Abstract
We designed a unique nanocapsule for efficient single CRISPR-Cas9 capsuling, noninvasive brain delivery and tumor cell targeting, demonstrating an effective and safe strategy for glioblastoma gene therapy. Our CRISPR-Cas9 nanocapsules can be simply fabricated by encapsulating the single Cas9/sgRNA complex within a glutathione-sensitive polymer shell incorporating a dual-action ligand that facilitates BBB penetration, tumor cell targeting, and Cas9/sgRNA selective release. Our encapsulating nanocapsules evidenced promising glioblastoma tissue targeting that led to high PLK1 gene editing efficiency in a brain tumor (up to 38.1%) with negligible (less than 0.5%) off-target gene editing in high-risk tissues. Treatment with nanocapsules extended median survival time (68 days versus 24 days in nonfunctional sgRNA-treated mice). Our new CRISPR-Cas9 delivery system thus addresses various delivery challenges to demonstrate safe and tumor-specific delivery of gene editing Cas9 ribonucleoprotein for improved glioblastoma treatment that may potentially be therapeutically useful in other brain diseases.
A unique brain delivery system transports CRISPR/Cas9 into the brain and inhibits orthotopic glioblastoma noninvasively.
INTRODUCTION
While CRISPR-Cas9 represents a revolutionary gene editing technology, its delivery to brain is fraught with challenges that current delivery systems do not completely address. Awarded the 2020 Nobel Prize in Chemistry, CRISPR-Cas9 combines Cas9 nuclease with single-guide RNA (sgRNA) to bind and cut target DNA for gene editing (1, 2) and applications in the treatment of genetic disorders (3, 4). However, most current CRISPR-Cas9 brain delivery systems use viral vectors, which have low packing capacity and pose safety concerns due to inducing undesirable genetic mutations and immunogenicity (5). Viral vectors are also limited to large-scale fabrication required for clinical translation (6), whereas nonviral nanoparticle-based vectors are often nonimmunogenic and permitted facile bulk production (7). Considering that the brain is a primary and sensitive site of human disease, much research has focused on applying CRISPR-Cas9–based gene therapy to brain-related diseases (8, 9). Preliminary work, while providing therapeutic proof of principle (10–12), involved undesirable invasive intrabrain injection of CRISPR-Cas9 complexes contained in viral vectors or nanoparticles, which often leads to serious side effects, including infection, inflammation swelling, and tissue injury (13, 14). Accordingly, noninvasive delivery of nanoparticle-encapsulated CRISPR-Cas9 complexes is urgently required to promote gene therapy of brain disorders (15). The key bottleneck challenge is bypassing the blood-brain barrier (BBB) whose phenotype and physical structure are variable according to the vascular environment and brain physiological conditions (16, 17). Therefore, the heterogeneity of BBB could be an advantage to take in designing a BBB-penetrating drug delivery system. Excessive nanoparticle surface positive charge that induces tissue toxicity and off-target effects are additional problems that have hampered further clinical translational (18, 19).
We addressed these challenges by developing a new CRISPR-Cas9 brain delivery platform that satisfies the following design criteria: ease of formulation, high loading content, small and uniform size, stability with a long plasma lifetime, BBB permeability, active targeting of the brain and brain tumor cells, rapid intracellular release, efficient gene editing, and negligible off-target effects. Our delivery platform comprises a thin, disulfide–cross-linked polymeric shell decorated with angiopep-2 peptide [a ligand that binds low-density lipoprotein receptor–related protein-1 (LRP-1) that is more highly expressed on BBB endothelial cells and glioblastoma (GBM) cells] (20, 21). This polymeric shell can encapsulate the single Cas9 ribonucleoprotein/sgRNA complexes into a small nanocapsule (~30 nm) with nearly neutral surface charge to protect the cargo from degradation by ribonuclease (RNase) and promote its blood stability and plasma lifetime, which is essential for noninvasive BBB penetration. The small size and angiopep-2 peptide functionalization further benefits BBB penetration and intracellular delivery in brain toward GBM-specific targeting. The single Cas9 ribonucleoprotein/sgRNA complex capsulation enables high (almost 100%) drug loading while disulfide–cross-linking exploits higher intracellular glutathione (GSH) conditions present in tumor cells to release cargo on-site by disulfide cleavage, leading to nanocapsule degradation for high-performance gene editing (22). We show that anti–Polo-like kinase 1 (PLK1) Cas9/sgRNA encapsulated within our newly developed nanocapsules significantly decreases expression of the cellular mitosis protein PLK1 in GBM with negligible off-target editing using two typical orthotopic GBM mouse models. Nanocapsules lacking disulfide–cross-linking or angiopep-2 functionalization are notably less efficient in reaching the brain and elicit less gene editing. To the best of our knowledge, our nanocapsules represent a foremost noninvasive and nonviral strategy for effective and safe brain delivery and GBM gene therapy.
RESULTS
Nanocapsule formulation
The angiopep-2–functionalized, disulfide–cross-linked nanocapsules containing Cas9 and sgRNA [“ANCSS(Cas9/sgRNA)”] were fabricated using a robust free-radical in situ polymerization, as we used previously to construct small interfering RNA (siRNA) single nanocapsules (23). Cas9/sgRNA complexes were coated with positively charged acrylate guanidine via electrostatic interactions and were cross-linked with N,N′-bis(acryloyl)cystamine and angiopep-2–decorated polyethylene glycol (PEG) with acrylate end groups (Fig. 1A). A nontargeting control lacking angiopep-2 [“NCSS(Cas9/sgRNA)”] was fabricated with acrylate-functionalized methoxy-PEG instead of angiopep-2–functionalized PEG. A nondegradable control lacking disulfide bonds [“ANC(Cas9/sgRNA)”] was prepared using the nondegradable cross-linker N,N′-methylene bisacrylamide.
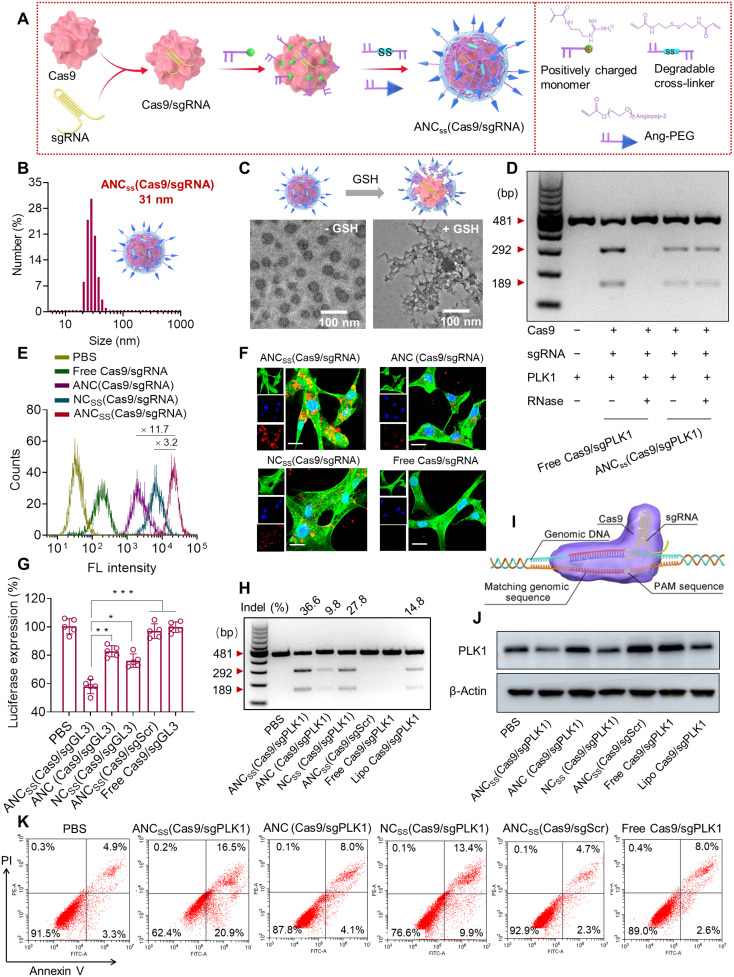
Fig. 1. Fabrication, physical properties, and cellular function of Cas9/sgRNA nanocapsules.
(A) In situ free-radical polymerization was used to synthesize disulfide–cross-linked nanocapsules containing Cas9/sgRNA and functionalized with angiopep-2 targeting ligand. (B) Size distribution of ANCSS(Cas9/sgRNA) nanocapsules determined by dynamic light scattering. (C) TEM images of ANCSS(Cas9/sgRNA) with or without GSH treatment. (D) Gel electrophoresis analysis of the ANCSS(Cas9/sgPLK1) or free Cas9/sgPLK1 with or without RNase treatment (1 mg/ml, 30 min). (E) Flow cytometry of U87MG cells following 4-hour incubation with ANCSS(Cas9/sgRNA) or controls. (F) Confocal laser scanning microscopy (CLSM) images of U87MG cells following 4-hour incubation with ANCSS(Cas9/sgRNA) or controls. Cas9 was labeled with Alexa Fluor 647 (AF647; red); the cytoskeleton was stained with Alexa Fluor 488 (green), and the nuclei was stained with Hoechst 33342 (blue). For (E) and (F), the AF647-Cas9 concentration was 20 nM. Scale bars, 20 μm. (G) Luciferase gene editing efficiency in U87MG-Luc cells incubated with ANCSS(Cas9/sgRNA) or controls for 72 hours. Data are presented as means ± SD (n = 5; *P < 0.05, **P < 0.01, and ***P < 0.001). (H) Indels of the PLK1 gene in U87MG cells transfected with ANCSS(Cas9/sgPLK1) or controls for 48 hours. (I) Schematic of gene editing in the nucleus. (J) Expression levels of PLK1 in U87MG cells after 72-hour incubation with ANCSS(Cas9/sgPLK1) or controls. (K) Apoptosis assay of U87MG cells after 72-hour incubation with ANCSS(Cas9/sgRNA) and other controls. For (G) to (K), the Cas9 concentration was 20 nM. bp, base pairs; PBS, phosphate-buffered saline.
The average hydrodynamic diameter of the ANCSS(Cas9/sgRNA) nanocapsules was 31 nm, while the size of naked Cas9/sgRNA was 17 nm (table S1), consistent with the previously reported thickness of the polymerization layer (24, 25), indicating successful surface coating and fabrication of single CRISPR-Cas9 nanocapsules (Fig. 1B and fig. S1A), where each of the nanocapsules likely only contains one Cas9/sgRNA molecule, consistent with reported results (24). Nontargeting NCSS(Cas9/sgRNA) and nonreducible ANC(Cas9/sgRNA) controls did not significantly vary in size, size distribution, or surface charge compared to ANCSS(Cas9/sgRNA (fig. S2). Hence, all nanocapsules exhibited physical parameters within ranges shown previously to be associated with excellent performance in vivo (26, 27). Transmission electron microscopy (TEM) images and dynamic light scattering revealed the spherical morphology (Fig. 1C) and nearly neutral surface charge (table S1) of ANCSS(Cas9/sgRNA), respectively, which confirmed particle size. However, free Cas9/sgRNA displayed obvious aggregation and poor dispersibility (fig. S1B). Notably, all nanocapsules quickly degraded and released Cas9/sgRNA in high GSH-mimicking intracellular reductive environment with the exception of the nonreducible ANC(Cas9/sgRNA) control (Fig. 1C).
To investigate whether the polymerization process causes protein denaturation, we performed circular dichroism spectroscopy analysis with bovine serum albumin as a model protein. Following polymerization, the protein structure remained intact after nanocapsule fabrication (fig. S3). We further evaluated the capability of our CRISPR-Cas9 nanocapsules to protect sgRNA by artificially introducing RNase into the in vitro gene editing evaluation system. In the presence of RNase, it was reassuring that ANCSS(Cas9/sgPLK1) gene-edited as efficiently as free Cas9/sgRNA in an RNase-free environment. However, the addition of RNase caused free Cas9/sgRNA to lose its ability to cleave target DNA (Fig. 1D). This result indicated that ANCSS(Cas9/sgPLK1) could shield Cas9/sgRNA complex from enzymatic hydrolysis, suggesting that ANCSS(Cas9/sgPLK1) would be a suitably protective delivery system for achieving in vivo gene editing.
Cell internalization of nanoparticles and intracellular release of Cas9/sgRNA
To examine cellular uptake of the CRISPR-Cas9 nanocapsules, we exposed GBM U87MG cells to Alexa Fluor 647 (AF647)–labeled Cas9 nanocapsules and analyzed the cell uptake efficiency using flow cytometry and confocal laser scanning microscopy (CLSM). Flow cytometry revealed 3.2- and 11.7-fold greater uptake of ANCSS(Cas9/sgRNA) than that achieved by NCSS(Cas9/sgRNA) or ANC(Cas9/sgRNA), respectively (Fig. 1E). Moreover, enhanced cellular uptake was observed for ANCSS(Cas9/sgRNA) as well when incubated with U251 human and CT2A mouse GBM cells (figs. S4 and fig. S5), supporting good targeting ability of these nanocapsules. CLSM demonstrated that U87MG cells treated with ANCSS(Cas9/sgRNA) showed a much stronger cellular fluorescence than cells receiving NCSS(Cas9/sgRNA) or ANC(Cas9/sgRNA) after 4 hours of incubation, while cells incubated with free Cas9/sgRNA exhibited negligible fluorescence (Fig. 1F). The significant improvement in cellular uptake of ANCSS(Cas9/sgRNA) compared to NCSS(Cas9/sgRNA) indicated that functionalization with angiopep-2 leads to the specific targeting on LRP-1–overexpressing GBM cells as confirmed by Western blotting (fig. S6). On the other hand, the much higher fluorescence intensity seen after treatment with ANCSS(Cas9/sgRNA) compared to ANC(Cas9/sgRNA) reflected GSH-responsive Cas9/sgRNA release from ANCSS(Cas9/sgRNA), thereby preventing the fluorescence quenching mediated by ANC(Cas9/sgRNA) (28, 29). Collectively, these results demonstrated the GBM-targeting capability of angiopep-2–functionalized nanocapsules and the fast intracellular release of Cas9/sgRNA triggered by the reducing intracellular environment.
Targeted delivery of Cas9/sgRNA-loaded nanocapsules into GBM cells for gene editing
To evaluate whether the nanocapsules can achieve specific gene disruption, we used luciferase as a model gene and encapsulated firefly luciferase guide RNA (sgLuc) as the targeting sequence in ANCSS(Cas9/sgLuc) nanocapsules. The Cell Counting Kit-8 (CCK-8) cell proliferation assay and a luciferase knockdown assay were used to assess the cell conditions after treating with the nanocapsules in luciferase-expressing U87MG cells constitutively (U87MG-Luc). The CCK-8 assay showed that the nanocapsules were nontoxic (fig. S7), which may be ascribed to their small size and nearly neutral surface charge (table S1). ANCSS(Cas9/sgLuc) treatment produced a 42.0% reduction in luciferase protein expression, a significantly larger reduction than that achieved by nonreducible nanocapsule ANC(Cas9/sgLuc) or by nontargeting nanocapsule NCSS(Cas9/sgLuc) at 17.2 and 25.1%, respectively (Fig. 1G). Nanocapsules containing Cas9 with scrambled guide RNA (sgScr) did not reduce luciferase expression, indicating that gene silencing by ANCSS(Cas9/sgLuc) was sgRNA sequence specific.
PLK1 plays a key role in cell mitosis and is overexpressed in a range of tumors (30, 31). PLK1 has emerged as an important GBM target, as PLK1 expression is associated with higher GBM tumor grade and a mesenchymal/proliferative GBM subtype. Inhibition of PLK1 suppresses GBM cell proliferation and results in apoptosis (32). PLK1 is highly overexpressed in GBM cells and has a much lower expression in normal brain cells including glial and astrocyte cells (fig. S6), suggesting that editing PLK1 gene in GBM is feasible. Therefore, we chose PLK1 as a model gene and incorporated sgPLK1 into nanocapsule as a model Cas9 capsule. To assess PLK1 gene editing efficiency of the nanocapsules, we performed T7 endonuclease I (T7E1) cleavage assays to measure the endogenous targeted disruption efficiency in U87MG cells treated with nanocapsules. Gene mutation occurred in the target PLK1 gene with mutation frequencies of 36.6, 9.8, and 27.8% for ANCSS(Cas9/sgPLK1), ANC(Cas9/sgPLK1), and NCSS(Cas9/sgPLK1), respectively (Fig. 1, H and I). The degree of gene editing was higher when using ANCSS(Cas9/sgPLK1) compared to Lipofectamine treatment (14.8% mutation frequency). Free Cas9/sgPLK1 and nanocapsules with scrambled sgRNA [ANCSS(Cas9/sgScr)] showed no PLK1 gene editing.
Indel frequencies at PLK1 target sites in cells treated with ANCSS(Cas9/sgPLK1) were evaluated by DNA sequencing. In 21 clones sequenced, 9 clones (42.9%) showed mutations near the target site, indicating efficient and specific gene disruption (fig. S8). Western blots indicated that PLK1 protein expression was reduced as much as 53% by ANCSS(Cas9/sgPLK1) nanocapsules (Fig. 1J and fig. S9), confirming PLK1 gene disruption. Together, these in vitro results indicate that the ANCSS(Cas9/sgRNA) nanocapsules achieve GBM tumor cell targeting with subsequent release of Cas9/sgRNA triggered by cytoplasmic GSH, resulting in specific gene editing in U87MG glioblastoma cells.
We assessed apoptosis in U87MG cells after incubation with nanocapsules for 72 hours. Treatment with ANCSS(Cas9/sgPLK1) resulted in 37.4% of U87MG cells entering late or early apoptosis, which was significantly higher than that of ANC(Cas9/sgPLK1) or NCSS(Cas9/sgPLK1) that induced 12.1 and 23.3% apoptosis, respectively (Fig. 1K and fig. S10). U87MG cells treated with nanocapsules containing Cas9 and scrambled RNA, ANCSS(Cas9/sgScr), showed a low level of apoptosis, similar to that in cells treated with free Cas9/sgPLK1 or phosphate-buffered saline (PBS). To evaluate whether the expression level of PLK1 has a potential influence on the efficacy of ANCSS(Cas9/sgPLK1), we knocked down the expression level of PLK1 gene in U87MG cells using siRNA followed by treatment with nanocapsules. The results showed that low PLK1 expression had little influence on ANCSS(Cas9/sgPLK1)-induced cell apoptosis (fig. S11), indicating that the level of PLK1 expression may have a negligible effect on the ANCSS(Cas9/sgPLK1)-induced antitumor efficacy. Notably, ANCSS(Cas9/sgRNA) nanocapsules demonstrated the similar apoptosis in U251 GBM cells as U87MG cells (fig. S12). Together, these results indicate that the increase in GBM cell apoptosis was the direct result of PLK1 gene editing.
BBB penetration, tumor permeability, and pharmacokinetics
We then investigated the BBB penetration of ANCSS(Cas9/sgRNA) nanocapsules in an in vitro BBB transwell model by seeding endothelial cells overexpressing LRP-1 (fig. S6). These results showed that ANCSS(Cas9/sgRNA) had significantly enhanced BBB penetration compared to nontargeting NCSS(Cas9/sgRNA) controls (Fig. 2A). We also evaluated whether ANCSS(Cas9/sgRNA) nanocapsules can penetrate into GBM tumor core using a well-established three-dimensional (3D) U87MG tumor spheroid model. These results showed that treatment with ANCSS(Cas9/sgRNA) produced a strong fluorescence in the tumor spheroids up to 100 μm deep (Fig. 2B and fig. S13). In pharmacokinetic studies, we quantified AF647-labeled Cas9 levels in circulating blood in tumor-free BALB/c mice at different intervals after intravenous administration of nanocapsules. ANCSS(Cas9/sgRNA) nanocapsules exhibited prolonged blood circulation with an elimination half-life (t1/2) of 57 min, comparable to that of the nontargeting control (53 min) and the nondegradable control (56 min). In contrast, free Cas9/sgRNA was eliminated quickly in the circulating blood (t1/2 = 5 min) (Fig. 2C), indicating that the polymeric shell with nearly neutral surface charge plays a vital role in protecting Cas9/sgRNA from enzymatic degradation.
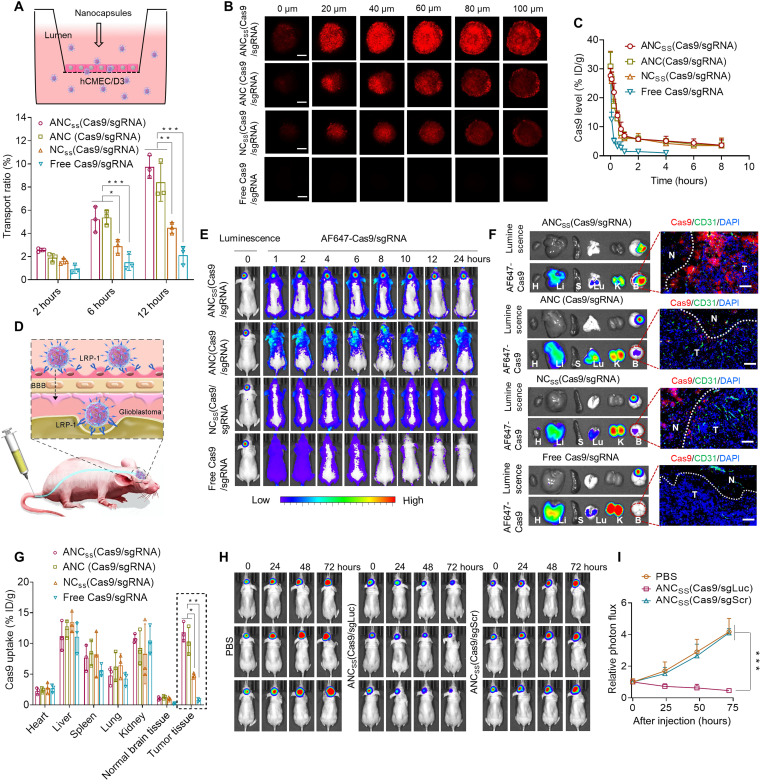
Fig. 2. BBB permeability, pharmacokinetics, deep tumor penetration, and biodistribution of Cas9/sgRNA nanocapsules.
(A) Cumulative transport ratio of ANCSS(Cas9/sgRNA) nanocapsules across the in vitro BBB barrier at 2, 6, and 12 hours (n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001). (B) Penetration of ANCSS(Cas9/sgRNA) nanocapsules into U87MG multicellular spheroids after 4 hours of incubation (AF647-Cas9 concentration was 20 nM). Scale bars, 200 μm. (C) Pharmacokinetics of ANCSS(Cas9/sgRNA) and controls in tumor-free mice (1.5 mg of Cas9 equiv./kg; n = 3). (D) Illustration of the nanocapsules follow intravenous injection specifically binding to LRP-1 that is overexpressed both on BBB endothelial cells and brain tumor cells. (E) Fluorescence images of orthotopic U87MG-Luc tumor-bearing nude mice following injection of ANCSS(Cas9/sgRNA) nanocapsules or controls (1.5 mg of Cas9 equiv./kg). (F) Luciferase luminescence and AF647-Cas9 fluorescence from major organs in nude mice bearing orthotopic U87MG-Luc 4 hours after intravenous injection of ANCSS(Cas9/sgRNA) nanocapsules or controls (1.5 mg of Cas9 equiv./kg). H, heart; Li, liver; S, spleen; Lu, lung; K, kidney; B, brain. Enlarged image: tumor penetration of ANCSS(Cas9/sgRNA) and controls observed by CLSM. Nuclei were stained with Nuclei were stained with DAPI (blue) and blood vessels with CD31 (green); AF647-Cas9 is red. Dotted lines indicate tumor boundary. N, normal brain tissue; T, tumor. Scale bars, 50 μm. (G) Quantitation of AF647-Cas9 accumulation in different organs (n = 3; *P < 0.05 and **P < 0.01). (H) Luciferase expression in glioblastoma in mice at 0, 24, 48, and 72 hours after injection of ANCSS(Cas9/sgLuc) or ANCSS(Cas9/sgScr) (1.5 mg of Cas9 equiv./kg). (I) Quantitation of luminescence intensity from U87MG-Luc tumor-bearing mice (n = 3; ***P < 0.001).
We then evaluated the ability of the nanocapsules to traverse the BBB in vivo following a single tail vein injection in the orthotopic U87MG-Luc tumor-bearing mouse model. Given that the LRP-1 is overexpressed by both endothelial cells of the BBB and U87MG brain cancer cells, LRP-1–targeting angiopep-2–functionalized ANCSS(Cas9/sgRNA) nanocapsules are expected to show enhanced BBB permeability via receptor-mediated transcytosis (Fig. 2D) and tumor accumulation. Orthotopic U87MG-Luc tumor-bearing immunocompromised mice injected with ANCSS(Cas9/sgRNA) exhibited strong AF647-Cas9 fluorescence in the tumor within 1 hour that steadily increased to a maximum at 4 hours with fluorescence maintained up to 24 hours, indicating efficient BBB penetration and tumor accumulation and retention (Fig. 2E and fig. S14A). Intravenous injection of NCSS(Cas9/sgRNA) resulted in weaker AF647 brain fluorescence, indicative of BBB disruption induced by GBM tumor (33–35). In contrast, almost no fluorescence was observed for the free Cas9/sgRNA treatment reflecting their short elimination half-life and poor BBB penetration. We further evaluated whether the developed nanocapsules work in infiltrative GBM stem cells (GSCs) bearing mice or not. We first established the infiltrative GSC-bearing mice model with LRP-1 receptor–expressed 83NS GSCs (fig. S15). Next, we tested the BBB penetration capability of our ANCSS(Cas9/sgRNA) with an in vivo imaging system. The results showed that ANCSS(Cas9/sgRNA) had excellent BBB penetration (fig. S16), implying that the developed nanocapsules have efficient BBB permeability in diffusely GSC mouse models as well.
These results highlight the role of both functionalization with the BBB-targeting peptide angiopep-2 and BBB disruption induced by GBM in facilitating BBB penetration and brain tumor accumulation of ANCSS(Cas9/sgRNA). Although the nonreducible ANC(Cas9/sgRNA) also has angiopep-2, treatment with ANC(Cas9/sgRNA) produced distinctly weaker tumor cell fluorescence compared to ANCSS(Cas9/sgRNA). Since ANC(Cas9/sgRNA) is nonreducible and thus cannot efficiently release its cargo, its lower fluorescence probably reflects self-quenching of encapsulated AF647-Cas9 by fluorescence resonance energy transfer (36, 37).
Nanocapsule distribution and organ accumulation were assessed by ex vivo organ luminescence in orthotopic U87MG–bearing mice followed by intravenously injecting nanocapsules via tail vein. Stronger fluorescence was observed in the brains of orthotopic U87MG-Luc tumor-bearing immunocompromised mice treated with ANCSS(Cas9/sgRNA) compared to mice treated with the NCSS(Cas9/sgRNA), ANC(Cas9/sgRNA), and free Cas9/sgRNA controls. The AF647-Cas9 signal in the brain colocalized with the luminescence of U87MG-Luc glioblastoma cells (Fig. 2F, left). The cellular distribution of nanocapsules in brain tissue sections was further assessed by CLSM (Fig. 2F, right, and fig. S14B), which confirmed that ANCSS(Cas9/sgRNA) nanocapsules showed better BBB penetration and brain tumor tissue accumulation compared to the three control treatments. Quantitation of fluorescence in the tumor and other organs showed that tumor tissue accumulation was 11.8% of the injected dose per gram of tissue (% ID/g) for ANCSS(Cas9/sgRNA), comparable to that achieved by ANC(Cas9/sgRNA) that was 2.5-fold more than that resulting from NCSS(Cas9/sgRNA) and 15.3-fold higher than the treatment with free Cas9/sgRNA (Fig. 2G).
To assess efficacy of gene disruption mediated by nanocapsules in vivo, transfection studies were performed with sgLuc in U87MG-Luc GBM–bearing nude mice. ANCSS(Cas9/sgLuc) treatment induced a large reduction of tumor bioluminescence, 37% at 48 hours after injection, which increased to 54% at 72 hours (Fig. 2, H and I), demonstrating efficient luciferase gene knockout in GBM in vivo.
To evaluate whether nanocapsules could deliver Cas9/sgRNA in immunocompetent mice, we first verified LRP-1 receptor expression in murine GL261 GBM cells (fig. S17) and then established an orthotopic GL261-bearing C57BL/6 mouse GBM model. The in vivo imaging results showed that treatment with AF647-labeled ANCSS(Cas9/sgRNA) exhibited strong fluorescence in the tumor by 1 hour that was maximal at 12 hours and observable up to 48 hours (fig. S18). Furthermore, the penetration results showed that ANCSS(Cas9/sgRNA) accumulated in the deep tumor sites but little in normal brain tissues (fig. S19A). Quantification of fluorescence in the tumor and other organs showed that tumor tissue accumulation of ANCSS(Cas9/sgRNA) was the highest (10.2% ID/g), which was comparable to nonreductive ANC (Cas9/sgRNA), and 3.4- and 10.2-fold higher than that achieved by NCSS(Cas9/sgRNA) or free Cas9/sgRNA control groups, respectively (fig. S19B). To evaluate whether the luciferase expression of GL261 has any influence on BBB penetration of nanocapsules, the in vivo imaging showed that ANCSS(Cas9/sgRNA) still had BBB penetration capability in luciferase-free GL261-bearing mice (fig. S20), indicating that the luciferase expression had little impact on the BBB crossing of ANCSS(Cas9/sgRNA). These results in GL261 GBM-bearing C57BL/6 mice agree with the results in U87MG GBM nude mice, showing that ANCSS(Cas9/sgRNA) has excellent BBB penetration and tumor accumulation in both immunocompromised and immunocompetent mouse models.
Assessment of the effect of CRISPR-Cas9 nanocapsules on orthotopic GBM xenografts
We evaluated the preclinical anti-GBM therapeutic efficacy of the CRISPR-Cas9 nanocapsules by examining their effects on the growth of orthotopic U87MG-Luc glioblastoma in mice (Fig. 3A). Mice were randomly assigned to groups and received intravenous tail-vein injections of ANCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), or PBS treatments every 2 days. Mice treated with ANCSS(Cas9/sgPLK1) nanocapsules exhibited remarkable tumor growth inhibition, indicated by the decrease in bioluminescence signal intensity (Fig. 3, B and D, middle). In contrast, mice treated with ANCSS(Cas9/sgScr) or PBS showed increased luminescence, indicating increased tumor growth (Fig. 3, B and D, left and right cohorts). Mice treated with ANCSS(Cas9/sgPLK1) showed negligible body weight loss, whereas ANCSS(Cas9/sgScr)- or PBS-treated mice exhibited a marked loss in body weight within 18 days, corresponding to rapid tumor growth and GBM infiltration into normal brain tissue (Fig. 3C), suggesting the superior safety profile of ANCSS(Cas9/sgPLK1) nanocapsules as well.
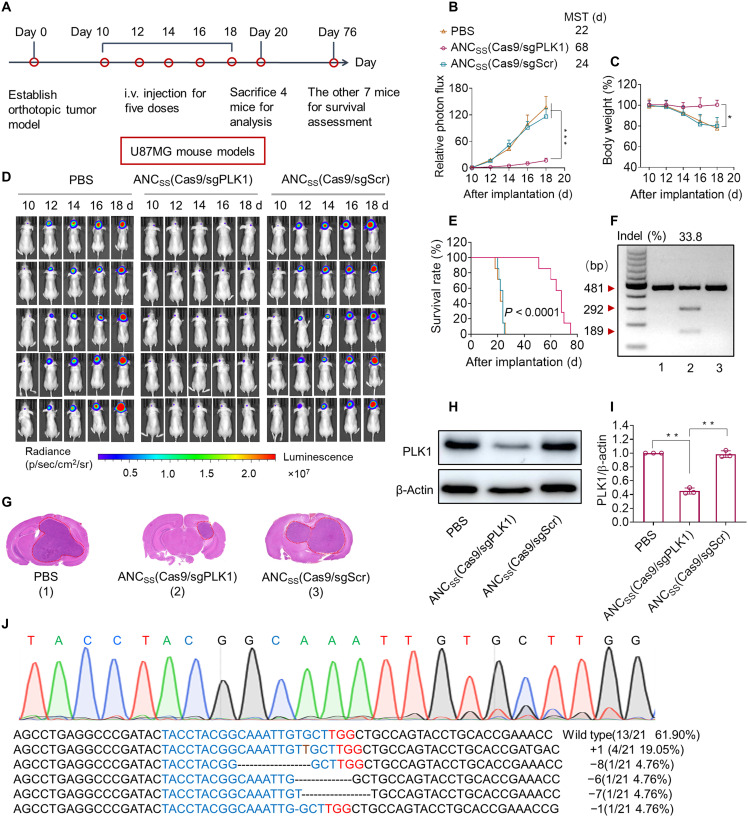
Fig. 3. Genome editing efficiency of CRISPR-Cas9 nanocapsule in orthotopic U87MG GBM xenografts.
(A) Schematic showing the timeline of the U87MG orthotopic tumor model study. (B) Quantified luminescence levels of mice using the Lumina IVIS III System following the indicated treatments. Data are means ± SD (***P < 0.001). (C) Body weight changes in mice following the indicated treatments. Data are means ± SD (*P < 0.05). (D) Luminescence images of orthotopic U87MG-Luc human glioblastoma tumor-bearing nude mice following treatment with PBS (left), ANCSS(Cas9/sgPLK1) (middle), or ANCSS(Cas9/sgScr) (right). Mice were intravenously injected at a dose of 1.5 mg of Cas9 equiv./kg on days 10, 12, 14, 16, and 18 after tumor implantation (n = 11). (E) Mice survival rates (n = 7). Statistical analysis: ANCSS(Cas9/sgPLK1) versus ANCSS(Cas9/sgScr) or PBS, (Kaplan-Meier analysis, log-rank test). (F) Indel frequency of PLK1 gene in tumor tissues excised from mice on day 20. (G) H&E staining of whole brain excised on day 20 from euthanized U87MG-Luc–bearing mice treated with different nanocapsule formulations as described above. (H) Western blot of PLK1 protein expression in tumor tissues excised on day 20. β-Actin was used as a reference. (I) Quantitation of Western blotting of PLK1 protein expression relative to β-actin. Data are means ± SD (n = 3; **P < 0.01). (J) Sequencing results of PLK1 gene editing in U87MG-bearing mice treated with ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg).
Survival curve analysis showed that treatment with ANCSS(Cas9/sgPLK1) remarkably extended median survival to 68 days versus 24 or 22 days following treatment with ANCSS(Cas9/sgScr) or PBS, respectively (Fig. 3E). Notably, the incomplete tumor retardation of ANCSS(Cas9/sgPLK1) is mainly attributed to the GBM heterogenetic, tumor microenvironment, complex pathogenetic mechanism, and potential resistance development. To confirm that tumor growth inhibition was due to PLK1 gene disruption and reduced PLK1 protein expression, the excised tumor tissues from mice treated with ANCSS(Cas9/sgPLK1) or the two control formulations were assessed on day 20 using a T7E1 mismatch detection assay and Western blotting. The indel frequency, a measure of gene editing efficiency, achieved by treatment with ANCSS(Cas9/sgPLK1) was 33.8%, whereas treatment with ANCSS(Cas9/sgScr) or PBS showed no observable cleavage of PLK1 (Fig. 3F). Hematoxylin and eosin (H&E) staining of whole-brain slices excised at day 20 showed that the tumor size of ANCSS(Cas9/sgPLK1)-treated mice was significantly smaller than that of the ANCSS(Cas9/sgScr) or PBS groups (Fig. 3G).
Western blotting demonstrated a significant reduction in PLK1 protein expression in the ANCSS(Cas9/sgPLK1)-treated cohort but not in control treatment groups (Fig. 3, H and I, and fig. S21). Sanger sequencing showed eight mutations at the target sequence in 21 clones, including T insertion single base substitutions, large deletions, and base deletions, providing an overall mutation frequency of 38.1% (Fig. 3J), which was consistent with the T7E1 results and results in U87MG cells (42.9%). To the best of our knowledge, this is the highest in vivo tumor genome editing efficiency observed using a noninvasive nanotechnology-based CRISPR-Cas9 system (38, 39).
Immunohistochemical analysis of tumor slices taken at the termination of treatment showed significantly fewer PLK1-positive tumor cells (brown) in ANCSS(Cas9/sgPLK1)-treated samples than in controls, which was consistent with Western blotting (fig. S22, A and B). Furthermore, tumor samples from ANCSS(Cas9/sgPLK1)-treated mice exhibited the highest expression of caspase-3, a marker of tumor cell apoptosis and the lowest expression of the Ki-67 proliferation marker (fig. S22, C and D). Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling assays confirmed that treatment with ANCSS(Cas9/sgPLK1) induced significant apoptosis (fig. S22E). H&E staining showed no obvious systemic toxicity or abnormality after ANCSS(Cas9/sgPLK1) treatment, with low nuclear-cytoplasmic ratios observed in major organs (heart, liver, and kidney) (fig. S23).
Assessment of the effect of CRISPR-Cas9 nanocapsules on patient-derived GSC xenografts
Before creating a patient-derived xenograft mouse model, we verified that the patient-derived Cancer Stem Cell-2 (CSC-2) GSCs expressed PLK1 at the gene and protein levels. DNA sequencing results showed that the PLK1 target sequence was present in the patient-derived GSCs, and the PLK1 protein expression level was comparable to those in U87MG cell line, confirming the suitability of the CSC2 GSC model (fig. S24). In addition, CSC2 cells overexpressed LRP-1 protein, which is necessary for enhanced recognition by targeting nanocapsules (fig. S6). We then validated the gene editing ability of ANCSS(Cas9/sgPLK1) nanocapsules using the CSC2 GSCs. We observed an indel frequency of 22.9% following ANCSS(Cas9/sgPLK1) treatment, which is significantly higher than that following treatment with NCSS(Cas9/sgPLK1) or ANC(Cas9/sgPLK1) (fig. S25). DNA sequencing analysis confirmed the high rate of PLK1 gene disruption (19.0%; fig. S26). Western blotting showed a large reduction in PLK1 protein expression following treatment with ANCSS(Cas9/sgPLK1) relative to controls (fig. S27).
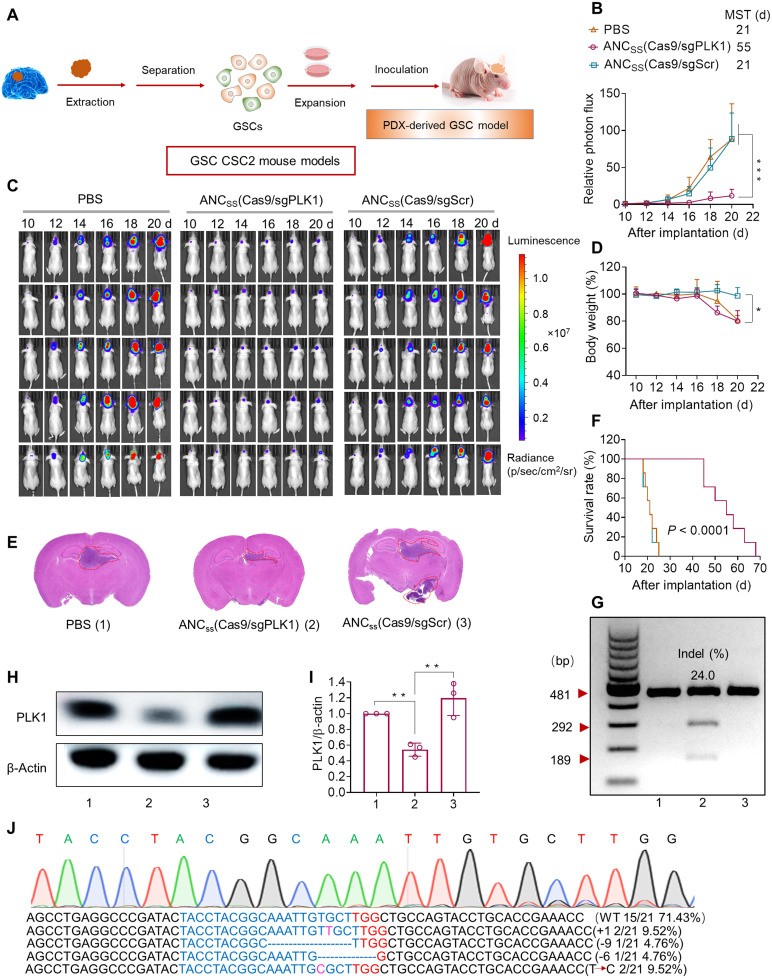
We then developed stable luciferase-expressing CSC2 cells (CSC2-Luc) to establish a facile bioluminescence-based orthotopic GSC mouse model to study the therapeutic effect of our CRISPR-Cas9 nanocapsules (Fig. 4A). As in the U87MG orthotopic GBM mouse model, ANCSS(Cas9/sgPLK1) treatment substantially inhibited tumor growth, as evidenced by reduced luminescence intensity in mice receiving ANCSS(Cas9/sgPLK1) treatment (Fig. 4, B and C). ANCSS(Cas9/sgPLK1) treatment did not cause marked decreases in body weight (Fig. 4D). In contrast, the obvious body weight loss was observed in mice treated with ANCSS(Cas9/sgScr) or PBS that resulted from the fast tumor proliferation and increased brain burden. Moreover, H&E staining of excised brain tissue confirmed the efficient antitumor efficacy of ANCSS(Cas9/sgPLK1) nanocapsules (Fig. 4E). Mice treated with ANCSS(Cas9/sgPLK1) also showed a marked extension in median survival to 55 days, which was significantly longer than the median survival of 21 days for mice receiving ANCSS(Cas9/sgScr) or PBS (Fig. 4F). T7E1 assays revealed that ANCSS(Cas9/sgPLK1) nanocapsules induced a high indel frequency of 24.0% (Fig. 4G), and PLK1 protein expression in GSC tumor tissue was also reduced (Fig. 4, H and I, and fig. S28), indicating PLK1 gene disruption. Sanger sequencing also verified PLK1 gene disruption (Fig. 4J), which showed a mutation rate of 28.6%, consistent with the T7E1 assay. Immunohistochemical analysis showed lower PLK1 and Ki-67 expression but higher caspase-3 apoptosis expression in tumor tissue slices excised from mice receiving ANCSS(Cas9/sgPLK1) treatment compared to control groups (fig. S29) and showed little damage to healthy tissues (fig. S30). These results were similar to those obtained in the U87MG xenograft model. Collectively, these in vitro and in vivo results demonstrate that ANCSS(Cas9/sgRNA) nanocapsules achieve effective BBB permeability, improved tumor accumulation and retention, active tumor cell uptake, and intracellular release of Cas9/sgRNA, resulting in high in vivo gene knockout efficiency and anti-GBM therapeutic efficacy.
Fig. 4. Genome editing efficiency of CRISPR-Cas9 nanocapsule in GSC CSC2 xenografts.
(A) Schematic of patient-derived xenograft (PDX)–derived GBM GSCs orthotopic model establishment. (B) Luminescence levels of mice over the 10-day treatment period measured with a Lumina IVIS III system. Data are means ± SD (***P < 0.001). (C) Luminescence images of orthotopic CSC2-Luc GSC tumor–bearing mice following treatment with ANCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), or PBS. Mice were intravenously injected at a dose of 1.5 mg of Cas9 equiv./kg on days 10, 12, 14, 16, and 18 after tumor implantation (n = 11). (D) Body weight changes over the 10-day treatment period in mice receiving ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg), ANCSS(Cas9/sgScr), or PBS. Data are means ± SD (*P < 0.05). (E) H&E staining of brain excised from CSC2-Luc GSC tumor–bearing mice treated with different nanocapsule formulations on day 20 after tumor implantation. (F) Mice survival rate curves (n = 7). Statistical analysis: ANCSS(Cas9/sgPLK1) versus ANCSS(Cas9/sgScr) or PBS, ****P < 0.0001 (Kaplan-Meier analysis, log-rank test). (G) Indel frequencies of PLK1 gene in tumor tissues from mice treated with ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg), ANCSS(Cas9/sgScr), or PBS on day 20 after tumor implantation. (H) PLK1 protein expression in tumor tissues excised from mice receiving different nanocapsule formulations on day 20 after tumor implantation. (I) Quantification of Western blotting of PLK1 expression relative to β-actin. Data are means ± SD (n = 3; **P < 0.01). (J) DNA sequencing results of PLK1 gene editing in GSC tumors excised from mice treated with ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg).
Safety evaluation of CRISPR-Cas9 nanocapsule in vivo
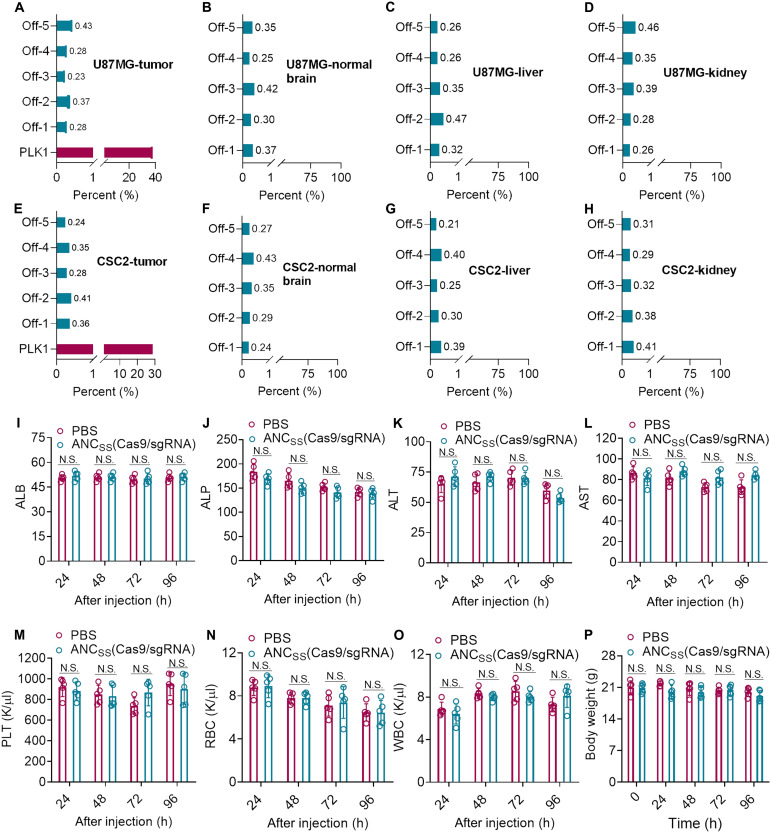
As genome editing by CRISPR-Cas9 may generate safety concerns due to off-target effects (40, 41), we therefore identified the riskiest off-target PLK1 sequence sites in tumor tissue (table S2) (42, 43). Deep sequencing assays revealed negligible gene disruption at these potential sites in tumor tissue in both U87MG- and CSC2 GSC–bearing mice following treatment with ANCSS(Cas9/sgPLK1). The mutation frequencies were below 0.5% at all five potential target sites in these models (Fig. 5, A and E). It is also important to evaluate potential off-target effects in nontumor brain tissue to ensure nanocapsule safety. Therefore, we investigated potential off-target effects in normal brain tissues. Again, deep sequencing results demonstrated that mutation frequencies were lower than 0.5% (Fig. 5, B and F), indicating the negligible impacts on these tissues. As PLK1 expression is much lower in endothelia, glia, and astrocytes and further considering that LRP-1 expression is also lower in glia and astrocytes cells, lower rates of off-target effects can be expected (fig. S6). As nanocapsules are eliminated mainly by the liver and kidney (Fig. 2F), we assessed these organs for potential off-target effects. Reassuringly, we found that mutation frequencies were below 0.5% at all potential target sites in liver and kidney in both U87MG- and CSC2 GSC–bearing mice (Fig. 5). The low off-targets of ANCSS(Cas9/sgRNA) nanocapsules may have resulted from the lower expression of LRP-1 receptor and PLK1 gene (fig. 31) and difficult Cas9/sgRNA release in normal cells due to low GSH concentration. Experiments assessing the effects of treatment on blood parameters and biochemistry indicated that mice treated with ANCSS(Cas9/sgPLK1) nanocapsules had similar profiles to those in mice treated with PBS over the treatment course. Furthermore, mice maintained their body weight, suggesting that ANCSS(Cas9/sgPLK1) nanocapsules had minimal impact on liver and kidney function or hematological parameters (Fig. 5, I to O). Together, these results indicate that the ANCSS(Cas9/sgPLK1) nanocapsules cause very limited safety concerns.
Fig. 5. Evaluation of the safety of CRISPR-Cas9 nanocapsules in vivo.
Mutation frequencies of off-target sites (tumor, normal brain tissue, liver, and kidney) in (A to D) U87MG and (E to H) CSC2 GSC tumor–bearing mice treated with ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg). Each value was determined from a single deep-sequencing library prepared from genomic DNA. Blood biochemistry analysis (I to L), blood parameter analysis (M to O), and body weight changes (P) of healthy BALB/c mice treated with ANCSS(Cas9/sgRNA) or PBS at 24, 48, 72, or 96 hours after nanocapsule injection. Data are presented as means ± SD (n = 5). N.S. represents nonsignificance. ALB, albumin; ALP, alkaline phosphatase; ALT, plasma alanine aminotransferase; AST, aspartate aminotransferase; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
DISCUSSION
CRISPR-Cas9 gene editing technology provides the means to rapidly create mutations, insertions, and deletions in the genome of targeted cells (44, 45). Although the technology has already been used for brain diseases via intrabrain injections (3, 46), further clinical applications of gene therapy in brain diseases has been hampered by the lack of noninvasive, effective, and safe brain delivery systems for transporting CRISPR-Cas9 across the BBB to target diseased cells in the brain (47). Existing approaches to deliver CRISPR-Cas9 to the brain include viral vector delivery (lentivirus and adeno-associated viruses) (45) and nonviral synthetic delivery (gold, lipid, and polymers) (48, 49). Although these approaches have provided valuable in vivo proof of principle, these methods are not ideal for human clinical applications. Viral vector delivery, for instance, may generate highly risky immune responses and complications due to off-targeting effects and is also difficult for large-batch production (50). On the other hand, current nonviral delivery systems are limited in their low loading efficiency, non–disease targeting, and problematic clearance out of the brain; risk of neuroinflammation; and lack of responsive drug release (51). They are both seriously limited in BBB penetration and lack specific brain disease site targeting (52). However, these properties are important, as they enable intravenous injection and thereby avoid risk of brain trauma arising from direct local brain administration or intracerebral delivery (14, 53).
In this study, we developed a nonviral CRISPR-Cas9 delivery system, ANCSS(Cas9/sgRNA), using angiopep-2–functionalized biodegradable nanocapsules that encapsulate and protect Cas9 protein and sgRNA for noninvasive, targeted gene knockdown. By design, our new ANCSS(Cas9/sgRNA) nanocapsules feature self-encapsulating single Cas9 ribonucleoprotein/sgRNA, which leads to high loading efficiency of nearly 100% (of Cas9 ribonucleoprotein/sgRNA complexes). Furthermore, single Cas9/sgRNA encapsulation led to a small uniform nanocapsule with a diameter of ~30 nm, which was beneficial for BBB penetration and subsequent deep transport into the brain and tumor. A further key design element was the incorporation of disulfide bonds (-SS-) cross-linked into the nanocapsule shell, which has two important functions: protection of the Cas9/sgRNA complex from enzymatic degradation in blood and facility for rapid Cas9/sgRNA release upon encountering the high intracellular reducing environment where the high concentration of GSH (2 to 10 mM) present in tumor cells is able to break the disulfide linkage to biodegrade the nanocapsule (Fig. 1C). The next key design element involved a “two birds, one stone” strategy by functionalizing the outer shell of Cas9/sgRNA nanocapsules with the angiopep-2 peptide, which specifically binds to LRP-1 receptor that is highly expressed on both the endothelial cells of the BBB and GBM cells, endowing ANCSS(Cas9/sgRNA) nanocapsules with both high BBB penetration and GBM-targeting capability.
To successfully reach glioblastoma target sites in the deep brain, therapeutic agents must remain intact in the bloodstream, long enough for sufficient BBB penetration and GBM accumulation. Our ANCSS(Cas9/sgRNA) nanocapsules exhibited a long circulation time in blood with an elimination half-life of 57 min that led to the BBB penetration via receptor-mediated transcytosis and exploited compromised BBB structure because of the BBB heterogeneity (54) in GBM to pronounced accumulation in the tumor, reaching a maximal concentration (11.8% of ID) 4 hours after injection. Considering that most other nanoparticle-based drug delivery systems only achieve an accumulation of only 1 to 5% of ID in the brain (55–57), our design strategy provides a clear advance, which was further demonstrated by achieving highly efficient PLK1 gene editing (up to 38.1% gene knockdown). To the best of our knowledge, this is the highest CRISPR-Cas9–based in vivo gene editing efficiency yet reported for a nonviral delivery system via intravenous injection. These encouraging results probably account for the marked inhibition of GBM tumor growth and the approximate trebling of median survival time mediated by the ANCSS(Cas9/sgPLK1) nanocapsules (68 days versus 24 days in the U87MG GBM model and 55 days versus 21 days in the CSC2 GSCs model).
Considering that off-target effects induced by CRISPR-Cas9 can lead to undesired chromosomal rearrangements and represent a major bottleneck limitation for CRISPR/Cas9 that seriously hinders clinical application, it was important to systematically evaluate the potential off-target effect of ANCSS(Cas9/sgPLK1) in both GBM and other normal tissue. Reassuringly, the noninvasive administration of ANCSS(Cas9/sgPLK1) caused negligible off-target side effects (<0.5%; Fig. 5), which mainly ascribed to the encapsulation of Cas9 ribonucleoprotein rather than plasmid or mRNA (58), neutral surface charge, specific targeting, and controlled Cas9/sgRNA intracellular release. Another factor working to reduce off-target effects is that expression of LRP-1 receptor and PLK1 target gene is much lower in normal brain tissue relative to GBM.
To promote clinical translation for GBM treatment, our nanocapsules could be further improved as follows: (i) Given that brain tumors have complex pathogenesis, single gene editing is unlikely to eradicate the tumor completely (as shown in this study). In future studies, we will encapsulate two or multiple sgRNAs with Cas9 for targeting and editing multiple pathogenetic genes simultaneously to achieve more effective brain tumor treatment. (ii) More specific targeting ligands could be developed to further increase the cellular uptake of GBM cells versus normal brain cells, thus reducing off-targets effects. (iii) These nanocapsules could be adapted for knocking in apoptosis genes [P53 (59) and PTEN (60)] into GBM cells for inducing tumor apoptosis.
In conclusion, we developed an angiopep-2–decorated, GSH-responsive single CRISPR-Cas9 nanocapsule [ANCSS(Cas9/sgRNA)] as a noninvasive brain delivery system and systemically demonstrated that it has the superior properties including high CRISPR-Cas9 loading and intracellular environment-responsive release capability, excellent brain and tumor targeting manners, outstanding gene editing efficiency, and negligible off-target effect with well-established animal models, successfully addressing the bottlenecks (low BBB penetration, weak diseased tissue targeting, low in vivo gene editing efficiency, and unwanted off-target effect) in CRISPR-Cas9 brain delivery toward an effective and safe approach for GBM gene therapy. This novel CRISPR-Cas9–brain delivery system is a versatile and potent platform for treating glioblastoma and other brain diseases.
MATERIALS AND METHODS
Analysis of RNase protection assay
To evaluate the stability of ANCSS(Cas9/sgPLK1) in the presence of RNase A, the free Cas9/sgPLK1 and ANCSS(Cas9/sgPLK1) (Cas9: 200 nM) was incubated in the RNase A (1 mg/ml) solution at 37°C for 30 min, followed by the addition of targeted DNA, and incubated at 37°C for 60 min to assess its ability to induce DNA double-stranded breaks in target DNA. The RNase protection effect was analyzed using gel electrophoresis.
Western blotting to determine the LRP-1 and PLK1 protein
U87MG human glioblastoma cells; GL261 mouse glioblastoma cells; CSC2 GSCs; 83NS GSCs; normal glial cell HA 1800; astrocytes cell BV2 and endothelial cell hCMEC/D3; and liver, lung, and kidney organs were lysed using lysis buffer (Beyotime, China). The protein concentrations were quantified by BCA Protein Assay. Lysates were separated by SDS–polyacrylamide gel electrophoresis and were transferred onto polyvinylidene difluoride (PVDF) membranes. PVDF membranes were incubated with primary antibody against PLK1 [mouse monoclonal antibody (mAb) 35-206; 1:1000; Abcam], LRP-1 [rabbit mAb (EPR3724); 1:50,000; Abcam], and secondary antibody (LI-COR IRDye 800CW). Protein bands were displayed by an ECL detection system and were analyzed using ImageJ software.
Gene editing and sequencing
U87MG cells were seeded in 24-well plates at 5 × 104 cells per well and were cultured for 24 hours. The cells were then incubated with ANCSS(Cas9/sgPLK1), ANC(Cas9/sgPLK1), NCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), Lipo(Cas9/sgPLK1), or free Cas9/sgPLK1 (Cas9, 20 nM) overnight, and the medium was replaced with fresh medium containing 10% fetal bovine serum (FBS). The cells were incubated at 37°C for another 48 hours and then were collected and processed to harvest genomic DNA using the Universal Genomic DNA Kit (CWBIO, China). The sgRNA-targeted genomic locus was amplified with High-Fidelity KOD-Plus-Neo (TOYOBO, Japan). After purification by gel extraction (CWBIO, China), T7E1 cleavage assays were conducted. Briefly, 200 ng of the purified polymerase chain reaction (PCR) product was denatured and reannealed in 2 μl of NEBuffer 2 (10×) using the following protocol: 95°C, 5 min; 95° to 85°C, −2°C/s; 85° to 25°C, −0.1°C/s; and then held at 4°C. Then, 1 μl of T7E1 (M0302S) was added to the annealed PCR products and incubated at 37°C for 1 hour. Products were analyzed on 2% agarose gels and imaged with a GelDoc imaging system (Bio-Rad). PCR products with mutations indicated by the T7E1 assay were subjected to DNA sequencing and subcloned into T-clone vectors (Vazyme Biotech, China). Colonies were picked randomly and further analyzed by Sanger sequencing using an M13F primer (Sangon Biotech). The in vivo gene editing and sequencing was similar to the previous procedure, except that the tumor genomic DNA samples was harvested using the Universal Genomic DNA Kit after treatment with ANCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), or PBS.
Western blotting to measure PLK1 protein expression in vitro and in vivo
In in vitro evaluation, U87MG cells were seeded in six-well plates at 1 × 106 cells per well, cultured for 24 hours, and incubated with ANCSS(Cas9/sgPLK1), ANC(Cas9/sgPLK1), NCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), Lipo(Cas9/sgPLK1), or free Cas9/sgPLK1 (Cas9, 20 nM) overnight; then, medium was replaced with fresh medium containing 10% FBS. The cells were incubated at 37°C for another 72 hours, and cells were treated with radioimmunoprecipitation assay lysis buffer (Beyotime, China). The concentrations of resulting proteins were quantified by BCA Protein Assay (Beyotime, China). The lysates were separated by SDS–polyacrylamide gel electrophoresis and were transferred onto PVDF membranes (Beyotime, China). PVDF membranes were incubated with primary antibody against PLK1 (mouse mAb 35-206; Abcam) at a dilution of 1:1000 and secondary antibody (LI-COR IRDye 800CW). Protein bands were visualized using an ECL detection system. Indel formation efficiencies were calculated using the value of β-actin as the denominator using ImageJ v.1.8.0. For in vivo evaluation, tumors from mice treated with ANCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), and PBS were lysed using lysis buffer (Beyotime, China). The following procedure was the same as the previous described.
Cell viability and in vitro luciferase assay
U87MG-Luc cells were seeded in 96-well plates at 2 × 103 cells per well and were incubated in 100 μl of Dulbecco’s modified Eagle’s medium containing 10% FBS for 24 hours. The culture medium was removed; the cells were incubated with ANCSS(Cas9/sgLuc), ANC(Cas9/sgLuc), NCSS(Cas9/sgLuc), or free Cas9/sgLuc (Cas9, 20 nM) overnight, and the medium was replaced with fresh medium containing 10% FBS. Then, after incubation for another 72 hours, CCK-8 solution (10/100 μl of medium) was added to each well, and after incubation for another 30 min, optical density at 450 nm (OD450) value (absorbance at 450 nm) was detected by a microplate reader (Devices/13×, Molecular Device, USA). Cell viability (%) was obtained by calculating the ratio of OD450 of the cells under targeted conditions to those of untreated control cells (n = 5). For the luciferase knockout assays, luminescence intensities of cell lysates were measured with the Firefly Luciferase Reporter Gene Assay Kit (Beyotime, China). Twenty microliters was used to quantify protein concentration with a BCA protein assay kit (Beyotime, China). The luciferase signal was divided by the amount of total protein for normalization (n = 5).
Apoptosis assay
To assess apoptosis, U87MG and U251 cells were seeded in 24-well plates at 5 × 104 cells per well and were cultured for 24 hours. The cells were then incubated with ANCSS(Cas9/sgPLK1), ANC(Cas9/sgPLK1), NCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), or free Cas9/sgPLK1 (Cas9, 20 nM) overnight, and the medium was replaced with fresh medium containing 10% FBS. The cells were incubated at 37°C for another 72 hours, after which the cells were incubated with Alexa Fluor 488–annexin V and propidium iodide (PI) (Beyotime, China). The stained cells were collected and washed with PBS, and fluorescence [fluorescein isothiocyanate: excitation (Ex.), 488 nm and emission (Em.), 520 nm; PI: Ex., 488 nm and Em., 630 nm] was measured following the manufacturer’s protocol using a flow cytometer (Becton Dickinson, USA) and FlowJo v10 software. The efficacy of ANCSS(Cas9/sgPLK1) in low PLK1-expressed U87MG cells was similar as above, except that the U87MG cells were pretreated with PLK1 siRNA (100 nM) using polyethyleneimine as a transfection agent for 48 hours.
In vitro BBB penetration evaluation
The in vitro BBB model was constructed with endothelial hCMEC/D3 cells using a transwell cell culture system. The transendothelial electrical resistance (TEER) instrument (World Precision Instruments Inc. Sarasota, FL, USA) was used to monitor the intactness of the cell monolayer. The following experiments were carried out only when the TEER value of the endothelial hCMEC/D3 cell monolayer was above 200 ohm·cm2. ANCSS(Cas9/sgRNA), ANC(Cas9/sgRNA), NCSS(Cas9/sgRNA), or free Cas9/sgRNA (AF647-Cas9: 20 nM) was added to the upper chamber. Then, FBS-free medium was added to the lower chamber. After 2, 6, and 12 hours of incubation, the supernatant in the upper chamber and the medium in the lower chamber were extracted with dimethyl sulfoxide and detected. The amounts of AF647-Cas9 in the supernatant and filtered compartments were determined using a standard microplate assay. The AF647-Cas9 ratio in each compartment was calculated compared with the feeding amount.
In vivo luminescence reduction in orthotopic U87MG-Luc glioblastoma–bearing nude mice
ANCSS(Cas9/sgLuc), ANCSS(Cas9/sgScr) (1.5 mg of Cas9 equiv./kg), or PBS was intravenously injected into the tail vein of U87MG-Luc orthotopic tumor-bearing nude mice (n = 3). Luminescence intensity in the brain was determined before injection and at 24, 48, and 72 hours after injection using an IVIS III instrument. Mice were anesthetized with isoflurane, and luciferin was injected intraperitoneally at a dosage of 150 mg/kg (100 μl). Measurements were performed at 10 min after luciferin injection. Photons emitted from the brain region were quantified using live imaging software.
Ex vivo imaging, penetration, and biodistribution
ANCSS(Cas9/sgRNA), ANC(Cas9/sgRNA), NCSS(Cas9/sgRNA), or free Cas9/sgRNA (1.5 mg of Cas9 equiv./kg) in PBS was administrated intravenously via the tail vein into orthotopic U87MG-Luc, 83NS GSC tumor–bearing nude mice, GL261 with or without luciferase expression–bearing C57BL/6 mice. Luminescence intensity in the brain was determined at different time points using an IVIS III instrument. At 4 hours after injection, the tumor-bearing mice were euthanized. The heart, liver, spleen, lung, kidney, brain, and tumors were collected, washed, and weighed. Fluorescence images were acquired with a Lumina IVIS III near-infrared fluorescence imaging system. To evaluate tumor penetration, cancerous brains were harvested, fixed in 4% formalin overnight, embedded in paraffin, and sliced for immunofluorescence staining analysis. Blood vessels were counterstained with an Alexa Fluor 488–donkey anti-rat secondary antibody (1:1000 dilution in PBS) in a humidified chamber at 37°C for 1 hour, followed by washing three times with PBS and staining with DAPI (5 μg/ml) for 10 min, and were observed with a CLSM imaging system (Zeiss 880). To quantify the amount of AF647-Cas9 delivered to the tumor and different organs, each tumor and organ was individually homogenized in 0.6 ml of 1% Triton X-100 with a homogenizer (70,000 Hz) for 6 min. The samples were then centrifuged at 15,000 rpm for 30 min. The content of AF647-Cas9 in the supernatant was determined by fluorometry (Ex., 649 nm; Em., 670 nm) based on a calibration curve. The penetration and biodistribution evaluation of ANCSS(Cas9/sgRNA) in GL261 mice was similar to that in U87MG mice except for the euthanasia of mice at 12 hours after injection of the nanocapsules.
Penetration evaluation in 3D spheroid tumor model
The 3D tumor spheroids of U87MG were established according to following steps. Briefly, U87MG cells (2 × 103 per well) were plated in PrimeSurface 96-well plates (Sumitomo Bakelite, Japan). After 3 days, the tumor spheroids were treated with the ANCSS(Cas9/sgRNA), NCSS(Cas9/sgRNA), ANCSS(Cas9/sgRNA), and free Cas9/sgRNA (the AF647-Cas9 concentration was 20 nM) for another 4 hours of incubation. Then, tumor spheroids were washed and fixed with 4% paraformaldehyde. The permeability of different nanoparticles into tumor spheroids was investigated by CLSM (Zeiss 880; ×110 magnification).
Effect of nanocapsules on the growth of GBM tumors in vivo
Tumor-bearing mice aged 6 to 8 weeks (n = 11; 7 for monitoring survival rate, 1 for histological analysis, and 3 for protein and gene editing evaluation) received an intravenous injection of ANCSS(Cas9/sgPLK1), ANCSS(Cas9/sgScr), or PBS every other day. At 48 hours after injection, the mice were anesthetized, and the Lumina IVIS III system was used to evaluate the tumor luminescence intensity. The relative photon flux was normalized to the initial intensity. On day 20, the treatment was terminated, and four mice from each group were euthanized. The brain tumors and organs, including heart, liver, spleen, lung, and kidney, were harvested, weighed, and fixed with 4% paraformaldehyde for immunohistochemical analysis, including staining for PLK1, cleaved caspase-3, and Ki-67, and hematoxylin and eosin (H&E) staining. Kaplan-Meier survival curves were determined for each treatment group, and the body weights of mice were measured individually.
In vivo DNA deep sequencing
To assess the off-target effects of nanocapsules, we predicted potential off-target sites based on the online database (https://cm.jefferson.edu/Off-Spotter/) according to the following acknowledged criteria (42, 43): (i) Cas9 tolerates single-base mismatches in the protospacer adjacent motif (PAM)-distal region to a greater extent than in the PAM-proximal region. (ii) Three or more mismatched base pairs eliminated detectable Cas9 cleavage in the vast majority of loci.
Briefly, after being treated with ANCSS(Cas9/sgPLK1), the genomic DNAs of tumor, normal brain tissue, liver, and kidney were harvested from mice bearing U87MG or CSC2 GSCs, using the Universal Genomic DNA Kit (CWBIO, China) according to the manufacturer’s instructions. One hundred nanograms of genomic DNA was used as a template to perform PCR using primers designed against on-target and off-target sites. Purified DNA was amplified again by PCR with primers containing sequencing adapters and then sequenced and analyzed by Sangon Biotechnology Company (Shanghai, China) to detect indels around target sites.
Safety evaluation
BALB/c mice were weighed and divided randomly into two groups (n = 5). ANCSS(Cas9/sgPLK1) (1.5 mg of Cas9 equiv./kg), or PBS was intravenously injected via the tail vein. Blood serum was collected at 24, 48, 72, and 96 hours and centrifuged at 800g for 5 min. The blood levels of alkaline phosphatase, aspartate aminotransferase, alanine aminotransferase, and serum albumin were determined by Wuhan Servicebio Technology Co. We also performed serial daily blood monitoring of white blood cell, platelet, and red blood cell levels. Daily body weights were recorded throughout the course of the nanocapsule and control treatments.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (NSFC 51803049, 31800841, 32071388, 52073079, U2004171, and U1804139), the National Key Technologies R&D Program of China (2018YFA0209800), NHMRC Investigator Grant (GNT1194825), and the Program for Science & Technology Innovation Talents in Universities of Henan Province (21HASTIT033). O.S. acknowledges the Australian Research Council and National Health and Medical Research Council (APP1101258 and IH150100028).
Author contributions: M.Z. and B.S. designed the experiments, supervised the project, and revised the manuscript. Y.Z., X.S., and Q.Y. prepared and characterized the nanocapsules and performed in vitro and in vivo experiments. W.R., Y.W., D.Z., and J.Y. analyzed the data. Y.Z. and X.S. wrote the manuscript and revised it with comments from M.Z., O.S., X.H., W.T., J.B.P., X-J.L., K.W.L., and B.S. All authors participated in discussions throughout the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 and S2
Figs. S1 to S31
REFERENCES AND NOTES
- 1.Shalem O., Sanjana N. E., Zhang F., High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleinstiver B. P., Pattanayak V., Prew M. S., Tsai S. Q., Nguyen N. T., Zheng Z., Joung J. K., High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park H., Oh J., Shim G., Cho B., Chang Y., Kim S., Baek S., Kim H., Shin J., Choi H., Yoo J., Kim J., Jun W., Lee M., Lengner C. J., Oh Y.-K., Kim J., In vivo neuronal gene editing via CRISPR–Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 22, 524–528 (2019). [DOI] [PubMed] [Google Scholar]
- 4.LaFleur M. W., Nguyen T. H., Coxe M. A., Yates K. B., Trombley J. D., Weiss S. A., Brown F. D., Gillis J. E., Coxe D. J., Doench J. G., Haining W. N., Sharpe A. H., A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 10, 1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin H., Kanasty R. L., Eltoukhy A. A., Vegas A. J., Dorkin J. R., Anderson D. G., Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Rybicki E. P., CRISPR–Cas9 strikes out in cassava. Nat. Biotechnol. 37, 727–728 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Roth T. L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P. J., Hiatt J., Saco J., Krystofinski P., Li H., Tobin V., Nguyen D. N., Lee M. R., Putnam A. L., Ferris A. L., Chen J. W., Schickel J.-N., Pellerin L., Carmody D., Alkorta-Aranburu G., del Gaudio D., Matsumoto H., Morell M., Mao Y., Cho M., Quadros R. M., Gurumurthy C. B., Smith B., Haugwitz M., Hughes S. H., Weissman J. S., Schumann K., Esensten J. H., May A. P., Ashworth A., Kupfer G. M., Greeley S. A. W., Bacchetta R., Meffre E., Roncarolo M. G., Romberg N., Herold K. C., Ribas A., Leonetti M. D., Marson A., Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernando M. B., Ahfeldt T., Brennand K. J., Modeling the complex genetic architectures of brain disease. Nat. Genet. 52, 363–369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pluvinage J. V., Haney M. S., Smith B. A. H., Sun J., Iram T., Bonanno L., Li L., Lee D. P., Morgens D. W., Yang A. C., Shuken S. R., Gate D., Scott M., Khatri P., Luo J., Bertozzi C. R., Bassik M. C., Wyss-Coray T., CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B., Lee K., Panda S., Gonzales-Rojas R., Chong A., Bugay V., Park H. M., Brenner R., Murthy N., Lee H. Y., Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2, 497–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staahl B. T., Benekareddy M., Coulon-Bainier C., Banfal A. A., Floor S. N., Sabo J. K., Urnes C., Munares G. A., Ghosh A., Doudna J. A., Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 35, 431–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei T., Cheng Q., Min Y.-L., Olson E. N., Siegwart D. J., Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rui Y., Wilson D. R., Green J. J., Non-viral delivery to enable genome editing. Trends Biotechnol. 37, 281–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozano A. M., Lipsman N., Bergman H., Brown P., Chabardes S., Chang J. W., Matthews K., McIntyre C. C., Schlaepfer T. E., Schulder M., Temel Y., Volkmann J., Krauss J. K., Deep brain stimulation: Current challenges and future directions. Nat. Rev. Neurol. 15, 148–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foldvari M., Chen D. W., Nafissi N., Calderon D., Narsineni L., Rafiee A., Non-viral gene therapy: Gains and challenges of non-invasive administration methods. J. Control. Release 240, 165–190 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Zheng M., Tao W., Zou Y., Farokhzad O. C., Shi B., Nanotechnology-based strategies for siRNA brain delivery for disease therapy. Trends Biotechnol. 36, 562–575 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Li J., Zheng M., Shimoni O., Banks W. A., Bush A. I., Gamble J. R., Shi B., Development of novel therapeutics targeting the blood–brain barrier: From barrier to carrier. Adv. Sci. 8, 2101090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trenkmann M., Gold rush to gene-editing in the brain. Nat. Rev. Genet. 19, 532–533 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., Liu J., Cheng C. J., Patel T. R., Weller C. E., Piepmeier J. M., Jiang Z., Saltzman W. M., Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater. 11, 82–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng M., Liu Y., Wang Y., Zhang D., Zou Y., Ruan W., Yin J., Tao W., Park J. B., Shi B., ROS-responsive polymeric siRNA nanomedicine stabilized by triple interactions for the robust glioblastoma combinational RNAi therapy. Adv. Mater. 31, 1903277 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y., Zhang J., Meng F., Zhong Z., Apolipoprotein E peptide-directed chimeric polymersomes mediate an ultrahigh-efficiency targeted protein therapy for glioblastoma. ACS Nano 12, 11070–11079 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Shimoni O., Postma A., Yan Y., Scott A. M., Heath J. K., Nice E. C., Zelikin A. N., Caruso F., Macromolecule functionalization of disulfide-bonded polymer hydrogel capsules and cancer cell targeting. ACS Nano 6, 1463–1472 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Zou Y., Sun X., Wang Y., Yan C., Liu Y., Li J., Zhang D., Zheng M., Chung R. S., Shi B., Single siRNA nanocapsules for effective siRNA brain delivery and glioblastoma treatment. Adv. Mater. 32, 2000416 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Chen G., Abdeen A. A., Wang Y., Shahi P. K., Robertson S., Xie R., Suzuki M., Pattnaik B. R., Saha K., Gong S., A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 14, 974–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian H., Du J., Wen J., Liu Y., Montgomery S. R., Scott T. P., Aghdasi B., Xiong C., Suzuki A., Hayashi T., Ruangchainikom M., Phan K., Weintraub G., Raed A., Murray S. S., Daubs M. D., Yang X., Yuan X.-b., Wang J. C., Lu Y., Growth-factor nanocapsules that enable tunable controlled release for bone regeneration. ACS Nano 10, 7362–7369 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Cabral H., Matsumoto Y., Mizuno K., Chen Q., Murakami M., Kimura M., Terada Y., Kano M. R., Miyazono K., Uesaka M., Nishiyama N., Kataoka K., Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 6, 815–823 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Mitchell M. J., Billingsley M. M., Haley R. M., Wechsler M. E., Peppas N. A., Langer R., Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y., Wang Y., Heath J. K., Nice E. C., Caruso F., Cellular association and cargo release of redox-responsive polymer capsules mediated by exofacial thiols. Adv. Mater. 23, 3916–3921 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Zou Y., Zheng M., Yang W., Meng F., Miyata K., Kim H. J., Kataoka K., Zhong Z., Virus-mimicking chimaeric polymersomes boost targeted cancer siRNA therapy in vivo. Adv. Mater. 29, 1703285 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Yao Y.-d., Sun T.-m., Huang S.-y., Dou S., Lin L., Chen J.-n., Ruan J.-b., Mao C.-q., Yu F.-y., Zeng M.-s., Zang J.-y., Liu Q., Su F.-x., Zhang P., Lieberman J., Wang J., Song E., Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci. Transl. Med. 4, 130ra148 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Gibori H., Eliyahu S., Krivitsky A., Ben-Shushan D., Epshtein Y., Tiram G., Blau R., Ofek P., Lee J. S., Ruppin E., Landsman L., Barshack I., Golan T., Merquiol E., Blum G., Satchi-Fainaro R., Amphiphilic nanocarrier-induced modulation of PLK1 and miR-34a leads to improved therapeutic response in pancreatic cancer. Nat. Commun. 9, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tandle A. T., Kramp T., Kil W. J., Halthore A., Gehlhaus K., Shankavaram U., Tofilon P. J., Caplen N. J., Camphausen K., Inhibition of Polo-like kinase 1 in glioblastoma multiforme induces mitotic catastrophe and enhances radiosensitisation. Eur. J. Cancer 49, 3020–3028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arvanitis C. D., Askoxylakis V., Guo Y., Datta M., Kloepper J., Ferraro G. B., Bernabeu M. O., Fukumura D., McDannold N., Jain R. K., Mechanisms of enhanced drug delivery in brain metastases with focused ultrasound-induced blood–tumor barrier disruption. Proc. Natl. Acad. Sci. U.S.A. 115, E8717–E8726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding S., Khan A. I., Cai X., Song Y., Lyu Z., Du D., Dutta P., Lin Y., Overcoming blood–brain barrier transport: Advances in nanoparticle-based drug delivery strategies. Mater. Today 37, 112–125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxwell K. G., Augsornworawat P., Velazco-Cruz L., Kim M. H., Asada R., Hogrebe N. J., Morikawa S., Urano F., Millman J. R., Gene-edited human stem cell–derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 12, eaax9106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T., He B., Tao J., He Y., Deng H., Wang X., Zheng Y., Application of Förster resonance energy transfer (FRET) technique to elucidate intracellular and in vivo biofate of nanomedicines. Adv. Drug Deliv. Rev. 143, 177–205 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Yang G., Jin S., Zhang R., Chen P., Tengjisi, Wang L., Chen D., Weitz D. A., Zhao C.-X., J-aggregate-based FRET monitoring of drug release from polymer nanoparticles with high drug loading. Angew. Chem. Int. Edit. 132, 20240–20249 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Wang P., Zhang L., Xie Y., Wang N., Tang R., Zheng W., Jiang X., Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core–shell nanocarrier. Adv. Sci. 4, 1700175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M., Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 31, 1902575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H.-X., Li M., Lee C. M., Chakraborty S., Kim H.-W., Bao G., Leong K. W., CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 117, 9874–9906 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Crudele J. M., Chamberlain J. S., Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 9, 3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L., Song L., Liu X., Yang X., Li X., He T., Wang N., Yang S., Yu C., Yin T., Wen Y., He Z., Wei X., Su W., Wu Q., Yao S., Gong C., Wei Y., Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 11, 95–111 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Li L., Yang Z., Zhu S., He L., Fan W., Tang W., Zou J., Shen Z., Zhang M., Tang L., Dai Y., Niu G., Hu S., Chen X., A rationally designed semiconducting polymer brush for NIR-II imaging-guided light-triggered remote control of CRISPR/Cas9 genome editing. Adv. Mater. 31, 1901187 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Dominguez A. A., Lim W. A., Qi L. S., Beyond editing: Repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platt R. J., Chen S., Zhou Y., Yim M. J., Swiech L., Kempton H. R., Dahlman J. E., Parnas O., Eisenhaure T. M., Jovanovic M., Graham D. B., Jhunjhunwala S., Heidenreich M., Xavier R. J., Langer R., Anderson D. G., Hacohen N., Regev A., Feng G., Sharp P. A., Zhang F., CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A. M., Schubert M. S., Friedmann-Morvinski D., Cohen Z. R., Behlke M. A., Lieberman J., Peer D., CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 6, eabc9450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F., In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng Y., Shen W., Zhang J., Yang B., Liu Y.-N., Qi H., Yu X., Lu S.-Y., Chen Y., Xu Y.-Z., Li Y., Gage F. H., Mi S., Yao J., CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci. 21, 447–454 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Wang H.-X., Song Z., Lao Y.-H., Xu X., Gong J., Cheng D., Chakraborty S., Park J. S., Li M., Huang D., Yin L., Cheng J., Leong K. W., Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. U.S.A. 115, 4903–4908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Loo J. C. M., Wright J. F., Progress and challenges in viral vector manufacturing. Hum. Mol. Genet. 25, R42–R52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin H., Song C.-Q., Dorkin J. R., Zhu L. J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A., Gupta A., Bolukbasi M. F., Walsh S., Bogorad R. L., Gao G., Weng Z., Dong Y., Koteliansky V., Wolfe S. A., Langer R., Xue W., Anderson D. G., Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L., Hu S., Chen X., Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 171, 207–218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller J. B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S. S., Zhu H., Siegwart D. J., Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Edit. 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang A. C., Stevens M. Y., Chen M. B., Lee D. P., Stähli D., Gate D., Contrepois K., Chen W., Iram T., Zhang L., Vest R. T., Chaney A., Lehallier B., Olsson N., du Bois H., Hsieh R., Cropper H. C., Berdnik D., Li L., Wang E. Y., Traber G. M., Bertozzi C. R., Luo J., Snyder M. P., Elias J. E., Quake S. R., James M. L., Wyss-Coray T., Physiological blood–brain transport is impaired with age by a shift in transcytosis. Nature 583, 425–430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patra J. K., Das G., Fraceto L. F., Campos E. V. R., del Pilar Rodriguez-Torres M., Acosta-Torres L. S., Diaz-Torres L. A., Grillo R., Swamy M. K., Sharma S., Habtemariam S., Shin H.-S., Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 16, 71 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banks W. A., From blood–brain barrier to blood–brain interface: New opportunities for CNS drug delivery. Nat. Rev. Drug Discov. 15, 275–292 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Pandit R., Chen L., Götz J., The blood-brain barrier: Physiology and strategies for drug delivery. Adv. Drug Deliv. Rev. 165-166, 1–14 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Kim S., Kim D., Cho S. W., Kim J., Kim J.-S., Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mai W. X., Gosa L., Daniels V. W., Ta L., Tsang J. E., Higgins B., Gilmore W. B., Bayley N. A., Harati M. D., Lee J. T., Yong W. H., Kornblum H. I., Bensinger S. J., Mischel P. S., Rao P. N., Clark P. M., Cloughesy T. F., Letai A., Nathanson D. A., Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat. Med. 23, 1342–1357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riquelme S. A., Lozano C., Moustafa A. M., Liimatta K., Tomlinson K. L., Britto C., Khanal S., Gill S. K., Narechania A., Azcona-Gutiérrez J. M., DiMango E., Saénz Y., Planet P., Prince A., CFTR-PTEN–dependent mitochondrial metabolic dysfunction promotes Pseudomonas aeruginosa airway infection. Sci. Transl. Med. 11, eaav4634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2
Figs. S1 to S31