Abstract
Aims
Current risk scores do not accurately identify patients at highest risk of recurrent atherosclerotic cardiovascular disease (ASCVD) in need of more intensive therapeutic interventions. Advances in high-throughput plasma proteomics, analysed with machine learning techniques, may offer new opportunities to further improve risk stratification in these patients.
Methods and results
Targeted plasma proteomics was performed in two secondary prevention cohorts: the Second Manifestations of ARTerial disease (SMART) cohort (n = 870) and the Athero-Express cohort (n = 700). The primary outcome was recurrent ASCVD (acute myocardial infarction, ischaemic stroke, and cardiovascular death). Machine learning techniques with extreme gradient boosting were used to construct a protein model in the derivation cohort (SMART), which was validated in the Athero-Express cohort and compared with a clinical risk model. Pathway analysis was performed to identify specific pathways in high and low C-reactive protein (CRP) patient subsets. The protein model outperformed the clinical model in both the derivation cohort [area under the curve (AUC): 0.810 vs. 0.750; P < 0.001] and validation cohort (AUC: 0.801 vs. 0.765; P < 0.001), provided significant net reclassification improvement (0.173 in validation cohort) and was well calibrated. In contrast to a clear interleukin-6 signal in high CRP patients, neutrophil-signalling-related proteins were associated with recurrent ASCVD in low CRP patients.
Conclusion
A proteome-based risk model is superior to a clinical risk model in predicting recurrent ASCVD events. Neutrophil-related pathways were found in low CRP patients, implying the presence of a residual inflammatory risk beyond traditional NLRP3 pathways. The observed net reclassification improvement illustrates the potential of proteomics when incorporated in a tailored therapeutic approach in secondary prevention patients.
Keywords: ASCVD, Risk score, Proteomics, Machine learning, NLRP3, C-reactive protein
Structured Graphical Abstract
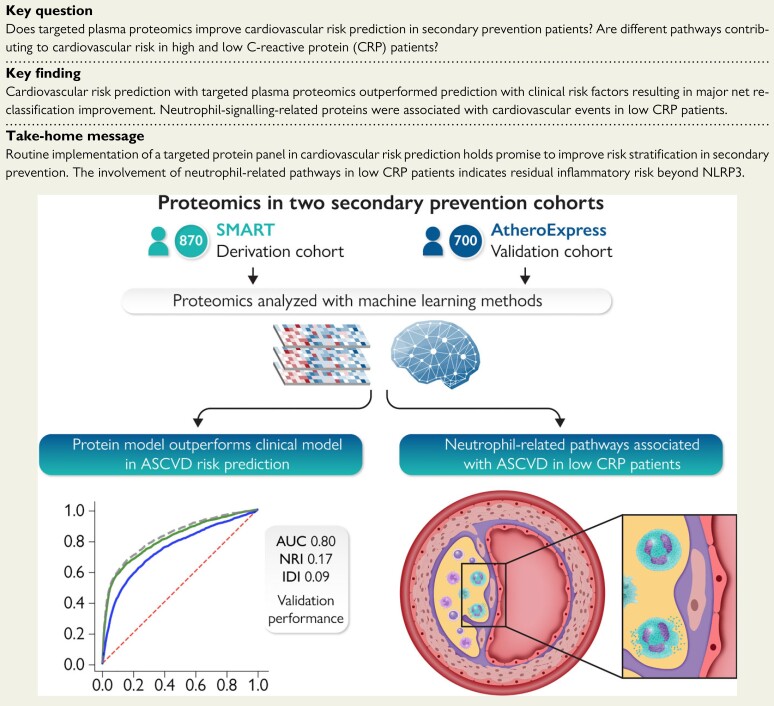
Structured Graphical Abstract.
Targeted proteomics in two secondary prevention cohorts outperforms a clinical risk model in terms of discrimination and reclassification. The involvement of neutrophil-related pathways was found in the subset of low C-reactive protein patients. ASCVD, atherosclerotic cardiovascular disease; AUC, area under the curve; NRI, net reclassification improvement; IDI, integrated discrimination index.
See the editorial comment for this article ‘Proteomics for the prediction and prevention of atherosclerotic disease’, by Paul M. Ridker, https://doi.org/10.1093/eurheartj/ehac036.
Introduction
The residual burden of atherosclerotic cardiovascular disease (ASCVD) remains large, despite the use of guideline-based preventive medication.1 The successful introduction of novel agents, comprising proprotein convertase subtilisin-like/kexin type 9 inhibitors,2,3 low-dose oral anticoagulants,4 sodium-glucose cotransporter 2 inhibitors,5 glucagon-like peptide-1 agonists,6,7 anti-inflammatory agents,8,9 and icosapent ethyl,10 offers an opportunity to further reduce the burden of recurrent ASCVD risk. However, the expanding choice of novel agents has also underscored the need to implement cost-effective therapeutic regimes, which mandates more accurate identification of patients at highest risk in order to solidify the highest absolute ASCVD benefit.11 Epidemiological surveys have demonstrated a highly variable residual risk in patients with established ASCVD ranging from <5% to a more than 40% 10-year recurrence risk.12 Clinical characteristics included in traditional risk prediction scores poorly discriminated individual recurrence of ASCVD,13 attested by the modest c-statistic of 0.64 [95% confidence interval (CI) 0.63–0.65] of the Second Manifestations of ARTerial disease (SMART) score in three independent secondary prevention cohorts.12 Ridker14,15 has argued to use C-reactive protein (CRP) as stratifying marker in order to identify residual inflammatory risk; however, it remains a matter of debate whether CRP reflects the entirety of inflammatory responses involved in atherogenesis.16 Therefore, improved methods to identify patients at highest recurrence risk are needed to help guide ASCVD risk-based therapeutic decisions.
Protein-based risk scores hold a major promise to improve ASCVD risk prediction, since proteins are not only influenced by the genetic background of an individual, but can also reflect adverse changes due to lifestyle alterations and specific pathways contributing to ASCVD risk.17,18 Improvements in machine learning techniques could allow clinical doctors to interpret the massive datasets emerging from proteomic analyses in an outpatient setting, which cannot be analysed using traditional statistical methods.17,19,20 Previously, we showed that the use of a targeted proteomics approach outperformed traditional ASCVD risk scores in a primary prevention setting.19 However, given their high recurrence risk, the most urgent need to identify highest-risk patients pertains to secondary prevention patients.11,21
In the present study, we evaluated the predictive value of targeted proteomics in a secondary prevention setting using advanced machine learning techniques. To this end, we performed plasma proteomics in two large secondary prevention cohorts. As a derivation cohort, we used a high-risk subset of secondary prevention patients included in the SMART cohort, followed by validation of these findings in an independent secondary prevention cohort; the Athero-Express.22,23 In an exploratory analysis, inflammatory pathways were assessed by dividing patients into high or low residual inflammatory risk profiles based on baseline CRP levels.
Methods
Selection of patients
The SMART cohort is an ongoing prospective single-centre cohort of the University Medical Center Utrecht.22 Patients younger than 80 years were included from 1996 onwards, if they had clinically manifest atherosclerotic disease or marked risk factors for atherosclerosis. Previously, a clinical risk model (SMART) was developed and validated to estimate the absolute risk for recurrent ASCVD events.24 We selected all subjects who entered the SMART cohort for myocardial infarction, stroke, or transient ischaemic attack with a 10-year SMART risk score above 15% and had blood samples available. A total of 870 participants were included as a derivation cohort.
The Athero-Express study was initiated in 2002, and included patients undergoing carotid and femoral endarterectomy for previous ischaemic cerebral events or peripheral artery disease.23 Patients were followed up until 3 years after the endarterectomy. We included 700 subjects who underwent a carotid endarterectomy following a stroke or transient ischaemic attack with plasma samples and complete follow-up data available as validation cohort.
Proteomic analyses
For both cohorts, the procedures for blood withdrawal and storage have been described previously.22,23 In short, plasma samples were collected fasting at baseline in the derivation cohort, whereas samples were collected non-fasting on the preoperative day in the validation cohort. In both cohorts, plasma samples were directly centrifuged and stored at −80°C for future analyses. For this study, frozen plasma samples of selected subjects from both cohorts were collected from storage and transferred to Olink proteomics AB (Utrecht, The Netherlands) on dry ice for Proximity Extension Assay analysis. We measured levels of 276 proteins from the Cardiovascular II, Cardiovascular III, and Cardiometabolic panels. These panels were selected based on known associations with ASCVD. All samples with a quality control warning or with ≥40% of measurements below the lower limit of detection (LOD) were excluded from the analysis; separately per proteomic panel. In addition, proteins with ≥90% of samples below the LOD were excluded from the model.
Statistical and machine learning methods
In both cohorts, we defined the primary outcome as the first recurrent ASCVD event, comprising acute myocardial infarction, ischaemic stroke, and cardiovascular death.
In the derivation cohort, we constructed three classification models: first, measured proteins passing quality control (267 proteins) were used to construct a protein-based model with 50 proteins with the highest predictive value. Second, to compare the protein model with current clinical practice, a clinical risk model was constructed and optimized using the same approach as the protein model, including parameters of different validated risk scores such as SMART, Reynolds Risk Score, and Framingham Risk Score.24–27 The clinical risk model comprised the following parameters: age, sex, body mass index, systolic blood pressure, total cholesterol, HDL cholesterol, CRP, smoking status, the presence of diabetes, the use of antihypertensive medication, and family history of cardiovascular disease. A third combined model was formed by stacking the clinical risk parameters with the protein parameters. For use in the validation cohort, all three models were recalibrated to allow an equal comparison and avoid miscalibration.28
All models were constructed using the same machine learning techniques. For the training and evaluation of the models as well as identification of the most reliable biomarker signature in our datasets (both proteomics and clinical), we used stability selection with extreme gradient boosting to predict a binary outcome (event/non-event).29,30 The model hyperparameters were selected using a Randomized Grid Search followed by classifier calibration using the Sigmoid method,31 both performed on the validation set. To prevent overfitting, ‘leave one out cross-validation’ was employed on a random subset with half the dimension of the original dataset. For increased confidence, this process was repeated 20 times. This method was coupled with a rigorous stability selection procedure to ensure the reliability and robustness of the obtained parameters. Finally, a permutation test (randomization test) was applied to evaluate the statistical validity of the results,32 since standard univariate significance tests cannot be applied to the used models due to the non-linear combination of feature functions.
To further explore the inflammatory pathways involved, we performed additional analyses by dividing the SMART cohort in a high CRP (>2 mg/L) and low CRP (≤2 mg/L) group. Patients with a suspected acute inflammatory episode (CRP > 20 mg/L) were excluded. In both groups, a model comprising 50 proteins was constructed to predict recurrent ASCVD events. Protein–protein association networks were assessed and graphically displayed using STRING v11 (string-db.org).33 Normalized protein expression (NPX) values (relative quantification on log2 scale) for interleukin-6 (IL-6) were compared between high and low CRP groups. To identify high or low CRP-specific proteins, the top 10 proteins from both groups were compared with the overall model.
Model performance was reported by means of discrimination, calibration, and reclassification. Discrimination was assessed using the receiver operating characteristic (ROC) curve with an area under the curve (AUC). Relative protein importance was reported in a bar plot.34 Calibration plots were constructed to display calibration performance. Reclassification performance was assessed using the category-free net reclassification improvement (NRI > 0) and integrated discrimination index (IDI).28 95% CI were reported using bootstrap intervals for point estimates of performance metrics when asymptotic intervals were not available.
Data are presented as mean ± standard deviation for normally distributed variables or median with interquartile range (IQR) for skewed data. Categorical variables are expressed as absolute numbers and percentages. Independent sample t-tests and Mann–Whitney U-tests were used where appropriate. Two-sided P-values of ≤0.05 were considered statistically significant. Data were analysed using Python version 3.7 (www.python.org) and RStudio version 3.6.1 (R Foundation, Vienna, Austria).
Results
Patient characteristics of both the derivation and validation cohort are listed in Table 1. In the derivation cohort, 263 (30.2%) participants experienced a recurrent ASCVD event during a median follow-up of 8.0 (4.6–12.2) years. The primary recurrent event consisted of myocardial infarction in 48 (5.5%) patients, ischaemic stroke in 105 (12.1%) patients, and 110 (12.6%) patients died of cardiovascular causes. In the validation cohort, 130 (18.6%) participants experienced a recurrent ASCVD event during a median follow-up of 3.0 (2.2–3.1) years. In this cohort, the primary recurrent ASCVD event was a myocardial infarction in 39 (5.6%) patients, whereas 53 (7.5%) patients had an ischaemic stroke and 38 (5.4%) patients died of cardiovascular causes. The final proteomic analysis included 267 unique proteins after exclusion of nine proteins with ≥90% of values below the LOD (see Supplementary material online, Table 1).
Table 1.
Patient characteristics
| Characteristic | Derivation cohort (SMART) | Validation cohort (Athero-Express) |
|---|---|---|
| Number of patients | 870 | 700 |
| Age (years) | 65 (9) | 70 (9) |
| Male sex | 657 (75.5) | 479 (68.4) |
| BMI (kg/m2) | 26.9 ± 3.9 | 26.2 ± 3.8 |
| Systolic blood pressure (mmHg) | 146 ± 22 | 152 ± 25 |
| Diastolic blood pressure (mmHg) | 82 ± 12 | 82 ± 31 |
| Active smoking | 299 (34.4) | 81 (20.2) |
| Total cholesterol (mmol/L) | 4.95 ± 1.22 | 4.31 ± 1.12 |
| HDL cholesterol (mmol/L) | 1.22 ± 0.36 | 1.10 ± 0.36 |
| LDL cholesterol (mmol/L) | 2.98 ± 1.07 | 2.43 ± 0.91 |
| Triglycerides (mmol/L) | 1.42 (1.00–2.10) | 1.49 (1.08–2.04) |
| C-reactive protein (mg/L) | 2.5 (1.2–5.2) | 2.0 (1.0–4.5) |
| Diabetes mellitus | 178 (20.5) | 163 (23.3) |
| Lipid-lowering therapy | 546 (62.8) | 541 (77.5) |
| Antihypertensive therapy | 578 (66.4) | 509 (72.9) |
| Follow-up time (years) | 7.98 (4.61–12.16) | 3.00 (2.17–3.10) |
| Recurrent ASCVD event | 263 (30.2) | 130 (18.6) |
| Myocardial infarction | 48 (5.5) | 39 (5.6) |
| Ischaemic stroke | 105 (12.1) | 53 (7.5) |
| Cardiovascular death | 110 (12.6) | 38 (5.4) |
Only primary recurrent ASCVD events are shown. Values are n (%), mean ± standard deviation, or median (IQR) for skewed data (triglycerides, C-reactive protein, and follow-up time). SMART, Second Manifestations of ARTerial disease; BMI, body mass index; ASCVD, atherosclerotic cardiovascular disease.
Discriminatory value of proteomic risk model
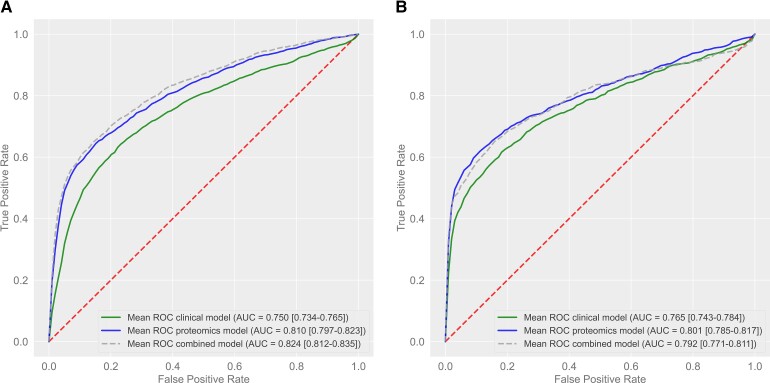
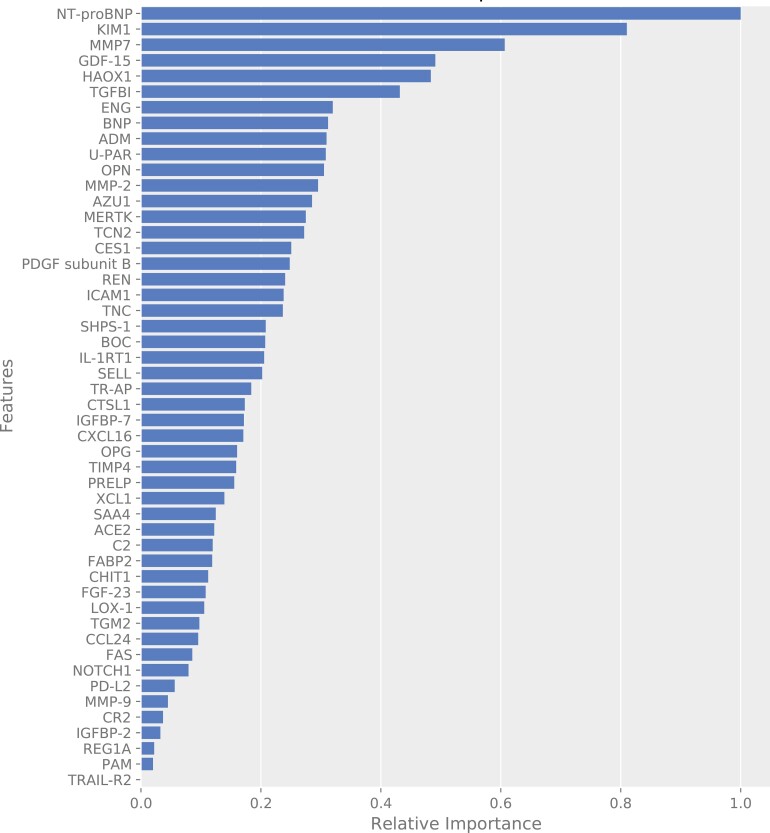
In the derivation cohort, prediction of recurrent ASCVD events using the protein model resulted in an ROC AUC of 0.810 (95% CI 0.797–0.823; Figure 1A and Table 2). The proteins with their relative importance are shown in Figure 2. In comparison, the clinical risk model resulted in an ROC AUC of 0.750 (95% CI 0.734–0.765; Figure 1A and Table 2). Combination of both models led to an ROC AUC of 0.824 (95% CI 0.812–0.835; Figure 1A and Table 2). The protein model performed significantly better than the clinical risk model (delta AUC 0.060, 95% CI 0.040–0.083, P < 0.001), whereas the combination of both models was only slightly superior to the protein model alone (delta AUC 0.014, 95% CI 0.009–0.019, P < 0.001).
Figure 1.
Discriminatory value in the derivation and validation cohort. Receiver operating characteristic curve of protein, clinical, and combined model in the derivation cohort (A) and in the validation cohort (B). The 95% confidence interval is shown between brackets. AUC, area under the curve.
Table 2.
Performance metrics
| Clinical model | Protein model | Combined model | |
|---|---|---|---|
| AUC | |||
| Derivation cohort | 0.750 (0.734–0.765) | 0.810 (0.797–0.823) | 0.824 (0.812–0.835) |
| Validation cohort | 0.765 (0.743–0.784) | 0.801 (0.785–0.817) | 0.792 (0.771–0.811) |
| NRI | |||
| Derivation cohort | Reference | 0.152 (0.110–0.196) | 0.174 (0.134–0.218) |
| Validation cohort | Reference | 0.173 (0.133–0.211) | 0.146 (0.099–0.188) |
| IDI | |||
| Derivation cohort | Reference | 0.098 (0.073–0.122) | 0.116 (0.094–0.139) |
| Validation cohort | Reference | 0.085 (0.068–0.101) | 0.070 (0.049–0.090) |
Summary statistics of performance: area under the curve (AUC), net reclassification improvement (NRI), and integrated discrimination index (IDI). 95% confidence interval is shown between parentheses.
Figure 2.
Importance plot of the protein model. Importance plot of the proteins in the protein model from the derivation cohort. The importance refers to the extent to which a model relies on a given protein. Shown is the relative importance of the 50 proteins in the model.
After recalibration of all models, the discriminatory value was tested in the validation cohort. Validation of the prediction of recurrent ASCVD events using the protein model resulted in an ROC AUC of 0.801 (95% CI 0.785–0.817; Figure 1B and Table 2). In comparison, the clinical risk model resulted in an ROC AUC of 0.765 (95% CI 0.743–0.784; Figure 1B and Table 2). Combination of both models led to an ROC AUC of 0.792 (95% CI 0.771–0.811; Figure 1B and Table 2). In the validation cohort, the protein model also outperformed the clinical risk model (delta AUC 0.036, 95% CI 0.020–0.051, P < 0.001), whereas a combination of both models was not superior to the protein model alone (delta AUC −0.007, 95% CI −0.023 to 0.004, P = 0.996).
Calibration and reclassification of the proteomic risk model
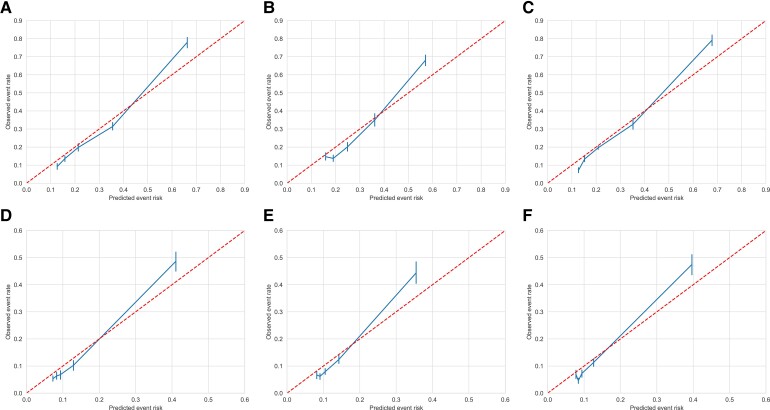
The calibration plots of the proteomic, clinical, and combined model for both the derivation cohort and validation cohort (after recalibration) are shown in Figure 3. The six models were well calibrated, although risk was slightly underestimated in the highest-risk categories. We calculated the NRI and IDI by comparing the protein model with the clinical risk model (Table 1). In the derivation cohort, the NRI was 0.152 (95% CI 0.110–0.196) and the IDI was 0.098 (95% CI 0.073–0.122), compared with an NRI of 0.173 (95% CI 0.133–0.211) and an IDI of 0.085 (95% CI 0.068–0.101) in the validation cohort.
Figure 3.
Calibration in the derivation and validation cohort. Calibration plots for the protein (A), clinical (B), and combined (C) model in the derivation cohort (SMART) and the protein (D), clinical (E), and combined (F) model in the validation cohort (Athero-Express). Predicted event risk vs. observed event rate per risk category quintiles.
Predictive value in high and low C-reactive protein subsets
In clinical practice, CRP is used to identify patients with ‘residual inflammatory risk’. To evaluate the impact of CRP on the performance of the proteomic panel, we divided patients based on CRP levels in the SMART cohort, resulting in 373 patients classified as low CRP (≤2 mg/L) vs. 463 patients classified as high CRP (>2 mg/L). Thirty-four patients with a suspected acute inflammatory episode (CRP > 20 mg/L) were excluded from the analysis. In the low CRP group, 27.3% of patients experienced an ASCVD event during follow-up, compared with 32.0% of patients in the high CRP group (P = 0.13). Interleukin-6 levels were much higher in the high CRP group compared with the low CRP group [NPX (log2 scale) 13.50, IQR 10.24–18.45 vs. 8.63, IQR 6.71–11.27]. The overview of the network pathway analysis in the high and low CRP group is depicted in Supplementary material online, Figure 1. The high CRP group showed a central role for IL-6, which was not present in the low CRP protein model. Conversely, four different inflammatory proteins, which were neither in the initial model nor in the high CRP group, were identified in the top 10 predicting proteins of the low CRP group (Table 3).
Table 3.
Most important proteins in the overall, high, and low C-reactive protein group
| Overall | High CRP subset | Low CRP subset |
|---|---|---|
| NT-proBNP | NT-proBNP | KIM1 |
| KIM1 | HAOX1 | BNP |
| MMP-7 | OPN | ADM |
| GDF-15 | KIM1 | AMBP |
| HAOX1 | PSGL-1 | NID1 |
| TGFBI | GDF-15 | TIMP4 |
| ENG | TIMD4 | FABP2 |
| BNP | MMP-2 | NT-proBNP |
| ADM | CTSL1 | VASN |
| U-PAR | XCL1 | TF |
Overview of the 10 most important proteins in the overall group as well as in the high and low CRP groups. Marked bold are proteins not in the overall 50-protein model. CRP, C-reactive protein; NT-proBNP, N-terminal prohormone brain natriuretic peptide; KIM-1, kidney injury molecule 1; MMP-7, matrix metalloproteinase 7; GDF-15, growth/differentiation factor 15; HAOX1, hydroxyacid oxidase 1; TGFBI, transforming growth factor-β-induced protein ig-h3; ENG, endoglin; BNP, brain natriuretic peptide; ADM, adrenomedullin; U-PAR, urokinase plasminogen activator surface receptor; OPN, osteopontin; PSGL-1, P-selectin glycoprotein ligand 1; TIMD4, T-cell immunoglobulin and mucin domain-containing protein 4; MMP-2, matrix metalloproteinase-2; CTSL1, cathepsin L1; XCL1, lymphotactin; AMBP, α1-microglobulin-bikunin precursor; NID1, nidogen-1; TIMP4, metalloproteinase inhibitor 4; FABP2, intestinal-type fatty acid-binding protein; VASN, vasorin; TF, tissue factor.
Discussion
Using targeted proteomics in two cohorts comprising 1570 patients with established arterial disease, we show that a panel of 50 proteins is superior to a clinical risk model in predicting recurrent ASCVD events. In both the derivation and the validation cohort, the proteomic model performed better in terms of discrimination, was similarly well calibrated and provided a significant NRI over the clinical risk model (Structured Graphical Abstract). Collectively, these data confirm the potential of improved, proteome-supported risk stratification in a secondary prevention setting.
Atherosclerotic cardiovascular disease risk prediction using clinical characteristics performs relatively poor in terms of discrimination.12,13 We previously showed that a targeted proteomics panel improves the prediction of ASCVD events in a primary prevention setting.19 Ganz et al. 35 illustrated that a nine-protein risk score also predicted recurrent ASCVD events in patients with coronary heart disease with modest discrimination (C-statistic 0.70 in validation). With improved proteomic and machine learning techniques, we now show that the use of proteomics significantly outperforms clinical risk prediction in two large secondary prevention cohorts (AUC of 0.801 in the validation cohort, delta AUC 0.036). Whereas in the highest-risk groups the models tended to underestimate ASCVD recurrence risk, the protein, clinical, and combined models were similarly and well calibrated.
Recurrent cardiovascular events: predictive proteins
A targeted proteomics panel was used comprising proteins related to ASCVD, metabolism, and inflammation. N-terminal pro-B-type natriuretic peptide (NT-proBNP), an established marker for heart failure,27 was the protein with the strongest predictive value. NT-proBNP was also found among the top proteins predicting primary ASCVD events in an earlier study.19 Kidney injury molecule-1 (KIM-1) was the second most predicting protein, and has been associated with cardiorenal syndrome.36 The top three proteins were completed by matrix metalloproteinase 7 (MMP-7), which was also found in the primary prevention population.19 MMP-7 and its family of matrix metalloproteinases, the main group of enzymes responsible for degradation of the extracellular matrix, are associated with plaque instability, through macrophage-related pathways.37 Lastly, growth differentiation factor 15 (GDF-15), as the top predictive protein in the earlier primary prevention cohort,19 was the fourth most predictive protein in this study. GDF-15 has been shown to play an important role in leucocyte integrin activation after myocardial infarction.38 The other proteins in the panel were primarily related to immune system involvement in atherosclerosis, including chemotaxis, migration, apoptosis, and angiogenesis.19,39
Residual inflammatory atherosclerotic cardiovascular disease risk
With respect to the residual inflammatory ASCVD risk, attention has primarily focused on the NLRP3 inflammasome with CRP as a reliable downstream marker.15 In a recent sub-study from low-dose colchicine for secondary prevention of cardiovascular disease (LoDoCo2),40 evaluating the impact of colchicine in secondary prevention, we observed colchicine-induced changes in a panel of 37 inflammatory proteins; the majority of which were, however, unrelated to CRP change. To evaluate the impact of CRP on the performance of a proteomic panel containing multiple inflammatory proteins, we compared the predictive value of our proteomic panel between patients with high (>2 mg/L) vs. low baseline (≤2 mg/L) CRP. As observed in Supplementary material online, Figure 2, the central protein in the high CRP group, linked to many other crucial proteins in the model, is IL-6 with much higher levels in the high CRP group compared with the low CRP group, substantiating the involvement of the NLRP3-IL6 pathway leading to CRP elevation. To further evaluate a potential role of inflammatory factors in patients with low CRP, we compared the 10 most important proteins in both high and low CRP groups with the overall 50 protein model. In contrast to the top 10 proteins in the high CRP protein model, which were all present in the overall protein model, the top 10 proteins in the low CRP group comprised four proteins not represented in the initial model nor in the high CRP model: α1-microglobulin-bikunin precursor (AMBP), nidogen-1 (NID1; also known as entactin), tissue factor (TF), and vasorin (VASN). All four proteins are related to neutrophil signalling, implying a role for pro-inflammatory innate immunity activation in the low CRP group independent from the NLRP3-IL6 inflammasome pathway.40 Thus, α1-microglobulin, which is a plasma and tissue protein derived from AMBP, has been shown to inhibit oxidation of LDL through the inhibition of myeloperoxidase (MPO).41 MPO, abundantly present in neutrophilic granules,42 has been shown to oxidize LDL, aggravating atherogenesis.43 NID-1 (entactin) is a component of basement membranes stimulating neutrophil adhesion and chemotaxis.44 Tissue factor has been shown to contribute to thrombosis at the site of plaque rupture via release from neutrophil extracellular traps and is critical in the formation of arterial thrombosis.45 Vasorin directly binds to and attenuates signalling of transforming growth factor beta (TGFß).46 TGFß, which can be produced by infiltrating cells such as neutrophils and macrophages, has been shown to have both atherogenic and atheroprotective properties.47,48 The preponderance of these neutrophil-related proteins in the model best predicting recurrent ASCVD risk in the low CRP group corresponds to our findings in LoDoCo2, where proteins related to neutrophil-activation such as MPO were reduced following colchicine treatment.40 Collectively, these findings imply a residual inflammatory risk also in secondary prevention patients with low CRP, with preliminary evidence pointing to the potential involvement of neutrophil-related pathways.
Strengths and limitations
The use of samples of two large, well-defined secondary prevention cohorts has supported a robust proteomic analysis. The use of state-of-the-art machine learning technology allows the discovery of non-linear relationships and interactions between proteins, which would not have been identified with traditional statistical methodology.
Several limitations to our study merit discussion. First, by using targeted proteomics, proteins not included in these panels which also predict recurrent ASCVD events may have been missed. However, the goal of this study was to evaluate the feasibility of a high-throughput, protein-based risk score for clinical use, rather than novel protein discovery. Nevertheless, we cannot exclude that the predictive value of a larger protein panel may be even better. Second, the derivation and validation cohort had selective and different enrolment criteria as well as different event risk distribution, which could complicate extrapolation to other risk groups. In the derivation cohort (SMART), patients were included following a myocardial infarction, ischaemic stroke, or transient ischaemic attack, whereas the patients from the validation cohort (Athero-Express) were included after carotid endarterectomy following an ischaemic stroke or transient ischaemic attack. Remarkably, while included after carotid endarterectomy, the relative proportion of patients with a myocardial infarction was higher in the validation cohort compared with the derivation cohort (30.0% vs. 18.3%). Despite these differences between the cohorts, the protein model performance in the validation cohort was comparable to the derivation cohort after recalibration, suggesting suitability for use in different populations. Yet, both cohorts primarily consisted of subjects from European ancestry, so extrapolation to other ethnicities remains to be determined. Lastly, in the derivation cohort, samples were collected after overnight fasting, in contrast to the validation cohort in which the samples were collected non-fasting.
Clinical relevance
Single plasma risk markers have failed to robustly improve ASCVD risk scores to date.49,50 Using a panel of 50 proteins, we show a significant improvement in discrimination and clinical value attested by the NRI and IDI in secondary prevention. The introduction of expensive novel therapeutics combined with the large variation in ASCVD recurrence risk in secondary prevention underscores the importance of reliable ASCVD risk stratification, which is essential when adhering to the ‘highest risk—highest benefit’ principle determining cost-efficacy of expensive novel medication.11 Routine implementation of a dedicated protein panel on top of clinical risk factors may therefore hold a promise to improve therapeutic decisions in secondary prevention.
C-reactive protein has been validated as a reliable marker of residual inflammatory risk,15 as well as a biomarker predicting therapeutic benefit from anti-inflammatory therapies.9 Conversely, colchicine treatment was recently reported to markedly reduce the residual ASCVD event rate in post-acute coronary syndrome patients, not selected for CRP elevation,8 whereas colchicine lowered CRP by only 10%.40 In the present study, we observe a preponderance of neutrophil-related proteins contributing to ASCVD risk prediction in patients with low CRP, implying another potential source of residual inflammatory risk independent of the IL6-CRP pathway.15 Collectively, these data lend further support to target specific pathways identified by proteomic analysis. The use of such a pathway-guided strategy instead of a single biomarker approach warrants prospective trials for further validation.
Propelled by expanding proteomic and machine learning technologies, optimal conditions for a high-throughput proteomic assay are approaching. As opposed to clinical risk scores or risk assessment based on genetic candidate genes,51 proteomic scores may more accurately mirror changes in lifestyle.17,18 The major NRI of ASCVD risk in secondary prevention heralds an important further step towards a tailored therapeutic approach in secondary prevention patients, aimed at introducing the use of effective novel medication in the highest-risk patients in a cost-effective manner.11
Conclusions
We show that a panel of 50 proteins is superior to a clinical risk model in predicting recurrent ASCVD events. In both the derivation and the validation cohort, the proteomic model performed better in terms of discrimination and provided significant NRI whereas calibration was comparable in comparison to the clinical risk model. In addition, we found involvement of neutrophil-related pathways in the subset of low CRP patients, indicating a residual inflammatory ASCVD risk beyond the traditional NLRP3 pathways. Further, large prospective studies will have to confirm the value of proteome-based risk scores in secondary prevention before routine clinical implementation can be advocated.
Supplementary material
Supplementary material is available at European Heart Journal online.
Supplementary Material
Acknowledgements
For the SMART cohort, we gratefully acknowledge the contribution of the research nurses; R. van Petersen (data-manager) and the members of the Utrecht Cardiovascular Cohort-Second Manifestations of ARTerial disease-Studygroup (UCC-SMART-Studygroup): F.W. Asselbergs and H.M. Nathoe, Department of Cardiology; G.J. de Borst, Department of Vascular Surgery; M.L. Bots and M.I. Geerlings, Julius Center for Health Sciences and Primary Care; M.H. Emmelot, Department of Geriatrics; P.A. de Jong and T. Leiner, Department of Radiology; A.T. Lely, Department of Obstetrics & Gynecology; N.P. van der Kaaij, Department of Cardiothoracic Surgery; L.J. Kappelle and Y.M. Ruigrok, Department of Neurology; M.C. Verhaar, Department of Nephrology & Hypertension, F.L.J. Visseren (chair) and J. Westerink, Department of Vascular Medicine, University Medical Center Utrecht and Utrecht University.
Contributor Information
Nick S. Nurmohamed, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands Department of Cardiology, Amsterdam University Medical Centers, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
João P. Belo Pereira, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands
Renate M. Hoogeveen, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands
Jeffrey Kroon, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands.
Jordan M. Kraaijenhof, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands
Farahnaz Waissi, Department of Vascular Surgery, Division of Surgical Specialties, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Nathalie Timmerman, Department of Vascular Surgery, Division of Surgical Specialties, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Michiel J. Bom, Department of Cardiology, Amsterdam University Medical Centers, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands
Imo E. Hoefer, Central Diagnostic Laboratory, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands
Paul Knaapen, Department of Cardiology, Amsterdam University Medical Centers, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Alberico L. Catapano, Department of Pharmacological and Biomolecular Sciences, University of Milan, Milano, Italy IRCCS Multimedica, Milano, Italy.
Wolfgang Koenig, Deutsches Herzzentrum München, Technische Universität München, Munich, Germany; German Centre for Cardiovascular Research (DZHK e.V.), Partner Site Munich Heart Alliance, Munich, Germany; Institute of Epidemiology and Medical Biometry, University of Ulm, Ulm, Germany.
Dominique de Kleijn, Department of Vascular Surgery, Division of Surgical Specialties, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Frank L.J. Visseren, Department of Vascular Medicine, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands
Evgeni Levin, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; HorAIzon BV, Delft, The Netherlands.
Erik S.G. Stroes, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands
Funding
This work was supported by an European Research Area Network on Cardiovascular Diseases (ERA-CVD) grant (ERA-CVD JTC2017) and the CVON-Dutch Heart Foundation (2017–20).
Conflict of interest
N.S.N. is the co-founder of Lipid Tools. A.L.C. reports consulting fees/lecturing fees from Akcea, Amgen, Amryt, Sanofi, Esperion, Kowa, Novartis, Ionis Pharmaceuticals, Mylan, Menarini, Merck, Recordati, Regeneron Daiichi Sankyo, Genzyme, Aegerion, and Sandoz. W.K. reports advisory board/lecturing fees from Novartis, The Medicines Company, DalCor, Kowa, Amgen, Corvidia, Daiichi-Sankyo, Genentech, Novo Nordisk, Esperion, OMEICOS, Sanofi, and Bristol-Myers Squibb, grants and non-financial support from Abbott, Roche Diagnostics, Beckmann, and Singulex, outside the submitted work. E.S.G.S. reports advisory board/lecturing fees paid to the institution of E.S.G.S. by Amgen, Sanofi, Regeneron, Esperion, and IONIS.
References
- 1. Jernberg T, Hasvold P, Henriksson M, Hjelm H, Thuresson M, Janzon M. Cardiovascular risk in post-myocardial infarction patients: nationwide real world data demonstrate the importance of a long-term perspective. Eur Heart J 2015;36:1163–1170. [DOI] [PubMed] [Google Scholar]
- 2. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 2017;376:1713–1722. [DOI] [PubMed] [Google Scholar]
- 3. Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, et al. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med 2018;379:2097–2107. [DOI] [PubMed] [Google Scholar]
- 4. Eikelboom JW, Connolly SJ, Bosch J, Dagenais GR, Hart RG, Shestakovska O, et al. Rivaroxaban with or without aspirin in stable cardiovascular disease. N Engl J Med 2017;377:1319–1330. [DOI] [PubMed] [Google Scholar]
- 5. Zinman B, Wanner C, Lachin JM, Fitchett D, Bluhmki E, Hantel S, et al. Empagliflozin, cardiovascular outcomes, and mortality in Type 2 diabetes. N Engl J Med 2015;373:2117–2128. [DOI] [PubMed] [Google Scholar]
- 6. Marso SP, Daniels GH, Frandsen KB, Kristensen P, Mann JFE, Nauck MA, et al. Liraglutide and cardiovascular outcomes in type 2 diabetes. N Engl J Med 2016;375:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marso SP, Bain SC, Consoli A, Eliaschewitz FG, Jódar E, Leiter LA, et al. Semaglutide and cardiovascular outcomes in patients with Type 2 diabetes. N Engl J Med 2016;375:1834–1844. [DOI] [PubMed] [Google Scholar]
- 8. Nidorf SM, Fiolet ATL, Mosterd A, Eikelboom JW, Schut A, Opstal TSJ, et al. Colchicine in patients with chronic coronary disease. N Engl J Med 2020;383:1838–1847. [DOI] [PubMed] [Google Scholar]
- 9. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 10. Bhatt DL, Steg PG, Miller M, Brinton EA, Jacobson TA, Ketchum SB, et al. Cardiovascular risk reduction with icosapent ethyl for hypertriglyceridemia. N Engl J Med 2019;380:11–22. [DOI] [PubMed] [Google Scholar]
- 11. Annemans L, Packard CJ, Briggs A, Ray KK. ‘Highest risk–highest benefit’ strategy: a pragmatic, cost-effective approach to targeting use of PCSK9 inhibitor therapies. Eur Heart J 2018;39:2546–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaasenbrood L, Boekholdt SM, Van Der Graaf Y, Ray KK, Peters RJG, Kastelein JJP, et al. Distribution of estimated 10-year risk of recurrent vascular events and residual risk in a secondary prevention population. Circulation 2016;134:1419–1429. [DOI] [PubMed] [Google Scholar]
- 13. Jensen JK. Risk prediction: are we there yet? Circulation 2016;134:1441–1443. [DOI] [PubMed] [Google Scholar]
- 14. Ridker PM. Residual inflammatory risk: addressing the obverse side of the atherosclerosis prevention coin. Eur Heart J 2016;37:1720–1722. [DOI] [PubMed] [Google Scholar]
- 15. Ridker PM. Clinician’s guide to reducing inflammation to reduce atherothrombotic risk: JACC review topic of the week. J Am Coll Cardiol 2018;72:3320–3331. [DOI] [PubMed] [Google Scholar]
- 16. Soehnlein O, Libby P. Targeting inflammation in atherosclerosis—from experimental insights to the clinic. Nat Rev Drug Discov 2021;20:589–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med 2019;25:1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindsey ML, Mayr M, Gomes AV, Delles C, Arrell DK, Murphy AM, et al. Transformative impact of proteomics on cardiovascular health and disease: a scientific statement from the American Heart Association. Circulation 2015;132:852–872. [DOI] [PubMed] [Google Scholar]
- 19. Hoogeveen RM, Pereira JPB, Nurmohamed NS, Zampoleri V, Bom MJ, Baragetti A, et al. Improved cardiovascular risk prediction using targeted plasma proteomics in primary prevention. Eur Heart J 2020;41:3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riley RD, Ensor J, Snell KIE, Harrell FE, Martin GP, Reitsma JB, et al. Calculating the sample size required for developing a clinical prediction model. BMJ 2020;368:m441. [DOI] [PubMed] [Google Scholar]
- 21. Ray KK, Molemans B, Schoonen WM, Giovas P, Bray S, Kiru G, et al. EU-wide cross-sectional observational study of lipid-modifying therapy use in secondary and primary care: the DA VINCI study. Eur J Prev Cardiol 2021;28:1279–1289. [DOI] [PubMed] [Google Scholar]
- 22. Simons PCG, Algra A, Van De Laak MF, Grobbee DE, Van Der Graaf Y. Second manifestations of ARTerial disease (SMART) study: rationale and design. Eur J Epidemiol 1999;15:773–781. [DOI] [PubMed] [Google Scholar]
- 23. Verhoeven BAN, Velema E, Schoneveld AH, de Vries JPPM, de Bruin P, Seldenrijk CA, et al. Athero-express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur J Epidemiol 2004;19:1127–1133. [DOI] [PubMed] [Google Scholar]
- 24. Dorresteijn JAN, Visseren FLJ, Wassink AMJ, Gondrie MJA, Steyerberg EW, Ridker PM, et al. Development and validation of a prediction rule for recurrent vascular events based on a cohort study of patients with arterial disease: the SMART risk score. Heart 2013;99:866–872. [DOI] [PubMed] [Google Scholar]
- 25. Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM Jr, Kastelein JJP, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 2008;359:2195–2207. [DOI] [PubMed] [Google Scholar]
- 26. Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA 2007;297:611–619. [DOI] [PubMed] [Google Scholar]
- 27. Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med 2006;355:2631–2639. [DOI] [PubMed] [Google Scholar]
- 28. Pencina MJ, D’Agostino RB, Pencina KM, Janssens ACJW, Greenland P. Interpreting incremental value of markers added to risk prediction models. Am J Epidemiol 2012;176:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caruana R, Niculescu-Mizil A, Crew G, Ksikes A. Ensemble selection from libraries of models. In: Proceedings, Twenty-First International Conference on Machine Learning, ICML 2004. Banff, AB, Canada: ACM; 2004. p137–144. [Google Scholar]
- 30. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. [Google Scholar]
- 31. Niculescu-Mizil A, Caruana R. Predicting good probabilities with supervised learning. In: ICML 2005—Proceedings of the 22nd International Conference on Machine Learning. [Google Scholar]
- 32. Ojala M, Garriga GC. Permutation tests for studying classifier performance. J Mach Learn Res 2010;11:1833–1863. [Google Scholar]
- 33. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hastie T, Tibshirani R, Friedman J. The elements of statistical learning, In: The Mathematical intelligencer, Springer Series in Statistics. New York, NY: Springer; 2009. p83–85. 10.1007/978-0-387-84858-7. [DOI] [Google Scholar]
- 35. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 2016;315:2532–2541. [DOI] [PubMed] [Google Scholar]
- 36. Figarska SM, Gustafsson S, Sundström J, Ärnlöv J, Mälarstig A, Elmståhl S, et al. Associations of circulating protein levels with lipid fractions in the general population. Arterioscler Thromb Vasc Biol 2018;38:2505–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abbas A, Aukrust P, Russell D, Krohg-Sørensen K, Almås T, Bundgaard D, et al. Matrix metalloproteinase 7 is associated with symptomatic lesions and adverse events in patients with carotid atherosclerosis. PLoS One 2014;9:e84935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kempf T, Zarbock A, Widera C, Butz S, Stadtmann A, Rossaint J, et al. GDF-15 is an inhibitor of leukocyte integrin activation required for survival after myocardial infarction in mice. Nat Med 2011;17:581–588. [DOI] [PubMed] [Google Scholar]
- 39. Bom MJ, Levin E, Driessen RS, Danad I, Van Kuijk CC, van Rossum AC, et al. Predictive value of targeted proteomics for coronary plaque morphology in patients with suspected coronary artery disease. EBioMedicine 2019;39:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Opstal TSJ, Hoogeveen RM, Fiolet ATL, Silvis MJM, The SHK, Bax WA, et al. Colchicine attenuates inflammation beyond the inflammasome in chronic coronary artery disease: a LoDoCo2 proteomic substudy. Circulation 2020;142:1996–1998. [DOI] [PubMed] [Google Scholar]
- 41. Cederlund M, Deronic A, Pallon J, Sørensen OE, Åkerström B. A1 M/α1-microglobulin is proteolytically activated by myeloperoxidase, binds its heme group and inhibits low density lipoprotein oxidation. Front Physiol 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aratani Y. Myeloperoxidase: its role for host defense, inflammation, and neutrophil function. Arch Biochem Biophys 2018;640:47–52. [DOI] [PubMed] [Google Scholar]
- 43. Delporte C, Boudjeltia KZ, Noyon C, Furtmüller PG, Nuyens V, Slomianny M-C, et al. Impact of myeloperoxidase–LDL interactions on enzyme activity and subsequent posttranslational oxidative modifications of apoB-100. J Lipid Res 2014;55:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Senior RM, Gresham HD, Griffin GL, Brown EJ, Chung AE. Entactin stimulates neutrophil adhesion and chemotaxis through interactions between its Arg–Gly–Asp (RGD) domain and the leukocyte response integrin. J Clin Invest 1992;90:2251–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stakos DA, Kambas K, Konstantinidis T, Mitroulis I, Apostolidou E, Arelaki S, et al. Expression of functional tissue factor by neutrophil extracellular traps in culprit artery of acute myocardial infarction. Eur Heart J 2015;36:1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikeda Y, Imai Y, Kumagai H, Nosaka T, Morikawa Y, Hisaoka T, et al. Vasorin, a transforming growth factor β-binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo. Proc Natl Acad Sci USA 2004;101:10732–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toma I, McCaffrey TA. Transforming growth factor-β and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell Tissue Res 2012;347:155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grainger DJ. Transforming growth factor β and atherosclerosis: so far, so good for the protective cytokine hypothesis. Arterioscler Thromb Vasc Biol 2004;24:399–404. [DOI] [PubMed] [Google Scholar]
- 49. Curry SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, Davidson KW, et al. Risk assessment for cardiovascular disease with nontraditional risk factors: US preventive services task force recommendation statement. JAMA 2018;320:272–280. [DOI] [PubMed] [Google Scholar]
- 50. Mortensen MB, Falk E, Li D, Nasir K, Blaha MJ, Sandfort V, et al. Statin trials, cardiovascular events, and coronary artery calcification: implications for a trial-based approach to statin therapy in MESA. JACC Cardiovasc Imaging 2018;11:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kessler T, Schunkert H. Coronary artery disease genetics enlightened by genome-wide association studies. JACC Basic Transl Sci 2021;6:610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.