered to represent the humoral response, and antigen-dependent T cell activation by an enzyme-linked immunosorbent spot (ELISpot) assay was evaluated for cellular immune responses. The functional activity of immune responses was evaluated in vitro by neutralization assays using pseudo-virion virus and live virus. The assays used for immunogenicity evaluation should be demonstrated to be suitable for their intended purpose. Furthermore, we conducted B cell epitope analysis for the induced antibodies. Finally, we conducted virus challenge test to evaluate vaccine protective efficacy in hamsters.
2. Methods
2.1. Animal protocol
Seven-week-old male and female Jcl:SD rats were purchased from Clea Japan Inc. (Tokyo, Japan), and housed with free access to food and water in a temperature and light cycle-controlled facility. All experiments were approved by the Ethical Committee for Animal Experiments of the Osaka University Graduate School of Medicine. For immunization, DNA vaccine (666.6 μg or 66.7 μg DNA plasmid with 66.7 μl of alum adjuvant in 400 μl) was intramuscularly injected at 0, 2, and 4 weeks with a needle and syringe (200 μl/injection x 2 sites/rat) [14,15]. The content of alum adjuvant was determined according to approved human vaccines [16]. 6 rats were used in vaccine group (Vaccine No.1-No.6). 3 rats were used in control group (Control No.1-No.3). Blood samples were collected every 2 weeks until 30 weeks after the first vaccination. At 7 weeks, the spleen, kidney, lung, and heart were collected for further analysis (spleen for ELISpot assay; kidney, lung, and heart for tissue toxicity).
2.2. DNA vaccine
pVAX1 was obtained from Invitrogen (USA). The virus RNA of SARS-CoV-2 (isolate Wuhan-Hu-1; MN_908,947.3) was obtained from the National Institute of Infectious Disease (Tokyo, Japan). A highly optimized DNA sequence encoding the SARS-CoV-2 Spike glycoprotein was created using an in silico gene optimization algorithm to enhance and immunogenicity. The synthesized sequences with Kozak consensus sequences were inserted between NheI and XbaI of the pVAX1 plasmid. Obtained plasmid sequences were confirmed by Sangar sequencing (Supplementary Information). Plasmid DNA was amplified and purified by GIGA prep (Qiagen, USA). Alum phosphate (5 mg/ml as aluminum content, Aluminium phosphate gel: vac-phos-250), as an adjuvant, was obtained from In Vivogen (USA).
2.3. Cells, DNA transfection, western blotting, and immunostaining
Human embryonic kidney 293 (HEK293) cells were cultured in DMEM containing 10% FBS plus penicillin/streptomycin and grown at 37 °C in a humidified 5% CO2 incubator.
For western blotting, plasmid DNA (pVAX1-SARS-CoV-2 Spike) was transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. After 48 h, the cells were washed and lysed with RIPA buffer (Sigma) containing protease inhibitor cocktail (Roche). After sonication and centrifugation, the supernatant was collected and stored at −80 °C until use. The protein concentration was determined by Bradford assay (Takara), and the cell lysate was mixed with 4x Laemmli loading buffer containing β mercapto-ethanol. The boiled samples were separated on gradient gels (4–20%, Bio-Rad) and transferred to PVDF membranes. After the membrane was blocked with PBS containing 5% skim milk for 1 h at room temperature (RT), the membrane was incubated with an antibody against SARS-CoV-2 Spike (GNX135356, GeneTex, Inc.) at 4 °C overnight. The washed membrane was incubated with a secondary antibody labeled with horseradish peroxidase (HRP) (GE healthcare) for 1 h at RT. After washing, the membrane was developed with a substrate (Chemi-Lumi One L, Nacalai Tesque). The bands were detected by ChemiDoc™ Touch (Bio-Rad).
For immunostaining, the cells were seeded on glass-bottomed dishes (Matsunami glass) and incubated for 24 h. At 80% confluence, plasmid DNA (pVAX1 or pVAX1-SARS-CoV-2 Spike) was transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. After 48 h, the cells were fixed with 4% paraformaldehyde and permealized with 0.2% Triton X-100. The cells were blocked with 5% skim milk-PBS for 1 h at RT and incubated with an antibody against SARS-CoV-2 spike (BLSN-005P, Beta Lifescience) or control IgG (Thermo Fisher) at 4 °C overnight. After washing the cells with PBS, the cells were incubated with a secondary antibody labeled with Alexa Fluor 488 (Molecular Probes) for 1 h at RT. Nuclei were stained with DAPI. The stained cells were observed by confocal microscopy (FV10i, Olympus).
2.4. Antibody titer determination by ELISA
In this study, we collected serum samples from the tail vein every 2 weeks and evaluated antibody titers by ELISA. Briefly, recombinant 2019-nCoV Spike S1+S2 protein (ECD: BLPSN-0986P, Beta Lifescience), recombinant 2019-nCoV Spike protein (RBD: BLPSN-0988P, Beta Lifescience), recombinant SARS-CoV-2 S protein, (SPN—C52H9, Acro Biosystems), recombinant SARS-CoV-2 S protein with mutations of B.1.1.7 variant(SPN—C52H6, Acro Biosystems), recombinant SARS-CoV-2 S protein with mutations of B.1.351 variant (SPN—C52Hk, Acro Biosystems), recombinant SARS-CoV-2 S protein with mutations of P.1 variant (SPN—C52Hg, Acro Biosystems), (1 μg/ml) were coated on 96-well plates on the first day. On the second day, wells were blocked with blocking buffer (PBS containing 5% skim milk) for 2 h at RT. The sera were diluted 10- to 31,250-fold in blocking buffer and incubated overnight at 4 °C. The next day, the wells were washed with PBS containing 0.05% Tween®20 (PBS-T) and incubated with HRP-conjugated IgG antibodies (NA935, GE healthcare; ab106753, ab106783, ab106750, abcam; 3075-05, Southern Biotech) for 3 h at RT. After washing with PBS-T, wells were incubated with the peroxidase chromogenic substrate 3,3′−5,5′-tetramethyl benzidine (Sigma-Aldrich) for 30 min at RT, and then the reaction was halted with 0.5 N sulfuric acid. The absorbance of the wells was immediately measured at 450 nm with a microplate reader (Bio-Rad). The value of the half-maximal antibody titer of each sample was calculated from the highest absorbance in the dilution range by using Prism 6 software.
2.5. Sandwich ELISA for detection of secreted spike protein
A diluted capture antibody for the spike protein (GTX632604, GeneTex) was coated on a 96-well plate and incubated at 4 °C overnight. After the plate was blocked with PBS containing 5% skim milk for 2 h at RT, diluted recombinant protein (S1+S2, Beta Lifescience) as a standard and samples were loaded and incubated at 4 °C overnight. The washed plate was incubated with a detection antibody (40,150-R007, Sino Biological) for 2 h at RT. After washing with PBS-T, a diluted secondary antibody labeled with HRP (GE healthcare) was added and incubated for 3 h at RT. After washing with PBS-T, HRP was developed by adding the peroxidase chromogenic substrate 3,3′−5,5′-tetramethyl benzidine (Sigma-Aldrich) for 30 min at RT, and then the reaction was halted with 0.5 N sulfuric acid. The absorbance at 450 nm was measured by a microplate reader (Bio-Rad). The concentration of the spike protein was calculated by a standard curve obtained from recombinant S1+S2 protein.
2.6. ELISpot assay
Cellular immune responses were measured by ELISpot assay, according to the manufacturer's instructions (UCT Biosciences). Ninety-six-well PVDF membrane-bottomed plates (Merck Millipore) were coated with an anti-rat IFNγ capture antibody or IL4 capture antibody and incubated at 4 °C overnight. After the plates were washed with PBS, the plates were blocked with blocking stock solution (UCT Biosciences) for 2 h at RT. Splenocytes from immunized or control rats were adjusted to 3 × 105 well and stimulated with recombinant 2019-nCoV Spike S1+S2 protein (ECD: Beta Lifescience) or recombinant 2019-nCoV-Spike protein (RBD; Beta Lifescience) at 37 °C for 48 h. The washed plates were incubated with a biotinylated polyclonal antibody specific for rat IFNγ or IL4 for 2 h at 4 °C. After washing with PBS-T, diluted streptavidin-HRP conjugate was added and incubated for 1 h at RT. HRP was developed with a substrate solution (AEC coloring system, UCT Biosciences), and then the reaction was stopped by rinsing both sides with demineralized water and air drying at RT in the dark. The colored spots were counted using a dissecting microscope (LMD6500, Leica).
2.7. Epitope array
Epitope mapping of vaccine-induced antibody was performed by using a CelluSpots peptide array (CelluSpots™ COVID19_HullB:98.301, Intavis), according to the manufacturer's instructions. After blocking the membrane with PBS containing 5% skim milk for 1 h at RT, diluted serum samples (1:10) were added and incubated overnight at 4 °C. The next day, the membrane was washed with PBS-T and then incubated with HRP-conjugated anti-Rat IgG (1:1000; NA935, GE healthcare) for 1 h at RT. After membrane washing, spots were developed by Chemi-Lumi One L (07,889–70, Nacalai Tesque). Signals were detected with a ChemiDoc Touch Imaging System (Bio-Rad) and analyzed with Image Lab software version 6.0.1 (Bio-Rad).
2.8. ACE2 binding assay
For the binding of RBD with ACE2, 96 well plate was coated with RBD recombinant protein (0.5 μg/ml, Fc tag; 40,592-V02H, Sino Biological) at 4 °C overnight. After the plate was blocked with PBS containing 5% skim milk for 2 h at RT, the plate was incubated with rat diluted serum 4 °C overnight. Next day, the plate was washed with PBS containing Tween 20 (0.05%) (PBS-T), and then human ACE2 recombinant protein (0.25 μg/ml, His-tag; 10,108-H08H, Sino Biological) was added to the wells and incubated for 2 h at RT. After washing with PBS-T, plate was incubated with anti-his antibody conjugated with HRP (ab1187, abcam) for 2 h at RT. After washing with PBS-T, wells were incubated with the peroxidase chromogenic substrate 3,3′−5,5′-tetramethyl benzidine (Sigma-Aldrich) for 30 min at room temperature, then reaction was halted with 0.5 N sulfuric acid. Absorbance of wells were immediately measured at 450 nm with a microplate reader (Bio-Rad).
For the binding of S1+S2 with ACE2, the 96 well plate was coated with human ACE2 recombinant protein (1 μg/ml, mFc tag; 83986S, Cell Signaling Technology). The plate was blocked with PBS containing 5% skim milk for 2 h at RT. After blocking, pre-incubated sample of serum with recombinant S1+S2 protein (2 μg/ml, His-tag; Beta Lifescience) was added to wells and incubated at 4 °C overnight. After the plate was washed with PBS-T, the wells was incubated with anti-his antibody conjugated with HRP (abcam) for 2 h at RT. After washing with PBS-T, the peroxidase chromogenic substrate 3,3′−5,5′-tetramethyl benzidine (Sigma-Aldrich) was added to wells and incubated for 30 min at RT. After stopped the reaction by adding 0.5 N sulfuric acid. The absorbance at 450 nm was measured by a microplate reader (Bio-rad).
2.9. Pseudo-virus neutralization assay for SARS-CoV-2
The neutralizing activity of vaccine-induced antibodies was analyzed with pseudo-typed vesicular stomatitis virus (VSVs), as previously described [17]. Briefly, Vero E6 cells stably expressing TMPRSS2 were seeded on 96-well plates and incubated at 37 °C for 24 h. Pseudo-viruses were incubated with a series of dilutions of inactivated rat serum for 1 h at 37 °C, and then added to Vero E6 cells. At 24 h after infection, the cells were lysed with cell culture lysis reagent (Promega), and luciferase activity was measured by a Centro XS3 LB 960 (Berthold).
2.10. Live virus neutralization assay for SARS-CoV-2
The neutralizing activity of vaccine-induced antibodies was analyzed by focus reduction neutralization test (FRNT) and tissue culture infectious dose (TCID) methods. SARS-CoV-2 JPN/TY/WK-521 strain obtained from the National Institute of Infectious Disease (Tokyo, Japan) was used in this study. The FRNT was carried out according to a previously described method with slight modification [18,19]. Briefly, neutralization assay was based on the reduction in focus forming units (FFU) after exposing a given amount of virus to the product to be characterized and comparing with the untreated control. VeroE6/TMPRSS2 cells (4 × 104 cells/well) were seeded on 96-well plates and incubated at 37 oC for 24 h. Serum samples were serially diluted (factor 2 dilutions: 1:10, 1:20, 1:40, 1:80, 1:160, 1:320 and 1:640) and incubated with equal volume of live SARS-CoV-2 (50 FFU/30 μl) at 37 °C for 30 min. Aliquots of 30 μl/well of each diluted serum-virus complex were added in VeroE6/TMPRSS2 cells and inoculation for 30 min (37 oC; 5% CO2). After inoculation, cells were added 50 μl/well of 1% carboxymethyl cellulose and 0.5% BSA supplemented Dulbecco's Modified Eagle Medium (DMEM) and incubated for 20 h (37 oC; 5% CO2). After incubation, plates were fixed with ethanol and virus infectious cells were stained by peroxidase-anti-peroxidase (PAP) method. As first antibody, 1 μg/ml of anti-SARS-CoV/SARS-CoV-2 nucleoprotein monoclonal antibody, HM1054 (East Coast Bio) was used. The TCID assay was carried out as previously described [20]. Briefly, Serum samples were serially diluted and incubated with equal volume of virus solution (100TCID50/50 μl) at 37 oC for 30 min. Aliquots of 50 μl of each diluted serum-virus complex were added in VeroE6/TMPRSS2 cells, seeded in 96-well plates and incubated for 2 days (37 oC; 5% CO2). The cells were fixed with 10% formaldehyde (Sigma-Aldrich), and stained with 0.1% methylene blue. Acute and convalescent sera of SARS-CoV-2 infected patients were used as negative and positive controls, respectively. The acute-phase serum showed no neutralization at all while the convalescent serum possessed the significant neutralization activity. The use of patients sera with written informed consent was approved by an Ethics Committee of Research Instituted for Microbial Diseases Osaka University (approval number 31–14–1).
2.11. Histological analysis
For histological analysis, tissues (kidney, liver, lung, and heart) were collected at 7 weeks after the first vaccination and fixed in 10% neutral buffered formalin. Fixed tissues were embedded in paraffin, cut into 5-μm-thick sections and stained with hematoxylin and eosin (HE staining). Stained tissue sections were observed using a BZ-X810 (Keyence).
2.12. Serum biochemical parameters
Rat serum biochemical parameters were measured by a Vetscan VS2 (Abaxis, Tokyo, Japan) analyzer with a multirotor II VCDP/VLA (Abaxis), according to the manufacturer's instructions. Biochemical parameters, such as albumin (ALB), alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), total bilirubin (TBIL), blood urea nitrogen (BUN), total calcium (CA), phosphorus (PHOS), creatinine (CRE), glucose (GLU), NA+, K+, total protein (TP), globulin (GLOB), and creatine kinase (CK) were analyzed.
2.13. SARS-CoV-2 viral challenge in golden Syrian hamsters
All experiments with hamsters were performed by Onco Design. Animal protocol was conducted under the French and European Regulations and the National Research Council Guide for the Care and Use of Laboratory Animals. All procedures were submitted the Institutional Animal Care and Use Committee of CEA approved by (CETEA DSV-no44). Hamsters were maintained in SPF and controlled environmental conditions. Wild-type (female, 6 week-old) Golden Syrian Hamsters were obtained from Janvier Labs (French). The body weight was daily taken through day 42 to 56.
Vaccine protocol: The hamsters were anesthetized by inhalation of vaporized isoflurane, and intramuscularly injected with total 100 μl volume of DNA vaccine (2 sites of 50 μl one shot) at day 0, day, 14 and day 28 [21]. The composition of DNA vaccine was mixed in 5:1 ratio (DNA vaccine; 175 μg and adjuvant; 83.3 μg), For control group, the mixture in 5:1 ratio (PBS and adjuvant 83.3 μg ) was intramuscularly injected.
Viral challenge: At 42 days after 1st vaccination, A dose of 105 pfu of SARS-CoV-2 virus (strain: Slovakia/SK-BMC5/2020) was inoculated via intranasal route under isoflurane-anesthetized condition.
Viral load determination in lung by quantitative realtime PCR using SARS-CoV-2 ORF1ab, and RNA dependent RNA polymerase (RdRp) genes. The lung samples were collected at 2, 4, 7, and 14 day after virus challenge. Viral RNA was extracted by using QIAamp Viral RNA Mini Kit (Qiagen), and was stored at −80 °C until RT-PCR. Complete RT-PCR was performed using SuperScript™ III One-step qRT-PCR System kit (Life Technologies) with ORF1ab primers: F_ CCGCAAGGTTCTTCTTCGTAAG, R_ TGCTATGTTTAGTGTTCCAGTTTTC, Probe_ AAGGATCAGTGCCAAGCTCGTCGCC, and RdRp Institut Pasteur (IP) 2 primers: F_ATGAGCTTAGTCCTGTTG, R_CCCCTTTGTTGTGTTGT, Probe_Hex-AGATGTCTTGTGCTGCCGGTA-BHQ-1, RdRp Institute Pasteur (IP)4 primers: F_GGTAACTGGTATGATTTCG, R_CTGGTCAAGGTTAATATAGG, Probe_Fam-TCATACAAACCACGCCAGG-BHQ-1, which were designed by Institut Pasteur, Paris [22,23]. Amplification of targeted sequences were performed by using a Bio-Rad CFX96™ or CFX384™ and adjoining software.
The expression of inflammatory cytokines (IFNγ, TNFα, IL6, and IL10) in lung was analyzed by quantitative realtime-PCR: The primers used for each target gene are shown as follows: IFNγ primers: F_ TGTTGCTCTGCCTCACTCAGG, R_ AAGACGAGGTCCCCTCCATTC, TNFα primers: F_ TGAGCCATCGTGCCAATG, R_ AGCCCGTCTGCTGGTATCAC, IL6 primers: F_ AGACAAAGCCAGAGTCATT, R_ TCGGTATGCTAAGGCACAG, IL10 primers: F_ GGTTGCCAAACCTTATCAGAAATG, R_ TTCACCTGTTCCACAGCCTTG. γ-actin primers: F_ ACAGAGAGAAGATGACGCAGATAATG, R_ GCCTGAATGGCCACGTACA.The expression of targeted genes were normalized with γ-actin.
Virus tissue culture infectious dose (TCID)50 determination in lung by immune-fluorescence plaque assay: Briefly, Vero6/TMPRSS2 cells was plated in 96-well plate at the density of 2.5 × 104 per well. Cells were infected with serial dilutions of each lung preparation after SARS-CoV-2 challenge for 1 h at 37 °C, followed by adding 100 μl of fresh growth medium to the cells. At 6 h after infection, cells were fixed with formalin, and permeabilized with 0.1% Triton-X. After blocking, SARS-CoV-2 nucleoprotein was stained with NP antibody (Sino Biological: 40,143-R019), and secondly antibody with Alexa Fluor 488 (Thermo Scientific, A-11,034). The cells were treated with Hoechst dye (Thermo Scientific, 33,342). Fluorescent images were obtained by using Operetta (Perkin Elmer).
Histological analysis of lung: Extracted lung tissues were embedded in paraffin and sectioned with 5 μm thickness. Sectioned tissues were stained with hematoxylin-phloxin (H&P) stain to visualize histomorphometric changes (inflammation). Slides were scanned using the NanoZoomer Digital Pathology System C9600–02, analyzed by using Definiens software.
Evaluation of SARS-CoV-2 antibody by ELISA: Anti-SARS-CoV-2 antibody (IgG) was measured by using V-PLEX SARS-CoV-2 Panel 2 plates (Meso Scale Discovery; K15383U), according to manufacturer's instruction. The antibodies for S, RBD, and N antigens of SARS-COV-2 were analyzed using anti-hamster IgG antibody conjugated with MSD SULFO-tag.
2.14. Statistical analysis
All values are presented as the mean±SEM. Student's t-test, One-way ANOVA followed by Bonferroni's multiple test was used to assess significant differences in each experiment using Prism 8 software (GraphPad Software). Differences were considered significant when the p value was less than 0.05.
3. Results
3.1. DNA vaccine design and in vitro expression
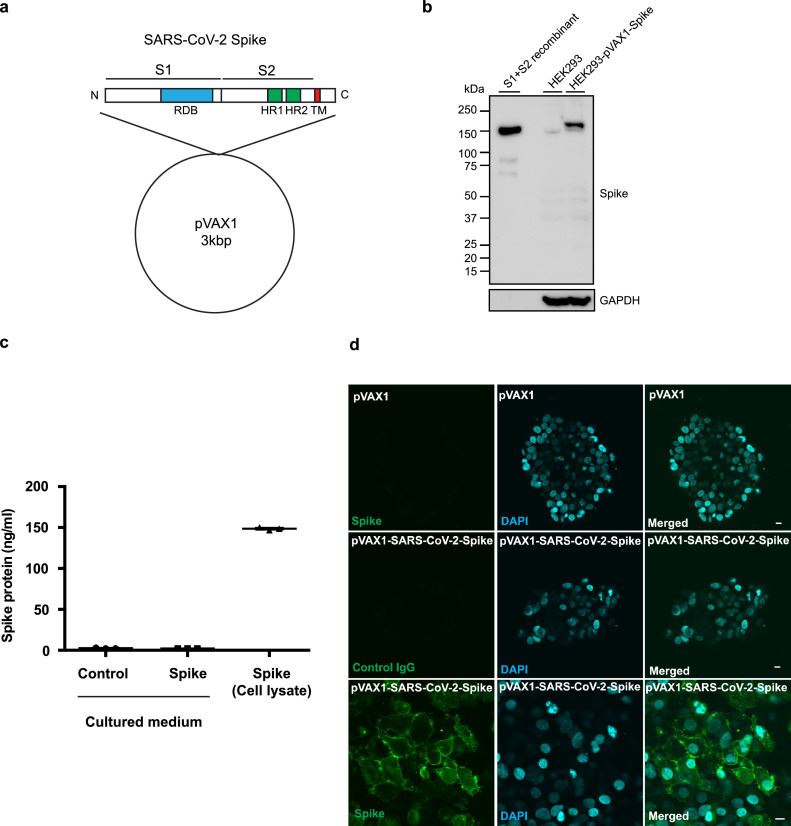
The optimized DNA sequence of the SARS-CoV-2 (isolate Wuhan-Hu-1; MN_908,947.3) spike glycoprotein was inserted into the pVAX1 plasmid (Fig. 1 a). The spike glycoprotein contains the RBD, heptad repeat 1 (HR1), heptad repeat 2 (HR2), the transmembrane domain, and the cytosolic domain. The expression of the spike glycoprotein was confirmed by western blot. The construct was transfected into HEK293 cells and incubated for 48 h. The spike glycoprotein was detected in HEK293 cells transfected with the DNA vaccine construct using a specific anti-Spike glycoprotein antibody, and the recombinant protein was almost the same size as the band of recombinant S1+S2 (extracellular domain: ECD, Fig. 1b). To quantify the expression level of the spike glycoprotein in the cells or in the supernatant, a sandwich ELISA was developed by using anti-Spike glycoprotein antibodies. It has been reported that SARS-CoV-2 infects host cells via ACE2 [6]. We are concerned with the possibility that the spike glycoprotein originating from DNA vaccine could bind to host ACE2 to regulate its function, including cardiopulmonary function [24,25]. Thus, we also evaluated the secretion of the spike glycoprotein in the culture medium of HEK293 cells transiently overexpressing the SARS-CoV-2 spike protein. Indeed, the expression of the spike glycoprotein was detected in the lysate of transfected cells, but not in the supernatant of transfected HEK293 cells (Fig. 1c). The localization of expressed spike glycoprotein was also analyzed by immunostaining in HEK293 cells. At 48 h after transfection, cells were fixed and stained with an anti-spike glycoprotein antibody. The spike protein was localized mainly on the plasma membrane (Fig. 1d, Fig. S1). These in vitro studies confirmed the expression of the spike glycoprotein from the DNA vaccine construct.
Fig. 1.
DNA vaccine design and expression of DNA plasmids in cells. (a) Design of DNA vaccine encoding SARS-CoV-2 spike protein based on pVAX1. The Spike glycoprotein is composed of an S1 subunit containing the RBD (receptor-binding domain), an S2 subunit containing HR1 (heptad repeat 1) and HR2 (heptad repeat 2), a transmembrane domain and a cytosolic domain. (b) In vitro expression of pVAX1-SARS-CoV-2 Spike in HEK293 cells as assessed by western blot. Transfected HEK293 cell lysate (HEK293-pVAX-Spike), non-transfected HEK293 cell lysate (HEK293) and recombinant spike glycoprotein(S1+S2 recombinant) were separated by SDS-PAGE and transferred to PVDF membrane. The Spike glycoprotein was detected by the polyclonal anti-spike antibody. GAPDH was used as loading control. (c) Quantification of pVAX1-SARS-CoV-2 Spike protein in HEK293 cells as assessed by sandwich ELISA. The Spike glycoprotein was detected in transfected HEK293 cell lysate (transfected HEK293 lysate), but not in the culture medium of transfected HEK293 cells (sup.). (d) Localization of pVAX1-SARS-CoV-2 Spike glycoprotein in HEK293 cells by immunostaining. The transfected spike protein was stained with the polyclonal spike antibody and the secondary antibody labeled with Alexa Fluor 488 (green). The nucleus was stained with DAPI (blue). Scale bar = 10 μm. Data are shown as mean ± SEM. See also Fig. S1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Animal protocol for DNA vaccine administration and evaluating humoral responses in rats
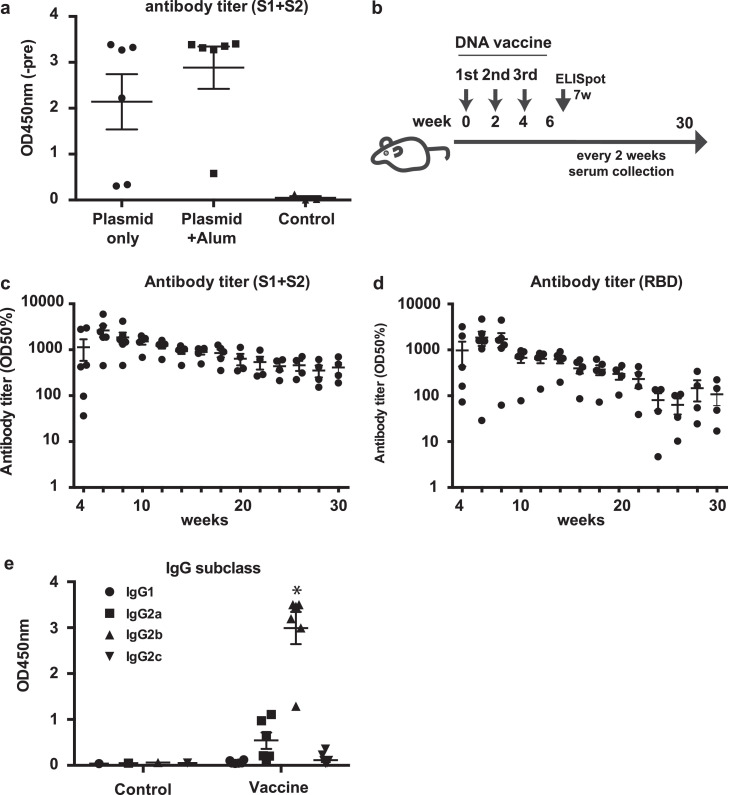
Although plasmid DNA itself induces innate immune responses leading to adjuvant action [26], previous reports suggest that the effect of DNA vaccines can be enhanced with alum adjuvant [27,28]. To determine whether co-administration of alum adjuvant with DNA vaccine for SARS-CoV-2 enhances antibody production, we compared the antibody titer induced by DNA vaccine with or without alum adjuvant at 2 weeks. Based on the formulation of several vaccines with alum adjuvants in humans, the dose of alum adjuvant in human clinical trials has been fixed at 0.2 mL (1 mg of aluminum). The compositions of the plasmid DNA and alum adjuvants in this study were calculated based on the clinical protocol (2 mg of DNA plasmid and 1 mg of aluminum). As a result, antibody production was enhanced by alum adjuvant (Fig. 2 a), and we selected the co-administration of alum adjuvant for further experiments. The DNA vaccine with alum adjuvant (666.6 μg of plasmid DNA with 66.7 μl of alum adjuvant/rat) was intramuscularly injected into SD rats three times at 2-week intervals (Fig. 2b). Anti-spike IgG was produced in DNA vaccine-dose dependent manner (Fig. S2). DNA vaccine-induced antibody production was followed up to 30 weeks after the 1st vaccination. The antibody titers (half maximum) for the spike glycoprotein and its RBD protein was elevated at 4 through 30 weeks (Fig. 2c, 2d, Fig. S3). Moreover, antibody titers for B.1.1.7, B.1.351, and P.1 were evaluated by ELISA using variant spike recombinant proteins. DNA vaccine-induced IgG titers for variants were slightly decreased against B.1.351 and P.1 spike recombinant protein (Fig. S4). At 4 weeks after the 1st vaccination, IgG subclasses (IgG1, IgG2a, IgG2b, and IgG2c) were analyzed by ELISA. Compared with IgG1, IgG2b (and IgG2a) was the main subclass produced by DNA vaccine, suggesting that the humoral immune response shifted toward Th1 response rather than Th2 (Fig. 2e). These data suggest that DNA vaccine effectively activated humoral immune responses.
Fig. 2.
DNA vaccine animal protocol and humoral response induced by the DNA vaccine. (a) Comparison of the effect of DNA vaccine with or without alum adjuvant. Antibody titers were compared at 2 weeks after vaccine administration. (b) Animal protocol for DNA vaccine administration. DNA vaccine was intramuscularly injected with alum adjuvant (666.6 μg of plasmid DNA with 66.7 μl of alum adjuvant) three times at 2-week intervals. Serum samples were collected every two weeks for antibody titer evaluation. (c) Antibody titer for recombinant S1 + S2 and (d) antibody titer for recombinant RBD assessed by ELISA from 4 to 16 weeks after the 1st vaccination. Serum dilution from 10x to x31250. The antibody titer is shown as the serum dilution exhibiting half maximum binding at optical density at 450 nm (OD50%). (e) IgG subclasses for recombinant S1 + S2 assessed by ELISA at 4 weeks. Serum dilution: 8x dilution. *p < 0.01 vs. IgG1, IgG2a, IgG2c, respectively. Data are shown as mean ± SEM. ANOVA followed by Bonferroni comparison. See also Fig. S2, S3 and S4.
3.3. The DNA vaccine effectively elicits a cellular immune response in rats
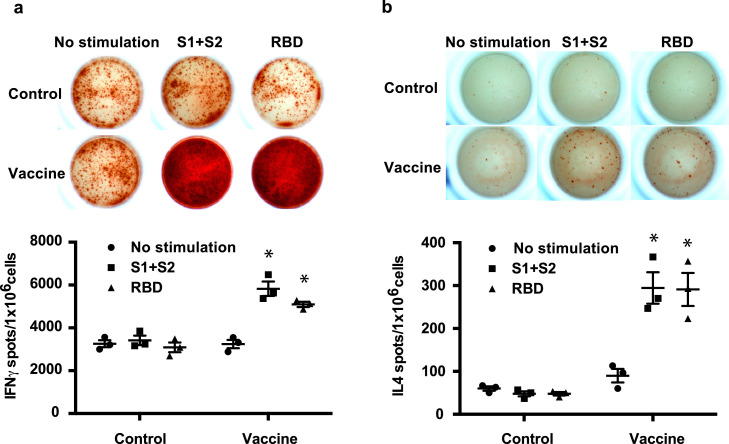
We also examined whether DNA vaccine would induce the cellular immune response by IFNγ and IL4 ELISpot assays at 7 weeks after the 1st vaccination. IFNγ spots were significantly increased by S1+S2 recombinant protein and recombinant RBD protein immunization in rats (Fig. 3 a). IL4 spots were slightly increased by S1+S2 recombinant and RBD recombinant protein administration (Fig. 3b). These results and the IgG subclass analysis (Fig. 2e) suggest that DNA vaccine would elicit the cellular immune response toward the Th1 type.
Fig. 3.
Cellular immune response to the DNA vaccine in rats. (a) IFNγ ELISpot assay and (b) IL4 ELISpot assay in immunized or control rats at 7 weeks after the first vaccination. Splenocytes were stimulated with recombinant S1 + S2 or recombinant RBD for 48 h. *P < 0.01 vs. control (splenocytes from untreated rats). Data are shown as mean ± SEM. ANOVA followed by Bonferroni comparison.
3.4. The DNA vaccine enhanced neutralizing antibodies
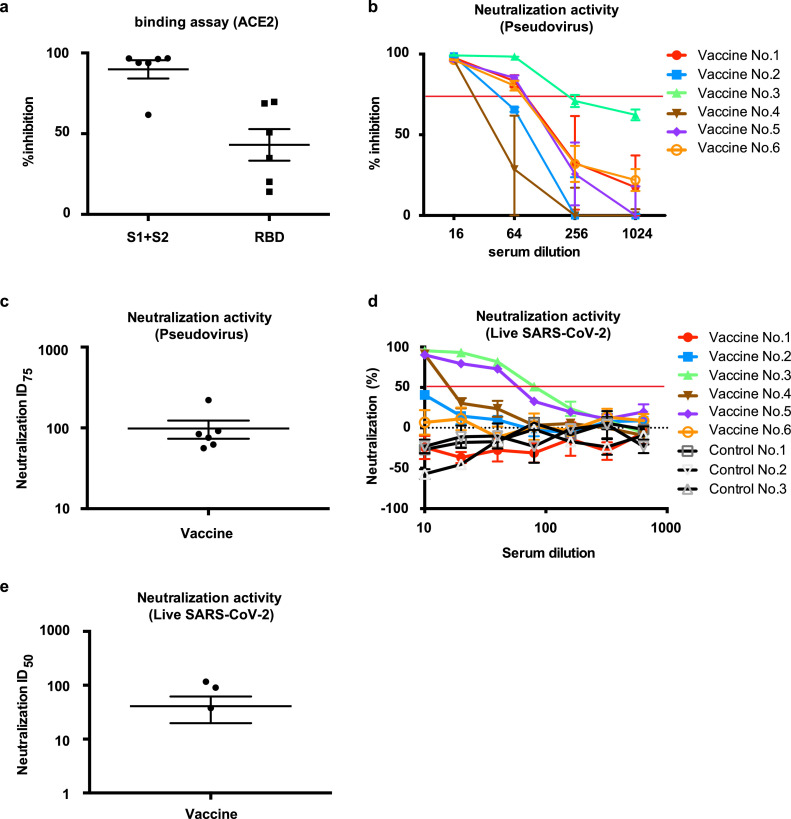
We further evaluated the neutralization activity of the vaccine-induced antibodies against SARS-CoV-2 with two different methods. Binding of human ACE2, a receptor of the SARS-CoV-2 spike glycoprotein [6], was evaluated by ELISA. The binding of ACE2 with S1+S2 recombinant protein was inhibited by a 5-fold dilution of immunized rat serum, and the binding of ACE2 and RBD protein was also decreased (Fig. 4 a). Moreover, neutralizing antibodies were tested by pseudo-typed VSV with the luciferase gene and Vero E6 cells stably expressing TMPRSS2 [6,29]. A series of dilutions of serum at 8 weeks after the first vaccination exhibited neutralizing activity on pseudo-virus infection (Fig. 4b), and neutralizing titers (75% inhibitory dose (ID75) shows the serum dilution that caused a 75% decrease in RLUs) were 98.4 on average at 8 weeks after the 1st administration of the DNA vaccine (Fig. 4c). We further evaluated the neutralizing activity by utilizing live SARS-CoV-2 in two different method (FRNT and TCID). The neutralizing titers (ID 50: 50% inhibitory dose) in FRNT method were 40.7 on average (Fig. 4d, 4e, Fig. S5), and 10–80 in TCID method (Table 1 , Fig. S6) 8 weeks after the 1st administration of the DNA vaccine.
Fig. 4.
Neutralizing activity of DNA vaccine-induced antibodies. (a) Inhibitory activity of vaccine-induced antibodies for the binding of ACE2 and SARS-CoV-2 Spike or RBD. The vaccinated sera at 8 weeks after 1st vaccination was used at 10-fold dilution for ELISA. “S1+S2” indicates the binding between ACE2 and SARS-CoV-2 Spike. “RDB” indicates the binding between ACE2 and SARS-CoV-2 RBD. (b) Dose-dependent pseudovirus neutralizing activity of vaccine-induced antibodies. Serial dilution (x16, x64, x256, and x1024) of vaccinated sera at 8 weeks after 1st vaccination was analyzed. The horizontal red line indicates 75% inhibition. Individual vaccinated rats were shown. (c) ID75 titers for pseudovirus neutralization activity of sera collected at 8 weeks after 1st vaccination from (b). (d) Dose-dependent live SARS-CoV-2 neutralization activity of vaccine-induced antibodies, analyzed by focus reduction neutralization test (FRNT). Serial dilution (x10, x20, x40, x80, x160, x320, and x640) of vaccinated sera at 8 weeks after 1st vaccination. The horizontal red line indicates 50% inhibition. Individual vaccinated rats were shown, (e) ID50 titers for live SARS-CoV-2 neutralization activity of sera collected at 8 weeks after 1st vaccination from (d). Vaccine No.3 and Control No.3: 7 weeks sample. Data are shown as mean ± SEM. See also Fig. S5 and S6. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
The result of neutralization activity of vaccinated serum by TCID.
The indicated values are maximum serum dilution that showed neutralization activity against live SARS-CoV-2. See also Fig. S6.
| No. | Neutralization activity | |
|---|---|---|
| Vaccine No.1 | <10 | |
| Vaccine No.2 | 10 | |
| Vaccine No.3 | 80 | |
| Vaccine No.4 | 20 | |
| Vaccine No.5 | 80 | |
| Vaccine No.6 | 10 | |
| Control No.1 | <10 | |
| Control No.2 | <10 | |
| Control No.3 | <10 |
3.5. Evaluation of epitopes in the SARS-CoV-2 spike protein
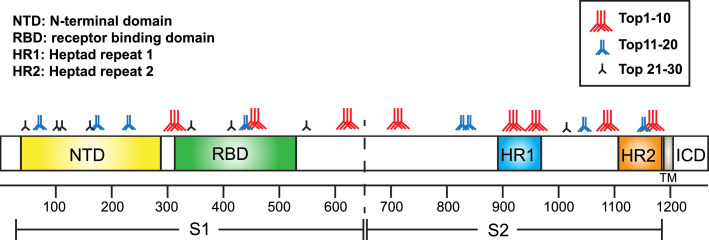
To identify the epitopes recognized by vaccine-induced antibodies, a peptide array based on the SARS-CoV-2 spike protein was performed with control serum or vaccine serum. The spike peptide-coated membrane treated with immunized rat serum showed many more dots than that treated with control serum (Fig. S7). Signals (dots) were ranked according to the strength of the signal for individual samples (Table S1), and the top 30 signals were determined from 6 rats (Fig. 5 and Table 2 ). The top 10 epitopes were localized in the RBD, HR1, HR2, and amino acids approximately 600–700 (near the S1/S2 cleaved site). These data suggest that mainly antibodies recognizing the RBD, HR1, and HR2 were produced by DNA vaccine with the SARS-CoV-2 spike glycoprotein.
Fig. 5.
Epitope mapping profiles of DNA vaccine-induced antibodies. The antigenic sites in the spike protein were ranked according to the results of epitope array. “Red three antibodies” indicate the top 10 epitopes. “Blue two antibodies” indicate the top 20 epitopes. “Black antibody” indicates the top 30 epitopes. See the details in Table 2, Table S1, and Fig. S7. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Top 30 strongest epitope recognized by vaccine-induced antibody: bold indicates RBD. See also Fig. 5, Fig. S7, and Table S1.
| Position | Amino Acid Sequence | |
|---|---|---|
| 1 | 1176 - 1190 | V-V-N-I-Q-K-E-I-D-R-L-N-E-V-A |
| 2 | 1256 - 1270 | F-D-E-D-D-S-E-P-V-L-K-G-V-K-L |
| 3 | 621 - 635 | P-V-A-I-H-A-D-Q-L-T-P-T-W-R-V |
| 4 | 1131 - 1145 | G-I-V-N-N-T-V-Y-D-P-L-Q-P-E-L |
| 5 | 971 - 985 | G-A-I-S-S-V-L-N-D-I-L-S-R-L-D |
| 6 | 851 - 865 | C-A-Q-K-F-N-G-L-T-V-L-P-P-L-L |
| 7 | 456 - 470 | F-R-K-S-N-L-K-P-F-E-R-D-I-S-T |
| 8 | 1251 - 1265 | G-S-C-C-K-F-D-E-D-D-S-E-P-V-L |
| 9 | 946 - 960 | G-K-L-Q-D-V-V-N-Q-N-A-Q-A-L-N |
| 10 | 311 - 325 | G-I-Y-Q-T-S-N-F-R-V-Q-P-T-E-S |
| 11 | 1211 - 1225 | K-W-P-W-Y-I-W-L-G-F-I-A-G-L-I |
| 12 | 811 - 825 | K-P-S-K-R-S-F-I-E-D-L-L-F-N-K |
| 13 | 451 - 465 | Y-L-Y-R-L-F-R-K-S-N-L-K-P-F-E |
| 14 | 221 - 235 | S-A-L-E-P-L-V-D-L-P-I-G-I-N-I |
| 15 | 181 - 195 | G-K-Q-G-N-F-K-N-L-R-E-F-V-F-K |
| 16 | 1096 - 1110 | V-S-N-G-T-H-W-F-V-T-Q-R-N-F-Y |
| 17 | 76 - 95 | T-K-R-F-D-N-P-V-L-P-F-N-D-G-V |
| 18 | 1171 - 1185 | G-I-N-A-S-V-V-N-I-Q-K-E-I-D-R |
| 19 | 1216 - 1230 | I-W-L-G-F-I-A-G-L-I-A-I-V-M-V |
| 20 | 846 - 860 | A-R-D-L-I-C-A-Q-K-F-N-G-L-T-V |
| 21 | 116 - 130 | S-L-L-I-V-N-N-A-T-N-V-V-I-K-V |
| 22 | 1221 - 1235 | I-A-G-L-I-A-I-V-M-V-T-I-M-L-C |
| 23 | 176 - 190 | L-M-D-L-E-G-K-Q-G-N-F-K-N-L-R |
| 24 | 556 - 570 | N-K-K-F-L-P-F-Q-Q-F-G-R-D-I-A |
| 25 | 106 - 120 | F-G-T-T-L-D-S-K-T-Q-S-L-L-I-V |
| 26 | 1056 - 1070 | A-P-H-G-V-V-F-L-H-V-T-Y-V-P-A |
| 27 | 326 - 340 | I-V-R-F-P-N-I-T-N-L-C-P-F-G-E |
| 28 | 31 - 50 | S-F-T-R-G-V-Y-Y-P-D-K-V-F-R-S |
| 29 | 416 - 430 | G-K-I-A-D-Y-N-Y-K-L-P-D-D-F-T |
| 30 | 1261 – 1275 | S-E-P-V-L-K-G-V-K-L-H-Y-T |
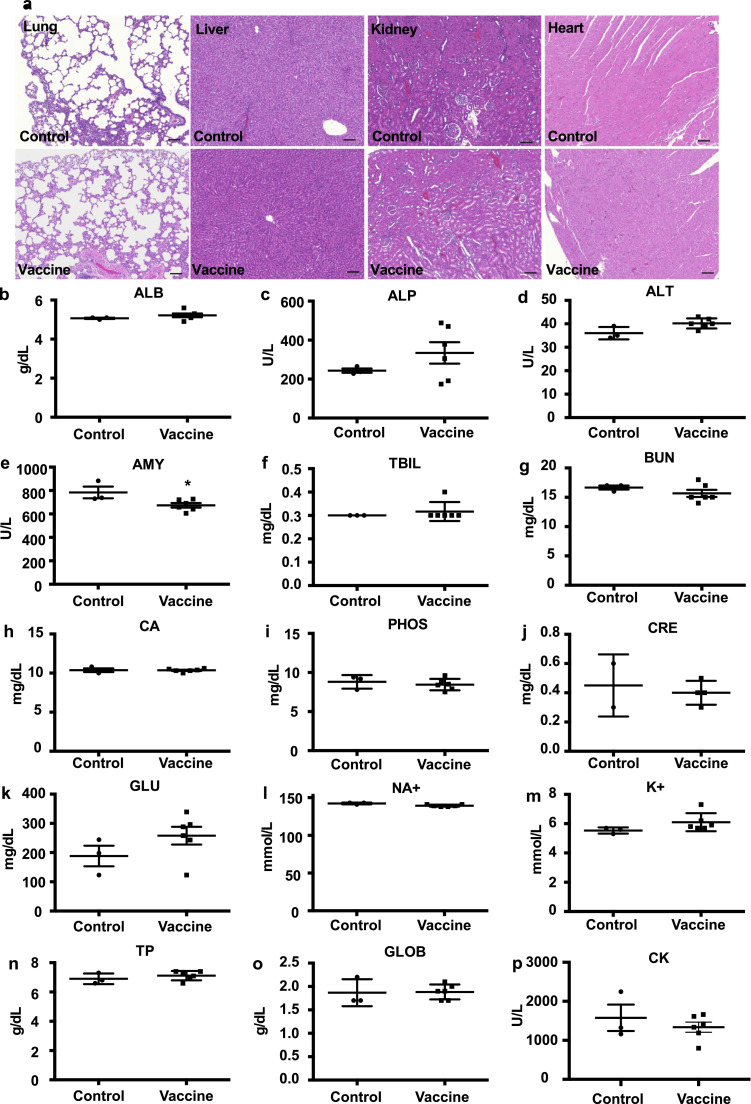
3.6. The DNA vaccine has no toxic effect
To evaluate the toxic effect of DNA vaccine on tissues, the lung, liver, kidney, and heart were collected from immunized rats at 7 weeks and analyzed by HE staining. No inflammatory cell infiltration, hemorrhage was detected in those tissues. The HE stained sections showed no tissue toxicity (Fig. 6 a). Additionally, serum biochemical parameters were evaluated to confirm the tissue toxic effect of the DNA vaccine. Serum biochemical data showed that amylase levels were slightly down-regulated by DNA vaccine compared with those in control serum. However, this variation was within the normal range. Other parameters were not changed (Fig. 6b-6p), suggesting that DNA vaccine does not have the toxic effects on tissues.
Fig. 6.
No tissue toxicity with DNA vaccine administration. (a) No tissue toxic effect with DNA vaccine administration assessed by histological analysis (HE staining; lung, liver, kidney, and heart). Scale bar = 100 μm.(b-p) Biochemical analysis of serum at 8 weeks after the 1st vaccination. ALB; albumin, ALP; alkaline phosphatase, ALT; alanine aminotransferase, AMY; amylase, TBIL; total bilirubin, BUN; blood urea nitrogen, CA; calcium, PHOS; phosphate, CRE; creatine, GLU; glucose, NA+; sodium, K+; potassium, TP; total protein, GLOB; globulin, CK; creatine kinase. In CRE analysis, values of 2 samples (1sample from control, 1 sample from vaccine) were below detection limit. Data are shown as mean ± SEM. t tests. *p < 0.05 vs control.
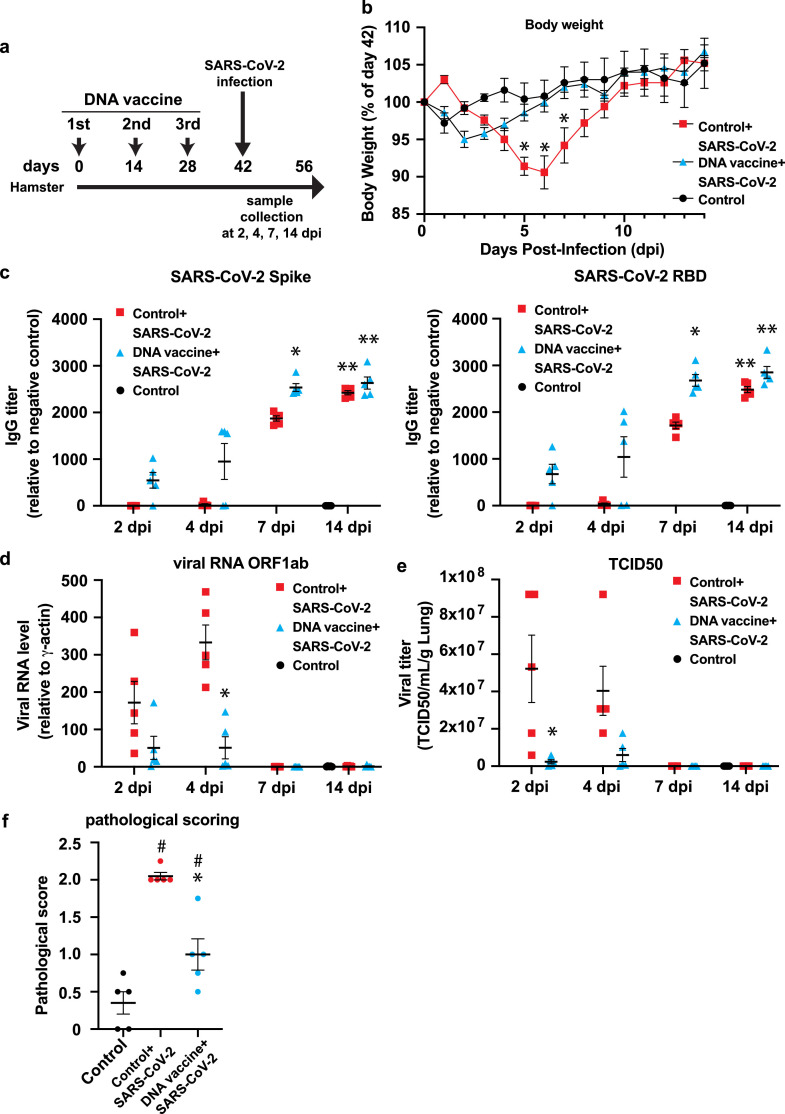
3.7. The DNA vaccine protects SARS-CoV-2 viral infection in hamsters
Finally, to evaluate the efficacy of vaccine for virus infection, the immunized Golden Syrian hamsters were challenged with SARS-CoV-2. Hamsters were immunized with DNA vaccine three times every 2 weeks (14 days), and challenged with SARS-CoV-2 via intranasal route (Fig. 7 a). After virus challenge, body weight was taken every day because body weight monitoring can be used as a parameter of severity of infection [30], [31], [32], [33]. In control group, hamsters lost body weight by 6 days after challenge, but not in DNA vaccinated group (Fig. 7b). Next, SARS-CoV-2 specific antibody (Spike, RBD, and nucleocapsid) was evaluated at 2, 4, 7, and 14 post-day infection (pdi) by ELISA. Anti-spike, RBD antibody was higher, compared with control with SARS-CoV-2 infection group at 7 pdi. Anti-nucleocapsid antibody was not different between control and vaccinated group (Fig. 7c,Fig. S8a). The amount of SARS-CoV-2 virus RNA (ORF1ab and IP2, IP4) in the lung was evaluated by quantitative realtime PCR. vRNA levels were increased at 2 and 4 pdi in control with SARS-CoV-2 infection group. However, DNA vaccine significantly suppressed vRNA level in the lung (Fig. 7d, Fig. S8b). Next, tissue viral loads were also measured in the lung at 4 pdi by tissue culture infection dose (TCID) assay. The viral load was suppressed in DNA vaccinated group, but not in control group (Fig. 7e). DNA vaccine suppressed inflammation, viral infection-related cytokines such as IL10 and IFNγ, and DNA vaccine tended to decrease inflammatory cytokines, TNFα, IL6 (Fig. S8c-S8f). Finally, pathological scoring (judged and scored by inflammation, edema, and hemorrhage) was also decreased in vaccinated group (Fig. 7f, Fig. S9) at 14 dpi. These data suggested that SARS-CoV-2 DNA vaccine protected the hamsters from COVID-19 infection.
Fig. 7.
SARS-CoV-2 viral protection by DNA vaccine in hamster. (a) Animal protocol for DNA vaccine administration in hamster. For DNA vaccine group, 100 μl of DNA vaccine was intramuscularly injected with alum adjuvant (175 μg of plasmid DNA with 83.3 μg of alum adjuvant/hamster) three times at 2-week intervals (two sites with 50 μl). For control group, 100 μl of PBS with alum adjuvant (83.3 μg of alum adjuvant/hamster) was intramuscularly injected. At day 42, hamsters were inoculated with 105 pfu (live SARS-CoV-2 virus) via intranasal route under a total volume of 70 μl (35 μl per nostril). (b) Body weight was monitored daily after viral challenge. (c) Antibody titer for the binding of antibody to recombinant spike protein S1+S2, or RBD recombinant. (d) Viral load determination in lung by RT-PCR. (e) Viral load determination by tissue culture infection dose assay (TCID). (f) At 14 day after viral challenge, pathological score was analyzed by lung section (Inflammation, edema, and hemorrhage). Inflammation, score 0: no pathological changes, score 1: 10% ≥ affected area, score 2: affected area > 10% to < 50%, score 3: affected area ≥ 50%; edema, score 0: absence, score 1, presence,;hemorrhage, score 0: absence, score 1, presence. *p < 0.05, vs. Control+DNA vaccine + SARS-CoV-2. # p < 0.05 vs. Control. Data are shown as mean ± SEM. ANOVA followed by Bonferroni comparison. See also Fig. S8 and S9.
4. Discussion
Here, we described the pre-clinical efficacy and safety studies of DNA vaccines for SARS-Cov-2. In this study, we confirmed the expression and immunogenicity of DNA vaccine in vitro and in vivo studies, and evaluated the humoral, cellular, and functional immune responses to support proceeding to initial human clinical trials.
DNA vaccine has several potential advantages, including the stimulation of both B and T cell responses, no observed integration of the vector into genomic DNA, rapid construction speed, and good thermostability during storage. DNA vaccine has been applied to many diseases, such as Alzheimer's disease, allergy, cancer and autoimmune diseases, as well as infections, such as those caused by HIV, hepatitis B, and West Nile virus (WNV) [34], [35], [36], [37], [38], [39], [40]. For example, DNA vaccine for WNV or Ebola/Marburg viruses efficiently induced immune responses in humans [41], [42], [43], [44], [45]. In the veterinary field, the protective immune responses have been observed against infectious agents in several target species, including fish, companion animals, and farm animals. DNA vaccines against WNV for use in horses and against infectious hematopoietic necrosis virus (IHNV) for use in salmon were licensed in the USA and in Canada, respectively. DNA vaccine against pancreatic disease was also licensed for use in farmed salmon in several countries [46]. Toward the clinical application in humans, many approaches have been conducted to enhance the immune response in clinical trials. Although optimization of the plasmid DNA vector (i.e., using strong promoters/enhancers or inserting CpG motifs to enhance adjuvant action) can potentially increase the immunogenicity and strength of gene expression, the plasmid backbone (pVAX) of DNA vaccine was employed in this study, since the pVAX plasmid has already been widely utilized for clinical use with good safety profile, and commercial launched as first gene therapy drug in Japan (Collategene) [47]. Instead, we optimized the formulation, including co-treatment of polymers, microparticles, utilizing the gene delivery system (intradermal injection, electroporation [48,49]), or co-administration of adjuvants [26], [27], [28]. For the rapid development of DNA vaccine, we selected the co-administration of alum adjuvant that has been clinically used in several vaccines. Although plasmid DNA itself induces innate immune responses leading to adjuvant action [26], co-administration of alum adjuvant with DNA vaccine for SARS-CoV-2 enhanced antibody production. The composition of DNA vaccine and alum adjuvants preferentially induced Th1-type T cell polarization based on ELISpot assay and IgG subclass analysis, which might be important to proceed to clinical trials. With model animals administered vaccine constructs against other coronaviruses, evidence of immunopathologic lung reactions characteristic of a Th-2 type hypersensitivity similar to enhanced respiratory disease (ERD) described with the respiratory syncytial virus (RSV) vaccine has been shown [26,[50], [51], [52], [53]].
In vivo animal experiments for SARS-CoV and MERS-CoV vaccines raised serious concerns of the potential risk for SARS-CoV-2 vaccine-associated antibody-dependent enhancement (ADE) [54,55]. In vitro studies of the effects of antibodies on viral infection have been used extensively to seek the correlates or predictors of ADE. These efforts are complicated by the fact that the antibody mechanisms for protection from viral disease for ADE are similar, and the effect of the administration of passive antibodies has been evaluated for the association with ADE. In small studies, patients infected with SARS-CoV or MERS-CoV received polyclonal antibodies without apparent worsening of their illness, and in the meta-analysis, early treatment with plasma from patients who had recovered from SARS-CoV infection correlated with an improved outcome [56], [57], [58], [59]. Moreover, a recent meta-analysis found no relationship between the kinetics of antibody responses to SARS-CoV, MERS-CoV or SARS-CoV-2 and clinical outcomes [60]. The current clinical experience is insufficient to implicate a role for ADE in the severity of COVID-19. To further investigate anti-Spike ADE, B cell epitope analysis was conducted using rat serum including DNA vaccine-induced antibody. Importantly, the antibodies produced by DNA vaccine greatly varied among the rats (Fig. 5). Thus, in the initial phase of clinical trials, B cell epitope analysis might be useful to evaluate the correlates or predictors of ADE with clinical symptoms, which include the magnitude and antigen specificity of antibodies, antibody subclasses and T cell subpopulations. To assess the risk of ADE, there is also value in available animal models for predicting the likelihood of such occurrence in humans. Therefore, post-vaccination animal challenge studies will be required in the near future for vaccine development. Currently, phase 1/2 clinical trial using this DNA vaccine has been tested in Japan. Preliminary data of this clinical trial demonstrated no serious adverse effects (data not shown), similar to the previous data of safety profile using DNA vaccines against various infectious disease.
In this study, our results suggests that DNA-based vaccine for SARS-CoV-2 effectively induced humoral and cellular immune responses, leading to protection from virus infection in animal model. Currently, circulatory five major variants as variant of concern (VOC) are spreading and presenting a growing thread to people worldwide and effectiveness of approved vaccines [61]. It has been shown that the approved vaccines-induced humoral immune responses were less effective against some VOCs, especially B.1.351 and P.1 [62,63]. DNA-based vaccine-induced IgG antibody titer specific for spikes of B.1.351 and P.1. were relatively decreased compared with original strain (Supplemental Figure 4). The result from epitope array using spike glycoprotein peptides showed that large portion of antibodies recognizing S2 domain were induced by DNA vaccine (Fig. 5, Table 2, and Supplemental Table 1). Many mutations has been accumulated in S1 domain in VOCs and others, affecting their infectivity [61,64], but not in S2 domain among coronaviruses [65], suggesting that antibodies derived by DNA vaccine are largely reactive with spike glycoprotein with original as well as VOC-related spikes. Moreover, it has been shown that monoclonal antibodies against HR1 and HR2 domains have potential to broadly neutralize coronaviruses through the inhibition of fusion process [65], [66], [67]. Additionally, cellular immune responses elicited by approved vaccines are equally effective among VOCs because CD4+ and CD8+ epitopes are almost conserved among VOCs and others [68]. Taken together, DNA-based vaccine-evoked immune activation, humoral and cellular immune responses, might be at least partly effective for VOCs and others. Though, the effectiveness of DNA vaccine developed in this study for emerging VOCs and others need to be further investigated.
So far, there are 2 papers available to compare with our DNA vaccine in small animal models, which are INO-4800 (intradermal injection with electroporation) from Inovio Pharmaceuticals [4], and Zydus-D (intradermal injection with PharmaJet®) from Zydus Cadila [69]. Both DNA vaccines proceeded to human clinical trial/application. The efficacy of vaccine: Their DNA vaccine also successfully and effectively induced anti-spike antibody production up to 6 weeks following immunization. Our result suggests anti-spike or RBD (receptor binding domain) antibody production, induced by our DNA vaccine, are durable, at least, up to 30 weeks (Fig. 2c and 2d). In our DNA vaccine system, plasmid DNA is intramuscularly injected with Alum adjuvant. Previous report suggested that alum adjuvant-containing vaccine promotes germinal center reaction, and increase follicular helper CD4 T (Tfh) cells, which are known as critical factors of long-lasting antibody production [70,71]. Our adjuvanted DNA vaccine is expected to induce more durable humoral immune responses, at least, in animal model. The safety issue: we evaluated the toxicity of DNA vaccine in two ways in this study. Histological observations suggested no abnormal alterations in lung, liver, kidney and heart (Fig. 6a). Also, there was no difference with control individuals in serum biochemical parameters (Fig. 6b-p). These indicated that our DNA vaccine is safe. Although the toxic profiles of their DNA vaccines have not been well investigated in their animal studies, there was no serious safety problem reported in clinical trials [72,73], suggesting that DNA vaccine is substantially safe way for immunization to fight against COVID-19.
Overall, these initial results describing the immunogenicity of DNA vaccine targeting S protein for SARS-CoV-2 will provide basal evidence toward to the clinical trials. Development of DNA vaccine against SARS-CoV-2 will lead to provide the novel vaccine for COVID-19 with high safety profile.
5. Contributors
HH, and HN contributed to conceptualization and writing original draft. HH, JS, YY, TO, RKK, CO, SY, RN, NJ, RI, AT, SK, TE, MS, SO, YA, HC, TK, YS contributed to visualization and formal analysis. TS, YM, HA, HT, MS, JM, HR, RM, and HN contributed to writing review & editing and Supervision. HA and HN contributed to Funding acquisition.
6. Research in context
6.1. Evidence before this study
The development of safer and more stable vaccine against SARS-CoV-2 causing COVID-19 pandemic worldwide is emerged. DNA vaccine is known to be effective, and more stable than other types of vaccine such as mRNA vaccines. So far, one DNA based-vaccine has been authorized for emergency use
6.2. Added value of this study
In this study, we developed a DNA-based vaccine targeting spike glycoprotein of SARS-CoV-2, which activated both humoral and cellular immune responses specific for SARS-CoV-2 without any tissue toxicity in rats. Furthermore, our DNA vaccine prevents hamsters from live SARS-CoV-2 infection.
6.3. Implications of all the available evidence
We successfully developed a DNA-based vaccine targeting SARS-CoV-2 in animal model, which has potential implications for proceeding this vaccine to clinical studies to combat the pandemic of COVID-19.
Declaration of Competing Interest
The Department of Health Development and Medicine is an endowed department supported by Anges, Daicel, and FunPep. The Department of Clinical Gene Therapy is financially supported by Novartis, AnGes, Shionogi, Boeringher, Fancl, Saisei Mirai Clinics, Rohto and Funpep. R.M. is a stockholder of FunPep and Anges. T.O. T.K. and Y.S. are employees of Anges. R.I, A.T, H.K, S.K, E.T, S.M, and H.T are employees of FunPep. R.M, H.T, and A.T. are FunPep stockholders. The funder provided support in the form of salaries for authors but did not have any additional role in the study design, data analysis, decision to publish, or preparation of the manuscript. All other authors declare no competing interests.
Acknowledgments
Acknowledgements
This study was supported by Project Promoting Support for Drug Discovery grants (JP20nk0101602 and JP21nf0101623h102) from the Japan Agency for Medical Research and Development and Panasonic Co. (Japan). We thank all the members of Department of Health Development and Medicine, Osaka University Graduate School of Medicine, for supporting this project, especially, Ms. Satoe Kitabata for secretarial support.
Data sharing statement
All data are available in this manuscript and supplementary files.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.retram.2022.103348.
Appendix. Supplementary materials
References
- 1.Corbett K.S., Edwards D.K., Leist S.R. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Q., Bao L., Mao H. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson L.A., Anderson E.J., Rouphael N.G. An mRNA Vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith T.R.F., Patel A., Ramos S. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J., Tostanoski L.H., Peter L. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020 doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahase E. Covid-19: where are we on vaccines and variants? BMJ. 2021;372 doi: 10.1136/bmj.n597. n597. [DOI] [PubMed] [Google Scholar]
- 8.Kutzler M.A., Weiner D.B. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9(10):776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanjuan R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J Virol. 2010;84(19):9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly J.J., Wahren B., Liu M.A. DNA vaccines: progress and challenges. J Immunol. 2005;175(2):633–639. doi: 10.4049/jimmunol.175.2.633. [DOI] [PubMed] [Google Scholar]
- 12.Hobernik D., Bros M. DNA vaccines-how far from clinical use? Int J Mol Sci. 2018;19(11) doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eusebio D., Neves A.R., Costa D. Methods to improve the immunogenicity of plasmid DNA vaccines. Drug Discov Today. 2021;26(11):2575–2592. doi: 10.1016/j.drudis.2021.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Franquesa M., Alperovich G., Herrero-Fresneda I. Direct electrotransfer of hHGF gene into kidney ameliorates ischemic acute renal failure. Gene Ther. 2005;12(21):1551–1558. doi: 10.1038/sj.gt.3302569. [DOI] [PubMed] [Google Scholar]
- 15.Herrero-Fresneda I., Torras J., Franquesa M. HGF gene therapy attenuates renal allograft scarring by preventing the profibrotic inflammatory-induced mechanisms. Kidney Int. 2006;70(2):265–274. doi: 10.1038/sj.ki.5001510. [DOI] [PubMed] [Google Scholar]
- 16.HogenEsch H., O'Hagan D.T., Fox C.B. Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. NPJ Vaccines. 2018;3:51. doi: 10.1038/s41541-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida S., Ono C., Hayashi H. SARS-CoV-2-induced humoral immunity through B cell epitope analysis in COVID-19 infected individuals. Sci Rep. 2021;11(1):5934. doi: 10.1038/s41598-021-85202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohshima N., Kubota-Koketsu R., Iba Y., Okuno Y., Kurosawa Y. Two types of antibodies are induced by vaccination with A/California/2009 pdm virus: binding near the sialic acid-binding pocket and neutralizing both H1N1 and H5N1 viruses. PLoS ONE. 2014;9(2):e87305. doi: 10.1371/journal.pone.0087305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno Y., Tanaka K., Baba K., Maeda A., Kunita N., Ueda S. Rapid focus reduction neutralization test of influenza A and B viruses in microtiter system. J Clin Microbiol. 1990;28(6):1308–1313. doi: 10.1128/JCM.28.6.1308-1313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota-Koketsu R., Terada Y., Yunoki M. Neutralizing and binding activities against SARS-CoV-1/2, MERS-CoV, and human coronaviruses 229E and OC43 by normal human intravenous immunoglobulin derived from healthy donors in Japan. Transfusion. 2021;61(2):356–360. doi: 10.1111/trf.16161. [DOI] [PubMed] [Google Scholar]
- 21.Maeda H., Kubo K., Sugita Y. DNA vaccine against hamster oral papillomavirus-associated oral cancer. J Int Med Res. 2005;33(6):647–653. doi: 10.1177/147323000503300606. [DOI] [PubMed] [Google Scholar]
- 22.Etievant S., Bal A., Escuret V. Performance Assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J. Clin. Med. 2020;9(6) doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pezzi L., Charrel R.N., Ninove L. Development and Evaluation of a duo SARS-CoV-2 RT-qPCR assay combining two assays approved by the world health organization targeting the envelope and the RNA-dependant RNA polymerase (RdRp) coding regions. Viruses. 2020;12(6) doi: 10.3390/v12060686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crackower M.A., Sarao R., Oudit G.Y. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 25.Imai Y., Kuba K., Rao S. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii K.J., Kawagoe T., Koyama S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 27.Grunwald T., Ulbert S. Improvement of DNA vaccination by adjuvants and sophisticated delivery devices: vaccine-platforms for the battle against infectious diseases. Clin Exp Vaccine Res. 2015;4(1):1–10. doi: 10.7774/cevr.2015.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Liu X., Fisher K. Enhanced type I immune response to a hepatitis B DNA vaccine by formulation with calcium- or aluminum phosphate. Vaccine. 2000;18(13) doi: 10.1016/s0264-410x(99)00391-6. 1227-35. [DOI] [PubMed] [Google Scholar]
- 29.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brocato R.L., Kwilas S.A., Kim R.K. Protective efficacy of a SARS-CoV-2 DNA vaccine in wild-type and immunosuppressed Syrian hamsters. NPJ Vaccines. 2021;6(1):16. doi: 10.1038/s41541-020-00279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan J.F., Zhang A.J., Yuan S. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai M., Iwatsuki-Horimoto K., Hatta M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117(28):16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tostanoski L.H., Wegmann F., Martinot A.J. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med. 2020;26(11):1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garren H., Robinson W.H., Krasulova E. Phase 2 trial of a DNA vaccine encoding myelin basic protein for multiple sclerosis. Ann Neurol. 2008;63(5):611–620. doi: 10.1002/ana.21370. [DOI] [PubMed] [Google Scholar]
- 35.Gottlieb P., Utz P.J., Robinson W., Steinman L. Clinical optimization of antigen specific modulation of type 1 diabetes with the plasmid DNA platform. Clin Immunol. 2013;149(3):297–306. doi: 10.1016/j.clim.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldonado L., Teague J.E., Morrow M.P. Intramuscular therapeutic vaccination targeting HPV16 induces T cell responses that localize in mucosal lesions. Sci Transl Med. 2014;6(221) doi: 10.1126/scitranslmed.3007323. 221ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierini S., Perales-Linares R., Uribe-Herranz M. Trial watch: dNA-based vaccines for oncological indications. Oncoimmunology. 2017;6(12) doi: 10.1080/2162402X.2017.1398878. e1398878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tebas P., Kraynyak K.A., Patel A. Intradermal SynCon(R) Ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers. J Infect Dis. 2019;220(3):400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 39.Trimble C.L., Morrow M.P., Kraynyak K.A. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386(10008) doi: 10.1016/S0140-6736(15)00239-1. 2078-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z., Yu J., Niu Y. Enhanced prophylactic and therapeutic effects of polylysine-modified Ara h 2 DNA vaccine in a mouse model of peanut allergy. Int Arch Allergy Immunol. 2016;171(3–4):241–250. doi: 10.1159/000453264. [DOI] [PubMed] [Google Scholar]
- 41.Barouch D.H., McKay P.F., Sumida S.M. Plasmid chemokines and colony-stimulating factors enhance the immunogenicity of DNA priming-viral vector boosting human immunodeficiency virus type 1 vaccines. J Virol. 2003;77(16):8729–8735. doi: 10.1128/jvi.77.16.8729-8735.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ledgerwood J.E., Pierson T.C., Hubka S.A. A West Nile virus DNA vaccine utilizing a modified promoter induces neutralizing antibody in younger and older healthy adults in a phase I clinical trial. J Infect Dis. 2011;203(10):1396–1404. doi: 10.1093/infdis/jir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin J.E., Pierson T.C., Hubka S. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J Infect Dis. 2007;196(12):1732–1740. doi: 10.1086/523650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin J.E., Sullivan N.J., Enama M.E. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin Vaccine Immunol. 2006;13(11):1267–1277. doi: 10.1128/CVI.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarwar U.N., Costner P., Enama M.E. Safety and immunogenicity of DNA vaccines encoding Ebolavirus and Marburgvirus wild-type glycoproteins in a phase I clinical trial. J Infect Dis. 2015;211(4):549–557. doi: 10.1093/infdis/jiu511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenler S., Blomberg P., Smith C.I. Safety and efficacy of DNA vaccines: plasmids vs. minicircles. Hum Vaccin Immunother. 2014;10(5):1306–1308. doi: 10.4161/hv.28077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suda H., Murakami A., Kaga T., Tomioka H., Morishita R. Beperminogene perplasmid for the treatment of critical limb ischemia. Expert Rev Cardiovasc Ther. 2014;12(10):1145–1156. doi: 10.1586/14779072.2014.955850. [DOI] [PubMed] [Google Scholar]
- 48.Jiang J., Ramos S.J., Bangalore P. Integration of needle-free jet injection with advanced electroporation delivery enhances the magnitude, kinetics, and persistence of engineered DNA vaccine induced immune responses. Vaccine. 2019;37(29):3832–3839. doi: 10.1016/j.vaccine.2019.05.054. [DOI] [PubMed] [Google Scholar]
- 49.Schommer N.N., Nguyen J., Yung B.S. Active Immunoprophylaxis and Vaccine Augmentations Mediated by a Novel Plasmid DNA Formulation. Hum Gene Ther. 2019;30(4):523–533. doi: 10.1089/hum.2018.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev Vaccines. 2016;15(3):313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marc M.A., Dominguez-Alvarez E., Gamazo C. Nucleic acid vaccination strategies against infectious diseases. Expert Opin Drug Deliv. 2015;12(12):1851–1865. doi: 10.1517/17425247.2015.1077559. [DOI] [PubMed] [Google Scholar]
- 52.Tregoning J.S., Kinnear E. Using plasmids as DNA vaccines for infectious diseases. Microbiol Spectr. 2014;2(6) doi: 10.1128/microbiolspec.PLAS-0028-2014. [DOI] [PubMed] [Google Scholar]
- 53.Williams J.A. Improving DNA vaccine performance through vector design. Curr Gene Ther. 2014;14(3):170–189. doi: 10.2174/156652321403140819122538. [DOI] [PubMed] [Google Scholar]
- 54.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20(6):339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S.F., Tseng S.P., Yen C.H. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451(2):208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Y., Wong R., Soo Y.O. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis. 2005;24(1):44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duan K., Liu B., Li C. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh K.M., Chiueh T.S., Siu L.K. Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J Antimicrob Chemother. 2005;56(5):919–922. doi: 10.1093/jac/dki346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang A.T., Garcia-Carreras B., Hitchings M.D.T. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11(1):4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krause P.R., Fleming T.R., Longini I.M. SARS-CoV-2 Variants and Vaccines. N Engl J Med. 2021 doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Beltran W.F., Lam E.C., St Denis K. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou D., Dejnirattisai W., Supasa P. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q., Wu J., Nie J. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah P., Canziani G.A., Carter E.P., Chaiken I. The Case for S2: the potential benefits of the S2 Subunit of the SARS-CoV-2 spike protein as an immunogen in fighting the COVID-19 Pandemic. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.637651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS ONE. 2012;7(11):e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sauer M.M., Tortorici M.A., Park Y.J. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol. 2021;28(6):478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- 68.Tarke A., Sidney J., Methot N. Negligible impact of SARS-CoV-2 variants on CD4 (+) and CD8 (+) T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [Google Scholar]
- 69.Dey A., Chozhavel Rajanathan T.M., Chandra H. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models. Vaccine. 2021;39(30):4108–4116. doi: 10.1016/j.vaccine.2021.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciabattini A., Pettini E., Fiorino F. Modulation of primary immune response by different vaccine adjuvants. Front Immunol. 2016;7:427. doi: 10.3389/fimmu.2016.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olatunde A.C., Hale J.S., Lamb T.J. Cytokine-skewed Tfh cells: functional consequences for B cell help. Trends Immunol. 2021;42(6):536–550. doi: 10.1016/j.it.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Momin T., Kansagra K., Patel H. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101020. 101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tebas P., Yang S., Boyer J.D. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine. 2021;31 doi: 10.1016/j.eclinm.2020.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.