Abstract
l-Nucleoside analogs are new therapeutic agents for treatment of chronic hepatitis B. However, their clinical application was limited by the emergence of viral resistance. It is important to develop a new system to evaluate drug cross-resistance and to test new agents that may overcome resistant virus. In this report, three cell lines HepG2-WT10, HepG2-SM1, and HepG2-DM2 are presented; these cell lines were established by transfection of HepG2 cells with unique fully functional 1.1× hepatitis B virus (HBV) genomes: wild-type HBV-adr and its L526M and L526MM550V variants, respectively. We have demonstrated that these genomes have different susceptibilities to lamivudine [l(−)SddC] and penciclovir (PCV). By examining HBV RNA transcription, antigen expression, progeny DNA replication, and viral susceptibilities to l(−)SddC, PCV, and other nucleoside analogs, it is concluded that the cell lines are able to stably produce l(−)SddC- and PCV-sensitive and -resistant HBV virions. In addition, the relative susceptibilities of the wild-type and mutant HBV produced from the stably transfected cell lines to several anti-HBV nucleoside analogs were also examined and found to be about the same as those found by using a transient infection system. PMEA [9-(2-phosphonylmethoxytehyl)-adenine] and QYL685 are able to suppress l(−)SddC- and PCV-resistant HBV. In conclusion, this cell culture system is a novel and useful tool for evaluating anti-HBV compounds and biologics.
Chronic hepatitis B virus (HBV) infection, one of the major causes of hepatocellular carcinoma and liver cirrhosis, affects more than 300 million people in the world. l(−)SddC (lamivudine) was demonstrated in our laboratory and in clinical trials by others to be an active anti-HBV drug (5, 6, 14). In clinical evaluations, rapid reduction of viral load in chronic HBV patient serum to undetectable levels could be achieved by l(−)SddC treatment. However, long-term treatment was required and drug resistance emerged during the prolonged l(−)SddC monotherapy (15, 21, 28, 32). Famciclovir (prodrug of penciclovir [PCV]) is another nucleoside analog in clinical trials for treatment of chronic HBV patients. Cases of resistance to famciclovir in patients were also reported (2, 38). Based on the clinical experience of using antiviral compounds, including lamivudine and famciclovir, for the treatment of patients with HBV and human immunodeficiency virus (HIV) infection (12, 18, 27, 30, 38), combinations of drugs with different mechanisms and spectra of anti-HBV activity for treatment will be needed to achieve the highest viral inhibition, minimize the possibility of developing resistance, and overcome HBV resistance. Therefore, establishment of an in vitro system for evaluating the activity of compounds against resistant virus is important.
Due to the lack of an efficient in vitro HBV cell culture system, it is not possible to continuously passage the virus to select for drug-resistant HBV variants as is specified by the protocol used for HIV study (36, 40). The stable HBV-producing cell lines, similar to 2.2.15 cells (27), which possess HBV DNA synthesis apparatus with different sensitivities to antiviral nucleoside analogs would be useful for selecting novel compounds against resistant HBV. The 2.2.15 cell line, established by G. Acs, is a stable HBV-producing cell line derived from the human hepatoma cell line HepG2. This cell line has been used extensively to assay the anti-HBV activities of compounds, including the original discovery of the use of l(−)SddC for the treatment of patients infected with HBV (6, 16, 17, 19). Recently, a tetracycline-inducible system producing wild-type HBV and an M550V variant was developed to study the susceptibility of the M550V variant to l(−)SddC and other nucleoside analogs (in the system used in the cited studies, the position of the amino acid change in the HBV polymerase is numbered M539) (20, 35).
The antiviral action of nucleoside analogs, such as l(−)SddC, against HBV and HIV is due to inhibition by their phosphorylated metabolites of viral-polymerase-catalyzed DNA chain elongation. The amino acid change of methionine, (M550) to either isoleucine or valine at the YMDD motif of HIV reverse transcriptase (RT) was demonstrated to be the key mechanism responsible for l(−)SddC resistance in AIDS patients. This motif is involved in nucleotide binding, which is also conserved in the HBV endogenous DNA polymerase RT domain. Based on clinical findings, a YMDD mutation (M550V or M550I) in the HBV RT is present in all the cases of l(−)SddC-resistant virus. Furthermore, it is found that M550V coupled with a leucine-to-methionine change at B domain position 526 (L526M) is the most common pattern, accounting for 80% of the cases (24, 25, 33). L526M also represents one of the most common changes in HBV polymerase identified in patients developing famciclovir resistance. Using in vitro transient transfection systems, it was demonstrated that these mutations were responsible for the development of HBV resistance to l(−)SddC and/or PCV (1, 3, 8, 9, 23, 26, 39). Therefore, we generated cell lines which can produce either wild-type HBV (adr substrain) or its L526M and L526MM550V mutants, by transfecting and integrating the HBV genomes into human hepatoma cell line HepG2. The availability of these cell lines will be useful for the discovery of new HBV drugs which are active against l(−)SddC- and/or PCV-resistant HBV.
MATERIALS AND METHODS
Chemicals.
l(−)SddC [l-(−)2′,3′-dideoxythiacytidine, 3TC] and l-FMAU (2′-fluoro-5-methyl-β-l-arabino-furanosyluracil) were provided by Chung K. Chu from the University of Georgia. l(−)Fd4C [2′,3′-dideoxy-2′,3′-didehydro-β-l(−)-5-fluorocytidine] was synthesized in the laboratory of the late Tai-Shun Lin at Yale University. PMEA [9-(2-phosphonylmethoxyethyl)-adenine] was provided by Gilead Sciences Inc. PCV was purchased from Moravek Biochemicals (Brea, Calif.). QYL685 was provided by Jiri Zemlicka from Wayne State University (33a).
Construction of HBV vectors.
HBV replication is via reverse transcription of 3.5-kb pregenomic RNA. The minimally sized functional HBV genome requires the entire transcriptional unit. The HBV genome construct used in this study was 1.1 times the length of the HBV genome, starting from enhancer II upstream of the core promoter, containing a 3′-terminal redundant region necessary for the transcription of all viral RNAs. This construct ensured that the HBV genome was able to express the viral RNAs, proteins, and virions driven by HBV autologous promoters and enhancers without help from any foreign regulation elements.
The recombinant retroviral vectors pG1Na-WT, pG1Na-SM, and pG1Na-DM were constructed by inserting wild-type and L526M and L526MM550V mutant constructs, 1.1 times the fully functional HBV genome, into the retroviral vector pG1Na (gift from Genetics Therapy Inc., Gaithersburg, Md.) downstream of the neomycin resistance gene. Figure 1 shows the genomic organization of the construct. The wild-type construct was derived from pHBV-adr, which was a gift from Yuan Wang, Shanghai Institute of Biochemistry, Academia Sinica (10, 11). The HBV mutants L526M and L526MM550V were generated by site-directed mutagenesis (8). All cloning procedures were performed by standard techniques (34).
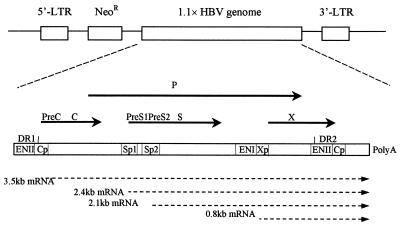
FIG. 1.
Genomic organization of retroviral vector pG1Na carrying the HBV genome 1.1 times the length of the fully functional HBV genome. A portion of HBV genome construction was adopted from a previous study (10). Solid arrows, HBV open reading frames; dashed arrows, mRNA transcripts driven by HBV promoters. Abbreviations: LTR, long terminal repeat sequence; NeoR, neomycin resistance gene; P, HBV DNA polymerase; C, HBV core antigen; S, HBsAg; X, HBV X protein; DR, direct repeat sequence; ENI and ENII, HBV enhancers I and II, respectively; Cp Sp, and Xp, HBV core gene, surface antigen gene, and X gene promoters, respectively.
Cells, transfection, and colony selection.
Human hepatoma HepG2 cells (from the American Type Culture Collection) were maintained in minimum essential medium (MEM) with 10% fetal bovine serum. Cells (2 × 105) were seeded into 100-mm-diameter culture dishes. Twenty micrograms of recombinant HBV vector plasmid was transfected by calcium precipitation. Twenty-four hours after transfection, the cells were split and cultured in G418 selection MEM (1 mg/ml). Culture medium was replaced every 3 days until colonies were formed. Individual colonies were isolated and seeded into six-well plates. After the cells grew to confluence, their ability to secrete HBV surface antigen (HBsAg) and e antigen (HBeAg) was tested by an immunoassay system (Abbott Laboratories). The cells capable of secreting both antigens were chosen for further characterization, as described below.
Analysis of HBV viral progeny DNA replication.
Cells (105) were seeded into six-well plates and allowed to grow to confluence. The medium was replaced every 3 days. After 12 days, the culture medium and cell lysate were harvested for analysis of the extracellular and intracellular viral progeny DNA by Southern hybridization. The viral DNA was prepared as follows.
(i) Extracellular.
The culture medium was harvested and centrifuged for 10 min at 2,000 × g to discard the floating cells and cell debris. The virus in the cleared culture medium was precipitated by polyethylene glycol 8000 (10%) in 0.5 M NaCl at 4°C overnight. After being centrifuged for 30 min at 16,000 × g, the virus was resuspended in 50 mM Tris-HCl (pH 8.0)–10 mM EDTA–100 mM NaCl–0.5% sodium dodecyl sulfate and digested with proteinase K (100 μg/ml) for 3 h at 50°C. After phenol extraction, the secreted virion DNA was recovered by ethanol precipitation.
(ii) Intracellular.
Cells were harvested and lysed in 1% NP-40–50 mM Tris-HCl at 37°C for 10 min followed by centrifuging for 2 min at 16,000 × g. The cleared cell lysate was treated by DNase I (100 μg/ml) with 10 mM MgCl2 and RNase A (100 μg/ml) at 37°C for 60 min. The digestion of viral capsid protein and recovery of the viral DNA were as described above.
Analysis of HBV RNA transcription.
Cells were grown in 100-mm-diameter dishes to confluence, and total RNAs were prepared by the RNeasy total RNA system (Qiagen). Northern analysis used 10 μg of total RNA per lane on a 1% agarose-formaldehyde gel. The RNA blot was hybridized with an HBV probe. Albumin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probes were used as internal controls.
Time course of HBV antigen secretion and viral production.
Cells were seeded into six-well plates at the density of 105 cells/well. Every 3 days one group of samples was harvested for the analysis of HBsAg and HBeAg secretion and viral production. The culture medium (200 μl) or a diluted solution of it was measured by the Abbott immunoassay system. Absorbance at 492 nm, representing the quantity of antigens in 200 μl of medium, was plotted against days of culture. Extracellular and intracellular viral progeny DNAs were detected by Southern hybridization as described above.
Assays of anti-HBV activity and cytotoxicity of drugs in the cell lines.
The effects of the compounds on HBV DNA replication were assessed as described previously with modification (6). Cells were seeded at the density of 125 cells/mm2. After 4 days of incubation, the medium was replaced with fresh medium containing drugs at various concentrations. Two days later, medium was again replaced with fresh medium containing the same drugs. After an additional 3-day treatment, half the volume of fresh medium containing the same drugs was added to continue treatment for three more days. After a total of 12 days, the cells were harvested and the intracellular progeny DNA replication was detected by Southern blot analysis. By using Molecular Dynamics personal densitometer SI with ImageQuaNT image analysis software, the HBV DNA bands were identified in the image. The quantitative analysis was done with this system.
For the cytotoxicity assay, cells were plated at a density of 5,000 cells/well in 24-well plates. Drug was added 24 h after seeding, and the cultures were maintained for 3 days in logarithmic growth. The methylene blue staining method was performed to determine the growth inhibition of the drug (7).
Statistical analysis.
Statistical differences between means were determined with a two-sided unpaired t test with the null hypothesis of no difference, and a P value of <0.05 was defined as a statistically significant difference.
RESULTS
Identification of cell colonies.
HepG2 cells were transfected by the vectors containing wild-type and L526M and M550V mutant HBV genomes and selected for resistance to G418. Twenty-four colonies from each transfected genome type were isolated and amplified for identification of viral production. The expression of HBV antigens HBsAg and HBeAg was detected. The cell clones, which were both HBsAg and HBeAg positive, were chosen to identify the extracellular virion DNA. There were six, two, and one positive virion producing colonies derived from the three groups of cells transfected by wild-type, single-mutation, and double-mutation HBV genomes, respectively. Among these cell clones, one from each group was chosen as the cell line for further characterization: HepG2-WT10, HepG2-SM1, and HepG2-DM2, respectively. The integrated HBV genomes in the chromosome DNA which encoded sequences including amino acids L526 and M550 in HBV polymerase were confirmed by sequencing (data not shown).
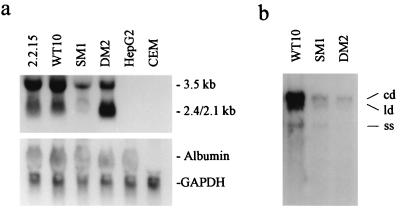
Selected cell lines HepG2-WT10, HepG2-SM1, and HepG2-DM2 were analyzed to determine the level of the expression of HBV RNA and the replication of virion DNA. As shown in Fig. 2a, HBV RNAs were transcribed in HepG2-WT10, HepG2-SM1, and HepG2-DM2 cells at different levels. HBV-transfected 2.2.15 cells were a positive control, and their parental cell line, HepG2, and leukemic CEM cells were used as negative controls. The RNA amount was normalized by using the housekeeping gene encoding GAPDH and the hepatocyte specific gene encoding albumin as internal controls. The Northern blot analysis indicated that all three HBV-transfected cell lines expressed viral transcripts. The Southern hybridization result of extracellular HBV DNA in the 12-day culture media showed that the cell lines secrete HBV in different amounts (Fig. 2b).
FIG. 2.
Characterization of HBV expression in the cell lines HepG2-WT10, HepG2-SM1, and HepG2-DM2, which were stably transfected by wild-type HBV and L526M and L526MM550V HBV mutants, respectively. (a) Transcription of HBV RNA (top) and albumin and GAPDH RNA (internal control) (bottom). (b) Extracellular HBV virion DNA. Abbreviations: cd, circular partial double-stranded HBV DNA; ld, linear partial double-stranded HBV DNA; ss, single-stranded HBV DNA.
Time courses of HBV antigen secretion and viral replication.
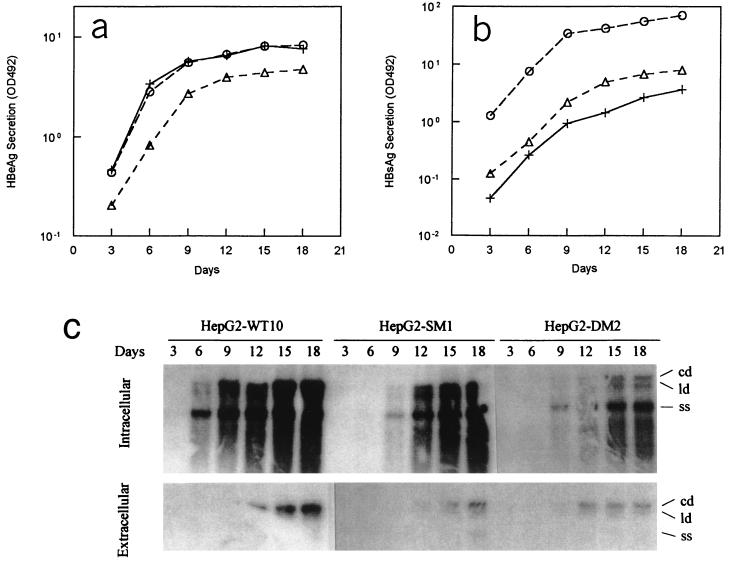
To further characterize these three cell lines, the time courses of HBV antigen secretion and progeny DNA replication were examined. As shown in Fig. 3a and b, both HBeAg and HBsAg were continually secreted. The levels of secreted HBeAg produced by HepG2-WT10 and HepG2-DM2 were similar and higher than that produced by HepG2-SM1, but for the secreted HBsAg levels, the lowest was produced by HepG2-WT10 and the highest was produced by HepG2-DM2. It was found that the amounts of extracellular and intracellular viral DNA were increased during the 18-day culture period (Fig. 3c). In any case, the amount of HBV DNA was highest in HepG2-WT10, lower in HepG2-SM1, and lowest in HepG2-DM2. This is consistent with previous observations by us (8) and others (23, 26).
FIG. 3.
Time course of viral antigen and progeny DNA production by HBV-producing cell lines. (a) Secretion of HBeAg. (b) Secretion of HBsAg. (c) Intracellular and extracellular production of HBV progeny DNA. Results shown in panels a and b represent the means of the results from triplicate independent experiments. The deviations from the means were ≤10% for all time points. Panel c shows results for one experiment representative of two. Abbreviations: cd, circular partial double-stranded HBV DNA; ld, linear partial double-stranded HBV DNA; ss, single-stranded HBV DNA; OD492, optical density at 492 nm. Symbols: +, HepG2-WT10; ▵, HepG2-SM1; ○, HepG2-DM2.
In addition, the stability of viral production in the cell lines also was examined. The three cell lines were cultured for 72 passages in the absence of G418 selection pressure. The HBsAg levels were monitored. The average variations of HBsAg level between the 15th passage and the 72nd passage in HepG2-WT10, -SM1, and -DM2 cells were 11.8, 17.1, and 5.4%, respectively. Also, it was observed that there was no significant change in the HBV progeny DNA level between the cell passages. Therefore, it was concluded that the expression of viral antigen and the production of virions were stable through 72 cell passages and that G418 selection was not required to maintain the integration.
Susceptibilities of wild-type HBV and its mutants produced from the cell lines to the anti-HBV compounds.
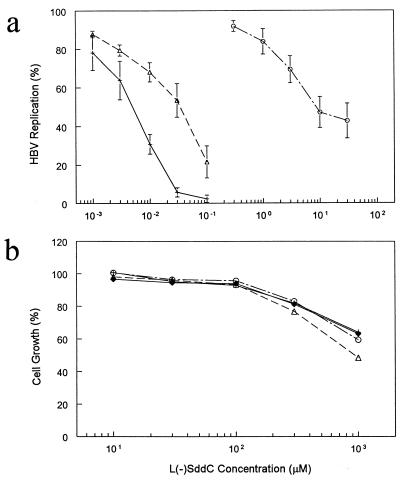
The HBV genomes used for the cell line construction were demonstrated to have different sensitivities to l(−)SddC (8). To determine whether the sensitivity of virion DNA synthesis to l(−)SddC in these cell lines has the same pattern as that in the transiently transfected cells, the cultures of HepG2-WT10, HepG2-SM1, and HepG2-DM2 cells were treated by l(−)SddC and the viral progeny DNA was observed by Southern hybridization. The dose-response relationship indicated that viral DNA synthesized in HepG2-SM1 was less sensitive to l(−)SddC than that in HepG2-WT10 but more sensitive than that in HepG2-DM2 (Fig. 4a). This result is consistent with the observation in a transient transfection system. The cytotoxicity assay of l(−)SddC in the HBV-producing cells and their parental HepG2 cells showed that the dose-response curve was about the same and that the concentrations required for 50% inhibition of cell growth for cell lines with different levels of genome integration were higher than 103 μM (Fig. 4b).
FIG. 4.
Antiviral activity and cytotoxicity of l(−)SddC. (a) Different susceptibilities of HBV variant to l(−)SddC. (b) Cell growth inhibition due to l(−)SddC of the generated HBV-producing cell lines and their parental cell line, HepG2. Values are means ± standard deviations of results from triplicate independent experiments. Symbols, +, HepG2-WT10; ▵, HepG2-SM1; ○, HepG2-DM2; ♦, HepG2.
It was observed that the L526M mutant was resistant to the treatment with PCV in transiently transfected HepG2 cells (9) and that L526M was a common mutation associated with famciclovir-resistant virion breakthrough in patients (25, 38). In the newly generated cell lines, the virus produced by HepG2-SM1 and HepG2-DM2 was also found to be resistant to PCV (Table 1). This confirmed that these cell lines could be used for evaluating compounds against the l(−)SddC- and PCV-resistant virus caused by the L526M or M550V mutation or both.
TABLE 1.
Anti-HBV nucleoside analogs against viral replication in the HBV producing cell lines
| Compound | IC50a (μM) for:
|
||
|---|---|---|---|
| HepG2-WT10 | HepG2-SM1 | HepG2-DM2 | |
| l(−)SddC | 0.005 ± 0.001 | 0.035 ± 0.004 | 8 ± 1.6 |
| PCV | 10 ± 0.9 | >200 | >200 |
| l(−)Fd4C | 0.0009 ± 0.0001 | 0.0027 ± 0.0003 | 0.21 ± 0.03 |
| l-FMAU | 0.02 ± 0.002 | 0.08 ± 0.01 | 50 ± 4.5 |
| PMEA | 0.028 ± 0.002 | 0.031 ± 0.003* | 0.07 ± 0.01 |
| QYL685 | 0.16 ± 0.02 | 0.16 ± 0.02* | 0.8 ± 0.1 |
Values are means ± standard deviations generated from the analysis of concentration dependence curves, which were plotted using results from triplicate independent experiments. IC50, 50% inhibitory concentration. ∗, P > 0.05 for IC50 of PMEA or QYL685 against HBV replication in HepG2-WT10 cells versus that in HepG2-SM1 cells. P values are <0.01 for all the other comparisons of IC50 of a compound against HBV replication in HepG2-WT10 cells versus that of the same compound in HepG2-SM1 or HepG2-DM2 cells.
As listed in Table 1, several compounds were tested in this new cell culture system; l(−)Fd4C, l-FMAU, and PMEA are well-identified anti-HBV compounds (3, 22, 31). The sensitivities of wild-type and mutated HBV DNA synthesis to these compounds were similar to those observed in the transient transfection system (9). Therefore, it can be concluded that l(−)SddC- and PCV-resistant viruses were cross-resistant to l(−)Fd4C and l-FMAU, but remained sensitive to PMEA. These observations made by using the new cell lines were consistent with results obtained by using transient transfections or an in vitro polymerase activity assay (9, 41).
QYL685, a purine analog, was recently identified to be an effective and promising anti-HBV compound by using 2.2.15 cells (33a). It was not known whether it was possible for QYL685 to inhibit the DNA synthesis of l(−)SddC-resistant virus. By using our new cell line system, it was found that the DNA synthesis of the L526M mutant, a l(−)SddC-resistant virus, in HepG2-SM1 cells, was as sensitive to QYL685 as that of wild-type virus in HepG2-WT10 cells whereas that of the L526MM550V mutant in HepG2-DM2 cells was five fold less sensitive than that of the wild type. It will be interesting to further study and develop this compound against l(−)SddC- and PCV-resistant virus.
DISCUSSION
Based on the clinical experience of antiviral chemotherapy for chronic hepatitis B and the lessons learned from the therapeutic management of HIV infection, using combination chemotherapy to maximize antiviral activity and decrease the rate of emergence of drug-resistant virus is the best strategy for future chronic hepatitis B therapy (24). Models for testing drug cross-resistance and screening new compounds with different targets of action against resistant HBV are urgently required. In this paper, we describe a new cell culture system, which is a group of three cell lines, HepG2-WT10, HepG2-SM1, and HepG2-DM2, stably transfected with wild-type, l(−)SddC- and PCV-resistant HBV genomes.
Viral production in the cell lines was studied. It was demonstrated that the cell lines were able to produce HBV RNAs, antigens, and virions. Furthermore, this ability was stable for at least 72 cell passages without requiring G418 selection pressure. This ensures that the cell culture is easy to perform and that the drug testing condition is consistent during a long culture period and avoids the complication caused by additional culture maintenance drugs.
Under our culture condition, it was observed that it was easier to detect intracellular viral DNA than extracellular viral DNA, due to the fact that more intracellular than extracellular HBV progeny DNA was present at the sampling time points. It was noticed that the HBV production was very low in HepG2-DM2 cells, although the RNA level was not dramatically lower than that in HepG2-WT10 cells and higher than that in HepG2-SM1 cells. This very low virion production could be due to defective replication of the HBV variant with two mutations, L526M and M550V, in the polymerase, which was demonstrated by previous studies (8, 23, 26).
The sensitivity of HBV DNA synthesis in these cell lines to l(−)SddC and PCV was studied, and it was confirmed that this system could be used for evaluating the activities of compounds against l(−)SddC- and PCV-resistant viruses. To further validate this cell-based model, several well-studied nucleoside analogs, l(−)Fd4C, l-FMAU, and PMEA, were tested in these cells. The results here were consistent with the observation obtained from other systems, such as transient transfection and an in vitro polymerase activity assay (9, 41).
Currently there are several systems for the study of antiviral activity against l(−)SddC-resistant HBV, such as a transient-transfection system, a system using isolated HBV polymerase from baculovirus, and stable virus-producing cell lines (1, 3, 8, 23, 26, 35, 39, 41). The transient-transfection system is suitable for initially identifying the role of mutations in the resistance phenotype. However, it is not easy to perform. Batch-to-batch variation, and lack of transfection efficiency and short-term stability are the major problems limiting its application for large-scale drug screening. The major application for the in vitro assay with a system using isolated HBV polymerase is the study of HBV replication at the HBV plus-strand DNA synthesis step, but the assay is not useful for studying the other steps of viral reproduction, such as regulation of HBV core particle assembly, HBV RNA processing (transcription and posttranscription), and cellular regulation (41). Our cell-based system is not only able to test the HBV DNA synthesis inhibitors but also is useful for the evaluation of HBV inhibitors that target other HBV regulation processes. With the cell lines presented here, we have found that a novel natural product could actively inhibit wild-type and resistant HBV by decreasing HBV RNA and HBV antigen levels without altering in vitro HBV DNA polymerase activity (L. Fu et al., unpublished data).
In conclusion, cell lines HepG2-WT, HepG2-SM1, and HepG2-DM2, established by introducing wild-type adr and L526M and L526MM550V mutant HBV genomes, respectively, into their parental HepG2 cells were capable of stably producing HBV virions with different susceptibilities to l(−)SddC and PCV. Using these cell lines, several compounds were tested, and the results were consistent with those from other testing systems (4, 9, 29, 41). In addition, purine analog QYL685 was identified as a promising inhibitor of resistant HBV. The establishment of these cell lines should be helpful in assessing new compounds against l(−)SddC- and PCV-resistant virus in a more controllable fashion and thus avoiding the complication of using a transient HBV DNA transfection system.
ACKNOWLEDGMENTS
We thank Yuan Wang, Shanghai Institute of Biochemistry, Academia Sinica, for her gift of the HBV genome (adr subtype), Gene Therapy Inc. for the gift of pG1Na vector; Chung K. Chu at the University of Georgia, Jiri Zemlicka at Wayne State University, and Gilead Sciences Inc. for providing compounds l(−)SddC, L-FMAU, QYL685, and PMEA; and Susan Grill and Susan Granata for critical reading of the manuscript.
This work was supported by NIH grants DK 34989, AI 38204, and AI 33655. Yung-Chi Cheng is a fellow of the National Foundation for Cancer Research.
REFERENCES
- 1.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L J, Brown N, Condreay L D for the Lamivudine Clinical Investigation Group. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Aye T T, Bartholomeusz A, Shaw T, Bowden S, Breschkin A, McMillan J, Angus P, Locarnini S. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J Hepatol. 1997;26:1148–1153. doi: 10.1016/s0168-8278(97)80125-0. [DOI] [PubMed] [Google Scholar]
- 3.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild-type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 4.De Clercq E. Perspectives for the treatment of hepatitis B virus infections. Int J Antimicrob Agents. 1999;12:81–95. doi: 10.1016/s0924-8579(99)00060-6. [DOI] [PubMed] [Google Scholar]
- 5.Dienstag J L, Perrillo R P, Schiff E R, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic viral hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 6.Doong S L, Tsai C H, Schinazi R F, Liotta D C, Cheng Y C. Inhibition of the replication of hepatitis B virus in vitro by 2′,3′-dideoxy-3′-thiacytidine and related analogues. Proc Natl Acad Sci USA. 1991;88:8495–8499. doi: 10.1073/pnas.88.19.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlay G J, Baguley B C, Wilson W R. A semiautomated microculture method for investigating exponentially growing carcinoma cells. Anal Biochem. 1984;139:272–277. doi: 10.1016/0003-2697(84)90002-2. [DOI] [PubMed] [Google Scholar]
- 8.Fu L, Cheng Y C. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(−)SddC (3TC) resistance. Biochem Pharmacol. 1998;55:1567–1572. doi: 10.1016/s0006-2952(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 9.Fu L, Liu S H, Cheng Y C. Sensitivity of L-(−)2′3′-dideoxythiacytidine resistant hepatitis B virus to other antiviral nucleoside analogues. Biochem Pharmacol. 1999;57:1351–1359. doi: 10.1016/s0006-2952(99)00073-8. [DOI] [PubMed] [Google Scholar]
- 10.Fu L, Wu X, Kong Y Y, Wang Y. Regulation of hepatitis B virus gene expression by its enhancers. Acta Biochim Biophys Sin. 1996;28:590–599. [PubMed] [Google Scholar]
- 11.Fu L, Wu X, Kong Y Y, Wang Y. Regulation of hepatitis B virus gene expression by core promoter and its upstream sequences. Chin J Virol. 1997;13:215–223. [Google Scholar]
- 12.Hammer S M, Katzenstein M D, Hughes M D, Gundacker H, Schooley R T, Haubrich R H, Henry W K, Lederman M M, Phair J P, Niu M, Hirsch M S, Merigan T C. A trial comparing nucleoside monotherapy with combination therapy in HIV-infected adults with CD4 cell counts from 200 to 500 per cubic millimeter. AIDS Clinical Trials Group Study 175 Study Team. N Engl J Med. 1996;335:1081–1090. doi: 10.1056/NEJM199610103351501. [DOI] [PubMed] [Google Scholar]
- 13.Heijtink R A, De Wilde G A, Kruining J, Berk L, Balzarini J, De Clercq E, Holy A, Schalm S W. Inhibitory effect of 9-(2-phosphonylmethoxyethyl)-adenine (PMEA) on human and duck hepatitis B virus infection. Antivir Res. 1993;21:141–153. doi: 10.1016/0166-3542(93)90050-s. [DOI] [PubMed] [Google Scholar]
- 14.Honkoop P, de Man R A, Zondervan P E, Schalm S W. Histological improvement in patients with chronic hepatitis B virus infection treated with lamivudine. Liver. 1997;17:103–106. doi: 10.1111/j.1600-0676.1997.tb00789.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoofnagle J H. Therapy of viral hepatitis. Digestion. 1998;59:563–578. doi: 10.1159/000007532. [DOI] [PubMed] [Google Scholar]
- 16.Innaimo S F, Seifer M, Bisacchi G S, Standring D N, Zahler R, Colonno R J. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob Agents Chemother. 1997;41:1444–1448. doi: 10.1128/aac.41.7.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen R W, Johnson L C, Averett D R. High-capacity in vitro assessment of anti-hepatitis B virus compound selectivity by a virion-specific polymerase chain reaction assay. Antimicrob Agents Chemother. 1993;37:441–447. doi: 10.1128/aac.37.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson V A, Merrill D P, Videler J A, Chou T C, Byington R E, Eron J J, D'Aquila R T, Hirsch M S. Two-drug combinations of zidovudine, didanosine, and recombinant interferon-α A inhibit replication of zidovudine-resistant human immunodeficiency virus type I synergistically in vitro. J Infect Dis. 1991;164:646–655. doi: 10.1093/infdis/164.4.646. [DOI] [PubMed] [Google Scholar]
- 19.Korba B, Boyd M. Penciclovir is a selective inhibitor of hepatitis B virus replication in cultured human hepatoblastoma cells. Antimicrob Agents Chemother. 1996;40:1282–1284. doi: 10.1128/aac.40.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladner S K, Miller T J, Otto M J, King R W. The hepatitis B virus M539V polymerase variation responsible for 3TC resistance also confers cross-resistance to other nucleoside analogues. Antivir Chem Chemother. 1998;9:65–72. [PubMed] [Google Scholar]
- 21.Lau G K, Carman W F, Locarnini S A, Okuda K, Lu Z M, Williams R, Lam S K. Treatment of chronic hepatitis B virus infection: an Asia-Pacific perspective. J Gastroenterol Hepatol. 1999;14:3–12. doi: 10.1046/j.1440-1746.1999.01812.x. [DOI] [PubMed] [Google Scholar]
- 22.Lin T S, Luo M Z, Liu M C, Zhu Y L, Gullen E, Dutschman G E, Cheng Y C. Design and synthesis of 2′,3′-dideoxy-2′,3′-didehydro-β-L-cytidine (β-L(-)d4C) and 2′,3′-dideoxy-2′,3′-didehydro-β-L-5-fluorocytidine(β-L(-)Fd4C), two exceptionally potent inhibitors of human hepatitis B virus (HBV) and potent inhibitors of human immunodeficiency virus (HIV) in vitro. J Med Chem. 1996;39:1757–1759. doi: 10.1021/jm950836q. [DOI] [PubMed] [Google Scholar]
- 23.Ling R, Harrison T J. Functional analysis of mutations conferring lamivudine resistance on hepatitis B virus. J Gen Virol. 1999;80:601–606. doi: 10.1099/0022-1317-80-3-601. [DOI] [PubMed] [Google Scholar]
- 24.Locarnini S, Birch C. Antiviral chemotherapy for chronic hepatitis B infection: lesions learned from treating HIV-infected patients. J Hepatol. 1999;30:536–550. doi: 10.1016/s0168-8278(99)80118-4. [DOI] [PubMed] [Google Scholar]
- 25.Locarnini S A. Hepatitis B virus surface antigen and polymerase gene variants: potential virological and clinical significance. Hepatology. 1998;27:294–297. doi: 10.1002/hep.510270144. [DOI] [PubMed] [Google Scholar]
- 26.Melegari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 27.Moyle G J. Use of viral resistance patterns to antiretroviral drugs in optimising selection of drug combinations and sequences. Drugs. 1996;52:168–185. doi: 10.2165/00003495-199652020-00002. [DOI] [PubMed] [Google Scholar]
- 28.Niesters H G M, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D M E. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 29.Ono-Nita S K, Kato N, Shiratori Y, Lan K H, Yoshida H, Carrilho F J, Omata M. Susceptibility of lamivudine-resistant hepatitis B virus to other reverse transcriptase inhibitors. J Clin Investig. 1999;103:1635–1640. doi: 10.1172/JCI5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oon C J, Chen W N, Lim N, Koh S, Lim G K, Leong A L, Tan G S. Hepatitis B virus variants with lamivudine-related mutations in the DNA polymerase and the ‘a’ epitope of the surface antigen are sensitive to ganciclovir. Antivir Res. 1999;41:113–118. doi: 10.1016/s0166-3542(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 31.Pai S B, Liu S H, Zhu Y L, Chu C K, Cheng Y C. Inhibition of hepatitis B virus by a novel l-nucleotide, 2′-fluoro-5-methyl-β-l-arabinofuranosyl uracil. Antimicrob Agents Chemother. 1996;40:380–386. doi: 10.1128/aac.40.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pessoa M G, Wright T L. Update on clinical trails in the treatment of hepatitis B. J Gastroenterol Hepatol. 1999;14(Suppl.):S6–S11. doi: 10.1046/j.1440-1746.1999.01877.x. [DOI] [PubMed] [Google Scholar]
- 33.Pillary D, Schinazi R F. Antivial drug resistance: from the laboratory to the patient. Antivir Ther. 1998;3:237. [PubMed] [Google Scholar]
- 33a.Qiu Y L, Ptak R G, Breitenbach J M, Lin J S, Cheng Y C, Drach J C, Kern E R, Zemlicka J. Synthesis and antiviral activity of phosphoralinate derivatives of methylenecyclopropane analogues of nucleosides. Antivir Res. 1999;43:37–53. doi: 10.1016/s0166-3542(99)00029-7. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schinazi R F, Ilan E, Black P L, Yao X, Dagan S. Cell-based and animal models for hepatitis B and C viruses. Antivir Chem Chemother. 1999;10:99–114. doi: 10.1177/095632029901000301. [DOI] [PubMed] [Google Scholar]
- 36.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A B. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 37.Sells M A, Chen M L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatits B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tillmann H L, Trautwein C, Bock T, Boker K H, Jackel E, Glowienka M, Oldhafer K, Bruns I, Gauthier J, Condreay L D, Raab H R, Manns M P. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology. 1999;30:244–256. doi: 10.1002/hep.510300141. [DOI] [PubMed] [Google Scholar]
- 39.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 40.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong X, Flores C, Yang H, Toole J J, Gibbs C S. Mutations in hepatitis B DNA polymerase associated with resistance to lamivudine do not confer resistance to adefovir in vitro. Hepatology. 1998;28:1669–1672. doi: 10.1002/hep.510280629. [DOI] [PubMed] [Google Scholar]