Graphical abstract

Keywords: Remdesivir derivatives, RNA polymerization inhibitor, RdRp, Binding mechanism, Molecular Dynamics Simulations
Abstract
The binding of the active form of Remdesivir (RTP) to RNA-dependent RNA Polymerase (RdRp) of SARS-CoV-2 was studied using molecular dynamics simulation. The RTP maintained the interactions observed in the experimental cryo-EM structure. Next, we designed new analogues of RTP, which not only binds to the RNA primer strand in a similar pose as that of RTP, but also binds more strongly than RTP does as predicted by MM-PBSA binding energy. This suggest that these analogues might be able to covalently link to the primer strand as RTP, but their 3′ modification would terminate the primer strand growth.
1. Introduction
The world today faces difficult times with the current coronavirus pandemic. The lack of effective therapeutics and rapid transmission of the virus has exacerbated the course of infection. There is some hope with SARS-CoV-2 vaccine development, however, due to the fact that not every individual may develop neutralizing antibodies, as well as the possibility of side effects which may emerge in the future, the development of small molecule antiviral drugs is urgently needed.
RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 has become an attractive drug target, as it exists only in viruses and not in humans, and it exhibits active site conservation around coronavirus. Remdesivir, an analogue of adenosine triphosphate (ATP) which targets RdRp [1], [2], has recently undergone emergency clinical authorization for the treatment of Covid-19. However, its clinical use in treating Covid-19 patients has been halted because it failed to show the significant improvement that was initially expected [3], [4].
Studies have indicated that the active form of Remdesivir (Remdesivir triphosphate, RTP) works by delayed chain termination mechanism, in which the RTP halt RNA primer strand extension at the i + 3 site after Remdesivir monophosphate (RMP) is incorporated into the RNA primer strand and translocated +3 steps [5], [6]. The nucleotide addition-inhibition cycle (NAC) consists of several sequential states, i.e., an open active site conformation without NTP substrate binding (S1, PDB ID 6M71) [7], initial binding mode of NTP substrate (S2), the conformational changes of the active site transition from an open state (S2) to a closed state (S3), the phosphoryl transfer reaction leading to a reaction product (S4, PDB ID 7BV2) [8], and finally i to i + 3 translocation of the primer strand (S5) [9], [10], [11], [12], [13]. More recently, Romero et al. (2021) studied the active-site open state of SARS-CoV-2 RdRp using apo form RdRp (PDB ID 7BTF) for the nucleotide initial binding and an closed active site of the polymerase using a reaction product (PDB ID 7BV2) for the stabilized nucleotide insertion [14], which were guided by base stacking and base pairing with the template nucleotide, respectively [14]. In this study, we used the reaction product structure (PDB ID 7BV2) as a starting structure for building the initial complex structure of the non-covalent binding step of RTP and its analogues to SARS-CoV-2 RdRp and monitoring their conformational stabilities using molecular dynamics simulations (MDS). It is worth to mention that the reactant complex structure (i.e. Remdesivir in the closed-active state of RdRP) is not yet available. Only 2 SARS-CoV-2 RdRp structures complexed with Remdesivir available in Protein Data Bank at present, i.e., 7BV2 and 7L1F, which are the reaction product and delayed translocation structures of RdRp-Remdesivir complexes, respectively.
Here we employed the quantum mechanics method in deriving ligand force field parameters. New analogues (R1T, R2T and R3T) of RTP were then designed not only to cap the RNA polymerization, but also to enhance the binding affinity toward RdRp. Indeed, we discovered that our designed compounds exhibited a similar binding mode to RTP but with a much higher affinity.
2. Materials and methods
The RdRp complex structure was retrieved from the RCSB data bank with PDB ID 7BV2 [8], which was the closed active site conformation [14]. From this structure, only protein chain A plus two RNA chains (chains P and T) were selected, which was then prepared using the Protein Preparation Wizard of Maestro software [15] using default settings.
The RTP structure was downloaded from the PubChem database, while RTP-like compounds were produced employing combinatorial synthesis of the Maestro CombiGlide module. Three compounds, i.e. R1T, R2T, and R3T, were selected based on RTP binding mode. The force field for RTP, R1T, R2T, and R3T molecules (Fig. S1) were developed in-house by generating charge models of nucleosides RN, R1N, R2N, and R3N (Fig. S2) using the standard AMBER protocol [16], [17], and merging them with the tri-phosphate force field developed by Carlson and coworkers (Fig. S1) [18]. In addition, 5′-form (R5), 3′-form (R3), and the middle-in-chain form (R) of Remdesivir nucleosides were created based on the AMBER protocol (Fig. S3 ). RNA OL3 and triphosphate parameters were assigned to each molecule and the GAFF force field was used to determine the remaining parameters [18], [19].
The molecular electrostatic potential (MEP) of the nucleosides (Fig. S2) were calculated at the HF/6-31G* methodology level using Gaussian 09. Then, geometry optimization was performed at the same level. MEP was then used to calculate the partial charges of all atoms in the nucleosides using the Restrained Electrostatic Potential (RESP) method with two-stage fitting and the use of multiple molecular orientations [16]. Partial charge constraints on each molecule is shown (Fig. S1-3), which retains necessary solved partial charges of RNA nucleosides [17]. After successful RESP calculations, the finalized nucleosides were merged with the triphosphate parameters from PARM94 and ATP to construct RTP, R1T, R2T, and R3T (Fig. S1).
Fig. 3.
The detailed interaction of the most dominant cluster of RTP with RdRp (A). The RTP interactions with two Magnesium ions (colored in pink), D618 (D249), D623 (D254), D760 (D391), and D761 (D392) of RdRp active site, in which distance was measured in Angstrom (B).
Five systems in complex were created for MDS (Table S1). While R3 was based on the solved crystal structure, RTP, R1T, R2T, and R3T were docked to i site (Fig. S13, S17) of crystal structure of RdRp employing Maestro’s extra precision (XP) docking procedure with the restraints to keep the R3 crystal conformation as much as possible. RTP pose is consistent with the pose of R3 (Fig. S13), as for R1T, R2T, and R3T. Therefore, our dock pose is a good starting pose for sampling the reactant state of the complex structure.
AMBER16 was employed to perform MDS following our previous protocol [20]. Three independent runs of 1 µs for the RTP-RdRp system were performed. Each system for R3, R1T, R2T, and R3T underwent MDS for 1 µs in addition to three independent runs of 200 ns for each R1T, R2T, and R3T; hence in total, 8.8 µs MDS were conducted for all systems. The run included a 1.0 ns MDS using the NPT ensemble mode to equilibrate the system’s density, following a 1000 ns dynamics in the equivalent NVT ensemble mode. The clustering analysis was performed using DBSCAN [21], in which the RMSD was chosen as the distance metric with epsilon and minpoints were 2.5 Å and 10, respectively [20]. The H-bond analysis was conducted using default setting (Donor-Acceptor distance:3.5 Å, and the Donor-Hydrogen-Acceptor angle > 90 degree). We further calculated binding free energy of each system using 250 snapshots taken from 0 to 1 µs simulation trajectories by employing MM-PBSA methods [22] and then predicted the ADME properties for ligands by using the SwissADME (https://www.swissadme.ch) [23].
3. Multiple sequence alignment
To identify critical residues for catalysis of SARS-CoV2 RdRP, its primary sequence (uniprotein ID: QHD43415_NSP11) was used as a query to search its closest protein family (PF00680/Viral RdRP family) on the protein families database (pfam 33.1) (https://pfam.xfam.org/) [24]. Next, a multiple sequence alignment using MAFFT method [25] in Jalview [26] was performed to identify conserved residues in PF00680 plus SARS-COV2 RdRP (See Fig. S14).
4. Results and discussion
4.1. Force field development for RTP, R1T, R2T and R3T and validation
Before developing the force fields for RTP, R1T, R2T and R3T, we took note of the structural similarities and differences that RTP and ATP share, which served as the basis for our strategy: Restrain the atom properties (atom name, type and partial charges) that RTP and ATP share, and change said atom properties where RTP and ATP differ. Fortunately, the AMBER RNA OL3 force field contains the full PARM94 parameters of ATP (Fig. S1 A) and its nucleosides A, A3, A5, and AN, in which the force fields of R, R3, R5 (Fig. S3), RN, R1N, R2N, R3N (Fig. S2), RTP, R1T, R2T, and R3T (Fig. S1 B-E) were derived.
Validation of the 7BV2 crystal structure with protein, RNA and R3 ligand covalently-bound to RNA is represented via a protein-RNA RMSD plot (Fig. S15) and a structure comparison figure between the reference and last snapshot structure complexes from a 1 μs MDS (Fig. S16), in which the protein, RNA and R3 ligand show significant overlap with one other; to note, since R3 is covalently-bound to RNA, it is incorporated in the RNA RMSD. The simulation system reached convergence quite early at ∼ 200 ns and maintained an RMSD value of ∼ 2 Å for the remainder of the simulation. This implies that the overall structure conformation of 7BV2 changed negligibly. Our simulation thus validates the stability of the crystal structure complex.
4.2. RTP binding to RdRp complex
RTP was docked at the i position (Fig. S17) of the RdRp catalytic site. The docking pose of RTP was similar to the crystal 7BV2 covalently bound R3, in which the phosphate group was positioned at the entry of the NTP channel formed by hydrophilic residues (K545, R553, and R555) [7], which would inhibit the entry of NTP to the RdRp active site. The reactant state (Fig. S18) of SARS-CoV-2 RdRp was indicated by the coordination of two Magnesium ions with RTP, D618, D760, and D761 of RdRp active site [27], [28]. Our multiple sequence alignment of SARS-CoV-2 RdRP to its protein family (PF00680) suggest D618, D623, D760 and D761 are 100% conserved in the family (Fig. S14), thus likely being key catalytic residues for the catalysis [29]. Fig. 1 shows the 3D conformation of RTP and its detailed interaction in the RdRp SARS-CoV-2, while Fig. S13 depicts the superimposed comparison of crystal 7BV2 covalently bound R3 to docked RTP.
Fig. 1.
The 3D surface representation of RdRp and conformation of RTP in the RdRp SARS-CoV-2 (A) and the detailed interaction of docked RTP with RdRp (B). Template and primer RNA strands are colored in cyan and red, respectively. The RTP interactions with two Magnesium ions (colored in pink), D618, D623, D760, and D761 of RdRp active site, in which distance was measured in Angstrom (C).
Gong and Peersen (2010) proposed the sequential catalytic cycle model of poliovirus polymerization [30], in which S1 is an apo form of initial structure without a bound NTP (PDB code: 3OL6, which is comparable with 6 M71 for SARS-CoV-2). S2 is an open active site with NTP non-covalently bound without Mg ions (PDB code: 3OLB). S3 is closed active site conformation with NTP non-covalently bound and presence of Mg ions. Of note, the crystal structure of S2 and S3 is not yet available for SARS-CoV-2. Table S2 depicts the comparison of RdRp structures of poliovirus, Norwalk virus, and SARS-CoV-2 virus in each state recorded in Protein Data Base (PDB).
Zamyatkin et al (2007) proposed the crystal structure of S3 of Norwalk virus (PDB ID: 3BSO), in which MnA and MnB ions were coordinated with D671 (D343) and D570 (D242), respectively, (distance each 2.15 Å) (Fig. S19 B) [31]. In our reactant state, the distances become longer, in which those between MgA and D760 and between MgB and D618 were 5.93 Å and 6.94 Å, respectively (Fig. 1 C). We showed the superimposition of the S3 of Norwalk virus (PDB ID: 3BSO) and the reaction product (S4) of SARS-CoV-2 (PDB code: 7BV2) in Fig. S19 A and Fig. S19 D.
S4 is a closed active site of post catalysis with NTP bound covalently and presence of Mg2 + ions (PDB code for poliovirus: 3OL7, comparable with 7BV2 structure in SARS-CoV-2), the superimposition of 7BV2 and 3OL7 was depicted in Fig. S20 A and Fig. S20 C. The distances between MgA ion and D328 and between MgB ion and D233 of poliovirus were 2.86 Å and 3.06 Å, respectively. However, the distances become longer in the SARS-CoV-2 virus, in which those between MgA ion and D760 and between MgB ion and D618 were 6.25 Å and 6.41 Å, respectively (Fig. S19 C). Thus, the longer distances between metal ions and key residues observed in our current study (Fig. 1 C), is consistent with the longer distances revealed in the experimental structure of reaction product (S4) of SARS-CoV-2 (PDB ID: 7BV2). It is clear that while MgB ion positions are conserved in Norwalk, poliovirus, and SARS-CoV-2 viruses, the MgA ion of SARS-CoV-2 is located at more upstream site than those found in Norwalk (differing by 3.37 Å) and poliovirus (differing by 2.99 Å) (Fig. S19 D and S20C). It is likely due to the protein sequence change from poliovirus and Norwalk virus to SAR-CoV-2 and is a hallmark for the latter. However, further experimental study is needed to clarify the issue.
Adenosine groups of RTP formed H-bond interactions with U10 of the template strand and U20 of the primer strand in addition to Pi-Pi orbital stacking interactions, which was also observed in the experimental structure [8], [32]. The ribose ring 3′-OH atom formed H-bond interactions with N691 and T680 in RTP, while the cyano group formed H-bond interactions with S759. The phosphate group of RTP formed salt bridge interactions with positively charged amino acid residues K621, R555, and R624. In brief, our docking pose is consistent with the experimental structures of RTP and RdRp complexes.
During the 1 µs MDS, several H-bond interactions were retained. The RTP H-bond interactions occurring with K621 (K252) and U10 (U542) of the template strand were preserved at high occupancies (79.6% and 64%, respectively), while the H-bond with R555 (R186) was maintained at modest occupancy (57.6%). Table S3 showed the H-bond occupancies of RTP during 1 µs MDS. Clearly, the RTP was able to maintain H-bond interactions with active site residues of RdRp.
The trajectory convergence during MDS was checked through the RMSD values averaged over three independent 1 µs MDS (Fig. S21). The receptor Cα achieved stability around 100 ns and remained stable towards the end of simulation time. The RMSD of RNA main atoms shows more fluctuation at around 2 Å and remains stable during MDS. The ligand RMSD of RTP did not change significantly during the simulation time. Fluctuations of RMSD values of ligand-heavy atoms were around 1 Å. The RMSD values of the RTP system were shown to be nearly constant during 1 µs MDS, which implied that the RTP attained a stable conformation in the RdRp active site. The RMSD values for the first, second, and third MDS of RTP show similar patterns (Fig. S22).
The fluctuation of protein amino acid residues during MDS was shown in the RMSF plot (Fig. S23-S24). The high peaks of residues were observed at V405, G432, T644, and G823, which corresponded to the protein loops, while E370 and L895 were the protein carboxy and amino ends. The residues R555, K621, D623, R624, T680, N691, S759, and D760, which directly interacted with RTP, were found to be stable.
The fluctuation of RNA O5′ and ligand heavy atoms was depicted in Fig. 2, S25-S26 , which were observed to be stable under 3 Å and 1 Å, respectively. High peaks were observed in G10, which is the primer strand end, as well as U8 and C21, which are the template strand ends. The U10 and A11 of template strands, which formed the H-bond interactions with RTP, were observed to be stable.
Fig. 2.
The RMSF plot of RNA O5′ atoms (black) and ligand heavy atoms (orange) (A) and labeled 2D structure of RTP (B).
4.3. Clustering analysis of RTP complex
Fig. 3A shows the representative structure of the only one cluster with 100% population extracted from MDS trajectories. The RTP interactions with two Magnesium ions, D618 (D249), D623 (D254), D760 (D391), and D761 (D392) of RdRp active site were depicted (Fig. 3 B). It was shown that RTP confirms the H-bond interactions with S759 through O4′ of the ribose ring, and with R555 through the triphosphate group while maintaining close distance with D618, D760, and D761. In addition, a 3′ hydroxyl group of ribose rings formed H-bond interactions with D623, which was also found in the previous experimental study [33]. Yin et al (2020) indicated that the S759, D760, and D761 residues comprise the catalytic active center of RdRp [8], while Gao et al., (2020) implied that R555, V557, and D618 were among the key binding residues [7]. Moreover, magnesium ions were located near the triphosphate group of RTP, and aspartate residues which indicated their role in stabilizing ligand conformation [8]. Prior studies have shown that incorporation of RTP at position i will block viral RNA synthesis through delayed chain termination mechanism at positions i + 3 or i + 5, which is responsible for antiviral activity of Remdesivir [33], [34], [35], as per present results.
4.4. Designing new analogues
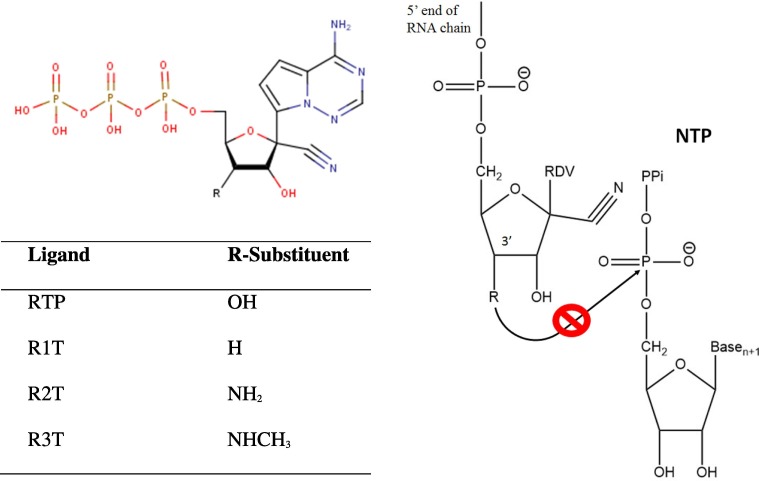
To improve the binding affinity of RTP, we performed combinatorial library enumeration which resulted in three RTP analogues, i.e. R1T, R2T, and R3T (Fig. 4 ).
Fig. 4.
The structures of designed RTP analogues (left) and schematic diagram showing the inhibition of 3′OH atom attack to the α-phosphate of NTP (right).
The docking conformations of R1T, R2T, and R3T were essentially similar to the RTP pose (Fig. 5 ). The H-bond and base stacking interactions were observed between the adenosine parts, template strand, and primer strands, respectively. The ribose ring hydroxyl group formed H-bond interactions with N691 in R1T, R2T, and R3T, while the phosphate groups interacted with positively charged amino acid residues K621 and R555. The aspartate residues and magnesium ions were observed to be close to phosphate groups of ligands.
Fig. 5.
The 3D surface representation of RdRp and conformation of R1T (A), R2T (B), and R3T (C) analogues in the SARS-CoV-2 RdRp. Template and primer RNA strands are colored in cyan and red, respectively. Magnesium ions are colored in pink.
We then conducted three independent runs of 200 ns for each R1T, R2T, and R3T to evaluate the complex stabilities and their binding affinities. We found out that the RTP analogues were all very stable during 3x 200 ns (Fig. S27-S36) and their binding affinities was also lower than that of RTP complex (Table S7). Further we performed individual MDS for 1 µs for each compound. Fig. S37 shows the RMSD values for protein Cα, RNA and ligand heavy atoms for 1 µs. The protein Cα RMSD values for R1T and RTP were nearly identical, while those for R2T and R3T were shown to be higher and lower than RTP, respectively. However, these two are stable enough despite fluctuation under 3 Å complexes. Some fluctuation was recorded in RMSD values of R3T RNA heavy atoms, while those for R1T and R2T were comparable to RTP. The RMSD values of ligand heavy atoms are quite stable in all complexes.
The protein RMSF profiles show that R2T generally enhances more flexibility in protein compared to RTP, which is consistent with the RMSD plot of protein Cα, as well as showing similar patterns between complexes. Fluctuation occurred at S384, T402, T644, and D824, which were protein loops (Fig. S38). Other residues were noticed to be stable.
The RMSF pattern of RNA O5′ atom of analogues were very similar to the RTP (Fig. 6 ). High atomic fluctuations were recorded in G10 (primer strand end), U8, and C21 (template strands ends), while U10 and A11 of the template strands were observed to be stable, as found in the RTP complex. The RMSF values of ligand main atoms were observed to be stable under 1.6 Å. The highest peak was observed in O2′ atoms of the ribose hydroxyl group of R3T. However, its fluctuation was considered to be stable.
Fig. 6.
The RMSF values for RNA O5′ and ligand heavy atoms for RTP (blue), R1T (green), R2T (orange), and R3T (purple) (A). Labeled 2D structure of R1T, R2T, and R3T (B).
4.5. Clustering analysis of analogues complexes
In the cluster analysis of analogues complexes, each R1T, R2T, and R3T, was able to reproduce the H-bond interactions with adenosine part and U10 (U542) of template strands. The R1T, R2T, and R3T compounds were also able to reproduce H-bond interactions with R555 (R186) (64.8%, 70.9%, 85.4%, respectively, for R1T, R2T, and R3T) and K621 (K252) (79.3%, 72.1%, 82.5%, respectively, for R1T, R2T, and R3T) (Table S3-S5) as well as pi-stacking interactions between adenosine motif and A11 (A543) of the template strand. All of those interactions were observed in RTP conformation.
The presence of amino groups in R2T and R3T would hypothetically block the nucleophilic attack on the α-phosphate of an incoming nucleotide as observed in RTP [33], [35]. As a result, we speculate that further nucleotide incorporation is still allowed which would lead to a delayed RNA synthesis inhibition [36]. In the case of R1T, further nucleotide addition would be prevented due to the absence of a ribose 3′OH group, and would eventually lead to classic chain termination [36], [37]. Fig. 4 presents the schematic diagram showing the inhibition of a nucleophilic attack of the 3′OH group on the α-phosphate atom of NTP, while Fig. 7 A-C shows the detailed interaction of the most dominant clusters of R1T, R2T, and R3T with RdRp.
Fig. 7.
The detailed interaction of the most dominant cluster of R1T (A), R2T (B), and R3T (C), each with RdRp.
The base pairing between RTP and U10 of template strand were maintained in 64% and 42.2% occupancies (Table S2), while those were 75.1% and 70.4% occurrences in R1T (Table S3), 70% and 47.6% occupancies in R2T (Table S4), and 78.6% and 45% occurrences in R3T (Table S5). It was clear that the RTP analogues maintain hbond base pairing better than RTP. In all ligand poses, D760 (D391) and Mg2+ ions were observed in close proximity to phosphate groups, indicating their important roles in ligand stabilization [38]. Clearly, the R1T, R2T, and R3T were able to reproduce the RTP interactions, while establishing more interactions with residues in the RdRp active site. Being that they have the ability to reproduce the RTP interactions, R2T and R3T are very likely to work in the same fashion as RTP in terms of inhibiting RNA polymerase specifically through chain delayed termination, while R1T would work through classic chain termination mechanism [39].
Next, using 1 µs MDS trajectory, each ligand was investigated for its free energy of binding using MM-PBSA protocol (Table 1 ). The electrostatic energies (ΔE ELE) were favorable for binding in each ligand, and although the polar contribution for solvation (ΔE PB) is unfavorable, the total electrostatic energy terms were still favorable. This is different from our previous work, which used a semi-empirical method to derive force field parameters, in which the total electrostatic contribution was unfavorable for RTP [20]. The present result is more reasonable in terms of the persistence of electrostatic interactions of nucleotides in the RdRp active site originating from interactions between the negatively charged ligand and positively charged residues, which was also previously reported [40]. This finding highlights the benefits of the quantum mechanics method over the semiempirical method in describing protein–ligand interaction [41]. Other favorable contributions originated from van der Waals (ΔE VDW) and non-polar contribution for solvation (ΔE PBSUR).
Table 1.
The binding energies and their standard deviations predicted by MM-PBSA protocol calculated at 1 µs trajectory*.
| Ligand | ΔEELE | ΔEVDW | ΔEPB | ΔEPBSUR | ΔEPBTOT | ΔΔEPBTOT |
|---|---|---|---|---|---|---|
| RTP | −121.87 ± 26.07 | −29.31 ± 7.46 | 87.19 ± 21.17 | −4.30 ± 0.12 | −68.24 ± 6.40 | 0.00 |
| R1T | −163.37 ± 19.68 | −31.40 ± 6.38 | 114.08 ± 14.16 | −4.35 ± 0.10 | −85.04 ± 6.21 | 16.80 ± 6.21 |
| R2T | −162.95 ± 12.50 | −29.49 ± 6.85 | 111.73 ± 8.14 | −4.33 ± 0.12 | −85.04 ± 7.14 | 16.80 ± 7.14 |
| R3T | −164.86 ± 14.51 | −36.69 ± 6.30 | 115.36 ± 10.52 | −4.65 ± 0.08 | −90.85 ± 6.98 | 22.61 ± 6.98 |
*All values are in kcal/mol. ΔΔEPBTOT is the relative binding energy with reference to the RTP.
It is worth noting that the R1T, R2T, and R3T showed higher affinities (−85.04 kcal/mol, −85.04 kcal/mol, −90.85 kcal/mol, respectively) toward RdRp as compared to RTP (−68.24 kcal/mol). The R1T, R2T, and R3T each enhanced the binding energy by 16.80 kcal/mol, 16.80 kcal/mol, and 22.61 kcal/mol, respectively. The more negative electrostatic contribution (ΔE ELE) of R1T, the clearer it becomes that R2T and R3T contributed to their higher affinities. Additionally, the van der Waals energy (ΔE VDW) of R3T is slightly more negative than that of R1T, R2T, and RTP, which explains its superior binding among other nucleotides. The data indicated that the replacement of the 3′ hydroxyl group of the ribose ring with the alkyl amino group would enhance the nucleotide binding toward RdRp. The alkyl amino which replaces the ribose 3′ hydroxyl group could function as a nucleophile to attack the α-phosphate of the incoming nucleotide and release a pyrophosphate molecule with the help of magnesium ion and aspartate residues around the triphosphate group [42], [43]. The entropy term was not calculated since it can be neglected for very similar molecules [44].
4.6. Prediction of ADME properties
The predicted ADME properties for ligands are shown in Table S8 and Fig. S39 A-D. All compounds show low intestinal absorption properties with no chance for distribution into the brain. They could not be inhibitors for the subtypes of cytochrome P450 enzymes (CYPs), which indicated that they most likely could not be metabolized. They also share the same violation of Lipinski’s rule of five, including molecular weight ((MW) > 500) and the number of H-bond acceptors ((NorO) > 10). Hence, the three analogues share the same ADME properties as RTP, which indicates their favorable use in prodrug form.
5. Conclusions
We developed the AMBER compatible ligand force fields of RTP, R1T, R2T, R3T, and three covalent forms (R5, R, and R3) of RTP by following the standard AMBER protocol and merging them with the existing tri-phosphate force field. The experimental structure was well maintained so that our force fields are well enough to be used with AMBER protein and nucleic acid force fields. Using MDS, we determined that RTP reproduced experimental structure interactions when binding to RdRp, and that S759, R555, D618, D760, and D761, were identified as key residues. The RTP analogues bind more strongly to the RdRp active site as compared to RTP based on MM-PBSA binding energy calculations and that electrostatic contribution is the dominant factor in enhancing binding affinity. These findings allude to our hypothesis that the analogues would potentially be better RdRp inhibitors than RTP, however further study on the inhibition mechanism of those RTP analogues are necessary.
CRediT authorship contribution statement
Muhammad Arba: Conceptualization, Methodology, Writing – original draft. Nicholas Paradis: Methodology. Setyanto T. Wahyudi: Methodology. Dylan J. Brunt: Methodology. Katherine R. Hausman: . Phillip M. Lakernick: Methodology. Mursalin Singh: Methodology. Chun Wu: Conceptualization, Software, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
MA and STW acknowledged the World Class Professor 2021 and Penelitian Dasar and Master Thesis Grants, respectively, from the Ministry of Education, Culture, Research and Technology Republic of Indonesia. CW thanks the support from the National Science Foundation of USA under Grants NSF RUI-1904797/ACI-1429467 and XSEDE MCB 170088.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cplett.2022.139638.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium, W.s.t., Repurposed Antiviral Drugs for Covid-19 — Interim WHO Solidarity Trial Results. 2020. [DOI] [PMC free article] [PubMed]
- 4.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y.i., Luo G., Wang K.e., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y.i., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y.i., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. The Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J., Wang H., Liu Q., Li R., Gao Y., Fang X., Zhong Y., Wang M., Wang Q., Rao Z., Gong P. Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex. Cell Reports. 2021;37(4):109882. doi: 10.1016/j.celrep.2021.109882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.) 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong P. Within and Beyond the Nucleotide Addition Cycle of Viral RNA-dependent RNA Polymerases. Frontiers in Molecular Biosciences. 2022;8 doi: 10.3389/fmolb.2021.822218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. 2016;113(28):E4005. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81(7):1548–1552.e4. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brueckner F., Ortiz J., Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr. Opin. Struct. Biol. 2009;19(3):294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero M.E., Long C., La Rocco D., Keerthi A.M., Xu D., Yu J. Probing remdesivir nucleotide analogue insertion to SARS-CoV-2 RNA dependent RNA polymerase in viral replication. Mol. Syst. Des. Eng. 2021;6(11):888–902. [Google Scholar]
- 15.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 16.Bayly C.I., Cieplak P., Cornell W., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. The Journal of Physical Chemistry. 1993;97(40):10269–10280. [Google Scholar]
- 17.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995;117(19):5179–5197. [Google Scholar]
- 18.Meagher K.L., Redman L.T., Carlson H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003;24(9):1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 20.Arba M., et al. Mechanistic insight on the remdesivir binding to RNA-Dependent RNA polymerase (RdRp) of SARS-cov-2. Comput. Biol. Med. 2021;129 doi: 10.1016/j.compbiomed.2020.104156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao J., Tanner S.W., Thompson N., Cheatham T.E. Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput. 2007;3(6):2312–2334. doi: 10.1021/ct700119m. [DOI] [PubMed] [Google Scholar]
- 22.Kollman P.A., et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 23.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1):42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Gebali, S., et al., The Pfam protein families database in 2019. Nucleic Acids Res, 2019. 47(D1): p. D427-D432. [DOI] [PMC free article] [PubMed]
- 25.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aranda, J. and M. Orozco, RNA-Dependent RNA Polymerase From SARS-CoV-2. Mechanism Of Reaction And Inhibition By Remdesivir. bioRxiv, 2020: p. 2020.06.21.163592.
- 28.Genna V., Vidossich P., Ippoliti E., Carloni P., De Vivo M. A Self-Activated Mechanism for Nucleic Acid Polymerization Catalyzed by DNA/RNA Polymerases. J. Am. Chem. Soc. 2016;138(44):14592–14598. doi: 10.1021/jacs.6b05475. [DOI] [PubMed] [Google Scholar]
- 29.Rungrotmongkol T., Mulholland A.J., Hannongbua S. QM/MM simulations indicate that Asp185 is the likely catalytic base in the enzymatic reaction of HIV-1 reverse transcriptase. Medchemcomm. 2014;5(5):593–596. [Google Scholar]
- 30.Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2010;107(52):22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamyatkin D.F., Parra F., Alonso J.M.M., Harki D.A., Peterson B.R., Grochulski P., Ng K.-S. Structural Insights into Mechanisms of Catalysis and Inhibition in Norwalk Virus Polymerase *. J. Biol. Chem. 2008;283(12):7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y.i., Yin W., Xu H.E. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A.n., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J.i., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;182(2):417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. The Journal of biological chemistry. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tchesnokov E., Feng J., Porter D., Götte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses-Basel. 2019;11(4):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Götte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. The Journal of biological chemistry. 2020;295(47):16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tchesnokov E.P., Obikhod A., Schinazi R.F., Gótte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. The Journal of biological chemistry. 2008;283(49):34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sgrignani J., Magistrato A. The Structural Role of Mg2+ Ions in a Class I RNA Polymerase Ribozyme: A Molecular Simulation Study. J. Phys. Chem. B. 2012;116(7):2259–2268. doi: 10.1021/jp206475d. [DOI] [PubMed] [Google Scholar]
- 39.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen H.L., Thai N.Q., Truong D.T., Li M.S. Remdesivir Strongly Binds to Both RNA-Dependent RNA Polymerase and Main Protease of SARS-CoV-2: Evidence from Molecular Simulations. J. Phys. Chem. B. 2020;124(50):11337–11348. doi: 10.1021/acs.jpcb.0c07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavasotto C.N., Adler N.S., Aucar M.G. Quantum Chemical Approaches in Structure-Based Virtual Screening and Lead Optimization. Front. Chem. 2018;6 doi: 10.3389/fchem.2018.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picarazzi F., Vicenti I., Saladini F., Zazzi M., Mori M. Targeting the RdRp of Emerging RNA Viruses: The Structure-Based Drug Design Challenge. Molecules. 2020;25(23):5695. doi: 10.3390/molecules25235695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho A.T.P., Fernandes P.A., Ramos M.J. The Catalytic Mechanism of RNA Polymerase II. J. Chem. Theory Comput. 2011;7(4):1177–1188. doi: 10.1021/ct100579w. [DOI] [PubMed] [Google Scholar]
- 44.Homeyer N., Gohlke H. Free Energy Calculations by the Molecular Mechanics Poisson−Boltzmann Surface Area Method. Mol. Inf. 2012;31(2):114–122. doi: 10.1002/minf.201100135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.