Abstract
Much of what is known and theorized concerning passive sampling techniques has been developed considering chemical analytes. Yet, historically, biological analytes, such as Salmonella typhi, have been collected from wastewater via passive sampling with Moore swabs. In response to the COVID-19 pandemic, passive sampling is re-emerging as a promising technique to monitor SARS-CoV-2 RNA in wastewater. Method comparisons and disease surveillance using composite, grab, and passive sampling for SARS-CoV-2 RNA detection have found passive sampling with a variety of materials routinely produced qualitative results superior to grab samples and useful for sub-sewershed surveillance of COVID-19. Among individual studies, SARS-CoV-2 RNA concentrations derived from passive samplers demonstrated heterogeneous correlation with concentrations from paired composite samples ranging from weak (R2 = 0.27, 0.31) to moderate (R2 = 0.59) to strong (R2 = 0.76). Among passive sampler materials, electronegative membranes have shown great promise with linear uptake of SARS-CoV-2 RNA observed for exposure durations of 24 to 48 h and in several cases RNA positivity on par with composite samples. Continuing development of passive sampling methods for the surveillance of infectious diseases via diverse forms of fecal waste should focus on optimizing sampler materials for the efficient uptake and recovery of biological analytes, kit-free extraction, and resource-efficient testing methods capable of rapidly producing qualitative or quantitative data. With such refinements passive sampling could prove to be a fundamental tool for scaling wastewater surveillance of infectious disease, especially among the 1.8 billion persons living in low-resource settings served by non-traditional wastewater collection infrastructure.
Keywords: COVID-19, SARS-CoV-2, Wastewater-based epidemiology, Passive sampling, Moore swab, Environmental surveillance, Wastewater surveillance
Graphical abstract
1. Wastewater surveillance of COVID-19
People infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shed the virus and its genetic material through their bodily excreta eventually ending up in wastewater (Crank et al., 2022). Therefore, the surveillance of wastewater at municipal wastewater treatment plants (WWTPs) has been instrumental to track coronavirus disease 2019 (COVID-19) dynamics at population levels (Graham et al., 2021; Feng et al., 2021; Peccia et al., 2020; Wu et al., 2020). Wastewater surveillance, also referred to as wastewater-based epidemiology (WBE), has been implemented at various scales in communities throughout the world (Sharara et al., 2021; Shah et al., 2022; Thompson et al., 2020; Naughton et al., 2021). One critical component of sensitive and accurate characterization of infectious disease status via WBE is the collection of a wastewater sample that is representative of the total wastewater volume produced by the community (Ahmed et al., 2022; George et al., 2022). Composite sampling, the pooling and mixing of serial subsamples collected from a wastewater stream into a single sample over a given time interval, is a frequently used approach to obtain a representative sample from WWTPs or other points in the wastewater collection system (Medema et al., 2020; Sherchan et al., 2020; Ahmed et al., 2020).
While wastewater samples can be composited manually, they are typically collected automatically using autosamplers. Autosamplers are expensive ($2300 to $7500 USD each) and can often require infrastructure modifications (i.e., housing, electrical power supply, insulation, conduits, access hole modifications, and security features) to accommodate the sampler and its associated accessories (Liu et al., 2022). Such expenses may be reasonable for wastewater surveillance at a single WWTP in well-resourced communities, but they greatly limit the application of wastewater surveillance at finer spatial scales (e.g., sub-sewershed or building-level) and in low-resource communities. For example, a large wastewater surveillance program at the University of California San Diego makes use of 70 Hach AS950 automatic samplers with a market-selling retail price (MSRP) of $4500 USD each and a total MSRP of $315,000 USD (Karthikeyan et al., 2021). Even with open-source in-house autosampler designs, such as implemented at the University of Colorado Boulder, autosamplers were still $800 USD each (Reeves et al., 2021). The large costs associated with best-practice sampling protocols and equipment could be prohibitively expensive for WBE efforts in low and middle-income countries (Naughton et al., 2021; Dzinamarira et al., 2022).
As an alternative to wastewater composite samples, some practitioners have used grab samples, discrete samples collected from the wastewater stream at a single location and time-point, to determine the presence of COVID-19 among populations (Vo et al., 2022; Brooks et al., 2021; Scott et al., 2021; Crowe et al., 2021). The reported success of wastewater surveillance in identifying COVID-19 cases at the building-level on university campuses highlights the usefulness of sub-sewershed sampling for directed public health response (Betancourt et al., 2021). Nonetheless, the collection of discrete small-volume samples at a single time-point increases the risk of erroneous results, especially as the number of individuals shedding SARS-CoV-2 RNA among a given population decreases (George et al., 2022). Thus, there is need for a robust and cost-efficient sampling approach capable of producing representative wastewater samples across a variety of contexts without the large expense of autosampling.
While interest in passive sampling of wastewater has been renewed during the COVID-19 pandemic, it is not an entirely novel approach. Herein, we briefly review the history of wastewater passive sampling, recent applications during the COVID-19 pandemic, including comparisons between passive, grab, and composite sampling, along with the potential benefits and constraints of passive sampling workflows. Finally, we propose a research agenda for continued refinement of passive sampling methodology and discuss the pressing need to scale wastewater surveillance to the sub-sewershed level in low-resource settings which are frequently not served by traditional wastewater infrastructure.
2. Passive sampling basics
Passive sampling is premised on the spontaneous exchange of an analyte between the bulk medium being sampled (i.e., wastewater) and a collecting medium (i.e., the passive sampler) (Salim and Górecki, 2019; Górecki and Namieśnik, 2002). This exchange is mediated by immersing the sampler within the sampled medium, hence the term “passive” sampling (without active response or resistance) although it has also been referred to as “trap” sampling (Matrajt et al., 2018). Because passive samplers are in continuous contact with the medium being sampled, they may collect analytes or molecules missed by discrete grab or composite sampling events (Habtewold et al., 2022). Passive samplers can be designed with a single-material as the sorption medium or with two materials: one as a rate-limiting barrier enclosing a second sorption medium (Salim and Górecki, 2019). In some cases, organisms such as plants, fish, and mussels have been used as passive samplers (Górecki and Namieśnik, 2002; Vrana et al., 2005). The earliest applications of passive sampling targeted chemicals in air, such as atmospheric ozone in 1873 and carbon monoxide in 1927 (Fox, 1873; Gordon and Lowe, 1927). Passive sampling has also been used extensively to monitor organic and inorganic chemical pollutants in various environmental compartments including air, water, wastewater, soil, groundwater, and pore water (Zabiegała et al., 2010; Ahrens et al., 2016; Petty et al., 2000). For this reason, much of the theoretical foundation for passive sampling has been derived and characterized using chemical pollutants (Salim and Górecki, 2019; Górecki and Namieśnik, 2002; Vrana et al., 2005). While microorganisms can be conceptualized as biocolloids, the development of theoretical frameworks for passive sampling of microorganisms remains limited (Tenney and Verhoff, 1973). Nonetheless, passive sampling has a rich history in the environmental surveillance of infectious disease.
3. History of passive sampling for infectious disease
Using passive samplers for disease surveillance via wastewater dates to the earliest days of bacteriology. Following the invention of selective media to isolate Salmonella typhi (“B. typhosus” or “S. typhi”) from wastewater, grab sampling of wastewater was used to assess typhoid fever epidemiology (Wilson, 1928; Gray, 1929; Wilson, 1933; Wilson, 1938). Many of these early efforts were focused on linking downstream cases of typhoid fever with upstream sources of fecal contamination. In the first account of passive sampling for S. typhi, Moore reported that the most successful sampling approach was a “strip of gauze, 4 feet by 6 inches in size, folded into 8 thicknesses, secured with string and immersed in the sewer for 48 hours” (Moore, 1948). This approach, called a “Moore swab”, allowed for more reliable detection by continuous sampling of sewage over two days rather than sporadic “catch sampling” (Kelly et al., 1955; Moore, 1950).
In the years following, Moore swabs were widely used to trace typhoid fever outbreaks back to the source of fecal contamination (Shinohara et al., 1981; Moore et al., 1952; Harvey and Phillips, 1955; Lendon and Mackenzie, 1951). Such swabs were even deployed directly into residential water closets to locate individual S. typhi carriers (Kwantes and Speedy, 1955). In the years since their invention, Moore swabs have been used to sample water and wastewater for Coxsackie viruses, Vibrio cholera, Escherichia coli O157:H7, norovirus, and poliovirus in sewage and environmental waters (Kelly et al., 1955; Isaäcson et al., 1974; Barrett et al., 1980; Sattar and Westwood, 1977; Pazzaglia et al., 1993; Sbodio et al., 2013; Cassemiro et al., 2016; Tian et al., 2018; Matrajt et al., 2018). More recently, Moore swabs have been suggested as a critical method to scale wastewater surveillance in response to the emergence of antibiotic-resistant S. typhi strains (Sikorski and Levine, 2020). Materials besides cotton gauze have also been tested for their effectiveness as passive samplers including glass beads for poliovirus from wastewater and a variety of polymers for norovirus from environmental waters (World Health Organization, 2003; Vincent-Hubert et al., 2017). However, passive samplers made from readily available materials, such as the Moore swab, remain attractive for their simplicity, availability, and affordability, especially in low-resource settings (Amereh et al., 2021).
4. SARS-CoV-2 wastewater passive sampling literature
The primary focus of this review is the application of passive sampling for wastewater surveillance of SARS-CoV-2 RNA and/or COVID-19, particularly studies reporting empirical observations. To identify relevant articles, we searched PubMed and Google Scholar using combinations of “SARS-CoV-2”, “passive sampler”, “Moore swab”, “swab”, “tampon”, and “wastewater” or “sewage” as summarized in Table S1. We identified 152 publications through the search, and after title and abstract screening 19 were categorized as relevant. We removed 3 preprints from the set after identifying their peer-reviewed version among the set yielding a total of 16 unique publications. We screened the text of these publications and removed an additional 5 publications that made mention of passive sampling but did not report empirical observations. Thus, the final set of manuscripts that reported primary datasets of SARS-CoV-2 wastewater surveillance via passive sampling totaled 11 publications as summarized in Table 1 .
Table 1.
Wastewater surveillance publications reporting the use of passive sampling for SARS-CoV-2 RNA or COVID-19.
| Setting | Location | Population | Quantitative or qualitative | Quantitative unit | Swab deployment frequency | Material | Swabs per deployment | Housing type | Exposure time | Portion used for analysis | Concentration method | Analytical platform | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Municipal wastewater system | Melbourne, Victoria, Australia | 260 to 2.6 M | Quantitative | GC/unit sample material | NR | Cotton gauze; electronegative membrane; cotton buds; | 3 replicates per material | Colander; boat; box; torpedo | 3 to 24 h | Direct extraction (cotton bud, electronegative filter); eluate (gauze) | Electronegative filter (gauze eluate) | RT-qPCR | Schang et al., 2021 |
| Office complex, university | Halifax, Nova Scotia, Canada | Unknown | Quantitative | GC/swab eluate | 15 events over 5 months | Cotton gauze; cheesecloth; cellulose sponge; electronegative membrane | 1 replicate per material | COSCa | 24 h 48 h 72 h |
Eluate | NA | RT-qPCR | Hayes et al., 2021 |
| Municipal | Guelph, Ontario, Canada | NR | Quantitative | GC/sampler | 3 events over 1 month | Cotton gauze; cotton bud; electronegative membrane | 6 replicates per material | Torpedo | 4 h 8 h 24 h 48 h 72 h 96 h |
Direct extraction (cotton bud, electronegative filter); eluate (gauze) | Electronegative filter (gauze eluate) | RT-qPCR | Habtewold et al., 2022 |
| University | Windsor-Essex, Ontario, Canada | ~200 | Quantitative | PMMoV-normalized | 3 days per week to daily | Tampon | 2 | None | 20 h | Swab sorbate | Ultrafiltration via CP Select | RT-qPCR | Corchis-Scott et al., 2021 |
| University | South Bend, Indiana, USA | 1627 | Qualitative | NA | 1 day per week | Tampon | 1 | None | 3 h | Swab sorbate + eluate solids fraction | NA | RT-LAMP | Bivins et al., 2021 |
| Municipal | Tehran, Iran | NR | Qualitative | NA | 2 events over 5 months | Cotton gauze | 1 | Stainless steel wire cage | 16 h | Swab sorbate + eluate liquid fraction | PEG precipitation | RT-qPCR | Rafiee et al., 2021 |
| University | Atlanta, Georgia, USA | NR | Qualitative | NA | 1 event per week | Cotton gauze | 1 | None | 24 to 72 h | Swab sorbate + eluate | Skimmed milk flocculation; PEG | RT-qPCR | Liu et al., 2022 |
| University | Atlanta, Georgia, USA | 91 to 600 | Qualitative | NA | 1 event per week | Cotton gauze | 1 | None | 24 to 72 h | Swab sorbate + eluate | Skimmed milk flocculation; PEG | RT-qPCR | Wang et al., 2022 |
| Olympic village | Tokyo, Japan | NR | Qualitative | NR | NR | NR | NR | NR | NR | NR | NR | RT-qPCR | Kitajima et al., 2022 |
| Bench scale; Residential area; Office complex; University | Halifax, Nova Scotia, Canada | Unknown | Quantitative | GC/swab eluate | 23 events over 15 weeks | Electronegative membrane | 1 | COSCa | 24 to 72 h | Eluate | NA | RT-qPCR | Hayes et al., 2022 |
| Municipal | Queensland, Australia | 13,000 to 231,000 | Quantitative | GC/sampler | 2 events | Electronegative membrane; cotton buds; cotton gauze; tampon | 1 | Hair roller (tampon); torpedo (others) | Up to 48 h | Sorbate (tampon); eluate (cotton gauze); direct extraction (membrane, cotton buds) | Ultrafiltration (tampon); electronegative filter (gauze eluate) | RT-qPCR | Li et al., 2022 |
PMMoV = pepper mild mottle virus.
PEG = polyethylene glycol.
Four publications reported paired data derived from passive samplers of various materials and more traditional grab or composite samples. Four publications reported on the use of passive samplers for building-level wastewater surveillance of COVID-19 in university residence halls and the Olympic Village in Tokyo, Japan. One study reported both a method comparison and COVID-19 surveillance with passive sampling of wastewater on a university campus. Lastly, two studies reported empirical observations of passive sampler kinetics and calibration for microbiological analytes from wastewater. Herein, we synthesize the observations from these studies into the following topical considerations: (i) passive sampling performance versus traditional sampling methods (composite and grab) for SARS-CoV-2 RNA in wastewater; (ii) passive sampling performance for COVID-19 surveillance; (iii) the theoretical and mechanical basis for passive sampling of microorganisms such as SARS-CoV-2 from wastewater; (iv) prospects of passive sampling to produce quantitative data; and (v) fabrication, deployment, and processing of passive samplers. We conclude with recommendations for a future research agenda to further improve passive sampling for microbiological analytes from wastewater and an outlook on the role of passive sampling to scale equitable application of wastewater surveillance.
5. SARS-CoV-2 wastewater passive sampling method comparisons
In the first comparison of composite, grab, and passive sampling across sewersheds of various populations and flows, Schang et al. assessed the performance of passive samplers constructed with electronegative membranes, medical gauze, and cotton buds (“Q-tips”) for the detection of SARS-CoV-2 RNA by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) (Schang et al., 2021). Overall, SARS-CoV-2 RNA was detected in 50% of grab and composite samples (92/183) and 31% of all passive samples (76/245), with electronegative membranes demonstrating the highest positive proportion (41%) among passive sampling materials, followed by gauze (31%) and cotton buds (25%) (Schang et al., 2021). When SARS-CoV-2 RNA concentrations were below 1.8 gene copies (GC)/mL of wastewater, passive samplers detected SARS-CoV-2 RNA more often than grab or composite sampling. In addition, there was a significant correlation between SARS-CoV-2 RNA concentrations on passive samplers and concentrations in the wastewater as measured by composite sampling (Schang et al., 2021).
In a second study, Habtewold et al. tested the same three passive sampler materials alongside traditional sampling methods for various exposure durations (4 to 96 h) to influent at a WWTP (Habtewold et al., 2022). They also reported superior performance of electronegative membranes with linear uptake of SARS-CoV-2 RNA between 4 and 48 h of exposure and a positive proportion of 79.6%, as high as traditional sampling methods, followed by cotton gauze pads (77.8%) (Habtewold et al., 2022) Cotton buds produced the lowest positivity rate (59%) among the three passive sampler materials (Habtewold et al., 2022). The authors theorized that this was because of a comparably smaller surface volume of cotton buds compared to the other materials. The proportion of samples positive increased with exposure duration for all three materials, and at 24 h exposures the proportion of gauze and membranes positive for SARS-CoV-2 RNA exceeded that of composite samples (Habtewold et al., 2022). These results along with those of Schang et al. (2021) suggest passive samplers constructed of certain materials may yield positivity similar to composite sampling for wastewater exposure durations of 24 h.
During passive and grab sampling at 17 maintenance access holes in Tehran, Iran, Rafiee et al. found the positivity of Moore swabs exposed to wastewater for 16 h achieved positivity equal to that of manually prepared composite samples (100%) from the same period. Moore swabs greatly outperformed grab sampling which demonstrated only 47% positivity over the same interval (Rafiee et al., 2021).
Unlike the three previous studies, Hayes et al. performed both bench-scale and in situ experiments to assess the performance of passive sampling compared to grab sampling. The authors tested passive samplers made of four different materials (cotton gauze, cheesecloth, cellulose sponges, and electronegative membranes) (Hayes et al., 2021). Among these materials, cheesecloth eluted with Tween 20 buffer recovered the greatest mean concentration of SARS-CoV-2 RNA during bench-scale experiments followed closely by electronegative filters eluted with Tween 20 buffer (Hayes et al., 2021). During in situ experiments, passive samplers were deployed within a novel 3D printed housing called the COVID-19 Sewer Cage (COSCa) (Hayes et al., 2021). During these experiments, cheesecloth, with 46.6% positivity, and electronegative filters, with 40.0% positivity, outperformed grab sampling for detecting SARS-CoV-2 RNA in wastewater (Hayes et al., 2021).
Across the four method comparisons, passive sampling with a variety of materials including cotton buds, cotton gauze, cheese cloth, and electronegative filters consistently outperformed grab sampling for detecting SARS-CoV-2 RNA in wastewater. In several instances, passive sampling with selected materials such as electronegative filters, cheese cloth, and cotton gauze was able to achieve RNA detection comparable to composite samples, although this was not observed across all studies. Electronegative membranes performed particularly well for passive sampling with promising linear uptake of SARS-CoV-2 RNA observed for exposure duration up to 48 h. While the strength of these results is limited by the small number of studies and the limited sample size, they are suggestive and should motivate efforts to replicate the findings. One fundamental limitation of the studies described above is their analyses consider SARS-CoV-2 RNA detection alone without consideration of COVID-19 incidence or prevalence among the relevant community. The prevalence of COVID-19 among the communities during two of the studies was 0.34 to 3.4 cases per 100,000 and 2 to 17 cases per 100,000 (Schang et al., 2021; Habtewold et al., 2022). The successful detection of SARS-CoV-2 RNA in wastewater by passive sampling with COVID-19 at such low prevalence suggests the approach could be sensitive for disease surveillance.
6. Wastewater passive sampling for COVID-19 surveillance
Several studies have leveraged passive sampling of wastewater for COVID-19 surveillance at the building-level. Liu et al. used Moore swabs made of cotton gauze to produce 442 wastewater samples from residential buildings, a hospital, and an isolation facility at Emory University, Georgia, USA. Over the study period the proportion of weekly positive Moore swabs ranged from 16.8 to 42.1% for wastewater effluent from residence halls with individual COVID-19 cases identified by follow up clinical testing in response to positive wastewater results (Liu et al., 2022). Paired grab and passive samples of hospital wastewater indicated that Moore swabs were more sensitive (24/26 positive; 92%) for the detection of SARS-CoV-2 RNA and COVID-19 cases than grab samples (18/26 positive; 69%) (Liu et al., 2022). Wastewater passive sampling results from the isolation facility indicated Moore swab positivity was 71.4% when there were one to four COVID-19 cases present (Liu et al., 2022). Despite these promising results, passive sampling yielded two false negative and two false positive results from the same isolation facility (Liu et al., 2022). However, even after accounting for these errors, accuracy in detecting COVID-19 cases in quarantine was 83% (Liu et al., 2022). During a second study using passive sampling for wastewater surveillance at the same university, the authors reported that weekly passive sampling was not sensitive (6/63, 9.5%) for the detection of single COVID-19 cases within 25 residential dormitories (Wang et al., 2022). This finding contradicts the observations from the isolation facility and suggests that there may be variation in performance from site to site. Wang et al. (2022) still found that passive sampling had better sensitivity than grab sampling and wastewater surveillance results from passive sampling identified a COVID-19 surge two weeks prior to clinical sampling.
Corchis-Scott et al., (2021) also reported superior performance of passive sampling compared to grab sampling to detect COVID-19 among residents of a university residence hall in Ontario, Canada. Over a period of seven weeks, thrice-weekly grab samples of wastewater from the hall were consistently negative for SARS-CoV-2 RNA; however, after deploying passive samplers made of tampon swabs, SARS-CoV-2 RNA was detected within two days (Corchis-Scott et al., 2021). When the RNA signal measured by passive sampling increased over four sampling days, clinical testing was triggered and a single infection was identified among 195 people (Corchis-Scott et al., 2021). In follow-up testing of close contacts, a second case was identified and subsequent testing identified the Alpha (B.1.1.7) variant of concern (VOC) in both wastewater and clinical samples (Corchis-Scott et al., 2021). Similar to the findings of Liu et al. (2022), these results suggest that passive sampling can be sensitive enough to detect single COVID-19 cases at the building level.
A fourth study reported the use of passive sampling with tampon swabs to monitor wastewater from nine university residence halls over six weeks at the University of Notre Dame, Indiana, USA (Bivins et al., 2021). The 1627 students residing in these halls were subject to weekly clinical testing for COVID-19 associated with the university's COVID-19 response. Unlike all other studies, Bivins et al. applied reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay to test wastewater for SARS-CoV-2 RNA. A comparison between the 143,884 clinical samples and 53 wastewater samples over the study period found that wastewater surveillance by passive sampling achieved a 75% three-day positive predictive value for incident COVID-19 cases and a same-day negative predictive value of 80%, despite RT-LAMP's decreased sensitivity compared to RT droplet digital PCR (Bivins et al., 2021).
A brief report of wastewater surveillance performed at the Olympic Village during the 2020 Olympics in Tokyo, Japan (which occurred in 2021) noted that passive sampling greatly increased the specificity of wastewater surveillance for incident COVID-19 cases compared to grab sampling but did not provide methodological details or empirical data (Kitajima et al., 2022). Finally, a publication evaluated the hypothetical application of passive samplers for wastewater surveillance during an outbreak of COVID-19 in Nanjing, China, although the study did not produce empirical data (Jiang et al., 2022). The results across the five studies reporting real-world implementation for disease surveillance suggest passive sampling can be a sensitive method for wastewater discharged from buildings. In some instances, passive sampling was sufficient to detect a single COVID-19 case among building populations up to 200 people. Despite these promising results, in three of five studies there were occasionally discordant results between wastewater testing results and the presence of COVID-19 cases in the relevant building (i.e., false positives and false negatives). However, such errors are possible even when using “best practice” wastewater surveillance methods including autosamplers, high-throughput workflows, and RT-qPCR (Karthikeyan et al., 2021). Despite these limitations, the collective experience indicates that well-designed passive surveillance programs could greatly inform the expenditure of clinical testing resources to manage COVID-19. However, four of the five studies included in this review were performed on university campuses. Replication efforts should focus on the use of passive surveillance for wastewater from other facilities and settings.
7. Passive sampling mechanisms and theory
The method comparisons and disease surveillance studies examined above establish a proof-of-concept for passive sampling of wastewater for SARS-CoV-2 surveillance. The results also emphasize the need for optimized methods to maximize the utility of the approach and further adapt it to other microbiological analytes. Such optimization will require an understanding and characterization of the fundamental mechanisms driving passive sampling of such analytes from wastewater.
Passive sampling is premised on the spontaneous enrichment of analyte molecules from the medium being sampled, in this case wastewater, to the receiving phase, the passive sampler, resulting from differences in chemical potential between the two (Górecki and Namieśnik, 2002; Zabiegała et al., 2010). Such samplers are considered “passive” because they are suspended within a medium to sorb the analyte of interest. Much of passive sampling theory has been derived with attention toward organic and inorganic chemical pollutants in air and water (Salim and Górecki, 2019; Vrana et al., 2005). The net flow of analyte from the sampled medium to the passive sampler continues until the sampler is removed from the sampled medium or equilibrium is reached. Thus passive sampling is conceptualized in two modes of operation: non-equilibrium and equilibrium (Górecki and Namieśnik, 2002; Salim and Górecki, 2019).
During non-equilibrium operation, the analyte accumulates in the passive sampler at a rate frequently modeled using first-order kinetics (Zabiegała et al., 2010; Salim and Górecki, 2019). Because the passive sampler is removed from the sampled medium prior to its saturation with the analyte, a time-weighted average of the analyte concentration in the sampled medium can be estimated using a calibrated kinetic model (Zabiegała et al., 2010; Vrana et al., 2005). Applying the linear uptake model depends on two key assumptions. First, the adsorption of the analyte to the passive sampler is assumed to be irreversible, referred to as the zero-sink assumption (Górecki and Namieśnik, 2002). This assumption can be problematic when the passive sampler relies on adsorption rather than absorption to uptake the analyte of interest and when sampling from complex mediums where the adsorption capacity could be exceeded (Górecki and Namieśnik, 2002). Second, the uptake rate is assumed to be constant throughout the exposure period, which could be dubious when the medium being sampled is heterogeneous (Zabiegała et al., 2010). To calibrate the kinetic model, the uptake rate for a particular passive sampler and sampled medium combination must be determined empirically (Salim and Górecki, 2019). After calibration, the appropriate exposure duration can be determined based on the uptake rate and the sorption capacity of the passive sampler (Zabiegała et al., 2010).
In the event the passive sampler is exposed for time periods exceeding the linear uptake duration, an equilibrium mode of operation must be used. When in equilibrium, the concentration of the analyte in the passive sampler can be used to estimate the sampled medium concentration via an empirically-determined partitioning coefficient (Vrana et al., 2005; Salim and Górecki, 2019). An important consideration for the equilibrium mode of operation is the equilibration time in relation to the temporal variation in the medium being sampled (Zabiegała et al., 2010). Rapid changes in the medium being sampled relative to the time required to reach equilibrium could lead to bias in the estimated concentrations. When passive samplers are used under equilibrium conditions, a time-weighted average cannot be calculated and instead the estimated analyte concentrations are comparable to a grab sample (Zabiegała et al., 2010).
Optimization of passive sampling from any medium must include considerations of the sampler fabrication and material, the partitioning behavior of the analyte between the sampled medium and the passive sampler, the concentration of the analyte in the sampled medium, and the composition of the sampled medium (Zabiegała et al., 2010). Passive sampling for wastewater surveillance of SARS-CoV-2 entails many challenges to the fundamental assumptions underlying both equilibrium and non-equilibrium theory. Wastewater is a complex mixture of chemical and biological constituents whose characteristics vary substantially. Wastewater flows in collection systems are often characterized by pulse inputs, heterogeneous mixtures, and large physicochemical variation. Such large variations in the sampled medium may invalidate constant uptake and zero-sink assumptions. Wastewater also contains many different suspended solids and biocolloids such that sorption of the analyte is likely to be competitive with other constituents and may saturate the sampler. The ability of passive samplers to capture and retain solids may enhance viral detection for enveloped viruses such as SARS-CoV-2. Enveloped viruses, such as HCoV and Phi 6 preferentially adsorbed to wastewater solids because of their hydrophobic viral envelope (Gundy et al., 2009; Yang et al., 2022). The hydrophobicity of the lipid surface of SARS-CoV-2 may also enhance recovery of intact virus present in aqueous wastewater onto hydrophobic samplers. However, if much of the viral RNA in wastewater is not intact, hydrophobic interactions may not be important. These complexities suggest calibration of passive samplers for wastewater surveillance will require in situ experiments with various combinations of passive sampler materials and configurations, wastewater composition, and sampling location.
The previously described studies provide preliminary data about passive sampling for wastewater surveillance of SARS-CoV-2. As might be expected, passive sampler positivity for SARS-CoV-2 RNA varied by material. During two different studies, electronegative membranes followed by cotton gauze demonstrated the highest positivity for SARS-CoV-2 RNA following exposure to wastewater (Schang et al., 2021; Habtewold et al., 2022). Habetwold et al. (2021) observed a linear accumulation of SARS-CoV-2 and total RNA on electronegative membranes exposed up to 48 h indicating linear uptake from wastewater. Passive sampling with gauze also demonstrated accumulation, but over a much shorter duration (Habtewold et al., 2022). During bench-scale experiments with distilled water and wastewater seeded with SARS-CoV-2, cheesecloth and electronegative membranes demonstrated the greatest accumulation of SARS-CoV-2 RNA whereas cellulose sponges performed poorly (Hayes et al., 2021). Another bench scale experiment performed using distilled water seeded with SARS-CoV-2 found that cotton gauze samplers were all positive for SARS-CoV-2 RNA when seeded concentrations exceeded 50 GC/mL whereas only half were positive at 5 GC/mL, although the low sample size precluded statistical comparisons (Liu et al., 2022). These findings indicate, as expected from theoretical underpinnings, passive sampler material is likely a critical consideration for accumulating SARS-CoV-2 RNA from wastewater. In addition to sorption mechanisms, mechanical impacts could also form a relevant source of SARS-CoV-2 RNA on passive samplers in wastewater collection systems. Schang et al. (2021) reported significant “ragging” of some passive sampler housings while they were deployed in the wastewater collection system, and Bivins et al. (2021) routinely encountered tampon swabs fouled with toilet paper upon retrieval.
Two publications investigated uptake kinetics and equilibrium of passive samplers for SARS-CoV-2 RNA from wastewater in more detail. The first by Hayes et al. (2022) found that the “adsorption” of seeded SARS-CoV-2 RNA from wastewater to electronegative membranes with low and medium total suspended solids was best characterized by Freundlich isotherms with a maximum adsorption capacity of 3.8 log10 GC/cm2 and pseudo-first-order kinetics with a first order rate constant of 0.05/h. The pseudo first-order kinetic model suggests a linear uptake range from 0 to 10 h with no increase in the adsorbed RNA quantity beyond 24 h (Hayes et al., 2022). Hayes et al. (2022) also observed a non-linear relationship between the total suspended solids present in the passive sampler eluate and the estimated maximum adsorption capacity, suggesting an optimal concentration of suspended solids to balance between increased adsorption and decreased recovery. Previous batch experiments have observed first-order partitioning kinetics between bacteriophages and sand and clay particles although these systems were greatly simplified compared to wastewater suspended solids (Syngouna and Chrysikopoulos, 2010; Grant et al., 1993). Since SARS-CoV-2 RNA has been observed to partition favorably to suspended solids, optimized solids retention and recovery are likely to be critical for passive sampling methods (Kostoglou et al., 2022; Kim et al., 2021; Hayes et al., 2022).
The second study, by Li et al. (2022), deployed passive samplers of various materials into wastewater collection systems for durations up to 48 h and assessed the uptake of pepper mild mottle virus (PMMoV), human adenovirus (HAdV 40/41), and enterovirus. Similar to the previously mentioned studies, they found a greater proportion of electronegative membranes (14/17; 82%) than tampon swabs (8/17; 47%) were positive for SARS-CoV-2 RNA; however, the low concentrations of SARS-CoV-2 RNA in the wastewater precluded a kinetic analysis (Li et al., 2022). For PMMoV, HAdV 40/41, and enterovirus, electronegative membranes demonstrated linear accumulation for 48 h of exposure with estimated sampling rates of 1 mL/h, 33.1 mL/h, and 0.3 mL/h, respectively (Li et al., 2022). Tampon swabs and cotton buds demonstrated non-linear uptake with saturation in 8 h or less whereas cotton gauze demonstrated rapid uptake followed by declines in viral analytes up to 48 h suggesting desorption or decreased recovery efficiency with increasing exposure duration (Li et al., 2022). Applying the linear uptake model derived for electronegative membranes to published SARS-CoV-2 RNA datasets indicated a sampling rate of 0.1 mL/h to 4.2 mL/h for SARS-CoV-2 in wastewater (Schang et al., 2021; Habtewold et al., 2022; Li et al., 2022).
These two studies highlight several important implications of passive sampling theory for SARS-CoV-2 and wastewater. Theory suggests that passive sampling performance will depend on the sampler material, the characteristics of the sampled medium, and the analyte. Both studies found that electronegative membranes demonstrated linear uptake of SARS-CoV-2 RNA from wastewater with linear range from 10 to 48 h of exposure. Cotton-based samplers demonstrated faster times to saturation and would therefore likely be more suited to exposure durations of 8 h or less. In the only study examining different materials, cotton gauze appeared to violate the zero-sink assumption with decreases in analyte concentration with increasing exposure duration (Li et al., 2022). For electronegative membranes, the concentration of total suspended solids in the sampled wastewater affected the sorption of SARS-CoV-2 RNA to the passive sampling material with high levels of suspended solids decreasing the observed RNA concentration. Importantly, the study designs could not distinguish between decreased sorption or decreased recovery during sampler processing, although the net effects would be similar. Other wastewater parameters such as temperature, pH, dissolved solids concentration and characteristics, will likely affect passive sampler performance in wastewater. Lastly, the characteristics of the biological analyte itself affect passive sampler performance since sampling rates varied between rod-shaped and icosahedral non-enveloped virus types suggesting differences in performance between virus morphologies as well (Li et al., 2022).
8. Passive sampling to generate quantitative SARS-CoV-2 RNA data
The uptake of SARS-CoV-2 and several other viruses from wastewater indicate passive samplers, especially electronegative membranes, may be capable of producing time-weighted average or semi-quantitative data. Despite the promising qualitative and kinetic results, studies considering the ability of passive samplers to produce accurate SARS-CoV-2 RNA quantitative data from wastewater are limited. Schang et al. (2021) found that SARS-CoV-2 RNA concentrations on passive samplers demonstrated a statistically significant correlation with concentrations in wastewater and suggested that passive samplers could yield semi-quantitative data, but replication is needed.
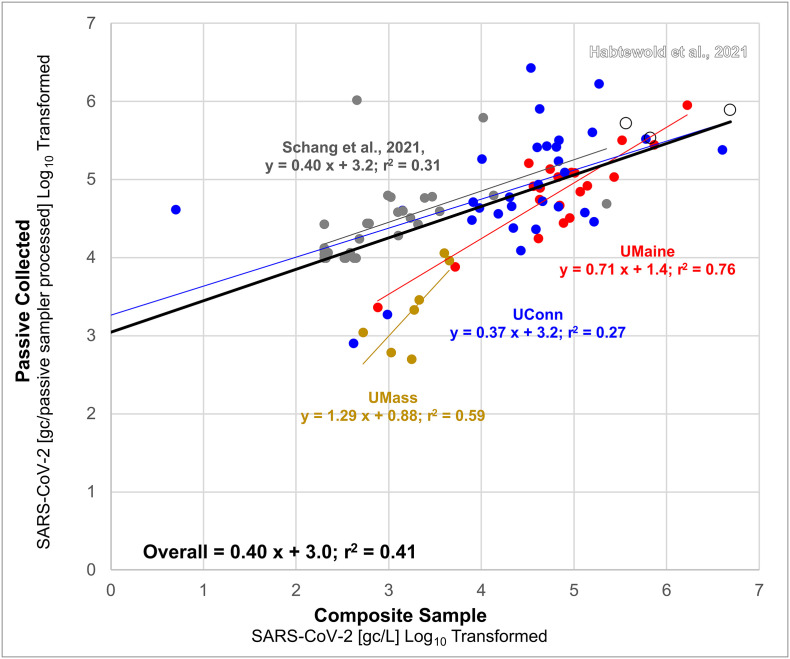
To assess the performance of passive sampling to produce quantitative data across five different settings, we considered SARS-CoV-2 RNA concentrations on various types of passive samplers as implemented on three university campuses in combination with datasets extracted from the work of Habtewold et al. (2022) and Schang et al. (2021). Universities have played a key role in piloting wastewater surveillance methods to manage COVID-19 among campus residents (Harris-Lovett et al., 2021). Wastewater surveillance programs at the University of Maine (UMaine), University of Massachusetts Amherst (UMass), and University of Connecticut (UConn) have used passive samplers to measure SARS-CoV-2 RNA in wastewater. The methodological details pertinent to each of these three programs are reported in the Supporting Information. From these five unique studies (UMaine n = 20; UMass n = 7; UConn n = 48; Schang et al. n = 33; Habtewold et al. n = 3) we compiled a dataset of paired measurements (n = 111) of SARS-CoV-2 RNA in wastewater (GC/L by composite sampling) and on passive samplers (GC/sampler), Fig. 1 . In the case of Habtewold et al. (2022), which compares three different passive sampler types to a single paired composite, we averaged across all three replicates of each passive sampler type to produce three paired samples. The concentration data was log transformed, with non-detects (n = 20) removed from the regression analyses. The non-detects reported in the datasets (not displayed on the graph) highlight a key consideration: a difference in the analytical sensitivity of passive samplers versus composite samplers. In the aggregated dataset, there were 17 instances in which passive samplers yielded non-detection while SARS-CoV-2 RNA was detected in the paired composite sample and three pairs consisting of passive sampler RNA detection and composite sample non-detection. These results suggest for qualitative data passive samples were 84% accurate in detecting SARS-CoV-2 RNA compared with composite samples.
Fig. 1.
Comparison of the reported SARS-CoV-2 concentrations in wastewater by composite sampling, genome copies per liter (GC/L), with paired passive samples, genome copies per passive sampler processed (GC/passive sampler processed) across five studies. Schang et al. (2021) (dark gray) and Habtewold et al. (2022) (open circles) data were extracted from the published supplemental. Habtewold et al. (2022) compares three different passive sampler types to a paired composite, therefore although nine points are represented, these represent three paired samples. The University of Massachusetts Amherst (UMass, gold), University of Maine (UMaine, red), and University of Connecticut (UConn, blue) datasets (unpublished) were provided by co-authors. Brief descriptions of these sampling campaigns are provided in the Supplemental Information. The concentrations were log transformed, and linear regressions were fit to each series individually (except for Habtewold et al., 2022) and to the aggregated data with the resulting fit and r2 shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When aggregating all five datasets, we observed a weak linear relationship between SARS-CoV-2 RNA concentrations from passive and composite samples (y = 0.40 x + 3.0, r2 = 0.41; Fig. 1). Since the passive sampling methods used by each sampling campaign were unique, we divided the analysis into individual campaigns to further examine the relationship across methodological approaches. The lowest r2 value for a linear model was observed with the UConn dataset (y = 0.37 x + 3.2; r2 = 0.27) while UMaine demonstrated the strongest linear fit (y = 0.71 x + 1.4; r2 = 0.76). The data from Schang et al. (y = 0.40 x + 3.2; r2 = 0.31) demonstrated a weak linear fit while the data from UMass demonstrated a moderate fit (y = 0.71 x + 1.4; r2 = 0.56). We did not fit a model to the data from Habtewold et al. due to the limited sample size. Overall, the results suggest a heterogeneous and weak quantitative relationship when passive sampling results are pooled across diverse settings. However, the UMaine and UMass data suggests that under some conditions passive sampling could yield semi-quantitative data, with consideration for the likely higher false negative rate. The heterogeneity of the results across studies is likely a function of the diverse methods currently used for wastewater surveillance via passive sampling.
9. Wastewater passive sampling methodologies
The first step in the process of any wastewater surveillance program is to define the relevant population, identify the available wastewater system access points, and formulate the desired public health response to the surveillance results. In most cases, passive sampling is being used to produce qualitative results at the building or sub-sewershed level such that COVID-19 clinical testing can be delivered in response to the wastewater testing results. Because such programs are often executed across multiple buildings and locations, it is important that the methodology is cost efficient and produces timely results for public health response. The passive samplers are typically deployed into the collection system via access holes for exposure durations from 4 to 96 h. Samplers are then collected, processed, and the resulting samples tested by molecular methods. There are many different types of sorbate removal procedures including mechanical squeezing, elution, or even direct extraction of the passive sampler itself. Recovered volumes of wastewater range from less than 1 mL up to 250 mL when combined with eluent. The resulting wastewater volume may be further concentrated or partitioned prior to extraction of the nucleic acids with a variety of commercial kits. Purified RNA is then tested for SARS-CoV-2 RNA presence and/or quantity via molecular methods.
9.1. Passive sampler sorbent materials and housings
A variety of sorbent materials have been used to construct passive samplers for wastewater surveillance. One of the most used materials is cotton gauze in the form of a Moore swab. The swab is made by cutting strips of readily available cotton medical gauze at roughly 120 cm long by 15 cm wide. The gauze strip is then folded multiple times and tied together in the middle with a nylon fishing line of sufficient length to submerge the sampler in wastewater at the bottom of an access hole (Rafiee et al., 2021; Liu et al., 2022). Another form of swab found in a conveniently available form is the tampon. Various brands of tampons have been tied to fishing line and deployed into sewers as passive samplers during COVID-19 surveillance efforts (Bivins et al., 2021; Li et al., 2022). Small cotton buds have also been deployed into wastewater using various containers; although, their performance was considered poor compared to cotton gauze (Habtewold et al., 2022; Schang et al., 2021). The performance of these cotton materials is similar to studies of fomite swab absorbance and release efficiencies where nylon or polyester swabs, designed to have an open structure, facilitate linear absorbance and release in contrast to cotton bud swabs which have fibers tightly wound around the shaft (Jansson et al., 2020). A single study found cheesecloth performed comparable to gauze whereas cellulose sponges performed poorly (Hayes et al., 2021). Electronegative membranes have demonstrated the highest positivity compared to composite sampling and linear uptake of SARS-CoV-2 RNA from wastewater over exposures lasting 24 to 48 h (Habtewold et al., 2022; Schang et al., 2021; Li et al., 2022; Habtewold et al., 2022). Cotton materials such as gauze have demonstrated rapid saturation and perhaps even loss of sorbed analyte with increasing exposure duration (Li et al., 2022).
In addition to sorption materials, a diverse array of housings has been used to protect passive samplers from debris while they are deployed in the wastewater collection system. Several teams have made use of custom 3D printed housings including shapes like colanders, boats, and boxes with the most popular shape being a torpedo, which demonstrated less “ragging” than other shapes (Schang et al., 2021; Habtewold et al., 2022; Li et al., 2022). One team invented a unique sphere with pore holes which they named the COSCa (Hayes et al., 2021). Most teams making use of Moore swabs or tampon swabs deployed them into the collection system without housing (Corchis-Scott et al., 2021; Bivins et al., 2021; Liu et al., 2022; Wang et al., 2022). One study made use of a stainless-steel wire cage for their Moore swabs while another used a plastic hair roller for a tampon swab (Rafiee et al., 2021; Li et al., 2022). No study has made a systematic comparison of SARS-CoV-2 RNA concentrations on passive sampling material with and without housing.
9.2. Sampler placement, exposure duration, and collection
Passive samplers are typically deployed into wastewater collection systems via maintenance access holes. The deployment locations are selected by examining the wastewater collection network design and selecting a downstream point from the facility of interest. These holes provide a convenient access point for lowering passive samplers tied to lengths of fishing line or cord of sufficient length to reach the manhole invert (Liu et al., 2022). These lines can be secured to the steps within the access hole or the access hole lid, but care must be taken as entering these spaces often constitutes confined space entry. To minimize the required ingress, two studies made use of fishing line tied to carabiners integrated with magnets (170 lb. swivel, WellZeer, Zhejiang P.R., China) that were then attached to the inside rim of the lid frame (Bivins et al., 2021; Corchis-Scott et al., 2021). Fishing line lengths can be made such that the passive sampler is washed just into the invert of the downstream pipe which may improve the passive sampler's contact with the passing wastewater volumes. If samplers are deployed directly into access hole inverts, the large diameter of the hole may allow wastewater to circumvent the sampler. The tensile strength of the fishing line or cord must be sufficient to prevent the passive sampler from breaking off, which has been reported in at least one case (Bivins et al., 2021). In sampling locations subject to high flow rate wastewater, the tensile strength of fishing line may not be sufficient and some teams have made use of materials such as Rexlace (Pepperell Brading Company, Pepperell, MA, USA), a sturdier plastic line, to deploy passive samplers. Passive samplers have been left deployed in wastewater systems for durations from 3 to 96 h with exposure durations of 24 to 48 h being most common. Samplers were collected from the access holes by lifting them out with the fishing line or string and depositing them into sterile bags or containers with proper aseptic and biosafety precautions. These containers are then transported on ice to laboratories for processing and analysis.
9.3. Passive sampler processing
Passive samplers have been processed with a wide variety of workflows and methodologies. In most cases Moore swabs have been processed via a combination of squeezing out the sorbate, with a potato ricer in one case, and then eluting the gauze with a variety of eluent solutions (Tween 80/Sodium polyphosphate/Antifoam Y-30, Tween 80, Tween 20) (Schang et al., 2021; Hayes et al., 2021; Habtewold et al., 2022; Liu et al. 2022; Wang et al., 2022; Rafiee et al., 2021). A single comparison of various eluents found that Tween 20 yielded higher SARS-CoV-2 RNA concentrations than a commercial lysis buffer and an acetonitrile-water solution (Hayes et al., 2021). The eluents from these gauze swabs have been handled in several different ways. Two studies used centrifugation to remove the solids fraction and then subjected the liquid fraction to polyethylene glycol precipitation and/or skim milk flocculation (Liu et al., 2022; Rafiee et al., 2021; Wang et al., 2022). In two other studies the eluate from the gauze swab was concentrated using electronegative membranes (Schang et al., 2021; Habtewold et al., 2022). In cases where electronegative membranes were used as passive samplers, nucleic acid was directly extracted from the membranes without intermediary elution or processing (Li et al., 2022; Schang et al., 2021; Habtewold et al., 2022; Hayes et al., 2022). Smaller passive samplers such as cotton buds were also subjected to direct nucleic acid extraction (Schang et al., 2021; Li et al., 2022). In studies that used tampons, sorbate was squeezed from the tampon using a large syringe followed by elution with Tween 20 solutions (Bivins et al., 2021; Corchis-Scott et al., 2021; Li et al., 2022). In one study the resulting tampon eluate was concentrated by ultrafiltration performed with a Concentrating Pipette (CP Select™) (Corchis-Scott et al., 2021). Whereas in another the solid fraction of the eluate was concentrated by centrifugation for further analysis while the liquid fraction was discarded (Bivins et al., 2021).
9.4. Molecular analysis of passive sampler-derived material
In every published study to date, nucleic acids were extracted from passive sampler-derived materials using commercial extraction kits. In most cases these kits have used silica columns for purification with satisfactory results (Schang et al., 2021; Habtewold et al., 2022; Corchis-Scott et al., 2021; Bivins et al., 2021; Liu et al., 2022; Wang et al., 2022; Li et al., 2022; Hayes et al., 2022). Two studies used a magnetic bead-based extraction, which also produced satisfactory results (Hayes et al., 2021; Hayes et al., 2022). Because these studies have made use of such kits, inhibition rates of passive sampler-derived extracts in most studies have been low and consistent with rates observed for RT-qPCR analysis of other wastewater samples. Bivins et al. (2021) attempted direct testing of materials without extraction and with heat extraction, but in all cases downstream RT-LAMP assays were inhibited by the resulting solution. In every case except one, purified nucleic acids were assayed for SARS-CoV-2 RNA by RT-qPCR using a variety of commercially available assays. A single study made use of a colorimetric RT-LAMP assay to produce qualitative results for SARS-CoV-2 RNA (Bivins et al., 2021).
10. Strengths and limitations of passive sampling for wastewater surveillance
Passive sampling for surveillance of wastewater affords some key advantages over other sampling methods. First, such methods are likely to be less expensive at scale, lacking any requirement for automated volumetric sampling for downstream molecular analysis, although concurrent measurement with other telemetry (e.g., flow rate) could offer added value (Schang et al., 2021; Bivins et al., 2021; Liu et al., 2022). Passive sampling may also reduce labor costs for installation and retrieval compared to longitudinal composite sampling. Once hardware is installed to affix passive samplers at a given site, little additional effort is needed to set up multiple samplers. A further advantage is that because passive samplers hang within collection pipes they are not dependent on persistent flow, provided that the water to be sampled does pass through the sampling point during the time the sampler is in place. This is especially important at small scales such as building-level surveillance where wastewater flow rates may be highly variable over time and therefore more difficult to collect via the serial sampling performed by autosamplers. In such scenarios the continuous exposure of the passive sampler may provide improved sensitivity.
The potential advantages of passive sampling approaches are accompanied by some important limitations. First, because the relationships between flow rate, target concentration in wastewater, and target loading of passive samplers remains poorly characterized, it is not currently possible to use passive sampling methods to estimate target concentrations reliably and consistently in wastewater. Semi-quantitative measurement may be possible via longitudinal sampling, but more work remains to establish sorption, desorption, and equilibria of targets on passive samplers as a function flow rate, target concentration in wastewater, target characteristics, sampler material, water chemistry, fouling, or additional variables affecting performance (Schang et al., 2021; Hayes et al., 2022; Li et al., 2022). These relationships may be context-specific (e.g., a function of concentrations for wastewater constituents), requiring experimental work of non-negligible complexity to establish empirical models for a given setting. These factors also contribute to the primary disadvantage of passive sampling methods. Because the capture efficiency of targets for many sorbent materials are currently uncharacterized, interpreting non-detects is further complicated. The absence of a biological analyte during analytical testing may reflect poor efficiency in sorbing that analyte rather than its absence from the wastewater. Further uncertainty stems from the observation that biological analyte sorption behavior may vary as a function of its morphology and physiology (Ye et al., 2016; Yang et al., 2022). Whereas a positive result does allow for an interpretation that the target was present in the wastewater during the interval that a passive sampler was in place, non-detection of the target on the sampler may not necessarily indicate that the target was absent, only that it was not captured or that it was not captured in sufficient quantities to be greater than the limit of detection of the analytical method used. This specificity problem is a primary constraint of all passive sampling methods and underscores the need for further experimental work to elucidate mechanisms, kinetics, and variability of target loading on passive samplers in context (Hayes et al., 2022). Further work is also needed on the development of simple, scalable, and appropriate methods for elution and extraction for specific targets, including the exploration of alternative analytical workflows beyond those typically used for SARS-CoV-2 (i.e., RT-qPCR) (Schang et al., 2021; Bivins et al., 2021). These could include simple culture-based methods traditionally used in surveillance of bacterial targets such as S. Typhi, phenotypic assays for antimicrobial resistance, novel applications using sequencing methods, or non-microbial methods suited to the nature of the analyte. Development and application of materials optimized for passive capture of targets of interest also holds promise, given that most passive sampling approaches have made use of simple materials such as cotton gauze that may or may not be ideal for specific targets of interest (Habtewold et al., 2022). Fouling and ragging of passive samplers deployed in wastewater collection systems may also present challenges to downstream processing including messiness in the lab and inhibition of molecular analyses.
11. Research opportunities for passive sampling of wastewater
The future of WBE would be greatly improved by the development of accessible and context-appropriate methods. Passive sampling constitutes an important opportunity to improve the spatial resolution of data and increase the footprint of wastewater surveillance to include low-resource settings (Schang et al., 2021). For SARS-CoV-2 wastewater surveillance using passive sampling, inferences remain constrained by the few studies published to date, which are limited in their size and scope. Beyond simple replication of these efforts in diverse settings, there are several opportunities to refine the passive sampling of wastewater. As previously mentioned, passive sampler performance is dependent on the interaction between the sampler material, the medium being sampled, and the analyte of interest. Development and examination of specialized materials for the uptake of microbial analytes from wastewater is critical. For example, nitrocellulose electronegative membranes have proven to perform quite well for passive sampling of SARS-CoV-2, but their performance may be limited by their structure and low recoveries when suspended solids concentrations are high (Hayes et al., 2022). Gun cotton, made by treating cotton with nitric and sulfuric acids, may afford nitrocellulose fibers in a more open structure that could potentially improve uptake and recovery (Abel, 1866). Other innovative materials, such as nanotraps and magnetic beads with conjugated receptors may afford the opportunity to fine tune passive samplers for optimal performance in wastewater (Xu et al., 2022; Oh et al., 2022).
For novel and existing passive sampling materials, sorption kinetics and calibrations should be performed for relevant combinations of materials and analytes of interest. These parameters are vital to inform the selection of exposure durations and translate counts from passive samplers into quantitative or semi-quantitative data. To date only two such studies have been performed using a limited number of materials and viruses (Hayes et al., 2022; Li et al., 2022). These examinations should be expanded to include other microorganisms such as bacteria, protozoa, and viruses of varying morphology and structure. The effect of wastewater characteristics on analyte accumulation should also be more thoroughly explored. Hayes et al. (2022) found that uptake and recovery were dependent on total suspended solids concentration. It is likely that other wastewater parameters also modulate the performance of passive samplers since they depend on chemical potential gradients for sorption of the target.
Beyond these fundamental research opportunities, there is also the need to develop optimized processing methods to maximize recovery while minimizing inhibition of downstream molecular analyses. This development should also include cost and resource-efficient processing methods such that passive surveillance of wastewater can be fully leveraged in resource-limited settings. One vital point of improvement is extraction, currently performed using various commercial kits, which can be slow and expensive. Rudimentary attempts at heat extraction failed to produce materials that could be amplified by RT-LAMP, but more exploration of simple extraction methods for passive sampling-derived material is warranted (Bivins et al., 2021). Another vital opportunity is the development of non-RT-qPCR analytical methods. The majority of passive sampling efforts have used RT-qPCR which can be slow, expensive, time and personnel intensive, and requires expensive specialized equipment, especially to produce data of reasonable quality. LAMP may offer a compelling alternative, but further development is needed particularly for LAMP integrated with passive sampling material such as electronegative membranes (Amoah et al., 2021; Zhu et al., 2022). Importantly, analytical tests, whether LAMP, RT-qPCR, or sequencing, must be appropriate for the detection and quantification of the relevant VOCs including novel VOCs which continue to emerge (Kirby et al., 2022). Published detections of specific VOCs by passive sampling remain limited with only a single study noting detection of the Alpha VOC via wastewater. The overall thrust of these research opportunities must be to improve the reliability of passive sampling of wastewater for scaling to achieve spatially resolved data across the resource availability gradient.
12. Scaling WBE with passive sampling
As of the time of this writing, wastewater surveillance for SARS-CoV-2 has primarily been reported in high-income settings that are well-served by modern wastewater infrastructure (Naughton et al., 2021). But wastewater surveillance has a long history of use in lower-income settings as well, most visibly in the context of polio eradication efforts where environmental surveillance has been widely deployed and in long-term surveillance for S. typhi and paratyphi (Pogka et al., 2017; Sikorski and Levine, 2020). Such systems, where extant, can be used in response to the ongoing COVID-19 pandemic as exemplified in Pakistan (Sharif et al., 2021). Wastewater surveillance systems for COVID-19 have been developed in at least 55 countries with demonstrated success across a range of climatic, economic, development, and logistical contexts (Tlhagale et al., 2022). While wastewater surveillance is not without limitations, it is increasingly clear that it can provide useful data concerning the epidemiology of an infectious disease, such as COVID-19, especially diseases that are characterized by asymptomatic infections or are inefficiently monitored through clinical surveillance (Safford et al., 2022; Hrudey and Conant, 2021). The wealth of experience gained during the COVID-19 pandemic can be leveraged to develop robust wastewater surveillance systems to monitor a wide-range of emerging and re-emerging pathogens, especially in settings where clinical surveillance is insufficient to capture spatial and temporal infection dynamics and inform public health response.
Because most WBE methods development and deployment have been in high-income settings served by centralized infrastructure, new methods and systems for WBE are needed in low-income settings where populations suffer from the greatest burden of disease. In these settings, wastewater surveillance techniques must be cost-efficient and adaptable to a wide range of wastewater infrastructure forms from centralized urban systems to decentralized ad hoc systems, to systems leveraging natural waterways (Shrestha et al., 2021; Basu et al., 2022). An estimated 1.8 billion people are served by onsite sanitation in low-income settings, and many high-risk settings in higher-income countries may also be unserved by wastewater collection and treatment, which suggests a major limitation of all waste surveillance approaches that focus on liquid wastewater alone (Berendes et al., 2017; Jones et al., 2021). Fecal sludges, for example, have shown to be promising matrices for surveillance and have been compared to stool sampling for a wide range of enteric pathogens in low-income communities (Capone et al., 2020). Wastewater surveillance methods that can be more broadly applied to other matrices containing human waste, such as surface waters or drains receiving human waste, fecal sludges in situ or in the fecal sludge management service chain, or aqueous effluent from septic tanks or in condominial sewerage.
Passive sampling offers a compelling combination of attributes that may make it especially suited to these settings: low cost and ease of use across a range of infrastructure types. Moore swabs have already been used to monitor for bacterial pathogens in a variety of hydraulic systems including irrigation canals, individual household drains, and surface waters (Giri et al., 2021; Sbodio et al., 2013; Sikorski and Levine, 2020). The applications of passive sampling for SARS-CoV-2 in wastewater reviewed herein suggests the value of the technique applied in these settings can be greatly increased through the use novel sampler materials, such as nitrocellulose membranes, and the development and optimization of resource-efficient processing methods including kit-free extractions and simple qualitative assays. Even in high-resource settings, cost-efficient surveillance methods could be used to greatly improve the spatial resolution of existing wastewater surveillance programs to better target public health interventions (Safford et al., 2022). In most applications to date, passive samplers have been used to produce binary wastewater testing results for COVID-19 surveillance. However, the findings of this review suggest passive samplers could produce semi-quantitative or, with finer tuning, quantitative data for public health response in low-resource settings. For example, at one university during wastewater surveillance of buildings with passive samplers, results were reported as non-detect, weak positive, moderate positive, or strong positive, depending on the proportion of RT-LAMP replicates positive for SARS-CoV-2 (Bivins et al., 2021). While at another university, building-level surveillance was structured such that follow up clinical testing was only triggered when genetic signals exceeded empirically determined thresholds. Even with the use of passive samplers, wastewater surveillance is unlikely to become a one-size-fits-all endeavor and will require institutional and professional support to determine the appropriate operational requirements and integrate such efforts into existing public health capacity (Kantor et al., 2022; Thompson et al., 2020).
Cost-efficient wastewater surveillance empowered by passive sampling could allow for the scaling of WBE to include low-resource settings, which to this point have been largely excluded. Paradoxically, settings that are not well served by wastewater infrastructure are likely to derive the most benefit from wastewater surveillance due to elevated burdens of infectious diseases (Prüss-Ustün et al., 2019). Refinement of passive sampling methods and their linkage to a broader environmental surveillance system, in conjunction with environmental laboratory systems, are needed to generate actionable public health data in such settings. Scaling wastewater surveillance to include low-resource settings is vital to secure a more equitable future in WBE.
CRediT authorship contribution statement
Aaron Bivins: Conceptualization, Formal analysis, Methodology, Project administration, Writing – original draft, Writing – review & editing. Devrim Kaya: Conceptualization, Data curation, Project administration, Writing - original draft, Writing - review & editing. Warish Ahmed: Conceptualization. Joe Brown: Writing – original draft, Writing – review & editing. Caitlyn Butler: Conceptualization, Data curation. Justin Greaves: Writing – original draft, Writing – review & editing. Raeann Leal: Writing – original draft, Writing – review & editing. Kendra Maas: Conceptualization, Data curation. Gouthami Rao: Writing – original draft, Writing – review & editing. Samendra Sherchan: Conceptualization, Writing – original draft, Writing – review & editing. Deborah Sills: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. Ryan Sinclair: Conceptualization, Writing – original draft, Writing – review & editing. Robert Wheeler: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. Cresten Mansfeldt: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.155347.
Appendix A. Supplementary data
Supplementary material
References
- Abel F.A. XIV. Researches on gun-cotton.—on the manufacture and composition of gun-cotton. Philos. Trans. R. Soc. Lond. 1866;156:269–308. doi: 10.1098/rstl.1866.0014. [DOI] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., D’Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Hamilton K.A., Hapuarachchi H.C., Harwood V.J., Haque R., Jackson G., Khan S.J., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B.A., Lipp E., McLellan S.L., McMinn B., Medema G., Metcalfe S., Meijer W.G., Mueller J.F., Murphy H., Naughton C.C., Noble R.T., Payyappat S., Petterson S., Pitkänen T., Rajal V.B., Reyneke B., Roman F.A., Rose J.B., Rusiñol M., Sadowsky M.J., Sala-Comorera L., Setoh Y.X., Sherchan S.P., Sirikanchana K., Smith W., Steele J.A., Sabburg R., Symonds E.M., Thai P., Thomas K.V., Tynan J., Toze S., Thompson J., Whiteley A.S., Wong J.C.C., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A.G., Shanks O.C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens L., Daneshvar A., Lau A.E., Kreuger J. Characterization and application of passive samplers for monitoring of pesticides in water. JoVE (J. Vis. Exp.) 2016 doi: 10.3791/54053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amereh F., Negahban-Azar M., Isazadeh S., Dabiri H., Masihi N., Jahangiri-rad M., Rafiee M. Sewage systems surveillance for SARS-CoV-2: identification of knowledge gaps, emerging threats, and future research needs. Pathogens. 2021;10:946. doi: 10.3390/pathogens10080946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Mthethwa N.P., Pillay L., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. RT-LAMP: a cheaper, simpler and faster alternative for the detection of SARS-CoV-2 in wastewater. Food Environ. Virol. 2021;13:447–456. doi: 10.1007/s12560-021-09489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T.J., Blake P.A., Morris G.K., Puhr N.D., Bradford H.B., Wells J.G. Use of Moore swabs for isolating Vibrio cholerae from sewage. J. Clin. Microbiol. 1980;11:385–388. doi: 10.1128/jcm.11.4.385-388.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu P., Choudhury S., Shridhar V., Huilgol P., Roychoudhury S., Nandi I., Chaudhuri A., Mitra A. Surveillance of SARS-CoV-2 RNA in open-water sewage canals contaminated with untreated wastewater in resource-constrained regions. Access Microbiol. 2022;4 doi: 10.1099/acmi.0.000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendes D.M., Sumner T.A., Brown J.M. Safely managed sanitation for all means fecal sludge management for at least 1.8 billion people in low and middle income countries. Environ. Sci. Technol. 2017;51:3074–3083. doi: 10.1021/acs.est.6b06019. [DOI] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Lott M., Shaffer M., Wu Z., North D., Lipp E.K., Bibby K. Building-level wastewater surveillance using tampon swabs and RT-LAMP for rapid SARS-CoV-2 RNA detection. Environ. Sci.: Water Res. Technol. 2021;8:173–183. doi: 10.1039/D1EW00496D. [DOI] [Google Scholar]
- Brooks Y.M., Gryskwicz B., Sheehan S., Piers S., Mahale P., McNeil S., Chase J., Webber D., Borys D., Hilton M., Robinson D., Sears S., Smith E., Lesher E.K., Wilson R., Goodwin M., Pardales M. Detection of SARS-CoV-2 in wastewater at residential college, Maine, USA, August–November 2020 - Volume 27, Number 12—December. Emerg. Infect. Dis. journal. 2021 doi: 10.3201/eid2712.211199. CDC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone D., Berendes D., Cumming O., Knee J., Nalá R., Risk B.B., Stauber C., Zhu K., Brown J. Analysis of fecal sludges reveals common enteric pathogens in urban MaputoMozambique. Environ. Sci. Technol. Lett. 2020;7:889–895. doi: 10.1021/acs.estlett.0c00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassemiro K.M.S.de M., Burlandy F.M., Barbosa M.R.F., Chen Q., Jorba J., Hachich E.M., Sato M.I.Z., Burns C.C., Silva E.E.da. Molecular and phenotypic characterization of a highly evolved type 2 vaccine-derived poliovirus isolated from seawater in Brazil, 2014. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0152251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchis-Scott R., Geng Q., Seth R., Ray R., Beg M., Biswas N., Charron L., Drouillard K.D., D’Souza R., Heath D.D., Houser C., Lawal F., McGinlay J., Menard S.L., Porter L.A., Rawlings D., Scholl M.L., Siu K.W.M., Tong Y., Weisener C.G., Wilhelm S.W., McKay R.M.L. Averting an outbreak of SARS-CoV-2 in a university residence hall through wastewater surveillance. Microbiol. Spectr. 2021 doi: 10.1128/Spectrum.00792-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank K., Chen W., Bivins A., Lowry S., Bibby K. Contribution of SARS-CoV-2 RNA shedding routes to RNA loads in wastewater. Sci. Total Environ. 2022;806 doi: 10.1016/j.scitotenv.2021.150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe J., Schnaubelt A.T., SchmidtBonne S., Angell K., Bai J., Eske T., Nicklin M., Pratt C., White B., Crotts-Hannibal B., Staffend N., Herrera V., Cobb J., Conner J., Carstens J., Tempero J., Bouda L., Ray M., Lawler J.V., Campbell W.S., Lowe J.-M., Santarpia J., Bartelt-Hunt S., Wiley M., Brett-Major D., Logan C., Broadhurst M.J. Assessment of a program for SARS-CoV-2 screening and environmental monitoring in an urban public school district. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.26447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzinamarira T., Murewanhema G., Iradukunda P.G., Madziva R., Herrera H., Cuadros D.F., Tungwarara N., Chitungo I., Musuka G. Utilization of SARS-CoV-2 wastewater surveillance in Africa—a rapid review. Int. J. Environ. Res. Public Health. 2022;19:969. doi: 10.3390/ijerph19020969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS EST Water. 2021;1:1955–1965. doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Fox C.B. 1873. Ozone and Antozone. [Google Scholar]
- George A.D., Kaya D., Layton B.A., Bailey K., Mansell S., Kelly C., Williamson K.J., Radniecki T.S. Impact of sampling type, frequency, and scale of the collection system on SARS-CoV-2 quantification fidelity. Environ. Sci. Technol. Lett. 2022;9:160–165. doi: 10.1021/acs.estlett.1c00882. [DOI] [PubMed] [Google Scholar]
- Giri S., Mohan V.R., Srinivasan M., Kumar N., Kumar V., Dhanapal P., Venkatesan J., Gunasekaran A., Abraham D., John J., Kang G. Case-control study of household and environmental transmission of typhoid fever in India. J. Infect. Dis. 2021;224:S584–S592. doi: 10.1093/infdis/jiab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.S., Lowe J.T. 1927. Carbon-monoxide Detector. 1,644,014A. [Google Scholar]
- Górecki T., Namieśnik J. Passive sampling. TrAC Trends Anal. Chem. 2002;21:276–291. doi: 10.1016/S0165-9936(02)00407-7. [DOI] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Grant S.B., List E.J., Lidstrom M.E. Kinetic analysis of virus adsorption and inactivation in batch experiments. Water Resour. Res. 1993;29:2067–2085. doi: 10.1029/93WR00757. [DOI] [Google Scholar]
- Gray J.D.A. The isolation of B. paratyphosus B from sewage. Br. Med. J. 1929;1:142–144. doi: 10.1136/bmj.1.3551.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Habtewold J., McCarthy D., McBean E., Law I., Goodridge L., Habash M., Murphy H.M. Passive sampling, a practical method for wastewater-based surveillance of SARS-CoV-2. Environ. Res. 2022;204 doi: 10.1016/j.envres.2021.112058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., Nelson K.L., Beamer P., Bischel H.N., Bivins A., Bruder A., Butler C., Camenisch T.D., De Long S.K., Karthikeyan S., Larsen D.A., Meierdiercks K., Mouser P.J., Pagsuyoin S., Prasek S.M., Radniecki T.S., Ram J.L., Roper D.K., Safford H., Sherchan S.P., Shuster W., Stalder T., Wheeler R.T., Korfmacher K.S. Wastewater surveillance for SARS-CoV-2 on college campuses: initial efforts, lessons learned, and research needs. Int. J. Environ. Res. Public Health. 2021;18:4455. doi: 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R.W.S., Phillips W.P. Survival of Salmonella paratyphi B in sewers. Its significance in the investigation of paratyphoid outbreaks. Lancet. 1955:137–139. doi: 10.1016/s0140-6736(55)92136-x. [DOI] [PubMed] [Google Scholar]
- Hayes E.K., Sweeney C.L., Anderson L.E., Li B., Erjavec G.B., Gouthro M.T., Krkosek W.H., Stoddart A.K., Gagnon G.A. A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19. Environ. Sci.: Water Res. Technol. 2021;7:1576–1586. doi: 10.1039/D1EW00207D. [DOI] [Google Scholar]
- Hayes E.K., Sweeney C.L., Fuller M., Erjavec G.B., Stoddart A.K., Gagnon G.A. Operational constraints of detecting SARS-CoV-2 on passive samplers using electronegative filters: a kinetic and equilibrium analysis. ACS EST Water. 2022 doi: 10.1021/acsestwater.1c00441. [DOI] [PubMed] [Google Scholar]
- Hrudey S.E., Conant B. The devil is in the details: emerging insights on the relevance of wastewater surveillance for SARS-CoV-2 to public health. J. Water Health. 2021;20:246–270. doi: 10.2166/wh.2021.186. [DOI] [PubMed] [Google Scholar]
- Isaäcson M., Clarke K.R., Ellacombe G.H., Smit W.A., Smit P., Koornhof H.J., Smith L.S., Kriel L.J. The recent cholera outbreak in the South African gold mining industry. A preliminary report. S. Afr. Med. J. 1974;48:2557–2560. [PubMed] [Google Scholar]
- Jansson L., Akel Y., Eriksson R., Lavander M., Hedman J. Impact of swab material on microbial surface sampling. J. Microbiol. Methods. 2020;176 doi: 10.1016/j.mimet.2020.106006. [DOI] [PubMed] [Google Scholar]
- Jiang A.Z., Nian F., Chen H., McBean E.A. Passive samplers, an important tool for continuous monitoring of the COVID-19 pandemic. Environ. Sci. Pollut. Res. 2022 doi: 10.1007/s11356-022-19073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.R., van Vliet M.T.H., Qadir M., Bierkens M.F.P. Country-level and gridded estimates of wastewater production, collection, treatment and reuse. Earth Syst. Sci. Data. 2021;13:237–254. doi: 10.5194/essd-13-237-2021. [DOI] [Google Scholar]
- Kantor R.S., Greenwald H.D., Kennedy L.C., Hinkle A., Harris-Lovett S., Metzger M., Thornton M.M., Paluba J.M., Nelson K.L. Operationalizing a routine wastewater monitoring laboratory for SARS-CoV-2. PLOS Water. 2022;1(2) doi: 10.1371/journal.pwat.0000007. [DOI] [Google Scholar]
- Karthikeyan S., Ronquillo N., Belda-Ferre P., Alvarado D., Javidi T., Longhurst C.A., Knight R. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego County. mSystems. 2021 doi: 10.1128/mSystems.00045-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S.M., Clark M.E., Coleman M.B. Demonstration of infectious agents in sewage. Am. J. Public Health Nations Health. 1955;45:1438–1446. doi: 10.2105/ajph.45.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kennedy L.C., Wolfe M.K., Criddle C.S., Duong D.H., Topol A., White B.J., Kantor R.S., Nelson K.L., Steele J.A., Langlois K., Griffith J.F., Zimmer-Faust A.G., McLellan S.L., Schussman M.K., Ammerman M., Wigginton K.R., Bakker K.M., Boehm A.B. 2021. SARS-CoV-2 RNA Is Enriched by Orders of Magnitude in Solid Relative to Liquid Wastewater at Publicly Owned Treatment Works. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A.E., Welsh R.M., Marsh Z.A., Yu A.T., Vugia D.J., Boehm A.B., Wolfe M.K., White B.J., Matzinger S.R., Wheeler A., Bankers L., Andresen K., Salatas C., New York Department of Environmental Protection. Gregory D.A., Johnson M.C., Trujillo M., Kannoly S., Smyth D.S., Dennehy J.J., Sapoval N., Ensor K., Treangen T., Stadler L.B., Hopkins L. Notes from the field: early evidence of the SARS-CoV-2 B.1.1.529 (Omicron) variant in community wastewater – United States, November – December 2021. MMWR Morb. Mortal. Wkly Rep. 2022;71:103–105. doi: 10.15585/mmwr.mm7103a5. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Murakami M., Iwamoto R., Katayama H., Imoto S. COVID-19 wastewater surveillance implemented in the Tokyo 2020 Olympic and Paralympic Village. J. Travel Med. 2022 doi: 10.1093/jtm/taac004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostoglou M., Petala M., Karapantsios T., Dovas C., Roilides E., Metallidis S., Papa A., Stylianidis E., Papadopoulos A., Papaioannou N. SARS-CoV-2 adsorption on suspended solids along a sewerage network: mathematical model formulation, sensitivity analysis, and parametric study. Environ. Sci. Pollut. Res. 2022;29:11304–11319. doi: 10.1007/s11356-021-16528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwantes W., Speedy W.J.Y. Monthly Bull. Ministry of Health and Pub. Health Lab. Service (directed by Med. Res. Council) Vol. 14. 1955. Detection of a paratyphoid carrier by sewer and water-closet swabs; pp. 120–123. [PubMed] [Google Scholar]
- Lendon N.C., Mackenzie R.D. Monthly Bull. Ministry of Health 6 Pub. Health Lab. Service (directed by Med. Res. Council) Vol. 10. 1951. Tracing a typhoid carrier by sewage examination; pp. 23–27. [Google Scholar]
- Li J., Verhagen R., Ahmed W., Metcalfe S., Thai P.K., Kaserzon S.L., Smith W.J.M., Schang C., Simpson S.L., Thomas K.V., Mueller J.F., Mccarthy D. In situ calibration of passive samplers for viruses in wastewater. ACS EST Water. 2022 doi: 10.1021/acsestwater.1c00406. [DOI] [Google Scholar]
- Liu P., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., Guo L., Moe C.L. A sensitive, simple, and low-cost method for COVID-19 wastewater surveillance at an institutional level. Sci. Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.151047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrajt G., Naughton B., Bandyopadhyay A.S., Meschke J.S. A review of the most commonly used methods for sample collection in environmental surveillance of poliovirus. Clin. Infect. Dis. 2018;67:S90–S97. doi: 10.1093/cid/ciy638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Moore B. Monthly Bull. Ministry of Health & Pub. Health Lab. Service. Vol. 7. 1948. The detection of paratyphoid carriers in towns by means of sewage examination; pp. 241–248. [Google Scholar]
- Moore B. Monthly Bull. Ministry of Health & PubHealth Lab. Service (directed by Med. Res. Council) Vol. 9. 1950. The detection of typhoid carriers in towns by means of sewage examination; pp. 72–78. [Google Scholar]
- Moore B., Perry C.E.L., Chard S.T. A Survey by the sewage swab method of latent enteric infection in an urban area. Epidemiol. Infect. 1952;50:137–156. doi: 10.1017/S0022172400019501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Ahmed W., Katsivelis P., Allan V., Sinclair R., Zhang Y., Kinyua M.N. 2021. Show Us the Data: Global COVID-19 Wastewater Monitoring Efforts, Equity, and Gaps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C., Kim K., Araud E., Wang L., Shisler J.L., Nguyen T.H. A novel approach to concentrate human and animal viruses from wastewater using receptors-conjugated magnetic beads. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118112. [DOI] [PubMed] [Google Scholar]
- Pazzaglia G., Lesmana M., Tjaniadi P., Subekti D., Kay B. Use of vaginal tampons in sewer surveys for non-O1. Vibrio cholerae. Appl. Environ. Microbiol. 1993 doi: 10.1128/aem.59.8.2740-2742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty J.D., Jones S.B., Huckins J.N., Cranor W.L., Parris J.T., McTague T.B., Boyle T.P. An approach for assessment of water quality using semipermeable membrane devices (SPMDs) and bioindicator tests. Chemosphere. 2000;41:311–321. doi: 10.1016/S0045-6535(99)00499-3. [DOI] [PubMed] [Google Scholar]
- Pogka V., Labropoulou S., Emmanouil M., Voulgari-Kokota A., Vernardaki A., Georgakopoulou T., Mentis A.F. Laboratory surveillance of polio and other enteroviruses in high-risk populations and environmental samples. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.02872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüss-Ustün A., Wolf J., Bartram J., Clasen T., Cumming O., Freeman M.C., Gordon B., Hunter P.R., Medlicott K., Johnston R. Burden of disease from inadequate water, sanitation and hygiene for selected adverse health outcomes: an updated analysis with a focus on low- and middle-income countries. Int. J. Hyg. Environ. Health. 2019;222:765–777. doi: 10.1016/j.ijheh.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M., Isazadeh S., Mohseni-Bandpei A., Mohebbi S.R., Jahangiri-rad M., Eslami A., Dabiri H., Roostaei K., Tanhaei M., Amereh F. Moore swab performs equal to composite and outperforms grab sampling for SARS-CoV-2 monitoring in wastewater. Sci. Total Environ. 2021;790 doi: 10.1016/j.scitotenv.2021.148205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves K., Liebig J., Feula A., Saldi T., Lasda E., Johnson W., Lilienfeld J., Maggi J., Pulley K., Wilkerson P.J., Real B., Zak G., Davis J., Fink M., Gonzales P., Hager C., Ozeroff C., Tat K., Alkire M., Butler C., Coe E., Darby J., Freeman N., Heuer H., Jones J.R., Karr M., Key S., Maxwell K., Nelson L., Saldana E., Shea R., Salveson L., Tomlinson K., Vargas-Barriga J., Vigil B., Brisson G., Parker R., Leinwand L.A., Bjorkman K., Mansfeldt C. High-resolution within-sewer SARS-CoV-2 surveillance facilitates informed intervention. Water Res. 2021;204 doi: 10.1016/j.watres.2021.117613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford H.R., Shapiro K., Bischel H.N. Opinion: wastewater analysis can be a powerful public health tool—if it's done sensibly. PNAS. 2022;119 doi: 10.1073/pnas.2119600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim F., Górecki T. Theory and modelling approaches to passive sampling. Environ. Sci.: Processes Impacts. 2019;21:1618–1641. doi: 10.1039/C9EM00215D. [DOI] [PubMed] [Google Scholar]
- Sattar S.A., Westwood J.C. Isolation of apparently wild strains of poliovirus type 1 from sewage in the Ottawa area. Can. Med. Assoc. J. 1977;116:25–27. [PMC free article] [PubMed] [Google Scholar]
- Sbodio A., Maeda S., Lopez-Velasco G., Suslow T.V. Modified Moore swab optimization and validation in capturing E. coli O157:H7 and Salmonella enterica in large volume field samples of irrigation water. Food Res. Int. 2013;51:654–662. doi: 10.1016/j.foodres.2013.01.011. [DOI] [Google Scholar]
- Schang C., Crosbie N.D., Nolan M., Poon R., Wang M., Jex A., John N., Baker L., Scales P., Schmidt J., Thorley B.R., Hill K., Zamyadi A., Tseng C.-W., Henry R., Kolotelo P., Langeveld J., Schilperoort R., Shi B., Einsiedel S., Thomas M., Black J., Wilson S., McCarthy D.T. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ. Sci. Technol. 2021;55:10432–10441. doi: 10.1021/acs.est.1c01530. [DOI] [PubMed] [Google Scholar]
- Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J., Pang J. Wastewater surveillance to infer COVID-19 transmission: a systematic review. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharara N., Endo N., Duvallet C., Ghaeli N., Matus M., Heussner J., Olesen S.W., Alm E.J., Chai P.R., Erickson T.B. Wastewater network infrastructure in public health: applications and learnings from the COVID-19 pandemic. PLOS Glob. Public Health. 2021;1 doi: 10.1371/journal.pgph.0000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmed J., Safdar R.M., Rehman L., Mujtaba G., Hussain J., Ali J., Angez M., Alam M.M., Akthar R., Malik M.W., Baig M.Z.I., Rana M.S., Usman M., Ali M.Q., Ahad A., Badar N., Umair M., Tamim S., Ashraf A., Tahir F., Ali N. Detection of SARs-CoV-2 in wastewater using the existing environmental surveillance network: a potential supplementary system for monitoring COVID-19 transmission. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0249568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N., Tanaka H., Saito T., Deguchi J., Kondo R., Soda K., Suzuki M. Detection of carriers of typhoid bacilli by sewerage-tracing surveillance in Matsuyama City. Jpn. J. Med. Sci. Biol. 1981;34:385–392. doi: 10.7883/yoken1952.34.385. [DOI] [PubMed] [Google Scholar]
- Shrestha S., Yoshinaga E., Chapagain S.K., Mohan G., Gasparatos A., Fukushi K. Wastewater-based epidemiology for cost-effective mass surveillance of COVID-19 in low- and middle-income countries: challenges and opportunities. Water. 2021;13:2897. doi: 10.3390/w13202897. [DOI] [Google Scholar]
- Sikorski M.J., Levine M.M. Reviving the “Moore swab”: a classic environmental surveillance tool involving filtration of flowing surface water and sewage water to recover typhoidal Salmonella bacteria. Appl. Environ. Microbiol. 2020 doi: 10.1128/AEM.00060-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syngouna V.I., Chrysikopoulos C.V. Interaction between viruses and clays in static and dynamic batch systems. Environ. Sci. Technol. 2010;44:4539–4544. doi: 10.1021/es100107a. [DOI] [PubMed] [Google Scholar]
- Tenney M.W., Verhoff F.H. Chemical and autoflocculation of microorganisms in biological wastewater treatment. Biotechnol. Bioeng. 1973;15:1045–1073. doi: 10.1002/bit.260150605. [DOI] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Yang D., Shan L., Li Q., Liu D., Wang D. Estimation of human norovirus infectivity from environmental water samples by in situ capture RT-qPCR method. Food Environ. Virol. 2018;10:29–38. doi: 10.1007/s12560-017-9317-1. [DOI] [PubMed] [Google Scholar]
- Tlhagale M., Liphadzi S., Bhagwan J., Naidoo V., Jonas K., van Vuuren L., Medema G., Andrews L., Béen F., Ferreira M.L., Saatci A.M., Alpaslan Kocamemi B., Hassard F., Singer A.C., Bunce J.T., Grimsley J.M.S., Brown M., Jones D.L. Establishment of local wastewater-based surveillance programmes in response to the spread and infection of COVID-19 – case studies from South Africa, the Netherlands, Turkey and England. J. Water Health. 2022 doi: 10.2166/wh.2022.185. jwh2022185. [DOI] [PubMed] [Google Scholar]
- Vincent-Hubert F., Morga B., Renault T., Le Guyader F.S. Adsorption of norovirus and ostreid herpesvirus type 1 to polymer membranes for the development of passive samplers. J. Appl. Microbiol. 2017;122:1039–1047. doi: 10.1111/jam.13394. [DOI] [PubMed] [Google Scholar]
- Vo V., Tillett R.L., Chang C.-L., Gerrity D., Betancourt W.Q., Oh E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana B., Allan I.J., Greenwood R., Mills G.A., Dominiak E., Svensson K., Knutsson J., Morrison G. Passive sampling techniques for monitoring pollutants in water. TrAC Trends Anal. Chem. 2005;24:845–868. doi: 10.1016/j.trac.2005.06.006. [DOI] [Google Scholar]
- Wang Y., Liu P., Zhang H., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., Saber L., Kraft C.S., Lane M., Shartar S., Moe C. Early warning of a COVID-19 surge on a university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Sci. Total Environ. 2022;821 doi: 10.1016/j.scitotenv.2022.153291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.J. Isolation of B. typhosus from sewage and shellfish. Br. Med. J. 1928;1:1061–1062. doi: 10.1136/bmj.1.3520.1061-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.J. Isolation of enteric bacilli from sewage and water and its bearing on epidemiology. Br. Med. J. 1933;2:560–562. doi: 10.1136/bmj.2.3794.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W.J. Isolation of Bact. typhosum by means of bismuth sulphite medium in water- and milk-borne epidemics. Epidemiol. Infect. 1938;38:507–519. doi: 10.1017/S0022172400011359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation (No. WHO/V&B/03.03) [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Xu N., Zhang M., Wang Y., Ling G., Yuan Y., Zhang P. Nanotraps based on multifunctional materials for trapping and enrichment. Acta Biomater. 2022;138:57–72. doi: 10.1016/j.actbio.2021.08.047. [DOI] [PubMed] [Google Scholar]
- Yang W., Cai C., Dai X. Interactions between virus surrogates and sewage sludge vary by viral analyte: recovery, persistence and sorption. Water Res. 2022;210 doi: 10.1016/j/waters.2021.117995. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zabiegała B., Kot-Wasik A., Urbanowicz M., Namieśnik J. Passive sampling as a tool for obtaining reliable analytical information in environmental quality monitoring. Anal. Bioanal. Chem. 2010;396:273–296. doi: 10.1007/s00216-009-3244-4. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wu X., Gu A., Dobelle L., Cid C.A., Li J., Hoffmann M.R. Membrane-based in-gel loop-mediated isothermal amplification (mgLAMP) system for SARS-CoV-2 quantification in environmental waters. Environ. Sci. Technol. 2022;56:862–873. doi: 10.1021/acs.est.1c04623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material