Abstract
The description of group-level, genotype- and phenotype-associated imaging traits is academically important, but the practical demands of clinical neurology centre on the accurate classification of individual patients into clinically relevant diagnostic, prognostic and phenotypic categories. Similarly, pharmaceutical trials require the precision stratification of participants based on quantitative measures. A single-centre study was conducted with a uniform imaging protocol to test the accuracy of an artificial neural network classification scheme on a cohort of 378 participants composed of patients with ALS, healthy subjects and disease controls. A comprehensive panel of cerebral volumetric measures, cortical indices and white matter integrity values were systematically retrieved from each participant and fed into a multilayer perceptron model. Data were partitioned into training and testing and receiver-operating characteristic curves were generated for the three study-groups. Area under the curve values were 0.930 for patients with ALS, 0.958 for disease controls, and 0.931 for healthy controls relying on all input imaging variables. The ranking of variables by classification importance revealed that white matter metrics were far more relevant than grey matter indices to classify single subjects. The model was further tested in a subset of patients scanned within 6 weeks of their diagnosis and an AUC of 0.915 was achieved. Our study indicates that individual subjects may be accurately categorised into diagnostic groups in an observer-independent classification framework based on multiparametric, spatially registered radiology data. The development and validation of viable computational models to interpret single imaging datasets are urgently required for a variety of clinical and clinical trial applications.
Keywords: Neuroradiology, Machine learning, Amyotrophic lateral sclerosis, Neuroimaging, Artificial neural networks
Introduction
Diagnostic delay in neurodegenerative conditions has a considerable literature. In ALS, the average interval from symptom onset to definite diagnosis is around 12 months [1, 2]. Patients often describe insidious symptom onset many months before medical advice is sought. The key milestones of the diagnostic journey in ALS include symptom manifestation, visit to a general practitioner, review in a general neurology clinic, diagnostic investigations, and assessment in a tertiary referral centre to confirm a suspected diagnosis [3–6]. The constellation of initial symptoms may be confounded by comorbid conditions, and misdiagnoses in the initial phase of ALS are not uncommon. The implications of diagnostic delay are considerable as it may delay recruitment into clinical trials, may have ramifications for genetic counselling, may increase the risk of misdiagnoses or potentially lead to unnecessary medical or surgical interventions such as spinal laminectomies, carpal tunnel surgery, and intravenous immunoglobulin (IVIg) treatment [1]. Recent imaging studies have revealed that by the time the diagnosis is confirmed, significant neurodegenerative changes have already occurred [7], limiting the potential of putative neuroprotective medications. Recent evidence also suggests that considerable presymptomatic disease burden can be readily detected long before symptom manifestation [8–11]. These observations would suggest that, the optimal window for clinical trials is not well into the ‘post-diagnostic’ phase of the disease, when widespread cerebral and spinal cord degeneration can already be detected, but as early as the diagnostic likelihood or mutation status permit. The role of neuroimaging in ALS has been extensively discussed [12], but the literature is dominated by papers describing group-level, phenotype- or genotype-associated imaging traits [13]. Various research consortia have invested considerable effort to increase cohort numbers, pool data from multiple centres to perform well-powered analyses and report radiological patterns representative of a particular phenotype [14]. The characterisation of stereotyped ‘signatures’ is academically interesting [12, 15], but the practical demands of clinical practice are markedly different [16]. As opposed to the scholarly pursuit of group-level descriptions, the priority of clinical neurology is the precision classification of a specific, single patient into diagnostic, phenotypic and prognostic categories through the quantitative interpretation of their biomarker profile. Relatively few studies have focussed on the classification of individual patient imaging data in ALS [17, 18]. A variety of innovative approaches have been explored [19] spanning from z-score based approaches, through support vector machine frameworks, discriminant function analyses, to regression models, with varying degree of classification accuracy [16, 20–24]. Several studies have reported excellent ‘area under the curve’ (AUC) values with reference to the discriminatory potential of a specific measure between patients and healthy controls, but binary classification into ‘ALS’ versus ‘healthy’ does not mirror real-life diagnostic dilemmas. In the clinical setting, the distinction between ‘ALS’ and ‘healthy’ is seldom challenging; instead, the dilemma is typically whether subtle clinical changes represent incipient ALS or rather, the harbinger of an alternative neurodegenerative condition. Another common shortcoming of classification studies is the a priori selection of anatomical regions, often referred to as ‘regions of interest’ (ROIs) which are known to be affected in ALS, rather than performing formal feature selection analyses or ranking variables based on their discriminatory potential. Finally, few studies have narrowed their analyses to a cohort of patients in their peri-diagnostic phase, which seems indispensable to scrutinise and validate proposed frameworks. The classification of cases with marked disability and long disease duration reveals relatively little about the efficacy of a specific model architecture. Accordingly, the objective of this study is the development of an observer-independent, multiclass (three-way) classification protocol to categorise multiparametric imaging data of a large cohort of subjects consisting of patients with amyotrophic lateral sclerosis (ALS), healthy controls (HC) and disease controls (DC). An additional objective of the study is to evaluate and rank the importance of imaging measures and anatomical foci for further model optimisation, and to test a proposed classification framework on subjects in their peri-diagnostic phase.
Methods
Participants
A total of 378 participants, 214 patients with ALS (‘ALS’), 37 disease controls (‘DC’) with a non-ALS neurodegenerative diagnosis and 127 healthy controls (‘HC’) were included in a prospective, single-centre imaging study. All participants gave informed consent in accordance with the Ethics Approval of this research project (Beaumont Hospital, Dublin, Ireland). Exclusion criteria included prior cerebrovascular events, known traumatic brain injury, comorbid neoplastic, paraneoplastic or neuroinflammatory diagnoses. Participating ALS patients were diagnosed according to the revised El Escorial criteria. Disease controls consisted of patients with FTD and were diagnosed based on the Rascovsky criteria. Participating patients had a uniform neurological assessment and key variables, such as disability scores, interval from diagnosis to scan, and handedness were recorded.
Magnetic resonance imaging
A standardised imaging protocol was implemented on a 3 Tesla Philips Achieva Magnetic resonance (MR) platform. A 3D Inversion Recovery prepared Spoiled Gradient Recalled echo (IR-SPGR) sequence was utilised to acquire T1-weighted (T1w) images with a field-of-view (FOV) of 256 × 256 × 160 mm, flip angle = 8°, spatial resolution of 1 mm3, SENSE factor = 1.5, TR/TE = 8.5/3.9 ms, TI = 1060 ms. A spin-echo echo planar imaging (SE-EPI) pulse sequence was used to acquire diffusion tensor images (DTI) using a 32-direction Stejskal-Tanner diffusion encoding scheme; FOV = 245 × 245 × 150 mm, 60 slices with no interslice gap, spatial resolution = 2.5 mm3, TR/TE = 7639/59 ms, SENSE factor = 2.5, b values = 0, 1100 s/mm2, dynamic stabilisation and spectral presaturation with inversion recovery (SPIR) fat suppression. To assess for comorbid inflammatory or vascular pathologies, fluid-attenuated inversion recovery (FLAIR) images were also acquired from each subject. An Inversion Recovery Turbo Spin Echo (IR-TSE) sequence was used for FLAIR imaging. Data were acquired in axial orientation: FOV = 230 × 183 × 150 mm, spatial resolution = 0.65 × 0.87 × 4 mm, 30 slices with 1 mm gap, TR/TE = 11,000/125 ms, TI = 2800 ms, 120° refocusing pulse, with flow compensation and motion smoothing and a saturation slab covering the neck region.
Imaging framework
Initial quality control steps included radiological review for incidental pathological findings, assessment for movement artefacts, and evaluation of white matter abnormalities on FLAIR. Following standardised pre-processing steps (described below), 28 volumetric metrics, 68 cortical thickness values and 120 white matter indices were uniformly retrieved from each subject’s imaging data; a total of 216 imaging measures were then appraised in each participant. These data were systematically analysed in post-hoc statistics.
Volume metrics
The standard anatomical reconstruction pipeline of the FreeSurfer image analysis suite [25], ‘recon-all’ was implemented, including non-parametric non-uniform intensity normalisation, affine registration to the MNI305 atlas, intensity normalisation, skull striping, automatic subcortical segmentation, linear volumetric registration, neck removal, tessellation of the grey matter-white matter boundary, surface smoothing, inflation to minimise metric distortion, and automated topology correction [26]. To segment the brainstem into the medulla oblongata, pons and midbrain, a Bayesian segmentation algorithm was utilised, which relies on a probabilistic atlas of the brainstem and its neighbouring anatomical structures generated based on 49 scans [27]. The following 28 cerebral volume values were uniformly retrieved from each pre-processed T1-weighted dataset: (1) left cerebellar white matter volume, (2) left cerebellar cortex volume, (3) left thalamus volume, (4) left caudate volume, (5) left putamen volume, (6) left pallidum volume, (7) left hippocampus volume, (8) left amygdala volume, (9) left accumbens volume, (10) right cerebellar white matter volume, (11) right cerebellar cortex volume, (12) right thalamus volume, (13) right caudate volume, (14) right putamen volume, (15) right pallidum volume, (16) right hippocampus volume, (17) right amygdala volume, (18) right accumbens volume, (19) posterior corpus callosum volume, (20) middle corpus callosum volume, (21) central corpus callosum volume, (22) mid-anterior corpus callosum volume, (23) anterior corpus callosum volume, (24) medulla volume, (25) pons volume, (26) superior cerebellar peduncle volume, (27) midbrain volume, and (28) total intracranial volume. Volumetric values of individual subjects were converted as percentage of the subject’s total intracranial volume (TIV) to account for individual TIV variations.
Cortical thickness values
Following pre-processing with ‘recon-all’, the labels of the Desikan–Killiany atlas were utilised to retrieve average cortical thickness values [20] from 34 cortical regions in the left and right cerebral hemispheres; (1) banks superior temporal sulcus, (2) caudal anterior-cingulate cortex, (3) caudal middle frontal gyrus, (4) cuneus cortex, (5) entorhinal cortex, (6) frontal pole, (7) fusiform gyrus, (8) inferior parietal cortex, (9) inferior temporal gyrus, (10) insula, (11) isthmus–cingulate cortex, (12) lateral occipital cortex, (13) lateral orbitofrontal cortex, (14) lingual gyrus, (15) medial orbital frontal cortex, (16) middle temporal gyrus, (17) parahippocampal gyrus, (18) paracentral lobule, (19) pars opercularis, (20) pars orbitalis, (21) pars triangularis, (22) pericalcarine cortex, (23) postcentral gyrus (24) posterior-cingulate cortex, (25) precentral gyrus, (26) precuneus cortex, (27) rostral anterior-cingulate cortex, (28) rostral middle frontal gyrus, (29) superior frontal gyrus, (30) superior parietal cortex, (31) superior temporal gyrus, (32) supramarginal gyrus, (33) temporal pole, and (34) transverse temporal cortex.
White matter indices
Pre-processing of diffusion tensor data were implemented using in FMRIB’s software library. Raw DTI data first underwent eddy current corrections and skull removal; a tensor model was then fitted to generate maps of axial diffusivity (AD), fractional anisotropy (FA), mean diffusivity (MD), and radial diffusivity (RD). FMRIB’s software library’s tract-based statistics (TBSS) module was utilised for non-linear registration and skeletonisation of individual DTI images. A mean FA mask was created and each subject’s individual AD, FA, MD and RD images were merged into 4-dimensional (4D) AD, FA, MD and RD image files. The study-specific white matter skeleton was masked by atlas-defined labels for the following 30 white matter regions of interests in MNI space: (1) left anterior thalamic radiation, (2) right anterior thalamic radiation, (3) left cerebellar white matter skeleton averaged, (4) right cerebellar white matter skeleton averaged, (5) left cingulum, (6) right cingulum, (7) left corticospinal tract, (8) right corticospinal tract, (9) left external capsule, (10) right external capsule, (11) forceps major, (12) forceps minor, (13) fornix, (14) left inferior cerebellar peduncle, (15) right inferior cerebellar peduncle, (16) left inferior fronto-occipital fasciculus, (17) right inferior fronto-occipital fasciculus, (18) left inferior longitudinal fasciculus, (19) right inferior longitudinal fasciculus, (20) left medial lemniscus, (21) right medial lemniscus, (22) middle cerebellar peduncle, (23) left posterior thalamic radiation, (24) right posterior thalamic radiation, (25) left superior cerebellar peduncle, (26) right superior cerebellar peduncle, (27) left superior longitudinal fasciculus, (28) right superior longitudinal fasciculus, (29) left uncinate fasciculus, and (30) right uncinate fasciculus. The labels of the standard-space ICBM-DTI-81 white matter atlas [28, 29] were utilised to create masks for the cerebellar peduncles, medial lemniscus, external capsule and posterior thalamic radiation. Labels of the JHU white matter tractography atlas [30, 31] were used to generate masks for the forceps major and minor, anterior thalamic radiation, uncinate, superior and inferior longitudinal fasciculi, cingulum, corticospinal tracts, inferior fronto-occipital fasciculi. The cerebellar label (label 2) of the MNI probabilistic atlas [32, 33] was used to generate a mask for averaged cerebellar diffusivity estimation. The FMRIB fornix template [34] was used to mask the study-specific white matter skeleton in MNI space. Four diffusivity metrics (AD, FA, MD, RD) were retrieved from 30 white matter regions in each subject, resulting in a white matter panel of 120 values.
Statistical analyses
An artificial neural network framework, a multilayer perceptron model was implemented with hyperbolic tangent as the hidden layer activation function. The diagnosis (ALS, HC, DC) was set as dependent variable, and the retrieved imaging measures as covariates. Imaging metrics were rescaled by standardisation; (x − mean)/s. The model architecture included one hidden layer with 6 units. Data were partitioned into a training sample (68%) and testing sample (32%). A batch-type training approach was utilised with a gradient descent optimisation algorithm; initial learning rate: 0.4, momentum: 0.9, interval centre: 0, interval offset: ± 0.5. Using the above model architecture, the following outputs were generated; synaptic weights, classification results, ROC curves, and AUC values. An independent variable importance analysis was also performed to rank the relevance of imaging metrics in determining group membership. To visually represent the accuracy of diagnostic classification, the predicted pseudo-probability of each diagnostic group was plotted in a bar chart. Based on the feature importance hierarchy, a streamlined classification model was tested using only the 20 most important imaging variables. Finally, to further scrutinise the classification framework, the model was tested on a subset of patients in their peri-diagnosis phase, who were scanned within 6 weeks of their formal diagnosis.
Results
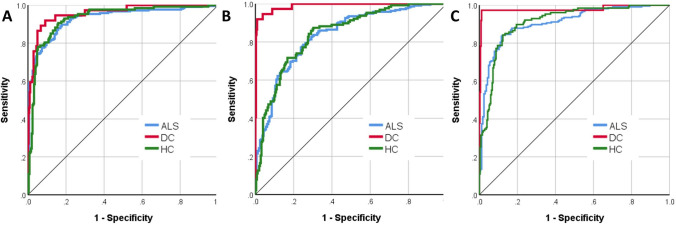
The three groups, ALS (n = 214, age: 60.97 ± 11.92, 140 male, 202 right handed), healthy controls (n = 127, age: 59.29 ± 10.95, 59 male, 112 right handed) and disease controls (n = 37, age: 62.4 ± 7.9, 20 male, 34 right handed) were matched for age (p = 0.23) and handedness (p = 0.12), but not for sex (p = 0.002). Of the 378 datasets, 256 (67.7%) was included in the training sample and 122 (32.3%) in the testing sample. Cross-entropy error was 73.49 in the training sample and 85.03 in the testing sample; incorrect predications were 10.9% in the training sample and 24.6% in the testing sample. Classification summary is presented in Table 1A. The predicted pseudo-probability of diagnosis in each cohort (confirmed diagnosis) is presented in Fig. 1. Receiver-operating characteristic (ROC) curves are presented in Fig. 2A. Area under the curve values were 0.930 for ALS, 0.958 for disease controls, and 0.931 for healthy controls relying on all input imaging variables. The normalised importance of the 20 most relevant imaging variables in predicting group membership is shown in Fig. 3 with their corresponding importance value. The ranked normalised importance of the 50 most relevant imaging metrics is presented in Table 2. The classification analyses were re-run with the 20 most important imaging metrics identified by the explorative analyses. Relying on only 20 imaging features, the classification accuracy was evaluated again (Table 1B). Area under the curve values based on only 20 core imaging features (Fig. 2B) were 0.835 for ALS, 0.990 for DC, and 0.842 for healthy controls. As 19 white matter metrics were ranked among the 20 most important imaging features (Fig. 3) and the vast majority (92%) of imaging metrics among the 50 diagnostically relevant variables (Table 2) were diffusivity metrics, a final post hoc analysis was conducted where only white matter diffusivity indices were included as covariates in the perceptron model; all 30 tracts and all four diffusivity metrics (120 variables in total). Area under the receiver-operating characteristic curves generated based on white matter features alone (Fig. 2C) were 0.907 for ALS, 0.979 for DC, and 0.911 for healthy controls. Classification outcomes using white measures alone are presented in Table 1C.
Table 1.
Classification outcomes in the training and testing samples using A, all imaging features B, the 20 most important variables only and C, white matter diffusivity metrics alone
| Sample | Observed | Predicted | |||
|---|---|---|---|---|---|
| ALS | DC | HC | % Correct | ||
| (A) All features included | |||||
| Training | ALS | 140 | 2 | 9 | 92.7 |
| DC | 3 | 19 | 2 | 79.2 | |
| HC | 11 | 1 | 69 | 85.2 | |
| Overall percent | 60.2% | 8.6% | 31.3% | 89.1 | |
| Testing | ALS | 52 | 6 | 5 | 82.5 |
| DC | 2 | 9 | 2 | 69.2 | |
| HC | 14 | 1 | 31 | 67.4 | |
| Overall percent | 55.7% | 13.1% | 31.1% | 75.4 | |
| (B) 20 core features included | |||||
| Training | ALS | 126 | 1 | 32 | 79.2 |
| DC | 3 | 23 | 1 | 85.2 | |
| HC | 24 | 0 | 63 | 72.4 | |
| Overall percent | 56.0% | 8.8% | 35.2% | 77.7 | |
| Testing | ALS | 46 | 0 | 9 | 83.6 |
| DC | 2 | 7 | 1 | 70.0 | |
| HC | 15 | 0 | 25 | 62.5 | |
| Overall percent | 60.0% | 6.7% | 33.3% | 74.3 | |
| (C) Only white matter metrics included | |||||
| Training | ALS | 128 | 1 | 18 | 87.1 |
| DC | 0 | 24 | 0 | 100.0 | |
| HC | 16 | 0 | 70 | 81.4 | |
| Overall percent | 56.0% | 9.7% | 34.2% | 86.4 | |
| Testing | ALS | 53 | 3 | 11 | 79.1 |
| DC | 0 | 12 | 1 | 92.3 | |
| HC | 11 | 0 | 30 | 73.2 | |
| Overall percent | 52.9% | 12.4% | 34.7% | 78.5 | |
ALS amyotrophic lateral sclerosis, DC disease control, HC healthy control
Fig. 1.
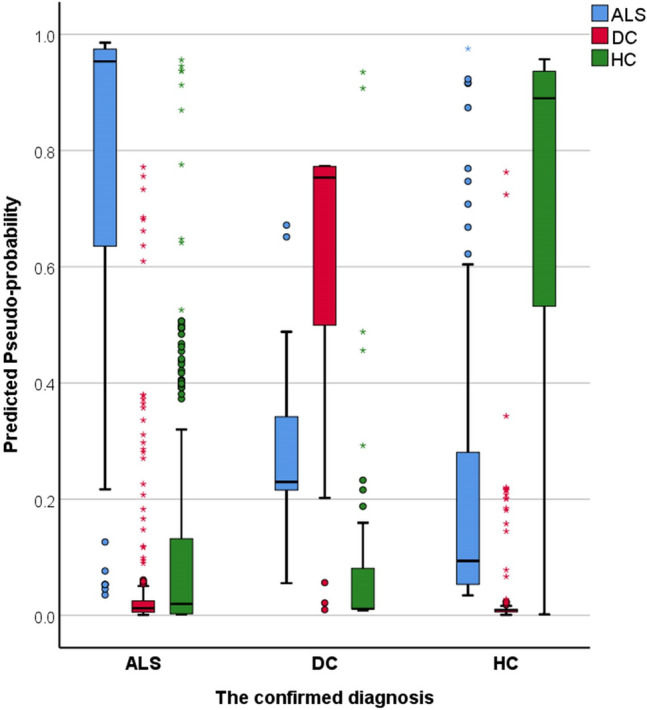
The predicted pseudo-probability profiles of subjects with an established diagnosis
Fig. 2.
Receiver-operating characteristic curve of patients with ALS, disease controls (DC) and healthy controls (HC) using all imaging features (A—left), the 20 most important variables only (B—middle) and white matter diffusivity metrics alone (C—right)
Fig. 3.
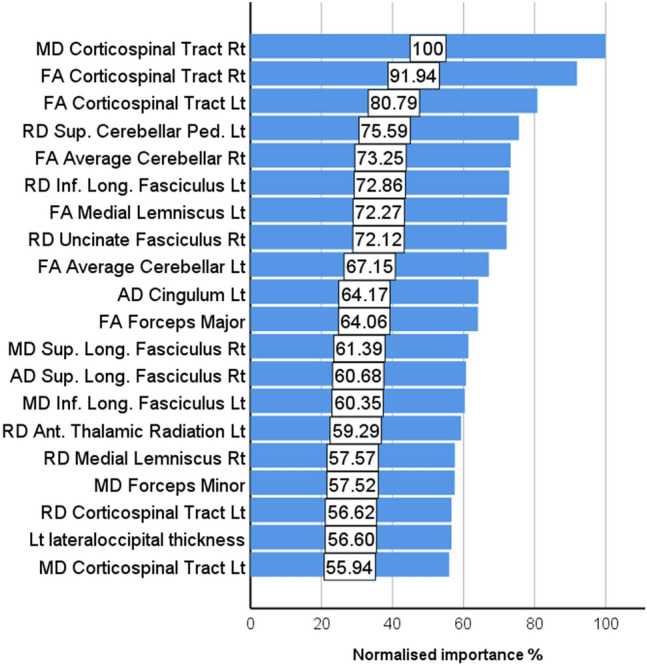
The hierarchy of normalised variable importance
Table 2.
The importance and normalised importance of the 50 most relevant imaging variables in predicting group membership
| Rank | Imaging metric | Importance | Normalised importance (%) |
|---|---|---|---|
| 1. | MD Corticospinal Tract Rt | 0.014 | 100.0 |
| 2. | FA Corticospinal Tract Rt | 0.013 | 91.9 |
| 3. | FA Corticospinal Tract Lt | 0.011 | 80.8 |
| 4. | RD Sup Cerebellar Ped Lt | 0.010 | 75.6 |
| 5. | FA Average Cerebellar Rt | 0.010 | 73.3 |
| 6. | RD Inf. Longitudinal_Fasciculus Lt | 0.010 | 72.9 |
| 7. | FA Medial Lemniscus Lt | 0.010 | 72.3 |
| 8. | RD Uncinate Fasciculus Rt | 0.010 | 72.1 |
| 9. | FA Average Cerebellar Lt | 0.009 | 67.1 |
| 10. | AD Cingulum Lt | 0.009 | 64.2 |
| 11. | FA Forceps Major | 0.009 | 64.1 |
| 12. | MD Sup. Longitudinal Fasciculus Rt | 0.008 | 61.4 |
| 13. | AD Sup. Longitudinal Fasciculus Rt | 0.008 | 60.7 |
| 14. | MD Inf. Longitudinal Fasciculus Lt | 0.008 | 60.4 |
| 15 | RD Anterior Thalamic Radiation Lt | 0.008 | 59.3 |
| 16. | RD Medial Lemniscus Rt | 0.008 | 57.6 |
| 17. | MD Forceps Minor | 0.008 | 57.5 |
| 18. | RD Corticospinal Tract Lt | 0.008 | 56.6 |
| 19. | Lt lateraloccipital thickness | 0.008 | 56.6 |
| 20. | MD Corticospinal Tract Lt | 0.008 | 55.9 |
| 21. | RD Medial Lemniscus Lt | 0.008 | 55.6 |
| 22. | AD External Capsule Rt | 0.008 | 54.6 |
| 23. | FA Uncinate Fasciculus Rt | 0.007 | 54.2 |
| 24. | RD Sup. Cerebellar Ped Rt | 0.007 | 54.0 |
| 25. | Lt posteriorcingulate thickness | 0.007 | 53.7 |
| 26. | AD Forceps Minor | 0.007 | 53.6 |
| 27. | RD Anterior Thalamic Radiation Rt | 0.007 | 53.5 |
| 28. | FA Inf. Cerebellar Peduncle Rt | 0.007 | 53.1 |
| 29. | FA Inf. Longitudinal Fasciculus Rt | 0.007 | 52.4 |
| 30. | Lt precentral thickness | 0.007 | 51.1 |
| 31. | RD Average Cerebellar Lt | 0.007 | 51.0 |
| 32. | RD Inf. Cerebellar Peduncle Lt | 0.007 | 51.0 |
| 33. | MD Middle Cerebellar Peduncle | 0.007 | 50.7 |
| 34. | RD Inf. Cerebellar Peduncle Rt | 0.007 | 50.1 |
| 35. | FA Inf. Cerebellar Peduncle Lt | 0.007 | 49.9 |
| 36. | MD Post. Thalamic Radiation Rt | 0.007 | 49.8 |
| 37. | MD Inf. Fronto-Occipital Fasciculus Rt | 0.007 | 49.7 |
| 38. | FA Inf. Longitudinal Fasciculus Lt | 0.007 | 49.3 |
| 39. | FA Sup. Cerebellar Peduncle Lt | 0.007 | 48.8 |
| 40. | MD Medial Lemniscus Lt | 0.007 | 48.3 |
| 41. | Lt Caudal middle frontal thickness | 0.007 | 48.2 |
| 42. | AD Medial Lemniscus Rt | 0.007 | 47.4 |
| 43. | Left-Amygdala Volume % | 0.007 | 47.1 |
| 44. | RD Middle Cerebellar Peduncle | 0.007 | 47.0 |
| 45. | MD Anterior Thalamic Radiation Rt | 0.006 | 46.8 |
| 46. | FA Uncinate Fasciculus Lt | 0.006 | 45.7 |
| 47. | FA Medial Lemniscus Rt | 0.006 | 45.4 |
| 48. | RD Forceps Major | 0.006 | 45.3 |
| 49. | RD Forceps Minor | 0.006 | 45.0 |
| 50. | MD External Capsule Rt | 0.006 | 45.0 |
AD axial diffusivity, ALS amyotrophic lateral sclerosis, DC disease control, FA fractional anisotropy, HC healthy control, Lt left, MD mean diffusivity, RD radial diffusivity, Rt right
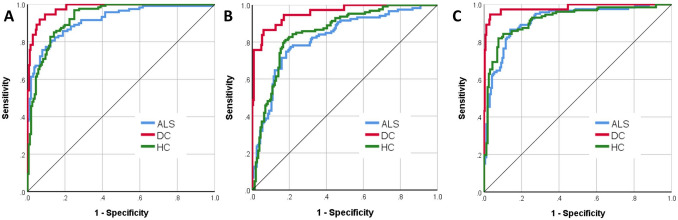
To scrutinise the validity of this classification strategy, the model was tested on a subset of patients (n = 119) who were scanned within 6 weeks of their diagnoses (‘peri-diagnosis cohort’). Using all imaging features AUC was 0.915 for ALS, 0.979 for DC and 0.929 for HC. Using the 20 most important imaging features alone, AUC was 0.822 for ALS, 0.958 for DC and 0.853 for HC. Using all WM metrics but no GM measures, AUC was 0.914 for ALS, 0.981 for DC and 0.92 for HC. Classification outcomes in the ‘peri-diagnosis’ cohort are presented in Table 3. Pseudo-probability profiles in the peri-diagnostic phase using all imaging features are presented in Fig. 4 and the three ROC curves are shown in Fig. 5. Model architecture is presented in Fig. 6 with 20 input variables.
Table 3.
Classification outcomes in the peri-diagnostic phase in the training and testing samples using A, all imaging features B, the 20 most important variables only and C, white matter diffusivity metrics alone
| Sample | Observed | Predicted | |||
|---|---|---|---|---|---|
| ALS | DC | HC | % Correct | ||
| (A) All features included | |||||
| Training | ALS | 76 | 5 | 9 | 84.4 |
| DC | 1 | 19 | 3 | 82.6 | |
| HC | 15 | 1 | 65 | 80.2 | |
| Overall percent | 47.4% | 12.9% | 39.7% | 82.5 | |
| Testing | ALS | 23 | 0 | 6 | 79.3 |
| DC | 2 | 11 | 1 | 78.6 | |
| HC | 11 | 0 | 35 | 76.1 | |
| Overall percent | 40.4% | 12.4% | 47.2% | 77.5 | |
| (B) 20 features included | |||||
| Training | ALS | 58 | 3 | 24 | 68.2 |
| DC | 3 | 22 | 1 | 84.6 | |
| HC | 15 | 1 | 68 | 81.0 | |
| Overall percent | 39.0% | 13.3% | 47.7% | 75.9 | |
| Testing | ALS | 26 | 4 | 4 | 76.5 |
| DC | 1 | 6 | 4 | 54.5 | |
| HC | 7 | 0 | 36 | 83.7 | |
| Overall percent | 38.6% | 11.4% | 50.0% | 77.3 | |
| (C) Only white matter metrics included | |||||
| Training | ALS | 62 | 3 | 13 | 79.5 |
| DC | 1 | 24 | 0 | 96.0 | |
| HC | 12 | 1 | 75 | 85.2 | |
| Overall percent | 39.3% | 14.7% | 46.1% | 84.3 | |
| Testing | ALS | 30 | 2 | 9 | 73.2 |
| DC | 2 | 9 | 1 | 75.0 | |
| HC | 5 | 1 | 33 | 84.6 | |
| Overall percent | 40.2% | 13.0% | 46.7% | 78.3 | |
ALS amyotrophic lateral sclerosis, DC disease control, HC healthy control
Fig. 4.
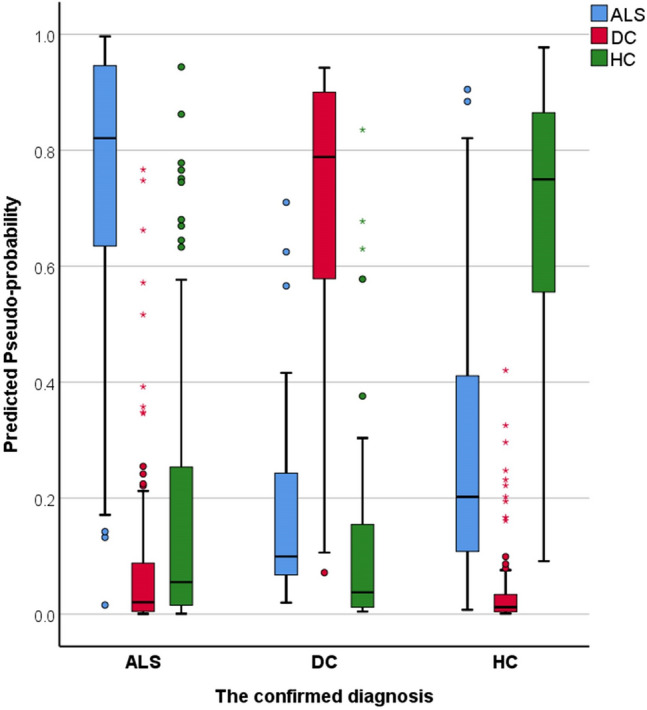
The predicted pseudo-probability profiles of ALS patients around the time of their diagnosis (< 6 weeks), disease controls (DC) and healthy controls (HC)
Fig. 5.
Receiver-operating characteristic curve of patients with ALS around the time of their diagnosis (< 6 weeks), disease controls (DC) and healthy controls (HC) using all imaging features (A—left), the 20 most important variables only (B—middle) and white matter diffusivity metrics alone (C—right)
Fig. 6.
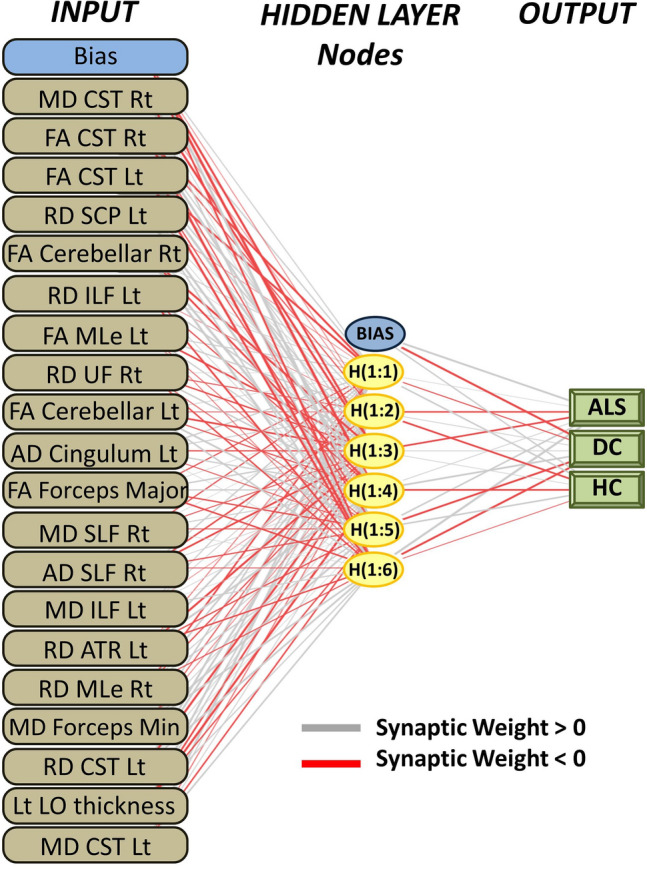
The multilayer perceptron model architecture. Input layer: 20 imaging metrics. Hidden layer: 6 nodes (units). Hidden layer activation function: hyperbolic tangent. Output layer: diagnostic label. CST corticospinal tract, SCP sup cerebellar ped, ILF inferior longitudinal fasciculus, MLe medial lemniscus, UF uncinate fasciculus, SLF superior longitudinal fasciculus, ATR anterior thalamic radiation, LO lateral occipital, Rt right, Lt left
Discussion
Our data indicate that quantitative imaging aids diagnostic classification and the systematic assessment of key anatomical regions may not only help to distinguish ALS from healthy controls, but also discriminates it from other neurodegenerative conditions. The presented framework operates in an observer-independent fashion and receiver-operating characteristic curves indicate excellent sensitivity/specificity profiles. In addition to the classification accuracy of the multilayer perceptron model utilised, the ranking of imaging features with respect to categorisation relevance offers valuable insights for the streamlining and optimisation of future models.
The utility of a variety of supervised and unsupervised machine-learning approaches have been explored in ALS, including support vector machines, regression-based approaches, random forests, discriminant function analyses, dimension reduction frameworks, but these are seldom applied to imaging data [17, 35, 36] due to challenges associated with MRI scanning, quality control, pre-processing, data acquisition costs and data harmonisation. Advanced neural network architectures have been successfully trialled in other conditions, including multilayer ‘deep-learning’ learning models and generative adversarial networks (GAN) [37–40]. The development of automated diagnostic frameworks based on radiology data in ALS are hampered by the scarcity of large, uniformly acquired training data sets. While the acquisition and recording of epidemiology data and clinical measures can be relatively easily harmonised, imaging data harmonisation requires considerable investment.
Our study consisted of an exploratory arm, where imaging metrics from the entire cerebrum were incorporated, without the a priori selection of anatomic regions considered relevant based on published imaging or post mortem evidence. While the predilection of disease burden to the corticospinal tracts, precentral gyrus and brainstem is well established, our strategy centred on the indiscriminate interrogation of imaging variables from across the entire cerebrum. To develop a truly observer-independent pipeline, individual imaging data were spatially co-registered to standard space, and only validated atlases were utilised to retrieve integrity variables from a range of anatomical regions. Cortical and subcortical, grey matter and white matter, supratentorial and infratentorial, left and right hemisphere structures were uniformly evaluated without prioritising potentially discriminatory anatomical regions a priori.
The ranking of variable importance revealed interesting trends. By large, integrity metrics of white matter regions discriminated the three groups better than grey matter measures. This is in line with previous observations that white matter degeneration is a relatively early feature of ALS [7, 41], while GM changes are less consistent, and may only become readily detectable in the later stages of the disease. A practical ramification of the recognition of the superior discriminatory power of white matter measures is that diffusion tensor protocols should be routinely incorporated into clinical and pharmacological trials protocols as opposed to only relying on T1-weighted, FLAIR and T2-weighted data sets which are classically used for clinical evaluation to rule out mimic conditions. Interestingly, there was only one grey matter variable among the first 20 diagnostically relevant imaging features and only 4 grey matter variables were ranked in the first 50 features. Our post hoc analyses also confirmed that excellent subject classification can be achieved relying on white matter measures alone (Figs. 2 and 5, Tables 1 and 3). These models provided accurate diagnostic classification without evaluating grey matter measures or volumes at all, and were solely based on measures derived from DTI. Furthermore, our results confirm the imperative of evaluating non-FA diffusivity measures. While FA is the most commonly evaluated white matter metric in descriptive analyses, RD, MD, and AD proved to be equally important discriminatory variables in our models. The review of ranked discriminating variables (Table 2.) is not only interesting from the perspective of biophysical measures, but also from an anatomical standpoint. The relative importance of key ALS-associated brain regions such as corticospinal tracts and precentral gyrus is not surprising given the ample evidence of the pathognomonic involvement of these structures in ALS. Conversely, the indices of some brain regions, such as the brainstem, ranked relatively low in the hierarchy of feature importance despite their archetypal involvement in ALS [42]. The discriminatory relevance of external capsule integrity is also of interest as ALS studies overwhelmingly emphasise internal capsule alterations [43]. It is also noteworthy that multiple cerebellar measures are among the most important discriminatory features, including intra-cerebellar white matter diffusivity metrics, volumetric values as well as cerebellar peduncle integrity measures. The recognition that cerebellar degeneration is an important facet of ALS biology is not new, but regional cerebellar disease burden has only been recently characterised in detail [44–50]. Our findings highlight the practical importance of systematically evaluating infratentorial indices in ALS and not only focussing on supratentorial variables. Several long association tracts (ILF, FOF) were also listed among the first 50 discriminatory regions, which are likely to aid discrimination from the disease-control group [51]. Frontotemporal dementia is a genetically, molecularly and clinically heterogeneous group of conditions, and specific subtypes are associated with specific imaging signatures [52, 53]. Our study illustrates the relevance of assessing brain regions which are not classically affected in ALS [54]. These regions may be preferentially affected in other conditions therefore the interrogation of imaging metrics from these anatomical foci is invaluable in discriminating ALS from alternative diagnoses. More broadly, our results support the importance of exploring imaging data without a priori anatomical assumptions.
Our approach illustrates that a multitude of metrics may be readily incorporated into complex classification models across a variety of anatomical regions. These models may be potentially further expanded to include additional measures such as wet biomarkers, additional imaging metrics or clinical measures [55–64]. In this application only cerebral measures were evaluated, despite the potential of spinal metrics [65–68]. Similar frameworks could potentially be utilised for the discrimination of other MND phenotypes such as PLS, PMA or flail-arm syndrome [69–71].
This study is not without limitations. A three-way classification scheme was implemented with a single disease-control group. The inclusion of a ‘mimic’ disease-control group would have been helpful to test the model further, but the definition of a true ALS mimic condition is contentious. Only total volumes of subcortical structures were explored as input variables in this study, even though the assessment of specific amygdalar nuclei, thalamic nuclei or hippocampal subfields may enhance the discrimination of ALS form other neurodegenerative conditions [72–75]. Moreover, our model only evaluated cerebral metrics, therefore LMN pathology is not accounted for and discrimination from LMN-predominant MNDs cannot be reliably assessed [76–82]. Additional validation of the model with presymptomatic mutation carriers would have tested the classification accuracy of the model further by evaluating subjects with limited disease burden [10, 83]. Model overfitting to a particular training cohort is invariably a significant risk and this study is no exception. Notwithstanding these limitations, our results indicate that subjects may be accurately classified into a diagnostic cohort, healthy control, or diseases control categories based on imaging data alone.
Conclusions
The meaningful interpretation of singe-subject imaging data is an urgent priority of clinical neuroradiology. Group-level descriptive analyses offer valuable academic insights, but the practical demands of clinical neuroradiology and clinical trial applications require accurate single-subject classification based on a core set of quantitative markers.
Acknowledgements
We are thankful for the participation of each patient and healthy control, and we also thank all the patients who expressed interest in this research study but were unable to participate for medical or logistical reasons. We also express our gratitude to the caregivers of patients for facilitating attendance at our neuroimaging centre. Without their generosity, this study would have not been possible.
Abbreviations
- AD
Axial diffusivity
- ALS
Amyotrophic lateral sclerosis
- ALSFRS-r
Revised amyotrophic lateral sclerosis functional rating scale
- ANN
Artificial neural network
- ATR
Anterior thalamic radiation
- AUC
Area under the curve
- C9orf72
Chromosome 9 open reading frame 72
- CST
Corticospinal tract
- CT
Cortical thickness
- DC
Disease control
- DTI
Diffusion tensor imaging
- EMM
Estimated marginal mean
- EPI
Echo-planar imaging
- FA
Fractional anisotropy
- FLAIR
Fluid-attenuated inversion recovery
- FOF
Fronto-occipital fasciculus
- FOV
Field of view
- FTD
Frontotemporal dementia
- FWE
Family-wise error
- GAN
Generative adversarial networks
- GM
Grey matter
- HC
Healthy control
- ICP
Inferior cerebellar peduncle
- ILF
Inferior longitudinal fasciculus
- IR-SPGR
Inversion recovery prepared spoiled gradient recalled echo
- IR-TSE
Inversion recovery turbo spin-echo sequence
- IVIG
Intravenous immunoglobulin
- LMN
Lower motor neuron
- LO
Lateral occipital
- Lt
Left
- MCP
Middle cerebellar peduncle
- MD
Mean diffusivity
- ML
Machine learning
- MLe
Medial lemniscus
- MND
Motor neuron disease
- MNI152
Montreal Neurological Institute 152 standard space
- MPM
Multilayer perceptron model
- NISALS
Neuroimaging Society in ALS
- PBA
Pseudobulbar affect
- PCC
Pathological crying and laughing
- PMC
Primary motor cortex
- QC
Quality control
- RD
Radial diffusivity
- ROC
Receiver-operating characteristic curve
- ROI
Region of interest
- Rt
Right
- SCP
Superior cerebellar peduncle
- SD
Standard deviation
- SE-EPI
Spin-echo echo planar imaging
- SENSE
Sensitivity encoding
- SLF
Superior longitudinal fasciculus
- SPIR
Spectral presaturation with inversion recovery
- T1w
T1-weighted imaging
- TBSS
Tract-based spatial statistics
- TE
Echo time
- TFCE
Threshold-free cluster enhancement
- TI
Inversion time
- TIV
Total intracranial volume
- TR
Repetition time
- UF
Uncinate fasciculus
- UMN
Upper motor neuron
- WM
White matter
Author contributions
Drafting the manuscript: PB. Neuroimaging analyses: PB and AM. Conceptualisation of the study: PB and AM. Revision of the manuscript for intellectual content: PB, AM, and OH.
Funding
Open Access funding provided by the IReL Consortium. Professor Peter Bede and the Computational Neuroimaging Group are supported by the Health Research Board (HRB EIA-2017-019), the Irish Institute of Clinical Neuroscience (IICN), the Spastic Paraplegia Foundation (SPF), the EU Joint Programme—Neurodegenerative Disease Research (JPND), the Andrew Lydon scholarship, and the Iris O'Brien Foundation.
Availability of data and material
Personal medical data are not publically available due to institutional data regulations.
Code availability
Statistical procedures are described in the methods section of the manuscript.
Declarations
Conflicts of interest
The authors have no competing interests to declare.
Ethics approval
This study was approved by the Ethics (Medical Research) Committee—Beaumont Hospital, Dublin, Ireland.
Consent to participate
All the participants had consented to participate.
Consent for publication
All the participants had consented to publish these research findings.
References
- 1.Cellura E, Spataro R, Taiello AC, La Bella V. Factors affecting the diagnostic delay in amyotrophic lateral sclerosis. Clin Neurol Neurosurg. 2012;114(6):550–554. doi: 10.1016/j.clineuro.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Chio A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, Traynor BG. Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler. 2009;10(5–6):310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czaplinski A, Yen AA, Appel SH. Amyotrophic lateral sclerosis: early predictors of prolonged survival. J Neurol. 2006;253(11):1428–1436. doi: 10.1007/s00415-006-0226-8. [DOI] [PubMed] [Google Scholar]
- 4.Donaghy C, Dick A, Hardiman O, Patterson V. Timeliness of diagnosis in motor neurone disease: a population-based study. Ulst Med J. 2008;77(1):18–21. [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster C, Elamin M, Hardiman O, Bede P. Presymptomatic and longitudinal neuroimaging in neurodegeneration–from snapshots to motion picture: a systematic review. J Neurol Neurosurg Psychiatry. 2015;86(10):1089–1096. doi: 10.1136/jnnp-2014-309888. [DOI] [PubMed] [Google Scholar]
- 6.Chipika RH, Finegan E, Li Hi Shing S, Hardiman O, Bede P. Tracking a fast-moving disease: longitudinal markers, monitoring, and clinical trial endpoints in ALS. Front Neurol. 2019;10:229. doi: 10.3389/fneur.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bede P, Hardiman O. Longitudinal structural changes in ALS: a three time-point imaging study of white and gray matter degeneration. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19(3–4):232–241. doi: 10.1080/21678421.2017.1407795. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand A, Wen J, Rinaldi D, Houot M, Sayah S, Camuzat A, Fournier C, Fontanella S, Routier A, Couratier P, Pasquier F, Habert MO, Hannequin D, Martinaud O, Caroppo P, Levy R, Dubois B, Brice A, Durrleman S, Colliot O, Le Ber I. Early cognitive, structural, and microstructural changes in presymptomatic C9orf72 carriers younger than 40 years. JAMA Neurol. 2018;75(2):236–245. doi: 10.1001/jamaneurol.2017.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen J, Zhang H, Alexander DC, Durrleman S, Routier A, Rinaldi D, Houot M, Couratier P, Hannequin D, Pasquier F, Zhang J, Colliot O, Le Ber I, Bertrand A. Neurite density is reduced in the presymptomatic phase of C9orf72 disease. J Neurol Neurosurg Psychiatry. 2019;90(4):387–394. doi: 10.1136/jnnp-2018-318994. [DOI] [PubMed] [Google Scholar]
- 10.Chipika RH, Siah WF, McKenna MC, Li Hi Shing S, Hardiman O, Bede P. The presymptomatic phase of amyotrophic lateral sclerosis: are we merely scratching the surface? J Neurol. 2020 doi: 10.1007/s00415-020-10289-5. [DOI] [PubMed] [Google Scholar]
- 11.Bede P, Siah WF, McKenna MC, Li Hi Shing S. Consideration of C9orf72-associated ALS-FTD as a neurodevelopmental disorder: insights from neuroimaging. J Neurol Neurosurg Psychiatry. 2020 doi: 10.1136/jnnp-2020-324416. [DOI] [PubMed] [Google Scholar]
- 12.Agosta F, Spinelli EG, Filippi M. Neuroimaging in amyotrophic lateral sclerosis: current and emerging uses. Expert Rev Neurother. 2018;18(5):395–406. doi: 10.1080/14737175.2018.1463160. [DOI] [PubMed] [Google Scholar]
- 13.Agosta F, Ferraro PM, Riva N, Spinelli EG, Chio A, Canu E, Valsasina P, Lunetta C, Iannaccone S, Copetti M, Prudente E, Comi G, Falini A, Filippi M. Structural brain correlates of cognitive and behavioral impairment in MND. Hum Brain Mapp. 2016;37(4):1614–1626. doi: 10.1002/hbm.23124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller HP, Turner MR, Grosskreutz J, Abrahams S, Bede P, Govind V, Prudlo J, Ludolph AC, Filippi M, Kassubek J. A large-scale multicentre cerebral diffusion tensor imaging study in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2016;87(6):570–579. doi: 10.1136/jnnp-2015-311952. [DOI] [PubMed] [Google Scholar]
- 15.Agosta F, Spinelli EG, Marjanovic IV, Stevic Z, Pagani E, Valsasina P, Salak-Djokic B, Jankovic M, Lavrnic D, Kostic VS, Filippi M. Unraveling ALS due to SOD1 mutation through the combination of brain and cervical cord MRI. Neurology. 2018 doi: 10.1212/wnl.0000000000005002. [DOI] [PubMed] [Google Scholar]
- 16.Ferraro PM, Agosta F, Riva N, Copetti M, Spinelli EG, Falzone Y, Sorarù G, Comi G, Chiò A, Filippi M. Multimodal structural MRI in the diagnosis of motor neuron diseases. NeuroImage Clin. 2017;16:240–247. doi: 10.1016/j.nicl.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grollemund V, Pradat PF, Querin G, Delbot F, Le Chat G, Pradat-Peyre JF, Bede P. Machine learning in amyotrophic lateral sclerosis: achievements, pitfalls, and future directions. Front Neurosci. 2019;13:135. doi: 10.3389/fnins.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grollemund V, Le Chat G, Secchi-Buhour MS, Delbot F, Pradat-Peyre JF, Bede P, Pradat PF. Manifold learning for amyotrophic lateral sclerosis functional loss assessment : Development and validation of a prognosis model. J Neurol. 2020 doi: 10.1007/s00415-020-10181-2. [DOI] [PubMed] [Google Scholar]
- 19.Bede P, Querin G, Pradat PF. The changing landscape of motor neuron disease imaging: the transition from descriptive studies to precision clinical tools. Curr Opin Neurol. 2018;31(4):431–438. doi: 10.1097/wco.0000000000000569. [DOI] [PubMed] [Google Scholar]
- 20.Schuster C, Hardiman O, Bede P. Development of an automated MRI-based diagnostic protocol for amyotrophic lateral sclerosis using disease-specific pathognomonic features: a quantitative disease-state classification study. PLoS ONE. 2016;11(12):e0167331. doi: 10.1371/journal.pone.0167331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsh RC, Jelsone-Swain LM, Foerster BR. The utility of independent component analysis and machine learning in the identification of the amyotrophic lateral sclerosis diseased brain. Front Hum Neurosci. 2013;7:251. doi: 10.3389/fnhum.2013.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bede P, Iyer PM, Finegan E, Omer T, Hardiman O. Virtual brain biopsies in amyotrophic lateral sclerosis: diagnostic classification based on in vivo pathological patterns. NeuroImage Clin. 2017;15:653–658. doi: 10.1016/j.nicl.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahedl M, Chipika RH, Lope J, Li Hi Shing S, Hardiman O, Bede P. Cortical progression patterns in individual ALS patients across multiple timepoints: a mosaic-based approach for clinical use. J Neurol. 2021;268(5):1913–1926. doi: 10.1007/s00415-020-10368-7. [DOI] [PubMed] [Google Scholar]
- 24.Tahedl M, Murad A, Lope J, Hardiman O, Bede P. Evaluation and categorisation of individual patients based on white matter profiles: single-patient diffusion data interpretation in neurodegeneration. J Neurol Sci. 2021;428:117584. doi: 10.1016/j.jns.2021.117584. [DOI] [PubMed] [Google Scholar]
- 25.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iglesias JE, Van Leemput K, Bhatt P, Casillas C, Dutt S, Schuff N, Truran-Sacrey D, Boxer A, Fischl B. Bayesian segmentation of brainstem structures in MRI. Neuroimage. 2015;113:184–195. doi: 10.1016/j.neuroimage.2015.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori S, Wakana S, Van Zijl P, Nagae-Poetscher L. MRI atlas of human white matter. The Netherlands: Elsevier; 2005. [Google Scholar]
- 30.Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage. 2007;36(3):630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage. 2008;39(1):336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;3(3):190–208. doi: 10.1002/hbm.460030304. [DOI] [Google Scholar]
- 33.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown CA, Johnson NF, Anderson-Mooney AJ, Jicha GA, Shaw LM, Trojanowski JQ, Van Eldik LJ, Schmitt FA, Smith CD, Gold BT. Development, validation and application of a new fornix template for studies of aging and preclinical Alzheimer's disease. NeuroImage Clin. 2017;13:106–115. doi: 10.1016/j.nicl.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grollemund V, Chat GL, Secchi-Buhour MS, Delbot F, Pradat-Peyre JF, Bede P, Pradat PF. Development and validation of a 1-year survival prognosis estimation model for Amyotrophic Lateral Sclerosis using manifold learning algorithm UMAP. Sci Rep. 2020;10(1):13378. doi: 10.1038/s41598-020-70125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster C, Hardiman O, Bede P. Survival prediction in Amyotrophic lateral sclerosis based on MRI measures and clinical characteristics. BMC Neurol. 2017;17(1):73. doi: 10.1186/s12883-017-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65(12):2720–2730. doi: 10.1109/tbme.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amato F, López A, Peña-Méndez E, VbHhara P, Hampl A, Havel J. Artificial neural networks in medical diagnosis. J Appl Biomed. 2013;11:47–58. doi: 10.2478/v10136-012-0031-x. [DOI] [Google Scholar]
- 39.Lisboa PJ, Taktak AF. The use of artificial neural networks in decision support in cancer: a systematic review. Neural Netw. 2006;19(4):408–415. doi: 10.1016/j.neunet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10(3):257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 41.Menke RAL, Proudfoot M, Talbot K, Turner MR. The two-year progression of structural and functional cerebral MRI in amyotrophic lateral sclerosis. NeuroImage Clin. 2018;17:953–961. doi: 10.1016/j.nicl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bede P, Chipika RH, Finegan E, Li Hi Shing S, Doherty MA, Hengeveld JC, Vajda A, Hutchinson S, Donaghy C, McLaughlin RL, Hardiman O. Brainstem pathology in amyotrophic lateral sclerosis and primary lateral sclerosis: a longitudinal neuroimaging study. NeuroImage Clin. 2019;24:102054. doi: 10.1016/j.nicl.2019.102054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster C, Elamin M, Hardiman O, Bede P. The segmental diffusivity profile of amyotrophic lateral sclerosis associated white matter degeneration. Eur J Neurol. 2016;23(8):1361–1371. doi: 10.1111/ene.13038. [DOI] [PubMed] [Google Scholar]
- 44.Abidi M, de Marco G, Couillandre A, Feron M, Mseddi E, Termoz N, Querin G, Pradat PF, Bede P. Adaptive functional reorganization in amyotrophic lateral sclerosis: coexisting degenerative and compensatory changes. Eur J Neurol. 2020;27(1):121–128. doi: 10.1111/ene.14042. [DOI] [PubMed] [Google Scholar]
- 45.Abidi M, de Marco G, Grami F, Termoz N, Couillandre A, Querin G, Bede P, Pradat PF. Neural correlates of motor imagery of gait in amyotrophic lateral sclerosis. J Magn Reson Imaging. 2021;53(1):223–233. doi: 10.1002/jmri.27335. [DOI] [PubMed] [Google Scholar]
- 46.Feron M, Couillandre A, Mseddi E, Termoz N, Abidi M, Bardinet E, Delgadillo D, Lenglet T, Querin G, Welter ML, Le Forestier N, Salachas F, Bruneteau G, Del Mar AM, Debs R, Lacomblez L, Meininger V, Pelegrini-Issac M, Bede P, Pradat PF, de Marco G. Extrapyramidal deficits in ALS: a combined biomechanical and neuroimaging study. J Neurol. 2018;265(9):2125–2136. doi: 10.1007/s00415-018-8964-y. [DOI] [PubMed] [Google Scholar]
- 47.McKenna MC, Chipika RH, Li Hi Shing S, Christidi F, Lope J, Doherty MA, Hengeveld JC, Vajda A, McLaughlin RL, Hardiman O, Hutchinson S, Bede P. Infratentorial pathology in frontotemporal dementia: cerebellar grey and white matter alterations in FTD phenotypes. J Neurol. 2021 doi: 10.1007/s00415-021-10575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bede P, Chipika RH, Christidi F, Hengeveld JC, Karavasilis E, Argyropoulos GD, Lope J, Li Hi Shing S, Velonakis G, Dupuis L, Doherty MA, Vajda A, McLaughlin RL, Hardiman O. Genotype-associated cerebellar profiles in ALS: focal cerebellar pathology and cerebro-cerebellar connectivity alterations. J Neurol Neurosurg Psychiatry. 2021 doi: 10.1136/jnnp-2021-326854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharti K, Khan M, Beaulieu C, Graham SJ, Briemberg H, Frayne R, Genge A, Korngut L, Zinman L, Kalra S. Involvement of the dentate nucleus in the pathophysiology of amyotrophic lateral sclerosis: a multi-center and multi-modal neuroimaging study. NeuroImage Clin. 2020;28:102385. doi: 10.1016/j.nicl.2020.102385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu S, Menke RAL, Talbot K, Kiernan MC, Turner MR. Cerebellar tract alterations in PLS and ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(3–4):281–284. doi: 10.1080/21678421.2018.1562554. [DOI] [PubMed] [Google Scholar]
- 51.Christidi F, Karavasilis E, Rentzos M, Kelekis N, Evdokimidis I, Bede P. Clinical and radiological markers of extra-motor deficits in amyotrophic lateral sclerosis. Front Neurol. 2018;9:1005. doi: 10.3389/fneur.2018.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agosta F, Galantucci S, Magnani G, Marcone A, Martinelli D, Antonietta Volonte M, Riva N, Iannaccone S, Ferraro PM, Caso F, Chio A, Comi G, Falini A, Filippi M. MRI signatures of the frontotemporal lobar degeneration continuum. Hum Brain Mapp. 2015;36(7):2602–2614. doi: 10.1002/hbm.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bede P, Omer T, Finegan E, Chipika RH, Iyer PM, Doherty MA, Vajda A, Pender N, McLaughlin RL, Hutchinson S, Hardiman O. Connectivity-based characterisation of subcortical grey matter pathology in frontotemporal dementia and ALS: a multimodal neuroimaging study. Brain Imaging Behav. 2018;12(6):1696–1707. doi: 10.1007/s11682-018-9837-9. [DOI] [PubMed] [Google Scholar]
- 54.Bede P, Iyer PM, Schuster C, Elamin M, McLaughlin RL, Kenna K, Hardiman O. The selective anatomical vulnerability of ALS: 'disease-defining' and 'disease-defying' brain regions. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17(7–8):561–570. doi: 10.3109/21678421.2016.1173702. [DOI] [PubMed] [Google Scholar]
- 55.Blasco H, Patin F, Descat A, Garcon G, Corcia P, Gele P, Lenglet T, Bede P, Meininger V, Devos D, Gossens JF, Pradat PF. A pharmaco-metabolomics approach in a clinical trial of ALS: identification of predictive markers of progression. PLoS ONE. 2018;13(6):e0198116. doi: 10.1371/journal.pone.0198116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devos D, Moreau C, Kyheng M, Garcon G, Rolland AS, Blasco H, Gele P, Timothee Lenglet T, Veyrat-Durebex C, Corcia P, Dutheil M, Bede P, Jeromin A, Oeckl P, Otto M, Meninger V, Danel-Brunaud V, Devedjian JC, Duce JA, Pradat PF. A ferroptosis-based panel of prognostic biomarkers for Amyotrophic Lateral Sclerosis. Sci Rep. 2019;9(1):2918. doi: 10.1038/s41598-019-39739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dukic S, McMackin R, Buxo T, Fasano A, Chipika R, Pinto-Grau M, Costello E, Schuster C, Hammond M, Heverin M, Coffey A, Broderick M, Iyer PM, Mohr K, Gavin B, Pender N, Bede P, Muthuraman M, Lalor EC, Hardiman O, Nasseroleslami B. Patterned functional network disruption in amyotrophic lateral sclerosis. Hum Brain Mapp. 2019;40(16):4827–4842. doi: 10.1002/hbm.24740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nasseroleslami B, Dukic S, Broderick M, Mohr K, Schuster C, Gavin B, McLaughlin R, Heverin M, Vajda A, Iyer PM, Pender N, Bede P, Lalor EC, Hardiman O. Characteristic increases in EEG connectivity correlate with changes of structural MRI in amyotrophic lateral sclerosis. Cereb Cortex. 2019;29(1):27–41. doi: 10.1093/cercor/bhx301. [DOI] [PubMed] [Google Scholar]
- 59.Iyer PM, Mohr K, Broderick M, Gavin B, Burke T, Bede P, Pinto-Grau M, Pender NP, McLaughlin R, Vajda A, Heverin M, Lalor EC, Hardiman O, Nasseroleslami B. Mismatch negativity as an indicator of cognitive sub-domain dysfunction in amyotrophic lateral sclerosis. Front Neurol. 2017;8:395. doi: 10.3389/fneur.2017.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Proudfoot M, Bede P, Turner MR. Imaging cerebral activity in amyotrophic lateral sclerosis. Front Neurol. 2018;9:1148. doi: 10.3389/fneur.2018.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verstraete E, Turner MR, Grosskreutz J, Filippi M, Benatar M. Mind the gap: the mismatch between clinical and imaging metrics in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(7–8):524–529. doi: 10.3109/21678421.2015.1051989. [DOI] [PubMed] [Google Scholar]
- 62.Burke T, Pinto-Grau M, Lonergan K, Elamin M, Bede P, Costello E, Hardiman O, Pender N. Measurement of social cognition in amyotrophic lateral sclerosis: a population based study. PLoS ONE. 2016;11(8):e0160850. doi: 10.1371/journal.pone.0160850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burke T, Elamin M, Bede P, Pinto-Grau M, Lonergan K, Hardiman O, Pender N. Discordant performance on the 'Reading the Mind in the Eyes' Test, based on disease onset in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2016 doi: 10.1080/21678421.2016.1177088. [DOI] [PubMed] [Google Scholar]
- 64.Finegan E, Chipika RH, Li Hi Shing S, Hardiman O, Bede P. Crying and laughing in motor neuron disease: pathobiology, screening, intervention. Front Neurol. 2019;10:260. doi: 10.3389/fneur.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Querin G, El Mendili MM, Bede P, Delphine S, Lenglet T, Marchand-Pauvert V, Pradat PF. Multimodal spinal cord MRI offers accurate diagnostic classification in ALS. J Neurol Neurosurg Psychiatry. 2018;89(11):1220–1221. doi: 10.1136/jnnp-2017-317214. [DOI] [PubMed] [Google Scholar]
- 66.El Mendili MM, Querin G, Bede P, Pradat PF. Spinal cord imaging in amyotrophic lateral sclerosis: historical concepts-novel techniques. Front Neurol. 2019;10:350. doi: 10.3389/fneur.2019.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Querin G, El Mendili MM, Lenglet T, Behin A, Stojkovic T, Salachas F, Devos D, Le Forestier N, Del Mar AM, Debs R, Lacomblez L, Meninger V, Bruneteau G, Cohen-Adad J, Lehericy S, Laforet P, Blancho S, Benali H, Catala M, Li M, Marchand-Pauvert V, Hogrel JY, Bede P, Pradat PF. The spinal and cerebral profile of adult spinal-muscular atrophy: a multimodal imaging study. NeuroImage Clin. 2019;21:101618. doi: 10.1016/j.nicl.2018.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valsasina P, Agosta F, Benedetti B, Caputo D, Perini M, Salvi F, Prelle A, Filippi M. Diffusion anisotropy of the cervical cord is strictly associated with disability in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2007;78(5):480–484. doi: 10.1136/jnnp.2006.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finegan E, Chipika RH, Shing SLH, Hardiman O, Bede P. Primary lateral sclerosis: a distinct entity or part of the ALS spectrum? Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(3–4):133–145. doi: 10.1080/21678421.2018.1550518. [DOI] [PubMed] [Google Scholar]
- 70.Finegan E, Li Hi Shing S, Siah WF, Chipika RH, Chang KM, McKenna MC, Doherty MA, Hengeveld JC, Vajda A, Donaghy C, Hutchinson S, McLaughlin RL, Hardiman O, Bede P. Evolving diagnostic criteria in primary lateral sclerosis: the clinical and radiological basis of “probable PLS”. J Neurol Sci. 2020;417:117052. doi: 10.1016/j.jns.2020.117052. [DOI] [PubMed] [Google Scholar]
- 71.Finegan E, Chipika RH, Li Hi Shing S, Doherty MA, Hengeveld JC, Vajda A, Donaghy C, McLaughlin RL, Pender N, Hardiman O, Bede P. The clinical and radiological profile of primary lateral sclerosis: a population-based study. J Neurol. 2019;266(11):2718–2733. doi: 10.1007/s00415-019-09473-z. [DOI] [PubMed] [Google Scholar]
- 72.Christidi F, Karavasilis E, Rentzos M, Velonakis G, Zouvelou V, Xirou S, Argyropoulos G, Papatriantafyllou I, Pantolewn V, Ferentinos P, Kelekis N, Seimenis I, Evdokimidis I, Bede P. Hippocampal pathology in amyotrophic lateral sclerosis: selective vulnerability of subfields and their associated projections. Neurobiol Aging. 2019;84:178–188. doi: 10.1016/j.neurobiolaging.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 73.Finegan E, Li Hi Shing S, Chipika RH, Doherty MA, Hengeveld JC, Vajda A, Donaghy C, Pender N, McLaughlin RL, Hardiman O, Bede P. Widespread subcortical grey matter degeneration in primary lateral sclerosis: a multimodal imaging study with genetic profiling. NeuroImage Clin. 2019;24:102089. doi: 10.1016/j.nicl.2019.102089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chipika RH, Finegan E, Li Hi Shing S, McKenna MC, Christidi F, Chang KM, Doherty MA, Hengeveld JC, Vajda A, Pender N, Hutchinson S, Donaghy C, McLaughlin RL, Hardiman O, Bede P. "Switchboard" malfunction in motor neuron diseases: selective pathology of thalamic nuclei in amyotrophic lateral sclerosis and primary lateral sclerosis. NeuroImage Clin. 2020;27:102300. doi: 10.1016/j.nicl.2020.102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chipika RH, Christidi F, Finegan E, Li Hi Shing S, McKenna MC, Chang KM, Karavasilis E, Doherty MA, Hengeveld JC, Vajda A, Pender N, Hutchinson S, Donaghy C, McLaughlin RL, Hardiman O, Bede P. Amygdala pathology in amyotrophic lateral sclerosis and primary lateral sclerosis. J Neurol Sci. 2020 doi: 10.1016/j.jns.2020.117039. [DOI] [PubMed] [Google Scholar]
- 76.Lebouteux MV, Franques J, Guillevin R, Delmont E, Lenglet T, Bede P, Desnuelle C, Pouget J, Pascal-Mousselard H, Pradat PF. Revisiting the spectrum of lower motor neuron diseases with snake eyes appearance on magnetic resonance imaging. Eur J Neurol. 2014;21(9):1233–1241. doi: 10.1111/ene.12465. [DOI] [PubMed] [Google Scholar]
- 77.Querin G, Lenglet T, Debs R, Stojkovic T, Behin A, Salachas F, Le Forestier N, Amador MDM, Bruneteau G, Laforêt P, Blancho S, Marchand-Pauvert V, Bede P, Hogrel JY, Pradat PF. Development of new outcome measures for adult SMA type III and IV: a multimodal longitudinal study. J Neurol. 2021 doi: 10.1007/s00415-020-10332-5. [DOI] [PubMed] [Google Scholar]
- 78.Querin G, Bede P, Marchand-Pauvert V, Pradat PF. Biomarkers of spinal and bulbar muscle atrophy (SBMA): a comprehensive review. Front Neurol. 2018;9:844. doi: 10.3389/fneur.2018.00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pradat PF, Bernard E, Corcia P, Couratier P, Jublanc C, Querin G, Morelot Panzini C, Salachas F, Vial C, Wahbi K, Bede P, Desnuelle C. The French national protocol for Kennedy's disease (SBMA): consensus diagnostic and management recommendations. Orphanet J Rare Dis. 2020;15(1):90. doi: 10.1186/s13023-020-01366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Hi Shing S, Chipika RH, Finegan E, Murray D, Hardiman O, Bede P. Post-polio syndrome: more than just a lower motor neuron disease. Front Neurol. 2019;10:773. doi: 10.3389/fneur.2019.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spinelli EG, Agosta F, Ferraro PM, Querin G, Riva N, Bertolin C, Martinelli I, Lunetta C, Fontana A, Sorarù G, Filippi M. Brain MRI shows white matter sparing in Kennedy's disease and slow-progressing lower motor neuron disease. Hum Brain Mapp. 2019;40(10):3102–3112. doi: 10.1002/hbm.24583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spinelli EG, Agosta F, Ferraro PM, Riva N, Lunetta C, Falzone YM, Comi G, Falini A, Filippi M. Brain MR imaging in patients with lower motor neuron-predominant disease. Radiology. 2016;280(2):545–556. doi: 10.1148/radiol.2016151846. [DOI] [PubMed] [Google Scholar]
- 83.Querin G, Bede P, El Mendili MM, Li M, Pelegrini-Issac M, Rinaldi D, Catala M, Saracino D, Salachas F, Camuzat A, Marchand-Pauvert V, Cohen-Adad J, Colliot O, Le Ber I, Pradat PF. Presymptomatic spinal cord pathology in c9orf72 mutation carriers: a longitudinal neuroimaging study. Ann Neurol. 2019;86(2):158–167. doi: 10.1002/ana.25520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Personal medical data are not publically available due to institutional data regulations.
Statistical procedures are described in the methods section of the manuscript.