Abstract
SARS-CoV-2 engages with human cells through the binding of its Spike receptor-binding domain (S-RBD) to the receptor ACE2. Molecular blocking of this engagement represents a proven strategy to treat COVID-19. Here, we report a single-chain antibody (nanobody, DL4) isolated from immunized alpaca with picomolar affinity to RBD. DL4 neutralizes SARS-CoV-2 pseudoviruses with an IC50 of 0.101 μg mL−1 (6.2 nM). A crystal structure of the DL4-RBD complex at 1.75-Å resolution unveils the interaction detail and reveals a direct competition mechanism for DL4's ACE2-blocking and hence neutralizing activity. The structural information allows us to rationally design a mutant with higher potency. Our work adds diversity of neutralizing nanobodies against SARS-CoV-2 and should encourage protein engineering to improve antibody affinities in general.
Keywords: Covid-19, Crystal structure, Neutralizing antibody, Protein engineering, Receptor-binding domain, Single-chain antibody
1. Introduction
An essential step for SARS-CoV-2 infection is its attachment to the human cells via a binding event between its Spike receptor-binding domain (S-RBD) and the human receptor angiotensin-converting enzyme 2 (ACE2) [[1], [2], [3], [4]]. S contains two subunits S1 and S2 that are generated by the cleavage of the proprotein [4]. The S1 subunit contains the RBD and is responsible for virus attachment, while the S2 subunit is responsible for a membrane fusion event that is triggered by the RBD-ACE2 engagement. S assembles into a trimer and is heavily decorated by glycosylation to escape immune surveillance. Suiting the role of molecular recognition, the RBD is relatively less glycosylated and therefore represents a hot spot for neutralizing antibodies [[5], [6], [7], [8], [9], [10], [11]] and vaccine development [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21]]. Much of the RBD and the receptor-binding motif (RBM, the ACE2-binding surface) are shielded by the N-terminal domain of S1 from adjacent protomers of the S trimer in the so-called more stable “closed conformation”; and such RBDs are referred to as “down”-RBD. In the “open-conformation”, the RBD ejects and exposes RBM for ACE2-binding [1,2,22,23] while also opening opportunities for neutralizing antibodies.
Naturally occurring single-chain antibodies (nanobodies) from camelids and sharks are increasingly recognized as “next-generation” therapeutics [24] owing to their unique advantages. Their small sizes (~14 kDa) allow convenient and high-yield production in microbial hosts, straightforward screening, and rapid directed evolution with various display platforms [25,26]. In the past months, dozens of neutralizing nanobodies against SARS-CoV-2 have been reported [8,[27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]], most of which target RBD with nanomolar affinities.
Here, we report the isolation of a neutralizing nanobody from an immunized alpaca with picomolar affinity to RBD. Using X-ray crystallography, we characterize its epitope and reveal the interaction details whereby all three complementarity-determination regions (CDRs) and a significant portion of the framework region participated in the antigen recognition. Guided by the high-resolution structure, we have designed a mutant that increases the neutralizing activity to 3-fold of the wild-type. Our work presents a high-affinity nanobody and a strategy to engineer antibodies to improve potencies in general.
2. Results
2.1. Isolation of a picomolar-affinity neutralizing nanobody from immunized alpaca
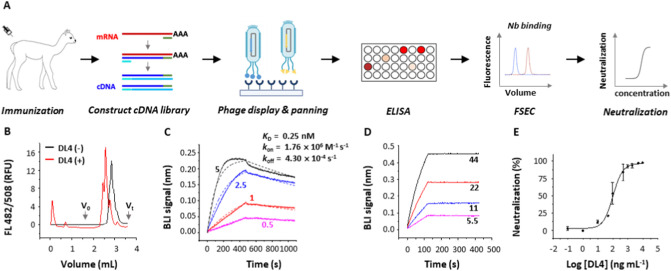
To obtain nanobodies against RBD, an adult female alpaca was immunized for four rounds with recombinant RBD expressed and purified from insect cells. The immunization effect was monitored by ELISA test which showed a high titer (~1 × 106) in comparison with the pre-immunization sera. A phage display library was constructed using mRNA isolated from peripheral blood lymphocytes. High-affinity nanobodies were enriched by three rounds of panning under increasingly stringent conditions (decreasing amount of the immobilized antigen RBD). Using ELISA, we identified 28 unique RBD binders and we focus on a nanobody named DL4 in this study.
On fluorescence-detection size exclusion chromatography, the elution volume of fluorescently labeled RBD [38] was decreased upon incubation with DL4 (Fig. 1B), suggesting a stable DL4-RBD complex in solution. Further binding kinetics measurements with a biolayer interferometry (BLI) assay confirmed a tight complex with picomolar affinity (K D = 0.25 nM) (Fig. 1C). The BLI assay also demonstrated DL4's ability to bind S (Fig. 1D). A neutralization assay using SARS-CoV-2 pseudotyped particles (pp) bearing the S from the first-reported strain from Wuhan displayed an IC50 of 0.101 μg mL−1 (6.23 nM) for DL4 (Fig. 1E).
Fig. 1.
Strategy and isolation of neutralizing nanobodies. (A) Flowchart for generation of neutralizing nanobodies (Nbs). A cDNA library was constructed using mRNA isolated from an immunized alpaca. The library was selected by rounds of panning and RBD-binders were screened by enzyme-linked immunosorbent assay (ELISA) and fluorescence-detector size exclusion chromatography (FSEC). Neutralizing nanobodies were identified by assays with SARS-CoV-2 pseudoviruses. (B) Unpurified DL4 causes earlier elution of the fluorescently labeled RBD on gel filtration. (C) Binding kinetics of DL4 to RBD using biolayer interferometry (BLI) with RBD immobilized and DL4 as analyte at indicated concentrations (nM). Solid lines indicate original data and dotted lines indicate fitted curves. (D) Evidence for binding between DL4 and Spike. Apparent binding kinetics are not fitted because of the likely existence of bridged complexes between immobilized DL4 and the trimeric analyte S. (E) Neutralization assay of DL4 against SARS-CoV-2 pseudoviruses. Mean and standard error of the mean (s.e.m., n = 3) are plotted.
2.2. Structural characterization of the DL4 epitope
To accurately characterize the epitope of DL4, we crystallized DL4 in complex with RBD in the space group of P22121 and solved its structure to 1.75-Å resolution by molecular replacement using previous RBD and nanobody structures as search models. The structure was refined to R work/R free of 0.1891/0.2174 with no geometry violations (Table 1 ). Each asymmetric unit contains two DL4-RBD complexes. Because the two copies are similar (Cα RMSD of 0.216 Å), we use chains A and B for structure description.
Table 1.
Data collection and refinement statistics.
| DL4-RBD | |
|---|---|
| Data collection | |
| Space group | P 2 21 21 |
| Cell dimensions | |
| a, b, c (Å) | 79.82, 95.04, 118.92 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.97915 |
| Resolution (Å) | 36.58–1.75 (1.78–1.75)a |
| Rmerge | 0.110 (1.183) |
| Rpim | 0.046 (0.522) |
| I/σI | 14.0 (2.1) |
| Completeness (%) | 100.0 (100.0) |
| Multiplicity | 12.8 (11.9) |
| CC*b | 0.999 (0.973) |
| Refinement | |
| Resolution (Å) | 33.66–1.75 |
| No. reflections | 91,696 |
| Rwork/Rfree | 0.1891/0.2174 |
| No. atoms | 5970 |
| Protein | 5127 |
| Ligands | 230 |
| Solvent | 613 |
| No. residues | 631 |
| B-factors (Å2) | 29.19 |
| Protein | 27.17 |
| Ligand/ion | 56.78 |
| Solvent | 35.72 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.880 |
| Ramachandran | |
| Favoured (%) | 96.31 |
| Allowed (%) | 3.69 |
| Outlier (%) | 0 |
| PDB ID | 7F5G |
Highest resolution shell is shown in parenthesis.
CC* = .
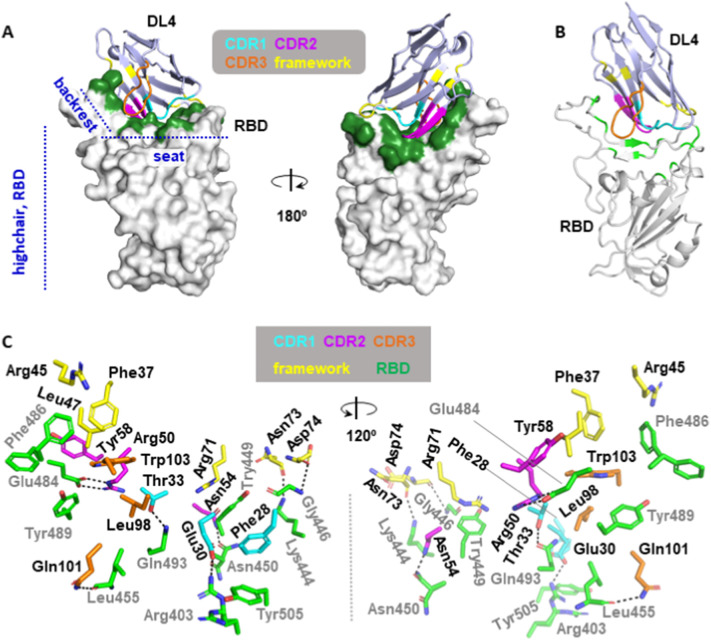
The RBD structure assembles a high-chair shape and DL4 binds RBD at the ‘seat’ and ‘backrest’ region with a buried surface area [39] of 1026.7 Å2 (Fig. 2A), with contributions of 205.6 Å2 from CDR1, 326.8 Å2 from CDR2, 257.5 Å2 from CDR3, and interestingly, 236.8 Å2 (23% of the total surface) from the framework region (Fig. 2B). For clarity, we label residues from RBD with a prime. The three CDRs interact with RBD via two salt bridge pairs (Glu30/Arg403′ and Arg50/Glu484′), three hydrogen bonds (Thr33/Gln493′, Asn54/Asn450′, Gln101/Leu 455′), and hydrophobic interactions by apolar residues or hydrocarbon potion of polar residues such as Glu484′ (Fig. 2C, Table S1). The framework loop contributed two hydrogen bonds (Asn73/Lys444′, Asp74/Gly446′) and a cation-π interaction (Arg71/Tyr449′) (Fig. 2C, Table S1).
Fig. 2.
Crystal structure of the DL4 in complex with the receptor-binding domain (RBD). (A) The overall structure of the DL4 (light blue) in complex with RBD (white). DL4 binds the highchair-shaped RBD at the ‘seat’ and ‘backrest’ region. The binding interface is colored green. Three CDRs and the framework residues involved in the binding are color-coded as indicated. (B) Cartoon representation of the overall DL4-RBD structure. The three CDRs are color-coded as in A. DL4-contacting sites in RBD are colored green. (C) Stick representation of the interaction residues from DL4 (cyan, magenta, orange, and yellow) and RBD (green). DL4 residues are labeled in black and RBD residues are labeled in grey. Dash lines indicate distances within 3.8 Å.
2.3. DL4 competes directly with ACE2 for RBD-binding
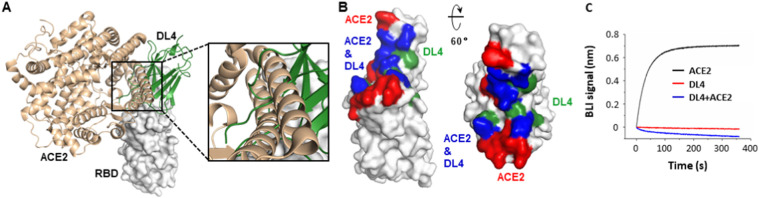
Aligning the DL4-RBD complex to the ACE2-RBD structure [40,41] reveals a large overlap between the DL4 epitope and the receptor-binding motif (RBM) (Fig. 3A, B). Specifically, the shared site includes 15 residues, some of which, such as Gln493′ and Glu484′ are key residues for both the receptor- and DL4-binding. Consistent with the structural observation, pre-incubation with DL4 completely blocked the binding between ACE2 and RBD (Fig. 3C). Aligning the DL4 structure to the S structures [1] showed that, although the epitope is shielded by Asn343-linked glycans and nearby residues in the ‘down’-RBD, it is well exposed on the ‘up’-RBD (Fig. S1). Taken together, DL4 neutralizes SARS-CoV-2 by directly blocking the receptor recognition.
Fig. 3.
Nanobody DL4 engages the receptor-binding domain (RBD) at the receptor-binding motif and directly competes with ACE2 for RBD-binding. (A) Aligning the DL4-RBD structure onto the ACE2-RBD structure (PDB ID 6M0J) [40] reveals clashes between ACE2 (wheat) and DL4 (green). Only the RBD from the DL4-binding structure is shown (white). (B) The overlap (blue) between the ACE2-binding site (red) and the DL4 epitope (green). (C) Pre-incubation of DL4 with RBD prevents ACE2 from binding to RBD. A sensor coated with RBD was first treated with 100 nM of DL4 (monovalent) before being incubated with a DL4-containing solution with (blue) or without (red) ACE2. As a control, the ACE2-RBD binding profile (black) was recorded using the same procedure without DL4 on a biolayer interferometry (BLI) system.
2.4. Structure-based design improved DL4's potency
Next, we set to engineer DL4 for higher neutralizing activity. Avidity effects are commonly exploited for nanobody engineering [8,42] and we also constructed the Fc version of DL4 (Fc-DL4). Unlike those in previous reports [8], however, the Fc fusion did not significantly increase neutralizing activity, displaying an IC50 of 0.142 μg mL−1/1.82 nM (0.101 μg mL−1/6.23 nM for DL4) (Fig. 4A).
Fig. 4.
Structure-based design for a gain-of-function DL4 triple mutant. (A) Neutralization assay of DL4 and the divalent form Fc-DL4. Data for DL4 are replotted from Fig. 1E. (B, C) The rationale for the design of H56Y (B) and G100E and Q101F (C). H56Y and Q101F may bind RBD tighter because of hydrophobic matching. G100E may bind RBD tighter by gaining a salt bridge. (D) Neutralization assay of the three single mutations and the triple mutant (3 m). The triple mutant displayed a 3-fold neutralizing activity compared to the wild-type DL4. In A and D, mean ± s.e.m. from three independent experiments are plotted.
Previously, we have designed gain-of-function nanobody mutants based on structural information to increase binding affinity and neutralizing activity [8]. This approach was used again for DL4. Analyzing the DL4-RBD structure reveals that His56 from CDR2 is located in a hydrophobic microenvironment (Fig. 4B) and does not contribute to hydrogen bonding (Fig. 2C). To match the hydrophobic patch, His56 was mutated to Phe, Tyr, and Trp. Similarly, Gln101 in CDR3 was also mutated to the three aromatic residues to match the hydrophobic patch on the RBD made by Tyr421′, Leu455′, Phe456′, Try473′, Tyr489′, and the hydrocarbon portion of Lys417′ (Fig. 4C). In addition, the G100E mutant was designed to introduce a possible salt bridge with Lys417′ or the nearby Arg403′. In neutralizing assays, H56Y, Q101F, and G100E inhibited viral entry with IC50 values of 0.088 μg mL−1 (1.13 nM), 0.163 μg mL−1 (2.09 nM), and 0.128 μg mL−1 (1.64 nM), respectively (the Fc-version was used, Fig. 4D). Although the single mutations behaved similarly to the wildtype (0.142 μg mL−1, 1.82 nM), the triple mutant (Fc-DL4(3m)) displayed a 3-fold neutralizing activity compared to the wild-type (IC50 = 0.046 μg mL−1, 0.59 nM) (Fig. 4D).
2.5. Structural interpretation of DL4's varying activity against variants
Next, the ability of Fc-DL4 (or Fc-DL4(3m)) against several variants of concern (VOCs) including the Alpha (B.1.1.17), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) was tested using pseudoviruses. DL4 remained effective against the Alpha strain, displaying a similar IC50 to the original Wuhan strain (Fig. 5A, B). Mutations in the Beta and Omicron strain almost escaped Fc-DL4, while the Gamma and Delta mutations compromised neutralization activity by 14 and 30 folds, respectively. Interestingly, despite the lack of neutralizing activity of Fc-DL4 against the Beta strain, the Fc-DL4(3m) showed weak inhibition with an IC50 of 3.23 μg mL−1 (Fig. 5A, B).
Fig. 5.
Structural interpretation of DL4's varying activity in neutralizing VOCs. (A) Neutralization assay of Fc-DL4 and Fc-DL4(3m) for the Alpha, Beta, Gamma, Delta, and Omicron pseudovirus. Mean ± s.e.m. from three independent experiments are plotted. (B) Summary of the strain information and neutralization results in A. Data for the original Wuhan strain (WT) are from Fig. 4A/D for comparison. (C–F) Structural interpretation of DL4's sensitivity for mutations from the Alpha (C), Beta/Gamma (D), Omicron (E), and Delta (F). The analysis was limited to the mutations occurring at the DL4's epitope. The overview and expanded view of the boxed region are shown. RBD residues are marked with a prime. Dash lines indicate H-bonds or salt bridges with distances shown in Å.
The atomic details of the DL4-RBD interactions provided possible explanations for DL4's varying activity against these variants. The Alpha strain contains a single mutation (N501Y) in the RBD. Although Asn501′ is in the vicinity of the CDR1, it does not form hydrogen bonds with DL4 (Fig. 5C). Therefore, mutation of Asn501′ is not expected to affect DL4-RBD binding, at least directly. In addition, a tyrosine replacement appeared to be compatible with the local hydrophobic patch consisting of Phe28/29/31; and Tyr501′ may even form a hydrogen bond with Glu30 (Fig. 5C), explaining DL4/DL4(3m)'s equal or slightly higher neutralizing activity against Alpha compared to the original Wuhan strain (Fig. 5A, B).
The Beta and Gama variant both contain a lysine replacement of Glu484′, a residue that forms a key salt bridge with Arg50 in CDR2 (Fig. 2C). The E484K mutation would not only eliminate the salt bridge but also introduce charge-charge repulsion with Arg50 (Fig. 5D). Similarly, the Omicron strain contains the E484A mutation. Although it alone may be less disruptive than E484K, its combination with other epitope mutations that either eliminate existing interactions (Q493R, Y505H) or distort backbone conformations (G446S, G496S) are expected to weaken the antibody-antigen interactions further (Fig. 5E). Finally, the structural reason for the weakened neutralizing activity against the Delta was less obvious. Although the Leu452′ was at the DL4's epitope, it is not immediately clear how the replacement with an arginine would affect the interactions, especially with the DL4 Asp55 in the vicinity for the formation of a possible salt bridge with Arg452′ (Fig. 5F). Possibly, L452R deforms the local region to prevent DL4 from accessing the epitope or to weaken its affinity with DL4.
3. Discussion
In this study, we report a high-affinity RBD binder isolated from immunized alpaca and its structural and biological characterization. Most monovalent RBD-targeting nanobodies bind S or RBD with K D in the nanomolar ranges [8,[27], [28], [29], [30], [31], [32],[35], [36], [37], [38]]. Structurally characterized RBM-type nanobodies with K D values in the low picomolar ranges include Nb20 (10.4 pM), Nb21 (<1 pM) [37], Huo-C5 (99 pM), Huo-F2 (40 pM) and Huo-H3 (25 pM) from immunized llama [43], and Fu2 (118 pM) [44]and Re5D06 from immunized alpaca (2 pM) [45]. With a K D of 245 pM, DL4 joins with these seven as ultra-high-affinity ACE2-blockers. This reinforces the notion that, despite their small sizes, nanobodies can bind antigens with comparable affinity with Fab which is four times in size. One of the reasons, as revealed in this study and previous structural reports [8,38,43,44,46], is that the framework region of the nanobodies can also participate in the antigen-binding, thus essentially expanding the binding surface and increasing the number of interactions. In the case of DL4, a concave surface of the nanobody framework aligns with the protruding “backrest” of RBD with good shape complementarity (Fig. 2A, B). This type of interaction has also been observed in the case of nanobodies against the KDEL receptor [47], the κ-opioid receptor [48], the folate transporter [49], and the histo-blood group antigen BabB [50].
It is noted that the IC50 (6.23 nM) of the monomeric DL4 is more than 20 folds than its K D (245 pM) to RBD. This would indicate epitope masking in the S trimer, as suggested by the structural superposition (Fig. S1). Thus, unlike in the case of the BLI binding experiment using isolated RBD, not all three RBMs are accessible to DL4 in the context of the S trimer on viral particles, reducing the apparent affinity of DL4 to S. Alternatively, the functional affinity between the S trimer and ACE2 dimer would be much higher than those reported which was measured using RBD and ACE2 monomers [40] due to avidity and positive cooperativity reasons [[51], [52], [53]]. To outcompete ACE2-RBD engagement, therefore, may require high concentrations of antibodies in the case of the monomeric DL4. A survey of the literature found this to be a general trend: most monomeric nanobodies (20 out of 24) show an IC50 value that is at least 5-fold higher than the K D value [37,[54], [55], [56]], and 8 of them report a >20 fold difference [8,44,46,54,[57], [58], [59]].
In the literature, increasing avidity generally improves potency, although the effect can vary from dozens to thousands of times [8,28]. Interestingly, the avidity effect for DL4 was not apparent (Fig. 4A). Mechanistically, fusing with Fc may introduce additional steric hindrance to prevent RBD-ACE2 binding. It may also tether two S trimers to restrict their conformational changes should the two nanobody entities bind to different S. More commonly, avidity is known to increase potency by boosting apparent binding affinity by increasing local concentration and hence a faster k on and a slower k off. In the case of DL4, the affinity may not be the limiting factor owing to its exceptional binding kinetics. Alternatively, the way DL4 binds RBD in the context of the S trimer may have prevented the avidity effects. Aligning the DL4 structure to the “two up-RBD” S structure [61] revealed a spacing of ~95 Å between the C-termini of the two DL4 molecules (Fig. S2). This would exceed the flexible limits of the disulfide-tethered Fc dimer (estimated distances of ~30–50 Å) [62]. Such arrangements could have two consequences (on a single S trimer): little avidity effects would occur, or the S trimer would collapse if both DL4 protomers bind S-RBD simultaneously. Based on the similar but slightly increased neutralizing activity after Fc-fusion (IC50 changed from 6.23 nM to 1.82 nM), both effects may exist only at a minor level. Nevertheless, because the Fc fusion can increase the potency in vivo by extending the serum half-life of nanobodies from several minutes to several days [8] and thus should be still be useful for therapeutic reasons.
The fact that the DL4(3m) is more potent than DL4 is worth discussing. Thus, despite multiple rounds of immunization, there was still space for rational design. Such practice may be applied to the existing antibodies although the effect of mutations on pharmacological behavior will have to be tested in the cases of therapeutic antibodies.
Accumulating evidence has suggested a general pattern regarding antibody characteristics and epitope. RBM-targeting antibodies are generally more potent than those targeting the RBD core [9,[63], [64], [65]] but are more susceptible to escape mutants [64,[66], [67], [68], [69], [70]]. Consistent with this trend, despite DL4's resistance to the mutations in the Alpha strain, its neutralizing activity is compromised or fully lost against variants with more RBM mutations. Although somewhat disappointing the results were not unexpected because the immunization was performed with RBD from the original Wuhan strain. Our results here highlight the lag-phase of antibody development in relation to the virus evolution and urge more rapid methods for developing antibodies with diverse epitopes and neutralizing mechanisms.
Owing to their minute sizes, nanobodies may bind surfaces that are inaccessible to conventional antibodies. DL4 may be able to bind to the ‘down’-RBD given the minor clashes with the ‘closed’ conformation of S, in addition to binding with the ‘up’-RBD (Fig. S1). This possibility remains to be experimentally investigated. The small size of nanobodies could also mean fewer chances for steric hindrance for the development of non-competing pairs that target different epitopes. Such pairs will allow the development of biparatopic nanobodies to increase tolerance to escape mutants, and DL4's ultra-high affinity could offer advantages in such applications.
4. Materials and methods
4.1. Protein expression and purification - Spike (S)
A DNA fragment encoding the polypeptide containing, from the N- to the C-terminus, residues Met1 – Gln1208 (without the C-terminal transmembrane domain, Uniprot P0DTC2) of the SARS-CoV-2 S with two stabilizing proline mutations K986P/V987P, a GSAS linker substituting the furin sites (Arg682-Arg685), a C-terminal T4 fibritin trimerization motif (GYIPEAPRDGQAYVRKDGEWVLLSTFL), a TEV protease cleavage site, a FLAG tag and a polyhistidine tag [22] was cloned into a pCDNA3.1 backbone vector and overexpressed in Expi293 cells by transient transfection using polyethylenimine (PEI). The supernatant of the cell culture after 3.5 days was harvested by filtration through a 0.22-μm membrane, and adjusted to contain 4 mM MgCl2, 20 mM imidazole, 200 mM NaCl, and 20 mM Tris-HCl pH 7.5. The filtrate was added with 3 mL of Ni-NTA beads and the mix was incubated at 4 °C for 2 h for batch binding. The beads were poured into a Bio-Rad gravity column, washed with 50 column volume (CV) of 20 mM imidazole, and eluted with 250 mM imidazole in 200 mM NaCl, 20 mM Tris-HCl pH 7.5. Fractions containing S were pooled, concentrated using a 100-kDa cut-off membrane concentrator, and further fractioned by size exclusion chromatography. Protein was quantified using a theoretical ε280 of 138,825 M−1 cm−1.
4.2. Protein expression and purification - RBD
A DNA fragment encoding the polypeptide containing, from the N- to C-terminus, the honey bee melittin signal peptide (KFLVNVALVFMVVYISYIYAA), a Gly-Ser linker, residues 330–531 of the SARS-CoV-2 S (Uniprot P0DTC2), a Gly-Thr linker, the 3C protease site (LEVLFQGP), a Gly-Ser linker, the Avi tag (GLNDIFEAQKIEWHE, for enzyme-based biotinylation), a Ser-Gly linker, and a deca-His tag was cloned in a pFastBac-backbone vector for overexpression in Trichoplusia ni High Five suspension cells. Cells at a density of 2 × 106 cells per milliliter were transfected with baculovirus produced using standard Bac-to-Bac procedures (Invitrogen) and the expression was carried out for 48–60 h at 27 °C in glass flasks. The medium from 1 L of culture was filtered through a 0.22-μm membrane and the filtrate was adjusted to contain 30 mM imidazole before being incubated with 3.0 mL of Ni-Sepharose Excel (Cat. 17-3712-03, GE Healthcare) resin for 2 h at 4 °C with mild agitation for batch binding. The beads were poured into a Bio-Rad gravity column, washed with 10 CV of 20 mM imidazole, and eluted with 300 mM of imidazole in 150 mM NaCl, 20 mM Tris HCl pH 8.0. For enzyme-catalyzed biotinylation, the Avi-tagged RBD at 0.8 mg mL−1 was incubated with 5 mM ATP, 10 mM magnesium acetate, 43.5 μM biotin, and 22 μg mL−1 home-purified BirA in a 3.2-mL reaction mix and incubated at 4 °C. After 16 h of reaction, biotinylated RBD was concentrated with a 10-kDa cut-off membrane to 3 mg mL−1 before being further fractioned on a Superdex Increase 200 10/300 GL gel filtration column. Fractions containing the RBD were pooled, aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C until use.
For crystallization, RBD eluted from the Ni-NTA column was desalted to remove imidazole using a desalting column, and digested with home-purified 3C protease to remove the C-terminal tags. After 16 h of digestion, the tag-free RBD was mixed with DL4 at a molar ratio of 1:1.3 and the complex was loaded onto a Superdex Increase 200 10/300 GL column for gel filtration. Fractions containing both RBD and DL4 complex were pooled, and concentrated to 10 mg mL−1 for crystallization.
4.3. Protein expression and purification - monovalent DL4 in Escherichia coli
Monovalent DL4 was expressed with a C-terminal Myc tag and a hexahistidine tag in the Escherichia coli strain MC1061. Briefly, cells carrying DL4-encoding pSb-init plasmids [25] were grown in Terrific Broth (TB, 0.017 M KH2PO4 and 0.072 M K2HPO4, 1.2% (w/v) tryptone, 2.4% (w/v) yeast extract, 0.5% (v/v) glycerol) supplemented with 25 mg L−1 chloramphenicol at 37 °C with vigorous shaking at 200 rpm. When OD600 reached 0.5 (typically ~2 h), the temperature was dropped to 22 °C and the cells were incubated for another 1.5 h before being added with 0.02% (w/v) arabinose to induce DL4 expression. After 17 h of induction, cells were collected by centrifugation and lysed by osmotic shock. Briefly, cells from 1 L of culture were first resuspended in 20 mL of TES-high Buffer (0.5 mM EDTA, 0.5 M sucrose, and 0.2 M Tris-HCl pH 8.0) and incubated at 4 °C for 30 min. The dehydrated cells were then abruptly rehydrated with 40 mL of ice-cold deionized water at 4 °C for 1 h. The periplasmic extract was separated from the cells by centrifugation at 20,000g at 4 °C for 30 min. The supernatant was added with 2 mM of MgCl2, 150 mM of NaCl, and 20 mM of imidazole. To the mix was added with Ni-NTA resin that had been pre-equilibrated with buffer containing 20 mM of imidazole, 150 mM NaCl, and 20 mM Tris HCl pH 8.0. After incubation for 2 h, the beads were washed with 30 mM imidazole and eluted with 300 mM imidazole, 150 mM NaCl, and 20 mM Tris HCl pH 8.0.
4.4. Protein expression and purification - divalent nanobodies in mammalian cells
Fc-DL4 (from N- to C-terminal: signal peptide, DL4, Fc) was transiently expressed in Expi293 suspension cells. Briefly, cells at a density of 2.5 × 106 cells per milliliter were transfected with a mix of plasmid and PEI. After 65 h at 37 °C, cells were separated from the medium by centrifugation at 1000g and filtration. The filtrate was mixed with rProtein A beads (Cat. SA012005, SmartLifesciences, China) for batch binding at 4 °C. After 3 h, the beads were poured into a gravity column, washed using 20 CV of PBS buffer, and eluted with an acidic buffer containing 0.1 M glycine pH 3.0. The elution was mixed immediately with 1 M Tris HCl pH 8.0. After neutralization, the buffer was exchanged to PBS using a Bio-Rad desalt column.
Nanobody mutants in this study were all generated on the Fc-fusion constructs with a standard PCR-based site-directed mutagenesis protocol. DNA sequences were verified by Sanger sequencing and the mutants were expressed and purified the same way as DL4.
4.5. Alpaca immunization and antibody titer determination
Purified RBD (2 mg mL−1, 0.5 mL) was emulsified with an equal volume of the Gerbu adjuvant (Cat. 3111) by vortexing. The emulsion was injected by the subcutaneous route at ten sites near the bow lymph node in the neck base of a 3-year old adult female alpaca. The immunization process was repeated for 3 rounds (a total of 4 injections) with 4 days between each injection.
To monitor antibody titer, 3 mL of blood samples before and after each injection were incubated at room temperature (RT, 20–25 °C) for 2 h. The clotted sample was then centrifuged at 3000g for 5 min at RT to harvest sera in the supernatant. A 96-well plate (Maxisorp, Nunc Thermo Fisher Scientific) was coated overnight at 4 °C with 100 μL of 2 μg mL−1 biotinylated RBD in TBS (150 mM NaCl, 20 mM Tris, pH 8.0), followed by blocking with 0.5% bovine serum albumin (BSA) in TBS. The plate was washed five times with TBS and incubated with serially diluted sera for 1 h. The plate was washed and the remaining nanobodies were detected by HRP-conjugated goat anti-alpaca IgG (Cat. S001P, NBbiolab) using Tetramethylbenzidine (TMB) (Cat. T2885, Merck) as an HRP substrate.
4.6. Phage display library and panning
Eighty milliliters of blood were drawn from the immunized alpaca into EDTA-coated tubes. Peripheral blood lymphocytes were isolated with Ficoll Plus (density of 1.077 g mL−1) following the manufacturer's instructions. mRNA was isolated from lymphocytes using RNAsio Plus (Takara). Reverse transcription of the mRNA was carried out using a commercial kit (Cat. R312-01, Vazyme). Subsequent PCR was performed with 50 ng of cDNA and the primer pair 5′-GTCCTGGCTGCTCTTCTACAAGG-3′ and 5′-GGTACGTGCTGTTGAACTGTTCC-3′. The PCR product was gel purified, and used as the template for a second PCR with the prime pair 5′-ATATGC TCTTCAAGTCAGGTGCAGCTGCAGGAGTCTGGRGGAGG-3′ and 5′-TATAGCTCTTCCTGCCGAGGAGACGGTGACCTGGGT-3′ which anneals with the framework 1 and framework 4 regions of nanobodies, respectively. These primers contained a site (italic) for the type IIs restriction enzyme BspQI.
One microgram of the gel-purified PCR product and 10 μg of the pDX_init vector [25] were digested separately with BspQI (Cat. R0712L, New England Biolabs) at 50 °C. After 1.5 h, the mixture was heated at 80 °C for 10 min to inactivate BspQI. The digested DNA fragments were gel-purified. For ligation, 0.3 μg of the digested PCR products were mixed with 1.2 μg of the digested vector. The mix was added with 10 units of T4 ligase in ligation buffer (Cat. B110041, Sangon Biotech, Shanghai, China) for 1.5 h. The ligation products were transformed into E. coli SS320 cells by electroporation in a 2-mm cuvette using a Gene Pulser Xcell (Bio-Rad) with the setting of 2400 V, 25 μF, and 750 Ω.
Cells treated above were grown in 225 mL of 2-YT broth (1.0% (w/v) yeast extract, 1.6% (w/v) tryptone, 0.5% (w/v) NaCl, pH 7.0) supplemented with 200 μg mL−1 ampicillin and 2% (w/v) glucose in a 37-°C incubator shaking at 220 rpm. To 10 mL of the overnight culture, 27 μL of the M13KO7 helper phage at 1012 plaque-forming units mL−1 were added. The mixture was incubated at 37 °C for 30 min for infection. Cells were harvested by centrifugation at 3200g for 10 min, resuspended in 2-YT broth supplemented with 200 μg mL−1 ampicillin and 25 μg mL−1 kanamycin, and cultured in an incubator at 37 °C shaking at 160 rpm. After 16 h, the medium from 50 mL of culture was separated from cells by centrifugation at 3200g for 30 min at 4 °C. The supernatant (40 mL) was transferred into a fresh tube and incubated with 10 mL of 20% (w/v) PEG 6000 and 2.5 M NaCl for 30 min on ice. Phage particles were harvested by centrifugation at 3200g for 30 min at 4 °C and washed with 1 mL of PBS buffer.
The first round of panning was carried out in a Nunc Maxisorp 96-well immunoplate that had been coated with 67 nM neutravidin (Cat. 31000, Thermo Fisher Scientific) overnight at 4 °C and blocked with TBS buffer supplemented with 0.5% (w/v) BSA for 30 min. Phage particles (4.9 mL) were incubated with biotinylated RBD (50 nM), aliquoted to the 96-well plate, washed, and released from the plate by tryptic digestion using 0.25 mg mL−1 trypsin in a buffer containing 150 mM NaCl and 20 mM Tris-HCl pH 7.4. Wells were treated with AEBSF and the selected phage particles were amplified in E. coli SS320 for the second round of panning which was performed essentially as the first one except that the plate was replaced with 12 μL of MyOne Streptavidin C1 beads (Cat. 65001, Invitrogen). In addition, the bound phage particles were challenged with 5 μM of non-biotinylated RBD to compete off weak binders. The third round of panning was carried out the same as the second round except that the biotinylated RBD concentration was kept at 5 nM. Enriched phagemid was sub-cloned into pSb_init vector by fragment-exchange (FX) cloning and transformed into E. coli MC1061 cells for nanobody expression and screening.
4.7. Enzyme-linked immunosorbent assay (ELISA)
Single colonies of the abovementioned E. coli MC1061cells were grown at 37 °C for 5 h in an incubator shaking at 300 rpm before 1:20 diluted into 1 mL of fresh TB supplemented with 25 μg mL−1 chloramphenicol and 0.02% (w/v) arabinose. After culturing at 22 °C for 17 h, cells were harvested by centrifugation at 3000g for 30 min. Pellets were resuspended in TES Buffer (0.5 mM EDTA, 20% (w/v) sucrose, 0.5 μg mL−1 lysozyme, 50 mM Tris-HCl pH 8.0) and lysed for 30 min at room temperature (RT, 20–25 °C). Cell lysate was added with 0.9 mL of TBS (150 mM NaCl, 20 mM Tris-HCl pH 7.4) supplemented with 1 mM MgCl2 and clarified by centrifugation at 3000g for 30 min at 4 °C. The supernatant containing extracted nanobodies was used for ELISA.
A 96-well plate (Cat. 442404, Thermo Fisher) was coated with Protein A at 4 °C for 16 h and blocked by 0.5% (w/v) BSA in TBS buffer for 30 min at RT, followed by washing three times with TBS. The plate was incubated with anti-Myc antibodies at 1:2000 dilution in TBS-BSA-T buffer (TBS supplemented with 0.5% (w/v) BSA and 0.05% (v/v) Tween 20) for 20 min at RT. After incubation, wells were washed three times using TBST (TBS supplemented with 0.05% Tween 20) to remove excess anti-Myc antibodies. The nanobody extracts prepared above were added to the wells and the plate was incubated for 20 min at RT. The plate was then washed trice with TBST followed by incubation with 50 nM of biotinylated RBD or the control protein MBP (the maltose-binding protein) for 20 min at RT. Wells were again washed trice followed by incubation with streptavidin-conjugated with horseradish peroxidase (HRP) (1:5000, Cat S2438, Sigma). After 30 min, the plate was washed trice and the ELISA signal was developed by incubating the wells with 100 μL of a mix containing 51 mM Na2HPO4, 24 mM citric acid, 0.006% (v/v) H2O2, and 0.1 mg mL−1 3,3′,5,5′-tetramethylbenzidine at RT. Absorbance at 650 nm was measured in a plate reader.
4.8. Fluorescence-detection size exclusion chromatography (FSEC)
The ability of nanobodies to shift the elution volume of a fluorescently labeled RBD was carried out as previously described [8]. Briefly, biotinylated RBD was mixed with streptavidin (Cat. 16955, AAT Bioquest) that was chemically labeled with fluorescein. The stable RBD-streptavidin complex (500 nM) was incubated with cell lysate containing nanobodies (a control nanobody against MBP was used as a control) and the mixture was applied onto an analytic gel filtration column (Cat. 9F16206, Sepax) on an HPLC system equipped with a fluorescence detector (RF-20A, Shimadzu) for FSEC. The elution profile was monitored by fluorescence (excitation/emission wavelength of 482/508 nm).
4.9. Biolayer interferometry
The binding kinetics were obtained using a bio-layer interferometry (BLI) assay on an Octet RED96 system (ForteBio). For DL4-RBD binding, biotinylated RBD (2 μg mL−1) was immobilized onto an SA sensor (streptavidin). The sensor was incubated with the monovalent DL4 at various concentrations (see figure legends) and the BLI signal was monitored for 480 s for association. The sensor was then transferred into DL4-free buffer (0.005% (v/v) Tween 20, 1 × phosphate-buffered saline) for dissociation.
For DL4 binding with S, an SA sensor was coated with 5 μg mL−1 biotinylated nanobodies for approximately 1 min. The sensor was equilibrated in a nanobody-free buffer for ~30 s, before bathing in various concentrations of S for 120 s. The dissociation was monitored for 300 s after transferring the sensor to the S-free buffer.
For competition between ACE2 and DL4, an SA sensor coated with biotinylated RBD was incubated with 100 nM of DL4 for 6 min. The sensor was then transferred into nanobody solutions with or without 100 nM of ACE2 (Cat. 10108-H08B, SinoBiological). The association of ACE2 was recorded for 360 s. As a control, the ACE2-RBD binding profile was monitored in the absence of nanobodies.
The data for DL4-RBD binding was fitted for a 1:1 stoichiometry for K D, k on, and k off calculations using the built-in software Data Analysis 10.0. The apparent binding kinetics for DL4-S interaction were not fitted owing to possible formation of bridged complexes.
4.10. Crystallization
Crystals were grown at 16 °C in a sitting drop plate which contained 1 μL of protein solution (10 mg mL−1) and 1 μL of precipitant solution (25% (w/v) polyethylene glycol 4000, 0.2 M ammonium sulfate, and 0.1 M sodium acetate pH 4.6) in a drop and 70 μL of reservoir solution in the well. Cryo protection was achieved by adding 20% (v/v) glycerol in the precipitant solution. Crystals were harvested using a MiTeGen loop, and flash-cooled in liquid nitrogen before X-ray diffraction data collection.
4.11. X-ray diffraction data collection and structure determination
Diffraction data were collected at beamline BL18U1 at Shanghai Synchrotron Radiation Facility with a 50 × 50 μm beam on a Pilatus detector with oscillation of 0.5° and a wavelength of 0.97915 Å. Data were processed using HKL2000 [71]. The structure was solved by molecular replacement using Phaser [72] with the RBD structure (PDB 6M0J) [40] and a nanobody structure (PDB 5M13) [25] as the search model. The model was manually adjusted according to 2F o-F c maps in Coot [73], and refined using Phenix [74]. Structures were visualized using PyMol.
4.12. Neutralization assay using SARS-CoV-2 pseudoviruses
Retroviral pseudotyped particles were generated by co-transfection of HEK293T cells using polyethylenimine with the expression vectors carrying the murine leukemia virus core/packaging components (MLV Gag-Pol), the various viral envelope glycoproteins, and a retroviral transfer vector containing the gene encoding the green fluorescent protein (GFP). The 19 amino-acids at the C-terminus of S were truncated. Supernatant containing pseudotyped particles were harvested 48 h post-transfection and filtered through a 0.45-μm membrane before neutralizing assays.
VeroE6-hACE2 cells were infected with 100 μL of virus supernatant in a final volume of 150 μL in a 48-well plate. Nanobodies were pre-incubated with the pseudotyped particles for 1 h at 37 °C prior to cell/virus co-incubation. After 6 h, the supernatant was removed and the cells were incubated with medium at 37 °C for 72 h. GFP expression was determined by fluorescence-activated cell sorting (FACS). The infectivity of pseudotyped particles incubated with nanobodies was normalized with the control (100%).
4.13. Animal experiment and ethics
The alpaca immunization procedures were conducted in conformity with the institutional guidelines for the care and use of laboratory animals, and the protocols were approved by the Institutional Committee of Ethics and Research of the Central Laboratory at Xinyang Agricultural and Forestry University.
CRediT authorship contribution statement
Tingting Li: Methodology, Validation, Formal analysis, Investigation, Writing-editing, Visualization.
Bingjie Zhou: Methodology, Validation, Formal analysis, Investigation, Writing-editing, Visualization.
Yaning Li: Methodology, Formal analysis.
Suqiong Huang: Methodology.
Zhipu Luo: Methodology, Visualization.
Yuanze Zhou: Methodology.
Yanling Lai: Methodology, Formal analysis, Investigation.
Anupriya Gautam: Methodology.
Salome Bourgeau: Methodology.
Shurui Wang: Methodology.
Juan Bao: Methodology.
Jingquan Tan: Methodology, Resources, Supervision.
Dimitri Lavillette: Conceptualization, Methodology, Resources, Formal analysis, Writing-editing, Supervision.
Dianfan Li: Conceptualization, Methodology, Resources, Formal analysis, Writing-editing, Visualization, Supervision.
Declaration of competing interest
A patent application for potential nanobody therapy for the treatment of COVID-19 has been submitted for DL4. Authors Y.Z., S.W., and J.T. are employed by the company Nanjing Crycision Biotech Co., Ltd., Nanjing, China.
Acknowledgments
Acknowledgement
We thank the staff scientists at the SSRF-BL18U1 beamline at National Facility for Protein Science (Shanghai) for technical support, and Dr. Peiling Li from Xinyang Agricultural and Forestry University for assistance in the alpaca immunization. This work has been supported by the National Natural Science Foundation of China (82151215, 31870726, D. Li; 31870153, D. La), the Strategic Priority Research Program of CAS (XDB37020204, D. Li), Key Program of CAS Frontier Science (QYZDB-SSW-SMC037, D. Li), CAS Facility-based Open Research Program, National Key Research and Development Program of China (2020YFC0845900, D. La), CAS president's international fellowship initiative (2020VBA0023, D. La), Science and Technology Commission of Shanghai Municipality (20ZR1466700, D. Li), Shanghai Municipal Science and Technology Major Project (20431900402, D. La).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2022.04.096.
Appendix A. Supplementary data
Supplementary Table 1 and Supplementary Figures 1 & 2.
PDB validation report for the DL4-RBD structure.
Data availability
The structure factors and coordinates are available through the protein data bank (PDB) under accession code 7F5G (DL4-RBD).
References
- 1.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang J., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 6.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 7.Kim C., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T., et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021;11:5588. doi: 10.1038/s41467-021-24905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T., et al. Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody. EMBO Mol Med. 2021;13 doi: 10.15252/emmm.202114544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes C.O., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min L., Sun Q. Antibodies and vaccines target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.671633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target Ther. 2020;5:282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. Journal of Thoracic Disease. 2013:S142–S148. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song S., et al. Sequential immunization with SARS-CoV-2 RBD vaccine induces potent and broad neutralization against variants in mice. Virol. J. 2022;19:2. doi: 10.1186/s12985-021-01737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng Q., et al. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdes-Balbin Y., et al. SARS-CoV-2 RBD-tetanus toxoid conjugate vaccine induces a strong neutralizing immunity in preclinical studies. ACS Chem. Biol. 2021;16:1223–1233. doi: 10.1021/acschembio.1c00272. [DOI] [PubMed] [Google Scholar]
- 19.Pino M., et al. A yeast-expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Science Immunology. 2021;6:eabh3634. doi: 10.1126/sciimmunol.abh3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalvie N.C., et al. Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2106845118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulton S., et al. Single-dose replicating poxvirus vector-based RBD vaccine drives robust humoral and T cell immune response against SARS-CoV-2 infection. Mol. Ther. 2021;30 doi: 10.1016/j.ymthe.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., et al. Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections. Nat. Commun. 2021;12:264. doi: 10.1038/s41467-020-20465-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu M., et al. Real-time conformational dynamics of SARS-CoV-2 spikes on virus particles. Cell Host Microbe. 2020;28:880–891 e8. doi: 10.1016/j.chom.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann I., et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife. 2018;7 doi: 10.7554/eLife.34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon C., et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018;25:289–296. doi: 10.1038/s41594-018-0028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pymm P., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X., et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021;37:109869. doi: 10.1016/j.celrep.2021.109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bracken C.J., et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat. Chem. Biol. 2021;17:113–121. doi: 10.1038/s41589-020-00679-1. 2020.04.16.045419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Custódio T.F., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mast F.D., et al. Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. eLife. 2021;10:e73027. doi: 10.7554/eLife.73027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huo J., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 33.Gai J., et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm (2020) 2021;2:101–113. doi: 10.1002/mco2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esparza T.J., Martin N.P., Anderson G.P., Goldman E.R., Brody D.L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020;10:22370. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nambulli S., et al. Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Gentili M., Hacohen N., Regev A., et al. A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing nanobodies. Nat. Commun. 2021;12:5506. doi: 10.1038/s41467-021-25777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Y., et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao H., et al. A high-affinity RBD-targeting nanobody improves fusion partner’s potency against SARS-CoV-2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 41.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma H., et al. Potent neutralization of SARS-CoV-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. J. Virol. 2021;95:e02438–20. doi: 10.1128/JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huo J., et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021;12:5469. doi: 10.1038/s41467-021-25480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanke L., et al. A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo. Nat. Commun. 2022;13:155. doi: 10.1038/s41467-021-27610-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guttler T., et al. Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies. EMBO J. 2021;40 doi: 10.15252/embj.2021107985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z., et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal Transduct Target Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bräuer P., et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science. 2019;363:1103–1107. doi: 10.1126/science.aaw2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Che T., et al. Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell. 2018;172:55–67.e15. doi: 10.1016/j.cell.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker J.L., et al. Structural basis of antifolate recognition and transport by PCFT. Nature. 2021;595:130–134. doi: 10.1038/s41586-021-03579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moonens K., et al. Structural insights into polymorphic ABO glycan binding by Helicobacter pylori. Cell Host Microbe. 2016;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caohuy H., et al. Common cardiac medications potently inhibit ACE2 binding to the SARS-CoV-2 Spike, and block virus penetration and infectivity in human lung cells. Sci. Rep. 2021;11:22195. doi: 10.1038/s41598-021-01690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand S.P., et al. Interaction of human ACE2 to membrane-bound SARS-CoV-1 and SARS-CoV-2 S glycoproteins. Viruses. 2020;12 doi: 10.3390/v12101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pak A.J., Yu A., Ke Z., Briggs J.A.G., Voth G.A. Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core. Nat. Commun. 2022;13:1002. doi: 10.1038/s41467-022-28654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter J.D., et al. Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance. EMBO Rep. 2022;e54199 doi: 10.15252/embr.202154199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Custodio T.F., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanke L., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoof M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koenig P.A., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371 doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrapp D., et al. Structural basis for potent neutralization of Betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1436–1441. doi: 10.1016/j.cell.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henderson R., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saphire E.O., et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 63.Dejnirattisai W., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Starr T.N., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piccoli L., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 Spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042 e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urdaniz I.F., et al. One-shot identification of SARS-CoV-2 S RBD escape mutants using yeast screening. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greaney A.J., et al. Complete mapping of mutations to the SARS-CoV-2 Spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57 e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hastie K.M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nabel K.G., et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2022;375:eabl6251. doi: 10.1126/science.abl6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen R.E., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Otwinowski Z., Minor W. vol. 276. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 72.McCoy A.J., Storoni L.C., Read R.J. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr. Sect. D. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 73.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 74.Adams P.D., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 and Supplementary Figures 1 & 2.
PDB validation report for the DL4-RBD structure.
Data Availability Statement
The structure factors and coordinates are available through the protein data bank (PDB) under accession code 7F5G (DL4-RBD).