Abstract
The bacterial phytopathogen Pseudomonas syringae causes disease on a wide array of plants, including the model plant Arabidopsis thaliana and its agronomically important relatives in the Brassicaceae family. To cause disease, P. syringae delivers effector proteins into plant cells through a type III secretion system. In response, plant nucleotide-binding leucine-rich repeat proteins recognize specific effectors and mount effector-triggered immunity (ETI). While ETI is pervasive across A. thaliana, with at least 19 families of P. syringae effectors recognized in this model species, the ETI landscapes of crop species have yet to be systematically studied. Here, we investigated the conservation of the A. thaliana ETI landscape in two closely related oilseed crops, Brassica napus (canola) and Camelina sativa (false flax). We show that the level of immune conservation is inversely related to the degree of evolutionary divergence from A. thaliana, with the more closely related C. sativa losing ETI responses to only one of the 19 P. syringae effectors tested, while the more distantly related B. napus loses ETI responses to four effectors. In contrast to the qualitative conservation of immune response, the quantitative rank order is not as well-maintained across the three species and diverges increasingly with evolutionary distance from A. thaliana. Overall, our results indicate that the A. thaliana ETI profile is qualitatively conserved in oilseed crops, but quantitatively distinct.
Subject terms: Natural variation in plants, Plant domestication, Plant immunity
Introduction
Gram-negative phytopathogenic bacteria, such as Pseudomonas syringae deliver type III secreted effector proteins (effectors hereafter) into host cells through a type III secretion system, where they function to suppress basal plant immunity and promote pathogen growth1,2. However, plant nucleotide-binding leucine-rich repeat (NLR) proteins can recognize the presence or activity of certain effectors and mount a robust immune response called effector-triggered immunity (ETI), which limits pathogen proliferation3,4. A systematic analysis of the ETI landscape of the model plant Arabidopsis thaliana against P. syringae revealed that 19 of 70 families of P. syringae effectors trigger ETI in the Col-0 ecotype of A. thaliana5. It was further revealed that ETI is remarkably pervasive in this plant-pathogen interaction, with nearly all analyzed P. syringae strains carrying at least one effector that has the potential to elicit ETI in A. thaliana. This suggests that ETI plays an important role in contributing to broad-spectrum disease resistance; however, it remains to be determined whether such a prominent ETI landscape exists beyond Arabidopsis.
A. thaliana is a small, annual weedy plant that belongs to the Brassicaceae, or mustard family, which also includes many important food crops such as radish, kale, and broccoli as well as oilseed crops such as Brassica napus (canola) and Camelina sativa (false flax)6. B. napus has an extensive breeding history as one of the world’s most important oilseeds, while C. sativa is an important emerging crop. C. sativa is in the tribe (i.e., subfamily) Camelineae, which includes A. thaliana, and these two species are believed to have diverged approximately 8 million years ago7. B. napus, on the other hand, is in the tribe Brassiceae, which is believed to have diverged from the ancestor of A. thaliana approximately 23 million years ago7. Consequently, these three species provide an interesting continuum, from the relatively divergent and highly cultivated B. napus, to the relatively closely related and recently cultivated C. sativa, to the ‘wild’ model species A. thaliana6,8–10. Despite both being crop species, B. napus and C. sativa offer similar advantages for researchers as the model plant A. thaliana, such as the availability of reference genomes11,12 and their amenability to genetic manipulation13–19.
Genome-wide comparative analyses of NLRs have revealed a high degree of NLR diversity within Brassicaceae20–27. For instance, compared to approximately 165 NLRs in the diploid A. thaliana, there are approximately 464 NLR-encoding genes in the tetraploid B. napus, which may reflect an expanded ETI landscape relative to A. thaliana24. The NLR repertoire of the hexaploid C. sativa has yet to be characterized. Nevertheless, orthologs of characterized NLRs from A. thaliana can be identified in both C. sativa and/or B. napus25. For instance, two copies of the NLR RPM1, which is required for the recognition of P. syringae effectors AvrB1 and AvrRpm1, are known to occur in B. napus28. Both B. napus and C. sativa also possess orthologs of ZAR129, which is required for the recognition of at least five distinct families of P. syringae effectors in A. thaliana: HopZ1, HopF1, HopBA1, HopO1, and HopX15,30,31. While the presence of NLR orthologs may indicate the conservation of ETI responses in close relatives of A. thaliana, this has yet to be systematically validated in Brassicaceous crops such as B. napus and C. sativa.
Here, we surveyed the conservation of ETI responses between A. thaliana and its two agronomically important Brassicaceous relatives, B. napus and C. sativa, by first establishing pathology assays on B. napus and C. sativa, and then screening these plant species with the 19 P. syringae effector families that elicit ETI in A. thaliana. We show that the P. syringae pathovar tomato DC3000 (PtoDC3000) can be adapted to study ETI on B. napus and C. sativa, and that 15 and 18 of the 19 ETI responses are conserved in B. napus and C. sativa, respectively. Our results indicate that the A. thaliana ETI responses are retained in oilseed crops, suggesting that domestication has not substantially compromised their ETI potential.
Results
Establishing P. syringae pathology assays on B. napus and C. sativa
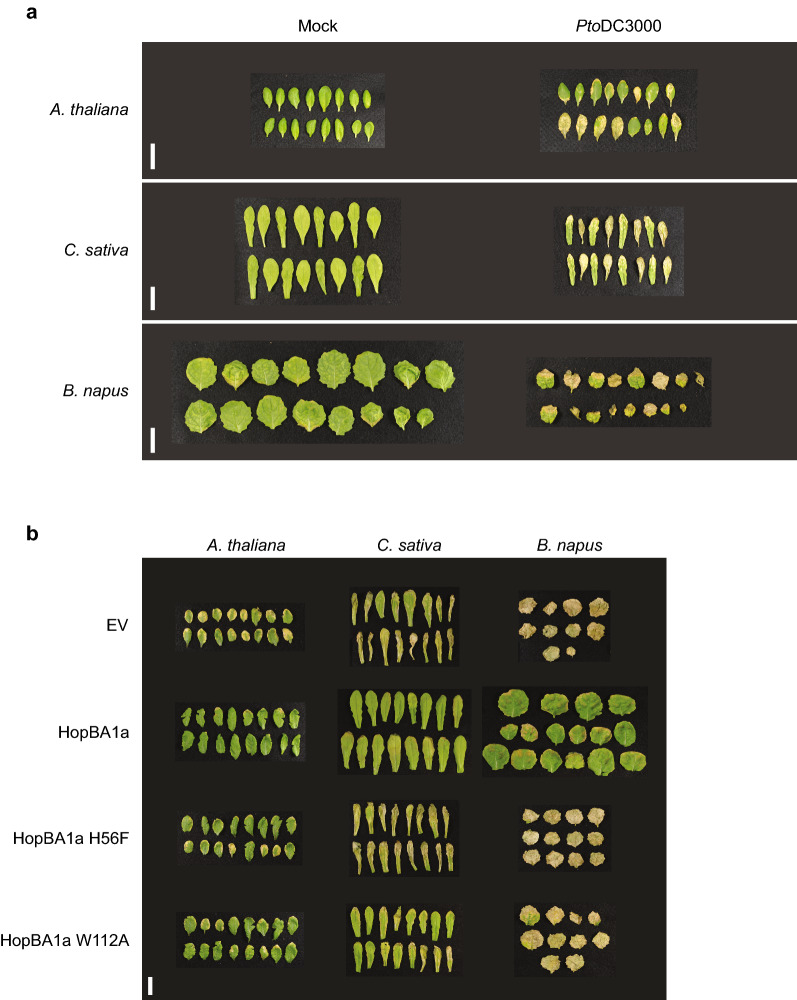
We first sought to identify a strain of P. syringae capable of causing disease on B. napus and C. sativa. In A. thaliana Col-0, spray inoculation of the highly virulent strain PtoDC3000 leads to chlorotic disease symptoms32–34. Similarly, spray inoculation of PtoDC3000 on B. napus and C. sativa resulted in distinctive and consistent chlorotic symptoms on both plant species to a similar extent as on A. thaliana Col-0 (Fig. 1a). Interestingly, disease symptoms on B. napus and C. sativa were associated with a more pronounced stunting of plant growth relative to A. thaliana (Fig. 1a).
Figure 1.
Establishing P. syringae pathology assays on B. napus and C. sativa. (a) P. syringae pv. tomato strain DC3000 (PtoDC3000) causes disease symptoms on B. napus and C. sativa similar to those on A. thaliana. A. thaliana Col-0, B. napus var. Topas, and C. sativa var. DH55 were spray inoculated with either a mock treatment of 10 mM MgSO4 0.04% Silwet (Mock) or PtoDC3000 at an OD600 = 1. A. thaliana was sprayed at 4 weeks old. B. napus and C. sativa were sprayed at 2 weeks old. Images show 2 leaves per plant. Photographs were taken at 6 days post-infection. Scale bar = 3 cm. (b) HopBA1a elicits ETI in B. napus and C. sativa. Representative photographs of chlorotic symptoms on A. thaliana Col-0, B. napus var. Topas, and C. sativa var. DH55 infected with PtoDC3000 expressing an empty vector (EV), HopBA1a, HopBA1a H56F, or HopBA1a W112A. Plants were spray inoculated as described above. Images show 2 leaves per plant. Photographs were taken at 6 days post-infection. Scale bar = 3 cm.
To determine whether B. napus and C. sativa could recognize P. syringae effectors and mount an ETI response, we spray inoculated B. napus and C. sativa with PtoDC3000 strains expressing an effector from each of the families that trigger ETI in A. thaliana Col-05 and assessed their ETI eliciting potential relative to a virulent negative control (PtoDC3000 carrying a pBBR1-MCS2 empty vector). In both B. napus and C. sativa, HopBA1a triggered a significant reduction in both disease symptoms and in planta bacterial growth compared to the empty vector control, indicating that it triggers a strong ETI response in these two plant species (Fig. 1b, Supplemental Figure 1). We further tested two point mutations in HopBA1a (H56F and W112A) that are known to abolish ETI-associated hypersensitive responses in the A. thaliana ecotype Ag-035 and found that they likewise restored disease symptoms and bacterial growth in B. napus and C. sativa, suggesting a similar recognition mechanism based on the interaction interfaces or catalytic activity that these residues confer to HopBA1a (Fig. 1b, Supplemental Figure 1). Overall, these results demonstrate that PtoDC3000 can grow and cause disease symptoms on B. napus and C. sativa and that ETI can suppress the virulence associated outcomes of P. syringae infection. This pathosystem could therefore be used to compare the ETI landscapes of these oilseed crops with that of A. thaliana.
Qualitative conservation of the A. thaliana ETI landscape across B. napus and C. sativa
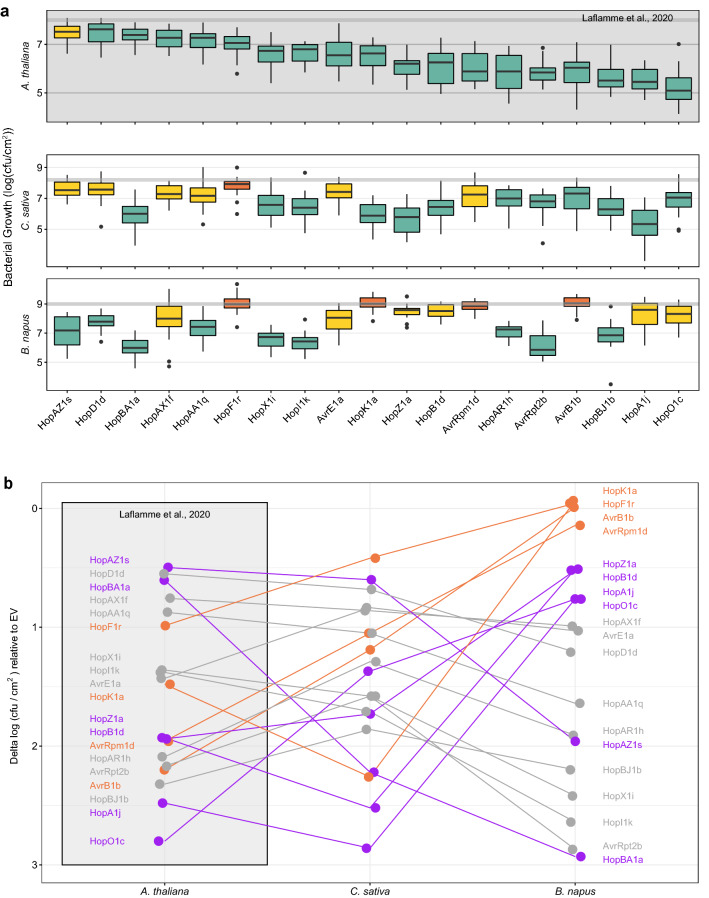
To assess the conservation of ETI responses between B. napus, C. sativa, and A. thaliana, we screened through the 19 ETI eliciting effector families previously identified in A. thaliana Col-05 (Table 1) by quantifying their ETI eliciting potential using bacterial growth assays (Fig. 2, Supplemental Figure 2). We normalized bacterial growth reductions induced by each effector to a virulent control (PtoDC3000::empty vector) and applied the same analysis to the A. thaliana Col-0 growth assay data from Laflamme et al. (2020) for comparison (Fig. 2a; Supplemental Figure 2)5. The C. sativa and B. napus spray inoculation resulted in a spectrum of immune phenotypes, which were broadly classified as: (1) strong ETIs, with a significant reduction in bacterial growth in all three replicates; (2) inconsistent ETIs, with a significant reduction in one or two of the three replicates; or (3) non-ETI, causing no significant reduction in bacterial growth in any replicate. In C. sativa, 12/19 effectors (63%) triggered strong ETI responses; 6/19 (32%) led to inconsistent ETI responses, and 1/19 effectors (5%) did not trigger ETI (Fig. 2a). In B. napus, 9/19 effectors (47%) triggered strong ETI responses, whereas 6/19 (32%) displayed inconsistent ETI phenotypes, and 4/19 effectors (21%) did not trigger an ETI response (Fig. 2a). ETI responses to the effectors AvrRpm1d, AvrB1b, and HopK1a, were lost in B. napus, while HopF1r-triggered ETI was lost in both B. napus and C. sativa (Fig. 2a). These results suggest that the majority of ETI responses are conserved between A. thaliana Col-0 and its two close relatives, though the robustness of these ETIs differ considerably across plant species.
Table 1.
ETI eliciting effectors used in this study.
| Effector family | Effector allele | Strain | Accession | Locus |
|---|---|---|---|---|
| AvrB | AvrB1b | PgyICMP807 | NZ_RBNZ01000352.1 | |
| AvrRpm1 | AvrRpm1d | PfiICMP7848 | NZ_LJQJ01000610.1 | |
| AvrRpt2 | AvrRpt2b | Pla1188_1 | NZ_RBPG01000101.1 | |
| HopK | HopK1a | PbrICMP13650 | NZ_LJPV01000582.1 | |
| HopAR | HopAR1h | PmeN6801 | NZ_LGLB01000021.1 | |
| HopA | HopA1j | PacICMP9850 | NZ_RBSM01000105.1 | |
| HopF | HopF1r | Pac302273 | NZ_GL385316.1 | |
| HopZ | HopZ1a | PssA2 | NZ_LGKU01000014.1 | |
| HopO | HopO1c | PsyUSA007 | NZ_AVDY02000338.1 | |
| HopX | HopX1i | PdpICMP13052 | NZ_RBRA01000108.1 | |
| HopBA | HopBA1a | PsfICMP4996 | NZ_RBSD01000159.1 | |
| HopB | HopB1d | PsyCC1466 | NZ_AVEM02000219.1 | |
| AvrE | AvrE1a | PsvICMP13519 | NZ_RBNW01000319.1 | |
| HopAA | HopAA1q | PsyCC1416 | NZ_AVEP02000280.1 | |
| HopD | HopD1d | PgyICMP2185 | NZ_RBRH01000243.1 | |
| HopI | HopI1k | PafICMP5011 | NZ_RBOK01000041.1 | |
| HopAX | HopAX1f | PcdICMP12341 | NZ_RBOV01000268.1 | |
| HopAZ | HopAZ1s | PhoICMP7847 | NZ_CP042804.1 | PSYTB_RS09780 |
| HopBJ | HopBJ1b | PsyCC1466 | NZ_AVEM02000066.1 |
Figure 2.
Conservation of the A. thaliana Col-0 ETI responses in C. sativa var. DH55 and B. napus var. Topas. (a) Growth assays of PtoDC3000 expressing 19 ETI eliciting effectors identified in5 normalized to the empty vector (EV) across assays for each species. A. thaliana Col-0 growth assay data is from Laflamme et al., 2020. The horizontal grey line across each plot represents the normalized mean of EV controls between assays. Green boxes represent the effectors that consistently caused a significant reduction in bacterial growth compared to the EV (ANOVA with post-hoc Tukey–Kramer HSD test, P < 0.05). Yellow boxes represent the effectors that led to inconsistent reductions in bacterial growth (not significantly different from EV in at least one experimental replicate). Orange boxes represent the effectors that were not significantly different from EV in any replicate. Box and whisker plots show pooled data from three experiments (n = 7 or 8 plants / experiment). Boxes show the first quartile, median, and third quartile. Whiskers extend to the smallest, and largest values no further than 1.5 × interquartile range from the first and third quartiles, respectively. Outlying points are plotted individually as solid circles. Raw growth assay data is presented in Supplemental Figure 2. (b) ETI intensity rank order profiles for A. thaliana Col-0, B. napus var. Topas, and C. sativa var. DH55. The delta log cfu/cm2 values of the normalized means of each effector relative to EV are plotted ranging from 0.0 logs (no reduction in bacterial growth relative to EV) to 3.0 logs (largest reduction in bacterial growth corresponding with the strongest ETI responses). Lines connect the means of each effector across the three plant species. Labels represent effector names. Orange represents effectors that do not trigger ETI in B. napus or C. sativa. Purple represents effectors of interest with very different responses between plants. Grey represents effectors that show similar responses between plants.
Quantitative conservation of the A. thaliana ETI landscape across B. napus and C. sativa
To assess the relative strengths of ETI in C. sativa and B. napus, we compared the rank order of normalized bacterial growth based on calculated delta log cfu/cm2 values relative to the EV and applied the same analysis to the A. thaliana Col-0 growth assay data from Laflamme et al. (2020) for comparison5 (Fig. 2b, Table 2). Notably, HopBA1a triggered strong decreases in bacterial growth in C. sativa (rank 4 of 19 ETI responses) and B. napus (rank 1) but was among the weakest ETI elicitors in A. thaliana (rank 17). HopAZ1s, which was among the weakest elicitors in A. thaliana (rank 19) and C. sativa (rank 18) led to a strong ETI response in B. napus (rank 6). In contrast, the ETI responses to HopA1j, HopB1d, and HopZ1a were among the strongest in A. thaliana (rank 2, rank 8, and rank 9 respectively) and C. sativa (rank 1, rank 6, and rank 2 respectively) but were weak in B. napus (rank 13, rank 14, and rank 15 respectively). HopO1c was the strongest ETI response in A. thaliana (rank 1) but was among the weak ETI elicitors in C. sativa (rank 10) and B. napus (rank 12). The overall ranked (Spearman) correlation between A. thaliana and C. sativa was 0.47, while the ranked correlation between A. thaliana and B. napus was − 0.24, indicating that the divergence of ETI profiles increases with evolutionary distance from A. thaliana (Supplemental Figure 3). Overall, these results emphasize that although the majority of ETI responses are conserved across the three Brassicaceous species, the quantitative nature of their ETI profiles are distinct.
Table 2.
Rank order of ETI responses in A. thaliana, B. napus, and C. sativa.
| Effector | Rank order of ETI responses based on delta log cfu/cm2 values relative to EVa | Delta log cfu/cm2 relative to EV | Significanceb | |||||
|---|---|---|---|---|---|---|---|---|
| At | Cs | Bn | At | Cs | Bn | At vs. Cs | At vs. Bn | |
| AvrB1b | 4 | 12 | 17 | 2.20 | 1.19 | − 0.010 | * | *** |
| AvrE1a | 11 | 16 | 10 | 1.43 | 0.835 | 1.03 | * | *** |
| AvrRpm1d | 7 | 14 | 16 | 1.96 | 1.05 | 0.143 | ** | *** |
| AvrRpt2b | 5 | 8 | 2 | 2.17 | 1.58 | 2.87 | ||
| HopA1j | 2 | 1 | 13 | 2.48 | 2.86 | 0.762 | *** | |
| HopAA1q | 15 | 13 | 8 | 0.873 | 1.05 | 1.64 | ||
| HopAR1h | 6 | 11 | 7 | 2.09 | 1.29 | 1.91 | * | *** |
| HopAX1f. | 16 | 15 | 11 | 0.755 | 0.861 | 0.991 | ||
| HopAZ1s | 19 | 18 | 6 | 0.496 | 0.600 | 1.96 | ||
| HopB1d | 8 | 6 | 14 | 1.94 | 1.73 | 0.520 | *** | |
| HopBA1a | 17 | 4 | 1 | 0.604 | 2.22 | 2.93 | *** | *** |
| HopBJ1b | 3 | 5 | 5 | 2.32 | 1.86 | 2.20 | ** | |
| HopD1d | 18 | 17 | 9 | 0.550 | 0.682 | 1.21 | ||
| HopF1r | 14 | 19 | 18 | 0.987 | 0.419 | − 0.044 | ** | *** |
| HopI1k | 12 | 7 | 3 | 1.38 | 1.71 | 2.64 | ||
| HopK1a | 10 | 3 | 19 | 1.48 | 2.26 | − 0.066 | *** | |
| HopO1c | 1 | 10 | 12 | 2.80 | 1.37 | 0.763 | *** | *** |
| HopX1i | 13 | 9 | 4 | 1.36 | 1.58 | 2.42 | ||
| HopZ1a | 9 | 2 | 15 | 1.93 | 2.52 | 0.511 | *** | |
At = A. thaliana, Cs = C. sativa, and Bn = B. napus.
aRank order is based on normalized bacterial growth assay data presented in Fig. 2, with rank 1 being the strongest ETI response (largest reduction in bacterial growth) and rank 19 being the weakest ETI response (smallest reduction in bacterial growth).
bSignificance is based on T-tests comparing normalized growth between A. thaliana and C. sativa and between A. thaliana and B. napus. T-test p-values were Bonferroni corrected for 19 × 2 = 38 tests. Bonferroni corrected p-values are indicated by 0.05 > * > 0.01 > ** > 0.001 > ***.
Discussion
We have established P. syringae pathology assays on B. napus and C. sativa and found that the A. thaliana ETI landscape is well conserved in these two Brassicaceous oilseed crop species. Out of 19 representative effectors that elicit ETI in A. thaliana Col-05, 18 elicited ETI in C. sativa and 15 elicited ETI in B. napus (Fig. 2a). Since A. thaliana is more closely related to C. sativa than B. napus7, the greater overlap between A. thaliana and C. sativa ETI responses may be reflective of a more similar arsenal of NLRs. In addition, the ETI profiles of B. napus and C. sativa may differ due to their different domestication histories. Further studies that leverage host diversity by surveying the ETI profiles across multiple accessions of the three plant species will be required to establish the full extent of ETI conservation.
We observed a difference in the patterns of qualitative and quantitative ETI responses across species that diverged with evolutionary distance from A. thaliana. While most of the ETI responses tested in our study are qualitatively conserved across A. thaliana, B. napus, and C. sativa, the quantitative magnitudes of these ETI responses differ considerably between the three species, with many weak and inconsistent responses in B. napus and C. sativa (Fig. 2b). From a qualitative perspective, the loss of ETI responses were nested and correlated with the evolutionary distance from A. thaliana (i.e., both species lost HopF1r, while only B. napus lost HopK1a, AvrRpm1d, and AvrB1b). The patterns are not nearly as clear from a quantitative perspective, with seven and ten inconsistent or absent ETI responses in C. sativa and B. napus, respectively. Of these, only four are shared (Jaccard similarity = 0.31), including two of the four lost ETI responses of B. napus. This pattern can also be seen in the extensive shuffling of ETI rank order between B. napus and C. sativa using bacterial growth assays (Fig. 2b). Nevertheless, the overall quantitative ETI profile of C. sativa was more similar to A. thaliana than B. napus (Supplemental Figure 3, Table 2). This suggests that there are more complex genomic differences that govern ETI responses than simple gain and loss of NLRs, leading to a continuum of disease and immune phenotypes across evolutionarily related plant species5.
Overall, our results suggest that effector recognition is broadly conserved across the Brassicaceae, indicating the possible functional conservation of several important NLRs. For example, the A. thaliana NLR ZAR1 is required for the recognition of at least five families of P. syringae effectors (HopZ1, HopF1, HopBA1, HopO1, and HopX1)5,30,31. ZAR1 is broadly conserved across angiosperms, and both B. napus and C. sativa possess several ZAR1 orthologs29. In both B. napus and C. sativa, four ZAR1-mediated ETI responses (HopX1i, HopZ1a, HopBA1a, and HopO1c) are conserved (Fig. 2a). However, HopF1r-mediated ETI is lost in both B. napus and C. sativa. ZAR1 associates with receptor-like cytoplasmic kinases (RLCKs) called ZED1-related kinases (ZRKs) and PBS1-like kinases (PBLs) to mediate ETI36. The loss of HopF1r-triggered ETI could indicate that its respective ZRK, ZRK331, is absent or non-functional in B. napus and C. sativa. However, ZRK3 is also required for ZAR1-mediated recognition of HopO1c in A. thaliana36, which can still elicit ETI in B. napus and C. sativa. The PBL kinase PBL27 is also required for HopF1r ETI, but not HopO1c ETI in A. thaliana37. We therefore hypothesize that PBL27 or another component of the ZAR1 ETI machinery necessary for HopF1r-triggered ETI in A. thaliana is absent or non-functional in B. napus and C. sativa. Alternatively, these ETI responses (or a subset of them) may be ZAR1-independent and mediated by convergent evolution of distinct NLR genes as observed in other crop species38–45.
While HopF1r-triggered ETI was the only ETI response lost in both B. napus and C. sativa, three additional ETI responses (to the effectors HopK1a, AvrRpm1d, and AvrB1b) were lost in B. napus (Fig. 2a). Our results agree with a previous study that found that B. napus does not recognize AvrB1 or AvrRpm1, despite possessing two copies of RPM1, the NLR responsible for recognizing these two effectors in A. thaliana28. In A. thaliana, HopK1a (also known as AvrRps446,47) is recognized by two distinct pairs of NLRs, RPS4/RRS148,49 and RPS4b/RRS1b50. B. napus possesses homologs of RPS4 and RRS1b, but not RRS1 or RPS4b. Improper pairings such as RPS4/RRS1b or RPS4b/RRS1 do not recognize HopK1a in A. thaliana50, perhaps explaining the lack of HopK1a recognition in B. napus.
While it is known that C. sativa possesses several orthologs of ZAR129, and that B. napus possesses copies of both ZAR1 and RPM128,29, the presence of other important NLRs that are known to recognize P. syringae effectors in A. thaliana have not yet been confirmed in these two species. A BLASTP51 analysis of RPM1, RPS2, RPS4, RRS1, RPS4b, RRS1b, RPS5, RPS6, ZAR1, CAR1, BAR1, and RBA1 in B. napus and C. sativa revealed the presence of putative homologs for all but three of these NLRs in B. napus and/or C. sativa (Supplemental Table 1), which may explain the conservation of ETI profiles in B. napus and C. sativa. Interestingly, RBA1, which recognizes HopBA1 in the Ag-0 ecotype of A. thaliana35, is the only NLR absent in both B. napus and C. sativa. As such, HopBA1a, which triggers strong ETI responses in both C. sativa and B. napus (Fig. 2) may be recognized by ZAR1 as observed in A. thaliana Col-05.
This study is the first example of a comprehensive investigation of ETI conservation between an important model plant and two closely related crop species. Most of the A. thaliana ETI responses are retained in B. napus and C. sativa, suggesting that domestication has not severely compromised ETI potential. Further studies testing the remaining alleles within ETI eliciting families will reveal whether A. thaliana, B. napus, and C. sativa possess differential allele specificity. Further, it will be interesting to comprehensively assess the ETI potential of the P. syringae Type III Effector Compendium in B. napus and C. sativa to determine whether the high extent of NLR diversity within Brassicaceous crops underlies an expansive ETI landscape beyond what is captured in A. thaliana. Nevertheless, our identification of ETI responses in B. napus and C. sativa provides insight into the conservation of crop immunodiversity and can be used to guide the development of crop protection strategies in these two important oilseed crops.
Materials and methods
Plant materials
A. thaliana Col-0, B. napus var. Topas and C. sativa var. DH55 plants were grown in Sunshine Mix 1 soil at constant 22 °C, a light intensity of 150 μmol/m2s, and a 12-h photoperiod. B. napus and C. sativa plants were sprayed at two weeks old, and A. thaliana plants were sprayed at three to four weeks old.
Spray infiltrations
P. syringae strains used in this study were previously described5. Prior to spray inoculation, bacterial strains were grown overnight at 28 °C on KB agar amended with 50 μg/ml rifampicin and 50 μg/ml kanamycin. Strains were re-suspended in 10 mM MgSO4 with 0.04% silwet surfactant L-77 and diluted to 1 × 109 CFU/ml (OD600 = 1). Individual plants were sprayed with approximately 3 ml of inoculum using Preval pressurized sprayers.
Growth assays
Bacterial growth assays were performed three days post-infection. Four leaf discs (1 cm2) from each plant were harvested and ground in 1 ml of 10 mM MgSO4 using a bead-beater. Serial dilutions were performed and 5 μL of each sample was plated on KB agar amended with 50 μg/ml rifampicin and incubated at 28 °C for 24 h, after which individual colonies were counted. To compare bacterial growth across individual assays, growth assay data was normalized to the empty vector (EV) for each species (mean of 8.0, 8.2, and 9.0 log cfu/cm2 for A. thaliana Col-0, C. sativa, and B. napus, respectively) (Fig. 2a). To determine the quantitative rank order of ETI responses in each species, the delta log cfu/cm2 values were calculated for each effector relative to the EV using the normalized growth assay data (mean EV log cfu/cm2 – mean effector log cfu/cm2) (Fig. 2b, Table 2).
Supplementary Information
Acknowledgements
We thank the members of the Desveaux and Guttman laboratories for their advice and feedback throughout this project, in particular Bradley Laflamme and Alexandre Martel for the HopBA1a mutants. We also thank Hossein Borhan and Isobel Parkin (Agriculture and Agri-Food Canada) as well as Tammy Sage, Eiji Nambara and Daphne Goring (University of Toronto) for C. sativa and/or B. napus seeds. This project is supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grants (DSG and DD), an NSERC postgraduate award (CB-M), an Ontario Graduate Scholarship (CB-M), and the Centre for the Analysis of Genome Evolution and Function (DSG and DD).
Author contributions
CB-M, DD, and DSG designed experiments and analyzed data. CB-M performed experiments. CB-M, DD, and DSG wrote the manuscript.
Funding
This project is supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grants (DSG and DD), an NSERC postgraduate award (CB-M), and an Ontario Graduate Scholarship (CB-M). All local, national or international guidelines and legislation were adhered to in the production of this study.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Darrell Desveaux and David S. Guttman.
Contributor Information
Darrell Desveaux, Email: darrell.desveaux@utoronto.ca.
David S. Guttman, Email: david.guttman@utoronto.ca
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-10410-w.
References
- 1.Xin XF, Kvitko B, He SY. Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 2018;16:316–328. doi: 10.1038/nrmicro.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buttner D. Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 2016;40:894–937. doi: 10.1093/femsre/fuw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 5.Laflamme B, et al. The pan-genome effector-triggered immunity landscape of a host–pathogen interaction. Science. 2020;367:763–768. doi: 10.1126/science.aax4079. [DOI] [PubMed] [Google Scholar]
- 6.Koenig D, Weigel D. Beyond the thale: Comparative genomics and genetics of Arabidopsis relatives. Nat. Rev. Genet. 2015;16:285–298. doi: 10.1038/nrg3883. [DOI] [PubMed] [Google Scholar]
- 7.Hohmann N, Wolf EM, Lysak MA, Koch MA. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu K, et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019;10:1154. doi: 10.1038/s41467-019-09134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolov LA, Tsiantis M. Using mustard genomes to explore the genetic basis of evolutionary change. Curr. Opin. Plant Biol. 2017;36:119–128. doi: 10.1016/j.pbi.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 2011;16:108–116. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Kagale S, et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014;5:3706. doi: 10.1038/ncomms4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalhoub B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 13.Morineau C, et al. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017;15:729–739. doi: 10.1111/pbi.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang WZ, et al. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 2017;15:648–657. doi: 10.1111/pbi.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aznar-Moreno JA, Durrett TP. Simultaneous targeting of multiple gene homeologs to alter seed oil production in Camelina sativa. Plant Cell Physiol. 2017;58:1260–1267. doi: 10.1093/pcp/pcx058. [DOI] [PubMed] [Google Scholar]
- 16.Lyzenga WJ, et al. CRISPR/Cas9 editing of three CRUCIFERIN C homoeologues alters the seed protein profile in Camelina sativa. BMC Plant Biol. 2019;19:292. doi: 10.1186/s12870-019-1873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng M, et al. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.) Plant Biotechnol. J. 2020;18:644–654. doi: 10.1111/pbi.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai Y, et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020;18:1153–1168. doi: 10.1111/pbi.13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Wu JJ, Tang T, Liu KD, Dai C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017;7:7489. doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peele HM, Guan N, Fogelqvist J, Dixelius C. Loss and retention of resistance genes in five species of the Brassicaceae family. BMC Plant Biol. 2014;14:298. doi: 10.1186/s12870-014-0298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics. 2014;15:3. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wroblewski T, Coulibaly S, Sadowski J, Quiros CF. Variation and phylogenetic utility of the Arabidopsis thaliana RPS2 homolog in various species of the tribe Brassiceae. Mol. Phylogenet. Evol. 2000;16:440–448. doi: 10.1006/mpev.2000.0781. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Han Z, Jiang H, Tian D, Yang S. Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J. Mol. Evol. 2010;70:137–148. doi: 10.1007/s00239-009-9316-4. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, et al. NBS-encoding genes in Brassica napus evolved rapidly after allopolyploidization and co-localize with known disease resistance loci. Front. Plant Sci. 2019;10:26. doi: 10.3389/fpls.2019.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YM, et al. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J. Integr. Plant Biol. 2016;58:165–177. doi: 10.1111/jipb.12365. [DOI] [PubMed] [Google Scholar]
- 26.Mun JH, Yu HJ, Park S, Park BS. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol. Genet. Genomics. 2009;282:617–631. doi: 10.1007/s00438-009-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alamery S, et al. Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop Pasture Sci. 2017;69:79–93. [Google Scholar]
- 28.Grant MR, et al. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;95:15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adachi H, Sakai T, Kourelis J, Maqbool A, Kamoun S. Jurassic NLR: Conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1. bioRxiv. 2020 doi: 10.1101/2020.10.12.333484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6:e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto D, et al. Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat. Plants. 2017;3:17027. doi: 10.1038/nplants.2017.27. [DOI] [PubMed] [Google Scholar]
- 32.Cuppels DA. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 1986;51:323–327. doi: 10.1128/AEM.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura MT, et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:E2053–E2062. doi: 10.1073/pnas.1620973114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martel A, et al. Immunodiversity of the Arabidopsis ZAR1 NLR Is conveyed by receptor-like cytoplasmic kinase sensors. Front. Plant Sci. 2020;11:1290. doi: 10.3389/fpls.2020.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seto D, et al. The small molecule Zaractin activates ZAR1-mediated immunity in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2021;118(47):e2116570118. doi: 10.1073/pnas.2116570118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter ME, et al. Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol. Plant. Microbe Interact. 2019;32:550–565. doi: 10.1094/mpmi-07-18-0202-fi. [DOI] [PubMed] [Google Scholar]
- 39.Fahrentrapp J, et al. A candidate gene for fire blight resistance in Malus ×robusta 5 is coding for a CC–NBS–LRR. Tree Genet. Genomes. 2013;9:237–251. doi: 10.1007/s11295-012-0550-3. [DOI] [Google Scholar]
- 40.Vogt I, et al. Gene-for-gene relationship in the host-pathogen system Malus × robusta 5-Erwinia amylovora. New Phytol. 2013;197:1262–1275. doi: 10.1111/nph.12094. [DOI] [PubMed] [Google Scholar]
- 41.Mazo-Molina C, et al. The Ptr1 locus of Solanum lycopersicoides confers resistance to race 1 strains of Pseudomonas syringae pv. tomato and to Ralstonia pseudosolanacearum by recognizing the type III effectors AvrRpt2 and RipBN. Mol. Plant Microbe Interact. 2019;32:949–960. doi: 10.1094/mpmi-01-19-0018-r. [DOI] [PubMed] [Google Scholar]
- 42.Prokchorchik M, et al. A host target of a bacterial cysteine protease virulence effector plays a key role in convergent evolution of plant innate immune system receptors. New Phytol. 2020;225:1327–1342. doi: 10.1111/nph.16218. [DOI] [PubMed] [Google Scholar]
- 43.Ashfield T, Keen NT, Buzzell RI, Innes RW. Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics. 1995;141:1597–1604. doi: 10.1093/genetics/141.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashfield T, et al. Evolutionary relationship of disease resistance genes in soybean and Arabidopsis specific for the Pseudomonas syringae effectors AvrB and AvrRpm1. Plant Physiol. 2014;166:235–251. doi: 10.1104/pp.114.244715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon MM, et al. Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant. Sci. 2019;10:418. doi: 10.3389/fpls.2019.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindeberg M, et al. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant Microbe Interact. 2005;18:275–282. doi: 10.1094/MPMI-18-0275. [DOI] [PubMed] [Google Scholar]
- 48.Birker D, et al. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 2009;60:602–613. doi: 10.1111/j.1365-313X.2009.03984.x. [DOI] [PubMed] [Google Scholar]
- 49.Narusaka M, et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- 50.Saucet SB, et al. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015;6:6338. doi: 10.1038/ncomms7338. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request.