Summary
Background
The increase in SARS-CoV-2 infections in December 2021 was driven primarily by the Omicron variant, which largely displaced the Delta over a three-week span. Outcomes from infection with Omicron remain uncertain. We evaluated whether clinical outcomes and viral loads differed between Delta and Omicron infections during the period when both variants were co-circulating.
Methods
In this retrospective observational cohort study, remnant clinical specimens, positive for SARS-CoV-2 after standard of care testing at the Johns Hopkins Microbiology Laboratory, between the last week of November and the end of December 2021, were used for whole viral genome sequencing. Cycle threshold values (Ct) for viral RNA, the presence of infectious virus, and levels of respiratory IgG were measured, and clinical outcomes were obtained. Differences in each measure were compared between variants stratified by vaccination status.
Findings
The Omicron variant displaced Delta during the study period and constituted 95% of the circulating lineages by the end of December 2021. Patients with Omicron infections (N = 1,119) were more likely to be vaccinated compared to patients with Delta (N = 908), but were less likely to be admitted (0.33 CI 0.21–0.52), require ICU level care (0.38 CI 0.17–0.87), or succumb to infection (0.26 CI 0.06–1.02) regardless of vaccination status. There was no statistically significant difference in Ct values based on the lineage regardless of the vaccination status. Recovery of infectious virus in cell culture was reduced in boosted patients compared to fully vaccinated without a booster and unvaccinated when infected with the Delta lineage. However, in patients with Omicron infections, recovery of infectious virus was not affected by vaccination.
Interpretation
Compared to Delta, Omicron was more likely to cause breakthrough infections of vaccinated individuals, yet admissions were less frequent. Admitted patients might develop severe disease comparable to Delta. Efforts for reducing Omicron transmission are required as, though the admission risk might be lower, the increased numbers of infections cause large numbers of hospitalizations.
Funding
NIH/NIAID Center of Excellence in Influenza Research and Surveillance contract HHS N2772201400007C, Johns Hopkins University, Maryland department of health, Centers for Disease Control and Prevention contract 75D30121C11061, and The Modeling Infectious Diseases in Healthcare Network (MInD) under awards U01CK000589.
Keyword: Omicron, Delta, Cycle thresholds, ELISA, Cell culture
Research in context.
Evidence before this study
The unprecedented increase in COVID-19 cases in the month of December 2021, associated with the displacement of the Delta variant with Omicron, triggered a lot of concerns. An understanding of the disease severity associated with infections with Omicron is essential as well as the virological determinants that contributed to its widespread predominance. We did a literature search on PubMed for articles published up to March 15th, 2022 using the search terms “Omicron”, “SARS-CoV-2”, and “Disease severity”, “Viral load” and “Cell culture”. Peer reviewed and preprint published studies showed that infection with Omicron is associated with lower likelihood of hospitalization, intensive care unit (ICU) level care, and mortality. In addition, peer reviewed and preprint studies that compared the infectious viral load or RNA loads in Omicron versus Delta showed that Omicron had equivalent infectious viral titers and duration of viral shedding.
Added value of this study
This study compared the clinical characteristics and outcomes after infection with the Omicron variant compared to Delta in the National Capital region (Maryland, Virginia, and D.C) using variants characterized by whole genome sequencing and a selective time frame when both variants co-circulated. The analysis was stratified by vaccination status to compare fully vaccinated patients who didn't receive a booster to patients who received a booster vaccination, and controlled for comorbidities and other known risk factors for more severe outcomes. In addition, we compared viral RNA and infectious virus load between Delta and Omicron samples from unvaccinated, fully vaccinated, and patients with booster vaccination.
Implications of all the available evidence
Omicron was more likely than Delta to cause breakthrough infections in fully vaccinated and booster vaccinated individuals, but these infections were less likely to result in admission or ICU level care. Admitted patients with Omicron infections had similar requirements for supplemental oxygen and ICU level care compared to patients admitted with Delta infections. Viral loads were similar in samples from Omicron and Delta infected patients regardless of vaccination status. The recovery of infectious virus on cell culture was reduced in samples from patients infected with Delta who received a booster dose, which was not the case with Omicron. In Omicron infected individuals, the recovery of infectious virus was equivalent in unvaccinated, fully vaccinated, and samples from patients who received booster vaccination.
Alt-text: Unlabelled box
Introduction
The SARS-CoV-2 Omicron variant was first identified in South Africa and reported to the World Health Organization (WHO) on November 24, 20211 and was designated as a variant of concern (VOC) on November 26, 2021.2,3 A large number of mutations and amino acid changes were noted across the Omicron genome, 15 of which are within the receptor binding domain (RBD) of the spike (S) protein.4 Some of the Omicron RBD characterized mutations raised concerns of a substantial impact on the transmissibility, immunity secondary to vaccination or prior infection, and efficacy of therapeutic monoclonal antibodies. Initial reports from South Africa5 and Europe6, 7–8 suggested that the Omicron variant may be more transmissible but cause less severe infection. However, the US population differs from both the South African and UK populations in multiple ways; most importantly, the percentage of the population that is vaccinated is lower in the US than the UK and prior infection is lower in the US compared to South Africa.9 Thus, important questions remain as to the regional impact of Omicron. Most importantly, how might outcomes, particularly hospitalization, differ between patients infected with Omicron compared to variants such as Delta. Additionally, there remains uncertainty as to whether enhanced evasion of pre-existing immunity or some other biological mechanisms drive the higher rate of Omicron transmission.
The Omicron variant was first identified in Maryland in the last week of November 2021, and became predominant in a matter of 3 weeks. In this study, we evaluated the biological differences between viral variants collected as a part of routine clinical care between November 22nd and December 31st in the Johns Hopkins Health System, as well as the clinical outcomes in patients infected with Omicron and Delta. We focused on this time frame as it witnessed the detection of the first Omicron case in our system in the last week of November, and the switch between Delta predominance in the beginning of December to Omicron predominance at the end of December 2021. We provide a comparison of clinical, demographic and virological load between Delta and Omicron infected individuals and stratify our results based on vaccination status.
Methods
Ethical considerations and data availability
Research was conducted under Johns Hopkins IRB protocol IRB00221396 with a waiver of consent. Remnant clinical specimens from patients that tested positive for SARS-CoV-2 after standard of care testing were used for whole genome sequencing. Whole genomes were made publicly available at GISAID.
Specimens and patient data
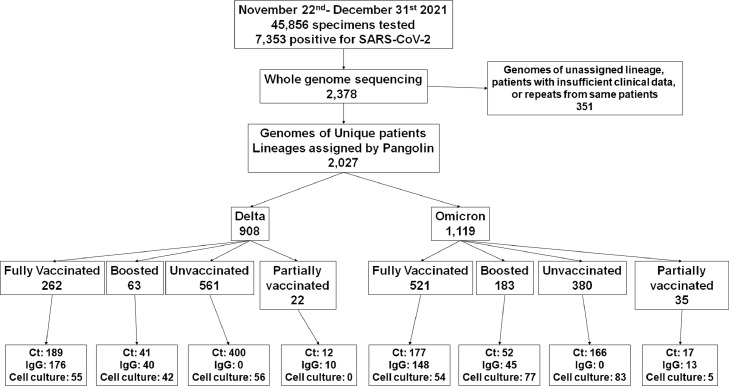
In this retrospective observational cohort study, nasopharyngeal (symptomatic) or lateral mid-turbinate nasal swabs (asymptomatic) after standard of care diagnostic or screening testing were collected and used for genome sequencing. At Johns Hopkins Medical System, SARS-CoV-2 clinical testing is performed for inpatients and outpatients (five acute care hospitals and more than 40 ambulatory care offices) as well as standard of care screening particularly prior to scheduled surgeries. Molecular assays used include primarily the NeuMoDx SARS-CoV-2 (Qiagen), Cobas SARS-CoV-2 (Roche), Xpert Xpress SARS-CoV-2/Flu/RSV (Cepheid), in addition to the RealStar® SARS-CoV-2 RT-PCR (Altona Diagnostics), ePlex Respiratory Pathogen Panel 2 (Roche), Aptima SARS-CoV-2 (Hologic), and Accula SARS-CoV-2 assays (ThermoFisher Scientific).10, 11, 12–13 Each sample in our cohort represents a unique patient as repeat tests from the same patient were excluded (samples with better quality genomes were used) as were uncharacterized genomes due to insufficient quality (Genomes with unassigned lineages, n = 347, Figure 1). Table 1 summarizes the numbers of patients and samples used for each part of the study.
Figure 1.
Flow chart of samples and patients used for the study.
Table 1.
Patients and samples used for the study.
| Omicron |
Delta |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total Patients | 1119 |
908 |
||||||
| Fully Vaccinated | With booster dose | Unvaccinated | Partially vaccinated | Fully Vaccinated | With booster dose | Unvaccinated | Partially vaccinated | |
| Patients(%) | 521 (46.6) | 183 (16.4) | 380 (33.96) | 35 (3.1) | 262 (28.9) | 63 (6.9) | 561 (61.8) | 22 (2.4) |
| Ct analysis(%) | 177 (33.97) | 52 (28.4) | 166 (43.7) | 17 (48.6) | 189 (72.1) | 41 (65.1) | 400 (71.3) | 12 (54.5) |
| Patients with known days from the onset of symptoms(%) | 154 (29.6) | 53 (28.96) | 135 (35.5) | 14 (40) | 105 (40.1) | 15 (23.8) | 222 (39.6) | 9 (40.9) |
| IgG(%) | 148 (28.4) | 45 (24.6) | 0 | 13 (37.1) | 176 (67.2) | 40 (63.5) | 0 | 10 (45.5) |
| Cell culture (total)(%) | 54 (10.4) | 77 (42.1) | 83 (21.8) | 5(14.3) | 55 (21) | 42 (66.7) | 56 (9.98) | 0 |
| Cell culture (Ct < 20) (%) | 45 (8.6) | 34 (18.6) | 51 (13.4) | 4 (11.4) | 51 (19.5) | 23 (36.5) | 39 (9.95) | 0 |
Full vaccination was based on the CDC definition of positive test results more than 14 days post the second shot for pfizer/BioNTech BNT162b2 and Moderna mRNA-1273 or 14 days after the J&J/Janssen, but no booster doses
Partially vaccinated include patients who received only one shot for pfizer/BioNTech BNT162b2 or Moderna mRNA-1273.
Sample size
A total of 1,119 Omicron and 908 Delta infected patients diagnosed between November 22nd 2021 and December 31st 2021 were included in the study. Patients’ sample size and cohorts were based on whole genome sequencing surveillance data. For whole genome sequencing, samples were collected randomly (daily, the first available 150- 200 samples with the same day's collection date) from the whole Johns Hopkins system. For Ct, ELISA and cell culture experiments, subsamples from the Omicron and Delta cohorts were selected based on availability (Table 1).
Clinical data analysis
Clinical and vaccination data for patients whose samples were characterized by whole genome sequencing was bulk extracted as previously detailed in.14 Cases with missing data were excluded (N = 4). The first Omicron infection was collected from a patient in the Johns Hopkins Medical System during the last week of November, 2021. COVID admission relatedness was determined based on presenting complaints, admission diagnoses, reason for testing, and timing of testing. Patients admitted without symptoms or whose primary reason for admission was not COVID were not counted as a COVID-related admission. Additionally, patients who developed symptoms after admission or who tested positive on regular asymptomatic surveillance of inpatients were also considered non-COVID-related admissions. In our vaccinated patients’ population, 68.6% received Pfizer/BioNTech, followed by the Moderna mRNA-1273 (26.6%), then the J&J/Janssen COVID-19 vaccines (4.8%). Full vaccination was based on the CDC definition of positive test results more than 14 days post the second shot for pfizer/BioNTech BNT162b2 and Moderna mRNA-1273 or 14 days after the J&J/Janssen.
Ct value analysis
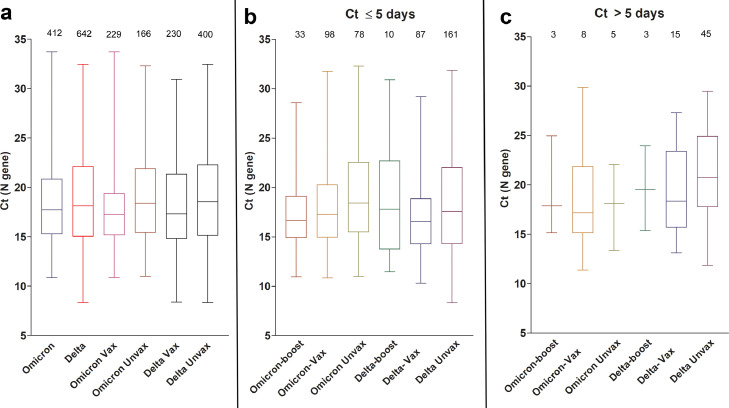
To ensure comparable Ct values for viral load analyses, samples were retested with the PerkinElmers SARS-CoV-2 kit (https://www.fda.gov/media/136410/download, Last accessed January 19, 2022) and Ct values of the N gene were used for comparisons. The distribution of nasopharyngeal (from symptomatic patients) and lateral mid-turbinate nasal swabs (from asymptomatic patients) were equivalent between the groups and the comprehensive analysis of all Ct values (Figure 3a) did not control for sample type.
Figure 3.
Omicron and Delta variants cycle threshold (Ct) values in upper respiratory samples.
(a) Ct values of Omicron and Delta from all samples with available Ct values (N gene) stratified by vaccination status. (b) Ct values of Omicron and Delta from samples collected 5 days or less from the onset of symptoms. For this analysis, samples from asymptomatic patients were not included. (c) Ct values of Omicron and Delta variants collected more than 5 days from the onset of symptoms. For this analysis, samples from asymptomatic patients were not included. Vax, fully vaccinated patients who didn't receive a booster dose (panel A only, Vax includes boosted patients); Unvax, unvaccinated; boost, patients with booster dose. Data shown as box and whisker plots, horizontal bars represent median Ct values, and total numbers per group are shown on top of each box.
Amplicon based sequencing
Specimen preparation, extractions, and sequencing were performed as described previously.15,16 Library preparation for this cohort was performed using the NEBNext® ARTIC SARS-CoV-2 Companion Kit (VarSkip Short SARS-CoV-2 # E7660-L). Sequencing was performed using the Nanopore GridION and reads were basecalled with MinKNOW, and demultiplexed with guppybarcoder that required barcodes at both ends. Alignment and variant calling were performed with the artic-ncov2019 medaka protocol. Only sequences with coverage >90% and mean depth >100 were submitted to GISAID database. Genomes with lineages assigned by Pangolin were included (coverage > 70%, Tables S1 and S2 detail the quality of the genomes, sequences with coverage >90% and mean depth >100: Delta 840/ 908 and Omicron 987/ 1,119).Query mutations were manually confirmed with Integrated Genomics viewer (IGV) (Version 2.8.10), clades were determined using Nextclade beta v 1.13.2 (clades.nextstrain.org, Last accessed March 30, 2022), and lineages were determined with Pangolin COVID-19 lineage Assigner.17
ELISA
Respiratory samples were tested, undiluted, with the EUROIMMUN Anti-SARS-CoV-2 ELISA (IgG) following the package insert (https://www.fda.gov/media/137609/download) as we described previously.14 The assay detects antibodies to the SARS-CoV-2 S1 domain of the spike protein. The value 1.1 was used as a cut off for positives.
Cell culture
VeroE6TMPRSS2 cells (RRID: CVCL_YQ49) were obtained from the cell repository of the National Institute of Infectious Diseases, Japan and are described in.18 One vial of frozen cells was received, thawed in the recommended media and expanded to generate approximately 20 vials of cells (seed stock). Subsequently, one vial of this batch of frozen cells is thawed, expanded for use and additional batches of frozen cell stocks (working stocks) are generated. All laboratory members are trained in good cell culture practice with annual refreshers. The original seed stock of cells is checked for mycoplasma using a real time quantitative PCR test (MycoSEQ Mycoplasma Detection kit, Thermo Fisher Scientific). VeroE6TMPRSS2 Cells were cultured and infected with aliquots of swab specimens (convenience sample, based on the availability) as previously described for VeroE6 cells.19 Cultures were incubated for 6 days and SARS-CoV-2 cytopathic effect (CPE) was confirmed by reverse transcriptase PCR.
Reagents validations
ELISA and PerkinElmers SARS-CoV-2 kits received the FDA emergency use authorization (EUA) and were clinically validated for diagnosis. SARS-CoV-2 whole genome sequencing was validated using reference materials and controls and each run includes positive and negative controls.
Statistical analysis
Fisher Exact test was used for categorical variable comparisons with confidence limits calculated by Wilson score interval method. One-way ANOVA was used for comparing continuous independent variables. Given the admission rate for Delta infections, we were powered to detect a 3-percentage point difference with 85% power at 5% statistical significance. The primary outcomes of interest were admission, ICU level care, and Death. To assess the association between variant type and outcomes, we conducted a multivariable logistic regression that controlled for noted risk factors for COVID-19 severe outcomes including, age, gender,20 race and ethnicity,21 and comorbidities (hypertension, pregnancy, lung disease, kidney disease, immunosuppression, diabetes, heart failure, atrial fibrillation, smoking, cerebrovascular diseases, cancer, and coronary artery disease). We further analyzed the association between strain type and Ct level controlling for the same factors using a linear regression analysis. All regression analyses were done in Stata (StataCorp. 2021. Stata Statistical Software: Release 17. College Station, TX: StataCorp LLC.), all other statistical analyses were conducted using GraphPad prism. There was no allowance for multiplicity in our statistical approaches.
Key resource table
| Reagent | Item Number |
| chemagic Viral DNA/RNA 300 Kit H96 | CMG-1033-S |
| Mag-Bind® TotalPure NGS | M1378 |
| NEBNextARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) - 96 reactions | E7660L |
| Qubit 1X dsDNA HS Assay Kit | Q33231 |
| Native Barcoding Expansion 96 | EXP-NBD196 |
| SFB Expansion | EXP-SFB001 |
| Adapter Mix II Expansion | EXP-AMII001 |
| Flow Cell Priming Kit XL | EXP-FLP002-XL |
| Sequencing Auxiliary Vials | EXP-AUX001 |
| Flow Cell (R9.4.1) | FLO-MIN106D |
| Software | Version |
| MinKnow | 21.11.6. |
| Nextclade | v1.13.2 |
| Pangolin | v3.0. |
Role of funders
The corresponding authors confirm that they had full access to all the data in the study and had final responsibility for the decision to submit for publication. The Funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
SARS-CoV-2 positivity and variants trends November–December 2021
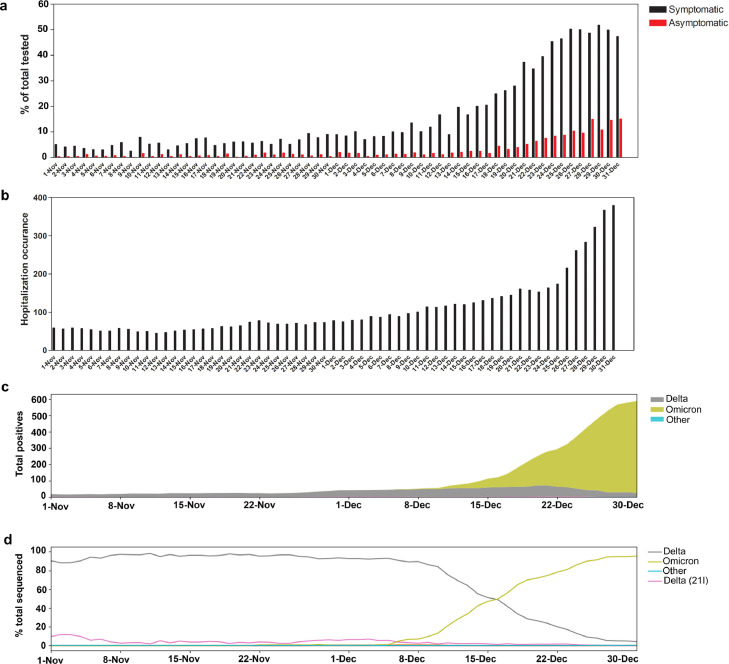
The SARS-CoV-2 positivity rate increased markedly in December 2021 (symptomatic, 25.5%, Table 2 and Figure 2a). The large increase in SARS-CoV-2 positivity in the month of December was the highest recorded since the beginning of the pandemic (Figure S1a and22). The spike in SARS-CoV-2 positivity was particularly evident during the last two weeks of December for both symptomatic and asymptomatic patients (Figure 2a). A systemwide increase in COVID-19 hospitalizations was notable in December 2021 (Figure 2b) and was also the highest recorded since the beginning of the pandemic (Figure S1b). The increase in the positivity correlated with an increase in the detection of the Omicron variant, that went from less than 1% of sequenced strains in the beginning of December to the dominant variant in less than 3 weeks (Figure 2c and d). Table 2 shows the total positive tests and samples sequenced in November and December 2021.
Table 2.
SARS-CoV-2 positive tests and positivity rates in November and December 2021 and total sequenced.
| November | December | November | December | |
|---|---|---|---|---|
| Positive tests | % | |||
| SARS-CoV-2 (All testing) | 726 | 7078 | 2.67 | 18.65 |
| SARS-CoV-2 (Symptomatic only) | 592 | 6236 | 5.7 | 25.5 |
| SARS-CoV-2 whole genome sequencing | ||||
| Sequenced | % | |||
| Total | 552 | 2179 | 76.03 | 30.79 |
| Delta | 492 | 827 | 89.13 | 37.95 |
| Omicron | 1 | 1208 | 0.18 | 55.44 |
| Other or low QC | 59 | 144 | 10.69 | 6.61 |
Sequence counts were up to the time of writing this manuscript.
Figure 2.
SARS-CoV-2 positivity, COVID-19 hospitalizations, and variants trends November- December 2021. (a) SARS-CoV-2 daily positivity rates between November and December 2021 for both symptomatic and asymptomatic testing. (b) COVID-19 related admissions between November and December 2021. (c) SARS-CoV-2 variant distribution between November and December 2021 relative to the 7 day rolling average positives from Johns Hopkins system. (d) Percent SARS-CoV-2 variants in November and December 2021.
Patient characteristics and infection outcomes in Omicron and Delta infections
A total of 7353 samples tested positive for SARS-CoV-2 of a total 45,856 tested in the Johns Hopkins Laboratories between November 22nd 2021 and December 31st 2021. Of these, 2378 were randomly selected for whole genome sequencing. After excluding repeat tests in patients and results that were unable to be characterized due to low quality, a total of 2027 patients, 1119 Omicron and 908 Delta, were included in the study. Patients infected with the Omicron variant were younger (median age 32 (IQR 22-46) vs 35 (IQR 17-55), Table 3) and more likely to be both fully vaccinated and boosted than patients with Delta infections (Table 3). A greater percentage of patients with Omicron infections were female compared to Delta infections, and a greater percentage of patients were non-White. Additionally, every comorbidity except cancer and pregnancy, were less frequent in Omicron patients.
Table 3.
Clinical and metadata of the Omicron and Delta infected patients.
| Variables, N (%) | Omicron | Delta |
|---|---|---|
| Total | 1119 | 908 |
| Collection range | 11/25/2021 to 12/31/21 | 11/22/21 to 12/31/21 |
| Fully Vaccinated (No booster) | 704 (62.9%) | 325 (35.8%) |
| Booster | 183 (16.4%) | 63 (6.9%) |
| Partially vaccinated | 35 (3.1%) | 22 (2.4%) |
| Symptomatic | 1036 (92.6%) | 836 (92.1%) |
| Gender | ||
| Female | 728 (65.1%) | 499 (55.0%) |
| Male | 391 (34.9%) | 409 (45.0%) |
| Age | ||
| median (IQR) | 32 (22-46) | 35 (17-55) |
| 0-44 | 809 (72.3%) | 577 (63.6%) |
| 45-64 | 255 (22.8%) | 215 (23.7%) |
| 65+ | 55 (4.9%) | 116 (12.8%) |
| Race/Ethnicity† | ||
| White | 360 (32.1%) | 381 (41.9%) |
| Black | 531 (47.4%) | 345 (37.9%) |
| Hispanic | 87 (7.8%) | 93 (10.2%) |
| Other | 143 (12.8%) | 91 (10.0%) |
| Comorbidities | ||
| Hypertension | 242 (21.6%) | 283 (31.2%) |
| Pregnancy | 84 (7.5%) | 53 (5.8%) |
| Lung Disease | 225 (20.1%) | 205 (22.6%) |
| Kidney Disease | 69 (6.2%) | 126 (13.9%) |
| Immunosuppression | 121 (10.8%) | 156 (17.2%) |
| Diabetes | 106 (9.5%) | 138 (15.2%) |
| Heart Failure | 29 (2.6%) | 62 (6.8%) |
| Atrial Fibrilation | 17 (1.5%) | 46 (5.1%) |
| Smoker | 115 (10.3%) | 142 (15.6%) |
| Cerebrovascular Disease | 46 (4.1%) | 69 (7.6%) |
| Cancer | 251 (22.4%) | 208 (22.9%) |
| Coronary Artery Disease | 85 (7.6%) | 151 (16.6%) |
| Outcome | ||
| COVID Related Admission* | 39 (3.5%) | 140 (15.4%) |
| ICU Level Care | 10 (0.9%) | 41 (4.5%) |
| COVID related Death | 3 (0.3%) | 21 (2.3%) |
| CT, N, (avg [StDev]) | 372 (18.7 [ 4.7]) | 640 (18.8 [ 4.9]) |
Patients admitted without symptoms or whose primary reason for admission was not COVID were not counted as a COVID-related admission
Race/Ethnicity separates out Hispanic as a separate group due to the higher noted incidence in this community.
Controlling for gender, age, race/ethnicity, and comorbidities, we found that, compared to patients with Delta, patients with Omicron were less likely to be admitted (0.34 (CI 0.21-0.53)), require ICU level care (0.39 (CI 0.17-0.89)), or die (0.22 (CI 0.05-0.91), Table 4). Regardless of variant, vaccinated patients were less likely to be admitted, require ICU level care, or die, however, admissions and mortality were more likely in older age groups, and men were more likely to die (Table 4). The only statistically significant comorbidities were kidney disease, immunosuppression, diabetes, atrial fibrillation, and coronary artery disease. Notably, while individuals that were boosted had a lower odds ratio of admission or ICU-level care, this was not statistically different from fully vaccinated individuals. Given the relatively low number of patients that died (24 overall, 21 Delta and 3 Omicron), none of whom had Omicron and were boosted, the difference between fully vaccinated and boosted is also not statistically significant. Evaluating patients separately by vaccination status, we found that patients with Omicron infections were less likely to be admitted regardless of vaccination status (Table S3). However, for unvaccinated older individuals, of which there were only 52, there was no difference in admission rates, as equal numbers of patients were admitted by virus type (22 of 41 Delta patients and 5 of 11 Omicron patients). This is in comparison to vaccinated elderly patients, of which there were 119 patients, where those with Omicron infections were less likely to be admitted (40% [30/75] for Delta (CI 0.297–0.513) and 20% [9/44] for Omicron (CI 0.112–0.345); Fisher Exact test, p = 0.04).
Table 4.
Multivariable regression.
| COVID Related Admission* | ICU Level Care | COVID related Death | Ct‡ | |
|---|---|---|---|---|
| Omicron | 0.34 (0.21-0.53) | 0.39 (0.17-0.89) | 0.22 (0.05-0.91) | 0.07 (-0.59-0.72) |
| Vaccine Status† | ||||
| Unvaccinated/Partially Vaccinated | Reference | Reference | Reference | Reference |
| Fully vaccinated | 0.30 (0.18-0.50) | 0.42 (0.19-0.90) | 0.17 (0.05-0.64) | -0.81 (-1.50–0.12) |
| Boosted | 0.18 (0.08-0.38) | 0.14 (0.04-0.51) | 0.35 (0.09-1.37) | -0.47 (-1.64-0.71) |
| Symptomatic | 5.26 (2.24-12.35) | 1.02 (0.40-2.57) | 0.22 (0.07-0.67) | 0.38 (-0.83-1.58) |
| Gender | ||||
| Female | Reference | Reference | Reference | Reference |
| Male | 1.32 (0.87-2.02) | 1.69 (0.84-3.38) | 3.43 (1.18-9.96) | 0.37 (-0.26-1.01) |
| Age | ||||
| 0-44 | Reference | Reference | Reference | Reference |
| 45-64 | 2.52 (1.49-4.28) | 0.96 (0.39-2.35) | 2.73 (0.54-13.89) | -0.33 (-1.22-0.56) |
| 65+ | 6.16 (3.20-11.86) | 0.64 (0.21-1.98) | 20.15 (4.06-100.03) | 0.07 (-1.30-1.43) |
| Race/Ethnicity⁎⁎ | ||||
| White | Reference | Reference | Reference | Reference |
| Black | 1.15 (0.72-1.84) | 0.63 (0.30-1.35) | 1.22 (0.36-4.08) | 1.12 (0.39-1.85) |
| Hispanic | 0.94 (0.42-2.11) | 0.43 (0.05-3.48) | 1.57 (0.16-15.21) | -0.39 (-1.57-0.78) |
| Other | 1.14 (0.54-2.39) | 1.22 (0.36-4.14) | 0.61 (0.07-5.51) | -0.67 (-1.69-0.34) |
| Comorbidities | ||||
| Hypertension | 0.83 (0.49-1.41) | 1.68 (0.64-4.39) | 0.61 (0.16-2.33) | -0.63 (-1.54-0.28) |
| Pregnancya | 2.14 (0.92-4.99) | 0.73 (0.09-6.29) | - | -0.40 (-1.67-0.88) |
| Lung Disease | 0.82 (0.51-1.33) | 1.82 (0.90-3.68) | 1.03 (0.35-3.00) | -0.37 (-1.11-0.37) |
| Kidney Disease | 4.76 (2.77-8.19) | 6.79 (2.63-17.51) | 3.32 (0.85-13.05) | 1.21 (-0.11-2.54) |
| Immunosuppression | 2.78 (1.74-4.46) | 1.06 (0.46-2.43) | 1.74 (0.57-5.28) | 1.02 (0.01-2.02) |
| Diabetes | 2.35 (1.41-3.91) | 2.13 (0.98-4.61) | 3.22 (1.11-9.39) | 1.17 (0.07-2.28) |
| Heart Failure | 0.60 (0.28-1.30) | 1.19 (0.46-3.08) | 0.51 (0.13-2.03) | -1.14 (-3.02-0.75) |
| Atrial Fibrilation | 3.41 (1.52-7.63) | 1.86 (0.72-4.82) | 1.34 (0.38-4.77) | 0.07 (-2.00-2.15) |
| Smoker | 0.93 (0.56-1.56) | 0.64 (0.29-1.42) | 0.56 (0.16-1.93) | 1.06 (0.11-2.00) |
| Cerebrovascular Disease | 0.88 (0.46-1.68) | 1.04 (0.44-2.44) | 0.36 (0.09-1.40) | -0.90 (-2.38-0.58) |
| Cancer | 1.34 (0.84-2.15) | 0.82 (0.38-1.74) | 1.47 (0.51-4.27) | 0.16 (-0.66-0.98) |
| Coronary Artery Disease | 2.84 (1.68-4.82) | 4.11 (1.64-10.31) | 2.15 (0.65-7.17) | -0.56 (-1.78-0.66) |
| N | 2027 | 2027 | 1890 | 1012 |
Patients admitted without symptoms or whose primary reason for admission was not COVID were not counted as a COVID-related admission;
Partially vaccinated were considered unvaccinated;
Regression results (see methods).
Race/Ethnicity separates out Hispanic as a separate group due to the higher noted incidence in this community.
Omicron and Delta variants cycle threshold (Ct) values in upper respiratory samples
To determine if the Ct values in upper respiratory samples were different in Omicron versus Delta infected individuals, we compared the Ct values collected for all the groups regardless of the days to the onset of symptoms or the status of the patients being symptomatic or asymptomatic (Omicron = 412, Delta N = 642). No difference in the mean or median Ct values were notable (Figure 3a). No differences were noted when Ct values were compared between vaccinated and unvaccinated patients from both groups as well (Omicron vaccinated = 229, unvaccinated = 166, Delta vaccinated = 230, unvaccinated = 400, Figure 3a). When the Ct analysis was correlated to the days from the onset of symptoms for symptomatic patients, no statistically significant differences were detected between Omicron and Delta fully vaccinated, boosted, or unvaccinated (Figures 3b and c and S2). Finally, a multivariable regression controlling for gender, age, race and ethnicity, and comorbidities found no statistically significant association between strain type and Ct-level (Table 4).
Recovery of infectious virus in Omicron versus Delta groups
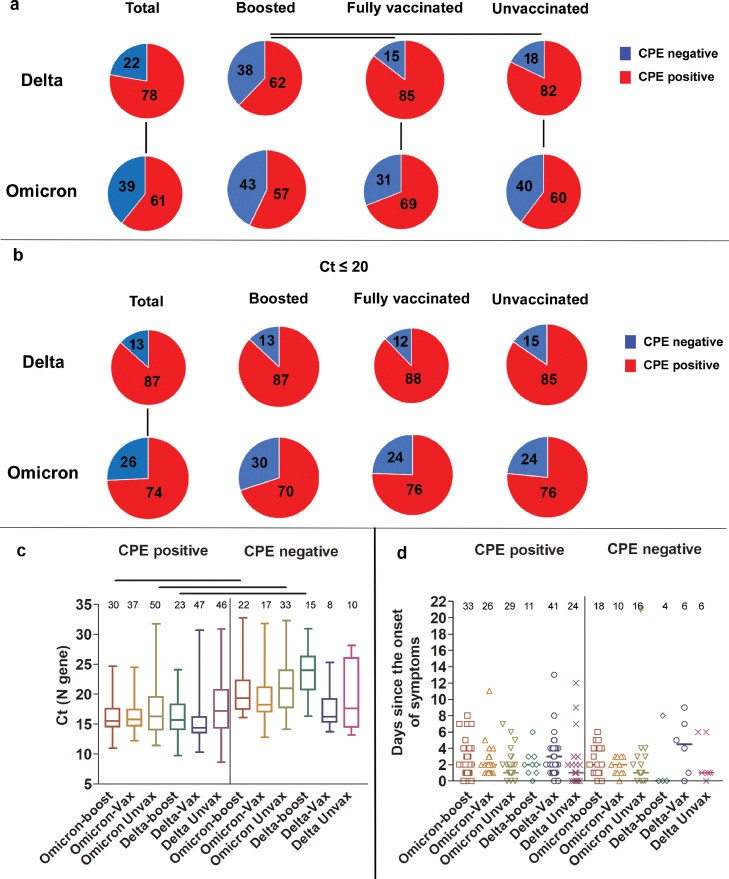
To assess the recovery of infectious virus from upper respiratory tract specimens of individuals infected with Omicron versus Delta variants, samples from 214 Omicron and 153 Delta (Table 5) infections were used to inoculate Vero-TMPRSS2 cells. Recovery of infectious virus (positive cytopathic effects; CPE) was noted from more specimens from the Delta group as compared to the Omicron group (Delta 78% (CI 0.71–0.84), Omicron 61% (CI 0.55–0.67); Figure 4a, Fisher Exact test, p = 0.0009). Specimens from the boosted Delta group showed statistically significant reduction in the recovery of infectious virus as compared to the fully vaccinated or the unvaccinated Delta groups (62% (CI 0.47–0.75) vs 85 (CI 0.74–0.92) and 82% (CI 0.7–0.9), Fisher Exact, p = 0.0096 and 0.037, Figure 4a). This was not the case in the Omicron groups which had no statistically significant differences in infectious virus recovery between the boosted, fully vaccinated and unvaccinated groups (57% (CI 0.46–0.68), 69% (CI 0.55–0.79), and 60% (CI 0.49–0.7), Figure 4a). A statistically significant increase in the recovery of infectious virus from specimens of patients infected with the Delta variant as compared to the Omicron was noted for both fully vaccinated (85% (CI 0.74–0.92) vs 69% (CI 0.55–0.79), Fisher Exact test, p < 0.042) and unvaccinated (82% (CI 0.7–0.9) vs 60% (CI 0.49–0.7), Fisher Exact test, p = 0.008) groups (Figure 4a). Consistent with our previously published reports (Figure 4c and14,19), and since lower Ct values have been associated with positive CPE, we compared samples with Ct values less than 20 for both Delta and Omicron groups (N, Table 5). Our analysis showed that the Delta infection was associated with a statistically significant increase in samples with positive CPE compared to Omicron (87% (CI 0.79–0.92) vs 74% (CI 0.66–0.81), Fisher Exact test, p = 0.0017) however, no statistically significant differences were seen between boosted, fully vaccinated, or unvaccinated groups (Figure 4b). Taken together, Omicron infections had lower numbers of samples with infectious virus when compared to Delta virus infections, indicating that a higher infectious virus load was not driving the higher transmissibility seen in Omicron infections. Notably, no statistically significant differences were noted in the specimen collection time frame in relation to the onset of symptoms in all groups (Figure 4d). Of note, 8.3% of samples with infectious virus were collected after 5 days from symptoms (14 of a total of 168 positives with known sample collection times, Tables S1 and S2).
Table 5.
Recovery of infectious virus from respiratory samples of patients infected with Delta or Omicron. P values, Fisher Exact test with Wilson 95% CI.
| Delta |
Omicron |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Positive | Negative | p value < 0.05 | Lower 95% CI | Upper 95% CI | Total | Positive | Negative | Lower 95% CI | Upper 95% CI | p value < 0.05 | p value < 0.05 (Omicron to Delta) | |
| Boosted | 42 | 26 | 16 | 0.47 | 0.75 | 77 | 44 | 33 | 0.46 | 0.68 | |||
| Fully vaccinated | 55 | 47 | 8 | 0.0096 | 0.74 | 0.92 | 54 | 37 | 17 | 0.55 | 0.79 | 0.042 | |
| Unvaccinated | 56 | 46 | 10 | 0.037 | 0.7 | 0.9 | 83 | 50 | 33 | 0.49 | 0.7 | 0.008 | |
| Total | 153 | 119 | 34 | 0.71 | 0.84 | 214 | 131 | 83 | 0.55 | 0.67 | 0.0009 | ||
| Ct < 20 |
Delta |
Omicron |
|||||||||||
| Total | Positive | Negative | p value < 0.05 | Lower 95% CI | Upper 95% CI | Total | Positive | Negative | Lower 95% CI | Upper 95% CI | p value < 0.05 | p value < 0.05 (Omicron to Delta) | |
| Boosted | 23 | 20 | 3 | 0.68 | 0.95 | 40 | 28 | 12 | 0.55 | 0.82 | |||
| Fully vaccinated | 51 | 45 | 6 | 0.77 | 0.94 | 45 | 34 | 11 | 0.61 | 0.86 | |||
| Unvaccinated | 39 | 33 | 6 | 0.7 | 0.93 | 51 | 39 | 12 | 0.63 | 0.86 | |||
| Total | 113 | 98 | 15 | 0.79 | 0.92 | 136 | 101 | 35 | 0.66 | 0.81 | 0.0017 | ||
Figure 4.
Recovery of infectious virus from respiratory samples of patients infected with Delta or Omicron. (a) Percent CPE positives and negatives for Delta and Omicron; total, patients who received a booster, fully vaccinated, and unvaccinated groups. Fisher Exact test (p, N, and CI, Table 5). (b) Percent CPE positives and negatives for Delta and Omicron; total, patients who received a booster, fully vaccinated, and unvaccinated groups with Ct values less than 20. Fisher Exact test (p, N, and CI, Table 5). (c) Box and whisker plots of Ct of Delta and Omicron samples, CPE positive and negative. Horizontal bars denote median Ct values. One-way ANOVA p < 0.05. (d) Distribution of sample collection time from each group in relation to days from the onset of symptoms. In c and d, total numbers per group are shown on top of each box.
Localized SARS-CoV-2 IgG in nasal specimens
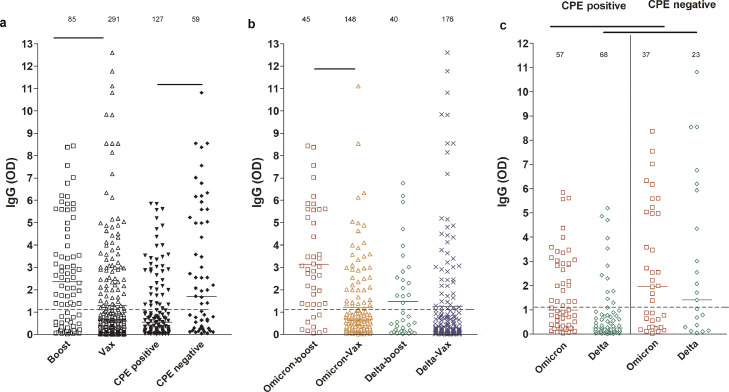
We previously showed that SARS-CoV-2 IgG levels in the upper respiratory tract are higher in samples from vaccinated individuals and correlate with less recovery of infectious virus on cell culture.14 To compare the anti-SARS-CoV-2 IgG levels between patients who received a booster and fully vaccinated patients, ELISA was performed on upper respiratory samples from the Omicron and Delta infected groups. As expected, a statistically significant increase in localized IgG levels was observed in patients who received a booster (Figure 5a, one-way ANOVA, p < 0.001 CI (0.3734 to 1.758)) and IgG levels were higher in samples with no detectable infectious virus compared to those with infectious virus (Figure 5a, one-way ANOVA, p < 0.001 CI (-2.447 to -0.6775)). The anti-SARS-CoV-2 IgG levels were higher in the boosted Omicron group compared to the Omicron- fully vaccinated group but this was not statistically significant in the Delta-infected groups (Figure 5b, one-way ANOVA, p < 0.001 (CI 0.9881 to 2.857). Both Omicron and Delta infected patients that had infectious virus showed a statistically significant decrease in anti-SARS-CoV-2 IgG levels when compared to specimens with no infectious virus (Figure 5c, one-way ANOVA, p < 0.001 (CI -3.348 to -0.8068). The data indicate that anti-SARS-CoV-2 antibody levels in the upper respiratory tract can be increased after booster vaccination but the presence of infectious virus with either Omicron or Delta infection is associated with lower local levels of vaccination induced antibodies.
Figure 5.
SARS-CoV-2 IgG levels in upper respiratory samples of infected vaccinated patients. Boost, patients with booster dose; Vax, fully vaccinated patients who didn't receive a booster dose. Dashed lines demarcate the limit of borderline and negative ELISA results as specified per assay's package insert. Statistical analyses, one-way ANOVA. Total numbers per group are shown on top of each scatter.
Discussion
In this study, we provide a comparison between Omicron and Delta infected patients from the transition from Delta to Omicron dominance. Using this tight time frame also controls for the timing of samples’ collection in relation to vaccinations and booster doses between the two groups in addition to the implemented community measures of infection control, including masking and social distancing. Our data showed that Omicron infected patients were associated with higher infection rates in vaccinated individuals and those who received booster vaccinations but admissions, ICU level care, and mortality were less likely. Additionally, even when examining only unvaccinated individuals, patients with Omicron infections were less likely to be admitted and though the low numbers precluded a statistically significant result, probably less likely to need ICU level care. Samples collected from Omicron infected patients had equivalent viral loads when compared to samples collected from Delta infected individuals regardless of the vaccination status. Recovery of infectious virus on cell culture was less frequent in the Omicron group with no statistically significant impact of vaccination with the exception of the Delta individuals who received a booster dose, this group showed a statistically significant reduced recovery of infectious virus when compared to the Delta fully vaccinated and unvaccinated groups. Consistent with our previously reported observations,14 samples with successful recovery of infectious virus on cell culture correlated with less IgG levels in the respiratory samples. In this study, we also report a statistically significant increase of IgG in samples collected from patients who received a booster dose.
The Omicron variant was first reported from South Africa early in November 2021. The first case was reported from the US on December 1st 2021, a few days after the WHO classified it as a variant of concern.23 The first case we identified as a part of our SARS-CoV-2 genomic surveillance was from a patient who developed symptoms during the last week of November 2021. A very quick increase in Omicron detection correlated with a marked increase in the overall SARS-CoV-2 positivity to reach an average of close to 50% in symptomatic patients in the last week of December. Notably, the Omicron detection and rapid increase occurred during a spike in Delta circulation. In contrast, the Delta displacement of Alpha variant occurred in a time of markedly low circulation of the latter (Figure S1 and14). The rapid increase in Omicron cases could be explained by either an increase in overall transmissibility of this variant or due to the enhanced immune evasion of Omicron through multiple mutations in the Spike protein. In a study that compared the household contacts of Delta and Omicron infected patients, the secondary attack rate was higher with Omicron, particularly in fully vaccinated and boosted contacts (preprint24). Notably, 53.1% of the Omicron infected patients from our cohort were fully vaccinated or received booster doses and there was no difference in the presence of Omicron infectious virus in either the unvaccinated, vaccinated or boosted groups, suggesting little effect of vaccination on infectious virus load. Multiple studies have shown that the neutralization of the Omicron variant is reduced compared to Delta or prior variants (preprint and peer reviewed25, 26, 27–28). Interestingly, there were fewer numbers of specimen containing infectious virus in the Omicron group compared to the Delta group, indicating that the presence of infectious virus alone may not explain the higher transmissibility of Omicron.
The omicron genome contains 32 amino acid changes in the spike protein, within its NTD, RBD, and close to the furin cleavage region, some of which are shared with prior variants of concern and were previously characterized (peer reviewed and preprint29,30). Interestingly, those changes are expected to impact the binding to the host receptor ACE2 and alter membrane fusion (preprint31). This likely explains the notable phenotypic changes of Omicron in cell culture and animal studies and the change in the viral tropism. Omicron was shown to cause a mild disease in animals and replicate less efficiently in the lungs.32 In addition, the use of the Vero-TMPRSS2 cell line which initially showed an enhanced sensitivity for the recovery of SARS-CoV-2,33 was also impacted by changes within Omicron. The slower viral growth might indicate an alternative entry pathway that is less dependent on TMPRSS2.34 This is consistent with our observations that samples from the Omicron infected group were associated with less recovery of infectious virus on this cell line, yet, using this cell line allowed the comparisons between different groups based on their vaccination status. Our data indicates that the recovery of infectious virus with Omicron is not impacted by booster vaccination, which was not the case with Delta infected patients. Even though, when we strictly limited our analysis to samples with Ct values less than 20, the recovery of infectious virus from the Delta infected, boosted group was equivalent to other groups. IgG levels were higher in samples from boosted, vaccinated patients and those with no infectious virus. Taken together, we believe that Omicron evasion of preexisting immunity contributes to lessen the impact of booster vaccination on the recovery of infectious virus, which might contribute to increased transmission, even in individuals who receive booster vaccination.
The most recent CDC guidelines for infection control and isolation indicates that in a contingency status, health care workers can quarantine for 5 days from the onset of symptoms if asymptomatic or with mild to moderate symptoms.35 Our study showed that 8.5% of CPE positive samples were collected after 5 days from symptoms’ onset from patients who did not require hospitalization. Our data indicates that it is not uncommon to recover infectious virus after 5 days from symptoms regardless of the vaccination status when patients are symptomatic. Hence care should be taken when making a release from quarantine decisions, especially when patients are showing symptoms.
The limitations of our study include the retrospective nature of data collections which doesn't allow the collection of baseline serum and respiratory IgG levels. Antibody neutralization assays and quantification of viruses from clinical samples were not conducted as a part of this study nor were Omicron or Delta specific IgG assays. Clinical data was compiled from patients tested and admitted to the Johns Hopkins Health System and the results might be impacted by regional differences. It is possible that patients who tested positive in our system sought additional clinical care, including admission, at a different hospital not affiliated with Johns Hopkins. While this could artificially lower our admission rates, we have no reason to think that this was different in the first half of December, when Delta was predominant, compared to the end of December. Additionally, the study was conducted in a highly vaccinated population, particularly in older individuals. Thus, while the results clearly suggest that vaccinated patients or relatively younger unvaccinated patients with Omicron infections were less likely to have severe disease, the impact on older individuals, while limited, suggests that Omicron remains highly dangerous for this community, particularly in those with no vaccination. This is consistent with the data from Hong Kong, where 95% of deaths in the recent Omicron wave were in the elderly.36
In conclusion, our data suggests that Omicron infected patients are less likely to develop severe disease when compared to Delta, yet we show that Omicron is associated with more infections of fully and booster vaccinated individuals likely due to immune evasion. It is important to note that the total numbers of admissions were higher and that admitted patients didn't show a statistically significant difference in the use of supplementary oxygen, or ICU level care. This highlights the importance of taking infection control precautions and raising an awareness that Omicron infections should not be underestimated.
Contributors
AF, RE, JS. data collection and data interpretation. CPM. data collection and analysis OA, JMN, NG, NS, ML, MF, DCG. data acquisition and collection. AP. cell culture, scientific and manuscript revision. EK. clinical data collection and analysis and scientific and manuscript revision. HHM. study design, data collection and analysis, data interpretation, writing, fund acquisition. EK and HHM verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing
Whole genome data were made available publicly (GISAID IDs, Tables S1 and S2) and raw genomic data requests could be directed to HHM.
Declaration of interests
We declare no relevant competing interests
Acknowledgements
This study was only possible with the unique efforts of the Johns Hopkins Clinical Microbiology Laboratory faculty and staff. HHM is supported by the HIV Prevention Trials Network (HPTN) sponsored by the National Institute of Allergy and Infectious Diseases (NIAID). Funding was provided by the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (UM1 AI068613), the NIH RADx-Tech program (3U54HL143541-02S2), National Institute of Health RADx-UP initiative (Grant R01 DA045556-04S1), Centers for Disease Control (contract 75D30121C11061), the Johns Hopkins University President's Fund Research Response, the Johns Hopkins department of Pathology, and the Maryland department of health. EK was supported by Centers for Disease Control and Prevention (CDC) MInD-Healthcare Program (Grant Number U01CK000589). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Institute of Biomedical Imaging and Bioengineering; the National Heart, Lung, and Blood Institute; the National Institutes of Health, or the U.S. Department of Health and Human Services.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104008.
Contributor Information
Andrew Pekosz, Email: apekosz1@jhu.edu.
Eili Y. Klein, Email: eklein@jhu.edu.
Heba H. Mostafa, Email: hmostaf2@jhmi.edu.
Appendix. Supplementary materials
References
- 1.Team CC-R SARS-CoV-2 B.1.1.529 (Omicron) variant - United States, December 1-8, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(50):1731–1734. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. World Health Organization . World Health Organization; Geneva, Switzerland: 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Accessed 3 December 2021. [Google Scholar]
- 3.Wolter N., Jassat W., Walaza S., et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022 doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kannan S., Shaik Syed Ali P., Sheeza A. Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci. 2021;25(24):8019–8022. doi: 10.26355/eurrev_202112_27653. [DOI] [PubMed] [Google Scholar]
- 5.Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA. 2021 doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Investigation of SARS-CoV-2 variants: technical briefings. UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England technical briefing 34. 14 January 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050236/technical-briefing-34-14-january-2022.pdf
- 7.Bouzid D., Visseaux B., Kassasseya C., et al. Comparison of patients infected with Delta versus Omicron COVID-19 variants presenting to Paris emergency departments : a retrospective cohort study. Ann Intern Med. 2022 doi: 10.7326/M22-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattiuzzi C., Henry B.M., Lippi G. COVID-19 vaccination and SARS-CoV-2 Omicron (B.1.1.529) variant: a light at the end of the tunnel? Int J Infect Dis. 2022 doi: 10.1016/j.ijid.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulliam J.R.C., van Schalkwyk C., Govender N., et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. MedRxiv. 2021 doi: 10.1126/science.abn4947. 2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarrett J., Uhteg K., Forman M.S., et al. Clinical performance of the GenMark Dx ePlex respiratory pathogen panels for upper and lower respiratory tract infections. J Clin Virol. 2021;135 doi: 10.1016/j.jcv.2021.104737. [DOI] [PubMed] [Google Scholar]
- 11.Mostafa H.H., Carroll K.C., Hicken R., et al. Multi-center evaluation of the Cepheid Xpert(R) Xpress SARS-CoV-2/Flu/RSV Test. J Clin Microbiol. 2020 doi: 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostafa H.H., Hardick J., Morehead E., Miller J.A., Gaydos C.A., Manabe Y.C. Comparison of the analytical sensitivity of seven commonly used commercial SARS-CoV-2 automated molecular assays. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhteg K., Jarrett J., Richards M., et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo C.H., Morris C.P., Sachithanandham J., et al. Infection with the SARS-CoV-2 Delta variant is associated with higher recovery of infectious virus compared to the Alpha variant in both unvaccinated and vaccinated individuals. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thielen P.M., Wohl S., Mehoke T., et al. Genomic diversity of SARS-CoV-2 during early introduction into the Baltimore-Washington metropolitan area. JCI Insight. 2021;6(6) doi: 10.1172/jci.insight.144350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris C.P., Luo C.H., Amadi A., et al. An update on SARS-CoV-2 diversity in the United States national capital region: evolution of novel and variants of concern. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Toole Á., Scher E., Underwood A., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evolution. 2021;7(2) doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuyama S., Nao N., Shirato K., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gniazdowski V., Morris C.P., Wohl S., et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully E.P., Schumock G., Fu M., et al. Sex and gender differences in testing, hospital admission, clinical presentation, and drivers of severe outcomes from COVID-19. Open Forum Infect Dis. 2021;8(9) doi: 10.1093/ofid/ofab448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez D.A., Hinson J.S., Klein E.Y., et al. SARS-CoV-2 positivity rate for Latinos in the Baltimore–Washington, DC region. JAMA. 2020;324(4):392–395. doi: 10.1001/jama.2020.11374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhteg K., Amadi A., Forman M., Mostafa H.H. Circulation of non- SARS-CoV-2 respiratory pathogens and coinfection with SARS-CoV-2 amid the COVID-19 pandemic. Open Forum Infect Dis. 2021 doi: 10.1093/ofid/ofab618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CDC Omicron Variant: What You Need to Know. 2021 Accessed 19 January 2022. [Google Scholar]
- 24.Lyngse F.P., Mortensen L.H., Denwood M.J., et al. SARS-CoV-2 Omicron VOC transmission in Danish households. medRxiv. 2021 2021.12.27.21268278. [Google Scholar]
- 25.Basile K., Rockett R.J., McPhie K., et al. Improved neutralization of the SARS-CoV-2 Omicron variant after Pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting. bioRxiv. 2021 doi: 10.3390/v14092023. 2021.12.12.472252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Planas D., Saunders N., Maes P., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt F., Muecksch F., Weisblum Y., et al. Plasma Neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2021 doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rössler A., Riepler L., Bante D., Dv L., Kimpel J. SARS-CoV-2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv. 2021 2021.12.08.21267491. [Google Scholar]
- 29.Starr T.N., Greaney A.J., Addetia A., et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19′s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Cai Y., Lavine C.L., Peng H., et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. bioRxiv. 2022 doi: 10.1016/j.celrep.2022.110729. 2022.01.11.475922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diamond M., Halfmann P., Maemura T., et al. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res Sq. 2021 [Google Scholar]
- 33.Matsuyama S., Nao N., Shirato K., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H., Lu L., Peng Z., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11(1):277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC . Centers for Disease Control and Prevention; 2021. Interim Guidance for Managing Healthcare Personnel with SARS-CoV-2 Infection or Exposure to SARS-CoV-2. Accessed 20 January 2022. [Google Scholar]
- 36.Taylor L. COVID-19: Hong Kong reports world's highest death rate as zero COVID strategy fails. BMJ. 2022;376:o707. doi: 10.1136/bmj.o707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.