Abstract
We present a genome assembly from an individual female Accipiter gentilis (the northern goshawk; Chordata; Aves; Accipitriformes; Accipitridae). The genome sequence is 1,398 megabases in span. The majority of the assembly (99.98%) is scaffolded into 40 chromosomal pseudomolecules, with the W and Z chromosomes assembled. The complete mitochondrial genome was also assembled and is 16.6 kilobases in length.
Keywords: Accipiter gentilis, northern goshawk, genome sequence, chromosomal, Aves
Species taxonomy
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda; Coelurosauria; Aves; Neognathae; Accipitriformes; Accipitridae; Accipitrinae; Accipiter; Accipiter gentilis (Linnaeus, 1758) (NCBI:txid8957).
Background
The northern goshawk, Accipiter gentilis, is a medium-sized, forest specialist, bird of prey inhabiting large parts of the Holarctic. The considerable morphological variation particularly within A. gentilis has resulted in the acknowledgement of 10 subspecies ( Del Hoyo & Collar, 2015; Dickson & Remsen, 2013), with the nominate European subspecies, A. gentilis gentilis found across Europe, except for the Iberian Peninsula, southern Italy, and Greece, and extending eastwards to the Carpathians and part of Russia. However, mitochondrial phylogenetic analyses suggest two monophyletic groups within the species, a Neartcic clade and a Palearctic clade ( Kunz et al., 2019).
In the UK, loss of woodland habitat and persecution drove the species to extinction by the end of the 19 th century, before it was reintroduced during the 1960s. Despite being legally protected, A. gentilis are still persecuted throughout Europe ( Rutz et al., 2006) and their nests are frequently robbed. For instance, in Scotland, illegal killing of birds of prey in general, and northern goshawks in particular, has not declined during the last 20 years ( RSPB, 2015), leading to the hypothesis that this might have contributed to the slow recovery of the population in the UK despite repeated reintroductions ( Kenward, 2006).
Genome sequence report
The genome was sequenced from a single female A. gentilis ( Figure 1) collected from Northumberland, UK. A total of 34-fold coverage in Pacific Biosciences single-molecule HiFi long reads and 31-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 113 missing/misjoins and removed 1 haplotypic duplication, reducing the assembly size by 0.04% and the scaffold number by 17.30%, and increasing the scaffold N50 by 9.56%.
Figure 1. Image of the Accipiter gentilis specimen from the previous breeding season, captured using a camera trap.
The final assembly has a total length of 1,398 Mb in 454 sequence scaffolds with a scaffold N50 of 34.0 Mb ( Table 1). The majority, 90.67%, of the assembly sequence was assigned to 40 chromosomal-level scaffolds, representing 38 autosomes (numbered by sequence length) and the W and Z chromosomes ( Figure 2– Figure 5; Table 2). Microchromosomes 35, 36, 37, and 38 were curated based on homology to microchromosomes found in Gallus gallus, Taeniopygia guttata, and Cuculus canorus.
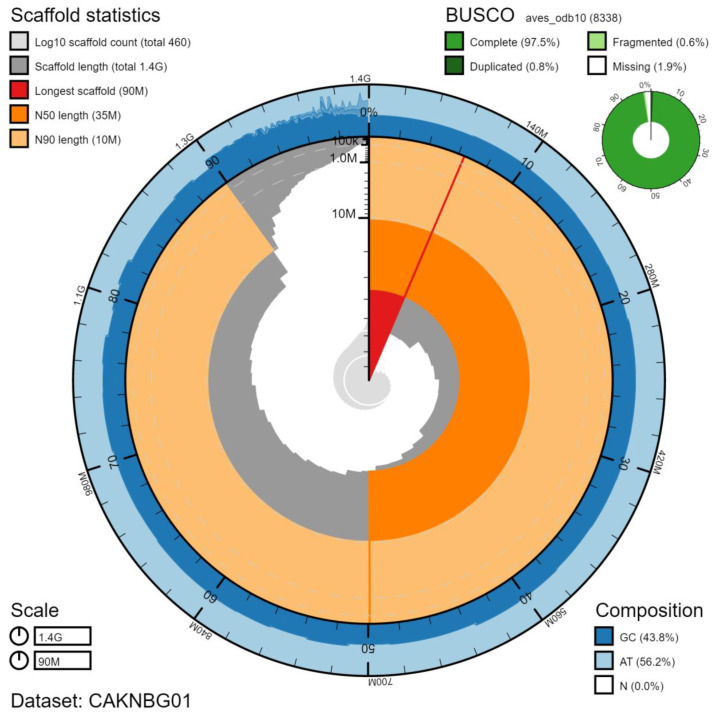
Figure 2. Genome assembly of Accipiter gentilis, bAccGen1.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 1,398,027,955 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (89,764,762 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (35,025,567 and 10,404,007 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the aves_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bAccGen1.1/dataset/CAKNBG01/snail.
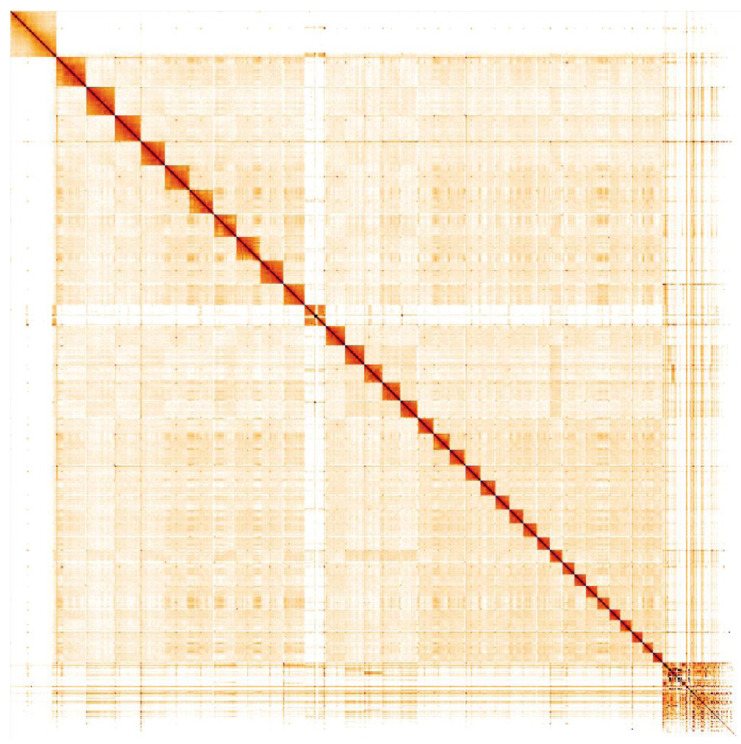
Figure 5. Genome assembly of Accipiter gentilis, bAccGen1.1: Hi-C contact map.
Hi-C contact map of the bAccGen1.1 assembly, visualised in HiGlass. Chromosomes are arranged in size order from left to right and top to bottom. The interactive Hi-C map can be viewed here.
Table 1. Genome data for Accipiter gentilis, bAccGen1.1.
| Project accession data | |
|---|---|
| Assembly identifier | bAccGen1.1 |
| Species | Accipiter gentilis |
| Specimen | bAccGen1 |
| NCBI taxonomy ID | 8957 |
| BioProject | PRJEB48396 |
| BioSample ID | SAMEA8235650 |
| Isolate information | female, heart tissue |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR7254635-ERR7254637 |
| 10X Genomics Illumina | ERR7220458-ERR7220461 |
| Hi-C Illumina | ERR7220461 |
| Genome assembly | |
| Assembly accession | GCA_929443795.1 |
| Accession of alternate haplotype | GCA_929447715.1 |
| Span (Mb) | 1,398 |
| Number of contigs | 637 |
| Contig N50 length (Mb) | 17.7 |
| Number of scaffolds | 454 |
| Scaffold N50 length (Mb) | 35.0 |
| Longest scaffold (Mb) | 55.8 |
| BUSCO * genome score | C:97.5%[S:96.7%,D:0.8%],
F:0.6%,M:1.9%,n:8338 |
*BUSCO scores based on the aves_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/bAccGen1.1/dataset/CAKNBG01/busco.
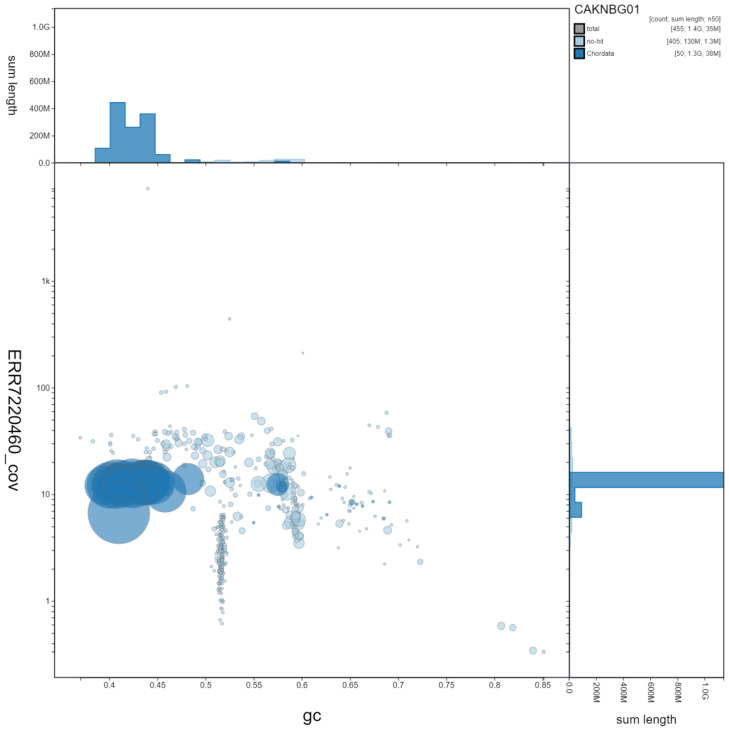
Figure 3. Genome assembly of Accipiter gentilis, bAccGen1.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bAccGen1.1/dataset/CAKNBG01/blob.
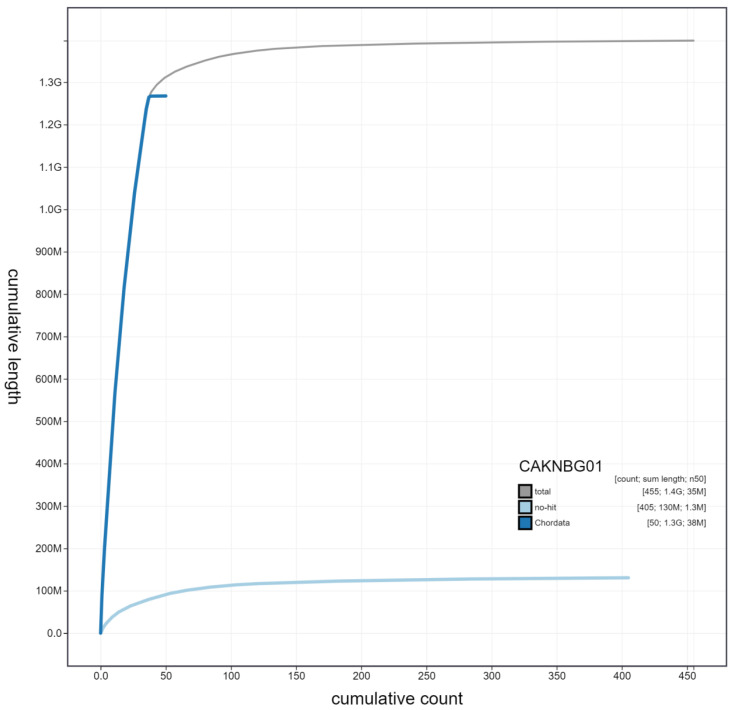
Figure 4. Genome assembly of Accipiter gentilis, bAccGen1.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bAccGen1.1/dataset/CAKNBG01/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Accipiter gentilis, bAccGen1.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| OV839361.1 | 1 | 55.81 | 42.4 |
| OV839362.1 | 2 | 55.41 | 40.8 |
| OV839363.1 | 3 | 49.24 | 40.1 |
| OV839364.1 | 4 | 48.36 | 41.4 |
| OV839365.1 | 5 | 47.46 | 43.7 |
| OV839366.1 | 6 | 45.22 | 43.7 |
| OV839367.1 | 7 | 44.90 | 44.0 |
| OV839368.1 | 8 | 44.73 | 43.6 |
| OV839369.1 | 9 | 44.71 | 42.6 |
| OV839370.1 | 10 | 41.28 | 44.5 |
| OV839372.1 | 11 | 37.57 | 41.0 |
| OV839373.1 | 12 | 37.31 | 40.9 |
| OV839374.1 | 13 | 35.03 | 39.5 |
| OV839375.1 | 14 | 34.43 | 43.4 |
| OV839376.1 | 15 | 31.97 | 40.4 |
| OV839377.1 | 16 | 31.42 | 42.9 |
| OV839378.1 | 17 | 31.04 | 44.2 |
| OV839379.1 | 18 | 29.66 | 42.7 |
| OV839380.1 | 19 | 28.75 | 42.3 |
| OV839381.1 | 20 | 28.03 | 40.6 |
| OV839382.1 | 21 | 27.96 | 41.1 |
| OV839383.1 | 22 | 27.20 | 41.1 |
| OV839384.1 | 23 | 25.48 | 43.2 |
| OV839385.1 | 24 | 25.38 | 43.5 |
| OV839386.1 | 25 | 23.93 | 42.7 |
| OV839387.1 | 26 | 23.47 | 44.6 |
| OV839388.1 | 27 | 22.46 | 39.8 |
| OV839389.1 | 28 | 21.93 | 43.3 |
| OV839390.1 | 29 | 21.76 | 48.2 |
| OV839391.1 | 30 | 21.43 | 41.5 |
| OV839392.1 | 31 | 21.27 | 41.3 |
| OV839393.1 | 32 | 21.17 | 41.8 |
| OV839394.1 | 33 | 21.08 | 45.0 |
| OV839395.1 | 34 | 17.59 | 40.8 |
| OV839396.1 | 35 | 10.40 | 57.5 |
| OV839397.1 | 36 | 2.11 | 57.8 |
| OV839398.1 | 37 | 0.65 | 59.8 |
| OV839399.1 | 38 | 0.43 | 65.1 |
| OV839371.1 | W | 39.88 | 45.8 |
| OV839360.1 | Z | 89.76 | 41.0 |
| OV839400.1 | MT | 0.02 | 44.2 |
| - | Unplaced | 130.34 | 55.7 |
The assembly has a BUSCO v5.1.2 ( Manni et al., 2021) completeness of 97.5% (single 96.7%, duplicated 0.8%) using the aves_odb10 reference set (n=8338). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
Methods
Sample acquisition and nucleic acid extraction
A single female A. gentilis specimen (bAccGen1) was found dead in coniferous woodland in Northumberland, UK by Katherine August (University of Aberdeen) and Martin Davison (Northumbria Ringing Group). The specimen was identified by Martin Davison and frozen at -20°C. Dissection of tissue samples occurred while the specimen was frozen, with the samples then stored at -80°C prior to sending to the Wellcome Sanger Institute on dry ice.
DNA was extracted at the Tree of Life laboratory, Wellcome Sanger Institute. The bAccGen1 sample was weighed and dissected on dry ice with tissue set aside for Hi-C and RNA sequencing. Heart tissue was cryogenically disrupted to a fine powder using a Covaris cryoPREP Automated Dry Pulveriser, receiving multiple impacts. Fragment size analysis of 0.01–0.5 ng of DNA was then performed using an Agilent FemtoPulse. High molecular weight (HMW) DNA was extracted using the Qiagen MagAttract HMW DNA extraction kit. Low molecular weight DNA was removed from a 200-ng aliquot of extracted DNA using 0.8X AMpure XP purification kit prior to 10X Chromium sequencing; a minimum of 50 ng DNA was submitted for 10X sequencing. HMW DNA was sheared into an average fragment size between 12–20 kb in a Megaruptor 3 system with speed setting 30. Sheared DNA was purified by solid-phase reversible immobilisation using AMPure PB beads with a 1.8X ratio of beads to sample to remove the shorter fragments and concentrate the DNA sample. The concentration of the sheared and purified DNA was assessed using a Nanodrop spectrophotometer and Qubit Fluorometer and Qubit dsDNA High Sensitivity Assay kit. Fragment size distribution was evaluated by running the sample on the FemtoPulse system.
Sequencing
Pacific Biosciences HiFi circular consensus and 10X Genomics Chromium read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Sequencing was performed by the Scientific Operations core at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II (HiFi) and Illumina NovaSeq 6000 instruments. Hi-C data were generated in the Tree of Life laboratory from remaining heart tissue of bAccGen1 using the Arima v2 kit and sequenced on a NovaSeq 6000 instrument.
Genome assembly
Assembly was carried out with Hifiasm ( Cheng et al., 2021); haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). One round of polishing was performed by aligning 10X Genomics read data to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using yahs. The assembly was checked for contamination as described previously ( Howe et al., 2021). Manual curation was performed using HiGlass ( Kerpedjiev et al., 2018) and Pretext. The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2021), which performs annotation using MitoFinder ( Allio et al., 2020). The genome was analysed and BUSCO scores generated within the BlobToolKit environment ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
Table 3. Software tools used.
| Software
tool |
Version | Source |
|---|---|---|
| Hifiasm | 0.15.3 | Cheng et al., 2021 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| yahs | 1.0 | https://github.com/c-zhou/yahs |
| longranger
align |
2.2.2 |
https://support.10xgenomics.com/
genome-exome/software/pipelines/ latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-
gaa2ace8 |
Garrison & Marth, 2012 |
| MitoHiFi | 2.0 | Uliano-Silva et al., 2021 |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.2.x |
https://github.com/wtsi-hpag/
PretextView |
| BlobToolKit | 3.0.5 | Challis et al., 2020 |
Ethics/compliance issues
The materials that have contributed to this genome note have been supplied by a Tree of Life collaborator. The Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible.
The overarching areas of consideration are:
Ethical review of provenance and sourcing of the material
Legality of collection, transfer and use (national and international)
Each transfer of samples is undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Tree of Life collaborator, Genome Research Limited (operating as the Wellcome Sanger Institute) and in some circumstances other Tree of Life collaborators.
Data availability
European Nucleotide Archive: Accipiter gentilis (northern goshawk). Accession number PRJEB48396; https://identifiers.org/ena.embl/PRJEB48396.
The genome sequence is released openly for reuse. The A. gentilis genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute [206194, <a href=https://doi.org/10.35802/206194>https://doi.org/10.35802/206194</a>] and the Darwin Tree of Life Discretionary Award [218328, <a href=https://doi.org/10.35802/218328>https://doi.org/10.35802/218328</a>].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
Author information
Members of the Wellcome Sanger Institute Tree of Life programme are listed here: https://doi.org/10.5281/zenodo.6125027.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.5746904.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.6125046.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.5638618.
References
- Allio R, Schomaker-Bastos A, Romiguier J, et al. : MitoFinder: Efficient Automated Large-Scale Extraction of Mitogenomic Data in Target Enrichment Phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat Methods. 2021;18(2):170–75. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Hoyo J, Collar NJ: HBW and BirdLife International Illustrated Checklist of the Birds of the World Volume 1: Non-Passerines.Lynx Edicions,2015. Reference Source [Google Scholar]
- Dickson EC, Remsen JR: The Howard and Moore Complete Checklist of the Birds of the World. Vol. 1. Non-Passerines.Aves Press, Eastbourne, UK,2013. Reference Source [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing.arXiv: 1207.3907.2012. 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenward R: The Goshawk.A&C Black,2006. [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz F, Gamauf A, Zachos FE, et al. : Mitochondrial phylogenetics of the goshawk Accipiter [ gentilis] superspecies. J Zool Syst Evol Res. 2019;57(4):942–958. 10.1111/jzs.12285 [DOI] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol Biol Evol. 2021;38(10):4647–54. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RSPB: The Illegal Killing of Birds of Prey in Scotland 1994–2014: A Review.2015. Reference Source [Google Scholar]
- Rutz C, Bijlsma RG, Marquiss M, et al. : Population Limitation in the Northern Goshawk in Europe: A Review with Case Studies. Studies in Avian Biology. 2006;31:158. Reference Source [Google Scholar]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, et al. : marcelauliano/MitoHiFi: mitohifi_v2.0.2021. 10.5281/zenodo.5205678 [DOI] [Google Scholar]