Abstract
Background
Natural killer (NK) cells play major roles in eliminating tumor cells. Preliminary studies have shown that NK cells and their receptors/ligands have prognostic value in malignant tumors. However, the relevance of NK cells and their receptors/ligands level to the prognosis of hepatocellular carcinoma (HCC) remains unclear.
Methods
Several electronic databases were searched from database inception to November 8, 2021. Random effects were introduced to this meta-analysis. The relevance of NK cells and their receptors/ligands level to the prognosis of HCC was evaluated using hazard ratios (HRs) with 95% confidence interval (95%CI).
Results
26 studies were included in the analysis. The pooled results showed that high NK cells levels were associated with better overall survival (HR=0.70, 95%CI 0.57–0.86, P=0.001) and disease-free survival (HR=0.61, 95%CI 0.40-0.93, P=0.022) of HCC patients. In subgroup analysis for overall survival, CD57+ NK cells (HR=0.70, 95%CI 0.55-0.89, P=0.004) had better prognostic value over CD56+ NK cells (HR=0.69, 95%CI 0.38-1.25, P=0.224), and intratumor NK cells had better prognostic value (HR=0.71, 95%CI 0.55-0.90, P=0.005) over peripheral NK cells (HR=0.66, 95%CI 0.41-1.06, P=0.088). In addition, high level of NK cell inhibitory receptors predicted increased recurrence of HCC, while the prognostic role of NK cell activating receptors remained unclear.
Conclusion
NK cells and their inhibitory receptors have prognostic value for HCC. The prognostic role of NK cell activating receptors is unclear and more high-quality prospective studies are essential to evaluate the prognostic value of NK cells and their receptors/ligands for HCC.
Keywords: natural killer cells, receptor, ligand, hepatocellular carcinoma, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignancy worldwide and the third leading cause of cancer-related mortality (1). The major risk factors for HCC involve chronic hepatitis B and hepatitis C infection, alcohol, and metabolic liver disease (2). Natural killer (NK) cells, characterized as CD3-CD56+ lymphocytes, are mainly involved in the early defense against virus infections and play major roles in eliminating tumor cells (3). NK cells account for only about 5–20% of the circulating lymphocytes in the peripheral blood. In contrast, NK cells are abundant in human liver, accounting for almost half of intrahepatic lymphocytes (4), which lays foundation for the powerful role of NK cells in the liver tumor microenvironment.
Human NK cells are divided into two major subpopulations based on the surface density of CD56 antigen (5). CD56dim NK cells display a mature phenotype, accounting for approximately 90% of all NK cells and mediating the cytolytic response, while immature CD56bright NK cells account for 5%-15% of total NK cells and are regarded as cytokine producers (6). Another surface marker is CD57, which is a marker for differentiated and highly cytotoxic NK cells, and is described as a phenotypically stable NK cells marker (7).
The regulation of NK cell function is mediated by a series of activated or inhibitory surface receptors. The major activated receptors involved in target cell killing are NK group 2 member D (NKG2D) and natural cytotoxic receptors (NCRs). NCRs mainly consist of NKp44, NKp46 and NKp30 (8), and can recognize ligands from different sources, including viral, parasitic, bacterial, as well as cellular ligands, such as HLA-B-associated transcript 3/Bcl-2-associated athanogene 6 (BAT3/BAG6), mixed lineage leukemia 5 (MLL5), proliferating cell nuclear antigen (PCNA) and B7 homolog 6 (B7-H6) (9, 10). In contrast, NKG2D mainly binds to the major histocompatibility complex class I chain-related protein A and B (MICA and MICB) and UL16-binding proteins (ULBPs). After binding, it can activate NK cells to produce cytotoxic substances to kill harmful and tumor cells (11). Other activated receptors include CD16, NKp88, CD244, CD226 and cytokine receptors such as interleukin (IL)-2R, IL-12R, IL-28R, IL-18R, IL-1R8, IL-15R, IL-10R, interferon receptor (IFNR) and tumor growth factor-β receptor (TGF-βR) (12–14).
The major inhibitory receptors involved in target cell killing are NKG2A, CD96, killer immunoglobulin-like receptors (KIRs), T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT) and T cell immunoglobulin domain and mucin domain-3 (TIM-3) (12). Other inhibitory receptors include programmed cell death-1 (PD-1), lymphocyte activation gene-3 (LAG3), leukocyte-associated immunoglobulin-like receptors (LAIRs), adenosine 2A receptor (A2AR) and immunoglobulin-like transcripts (ILTs) (15). PD-1 is primarily expressed by activated T lymphocytes, but may also be expressed by NK cells in tumor patients. PD-1/Programmed cell death ligand-1 (PD-L1) interactions can inactivate T cells and NK cells, allowing tumor cells to escape immune surveillance (16). Human histocompatibility leucocyte antigen E (HLA-E) is the main ligand of NKG2A, and is generally upregulated in cancer patients and predicts poor prognosis (17–19). The major histocompatibility complex class I (MHC-I) is the main ligand of KIRs, and is expressed on healthy hepatocytes. It interacts with inhibitory receptors on NK cells to prevent the activation of NK cells (14).
Until now, the correlation of NK cells and their receptors/ligands with the prognosis of HCC remains controversial (20–28). The purpose of this meta-analysis and review is to evaluate the prognostic value of NK cells and their receptors/ligands in HCC.
Materials and Methods
Search Strategy and Study Selection Criteria
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines (supplementary PRISMA Checklist) (29), and inclusion criteria were based on the PICOS model.
Relevant studies were independently searched by two authors (JSX, ZND) from the PubMed, Embase, Web of Science and Cochrane Library literature databases from the beginning of the database until November 8, 2021. Detailed search strategy was as described in the supplement. Additional articles were identified by a manual search of the references of eligible articles.
Studies were included if they met the following criteria. (1) all patients were identified as having HCC; (2) studies revealed the expression of NK cells and their receptors/ligands and obtained their levels by assaying; (3) studies provided adequate information to evaluate the hazard ratio (HR) and 95% confidence interval (95% CI); (4) the prognostic indexes such as overall survival (OS), cancer-specific survival (CSS), disease-free survival (DFS), recurrence-free survival (RFS), time-to recurrence (TTR), and progression-free survival (PFS) were evaluated; (5) Anti-tumor treatments were not conducted; (6) studies must be published in English. Studies were excluded if they met the following criteria. (1) review, meta-analysis and case report; (2) basic experimental researches of HCC and studies unrelated to the NK cells and their receptors/ligands; (3) studies provided inadequate data to evaluate the correlation of NK cells and their receptors/ligands with prognosis. For republished studies, only the studies with the largest sample size were selected; alternatively, the most recent literature and relevant data were collected.
Data Extraction and Quality Assessment
Eligible study data were extracted independently by two investigators (JSX, ZND). Disagreements could be discussed and resolved with a third investigator (GXM). Baseline characteristics were extracted from the included studies. Only one study had outcome indicator for CSS, which we uniformly classified as OS. OS and DFS/RFS/TTR/PFS were used as endpoints for the meta-analysis. The quality of eligible studies was assessed by the Newcastle–Ottawa Scale (NOS) criteria (30).
Statistical Analysis
Most of the relevant data from the studies could be directly collected. However, for those studies that did not provide hazard ratios (HRs) and 95% confidence intervals (95% CIs), we obtained estimates from known information using the method of Altman and Tierney (31, 32). Random effects models were applied. P < 0.05 was considered statistically significant. The pooled HR and 95% CI were used to assess the relevance between NK cell level and prognosis of HCC patients. Cochran’s Q test and Higgins’ I2 statistic were used to assess the heterogeneity. P-value of heterogeneity > 0.10 and I2 < 50% were considered as no significant heterogeneity. At the same time, subgroup analyses were performed by surface marker, source of NK cell, and outcome of patients. Sensitivity analysis was performed by removing each study to test the stability and reliability of the results. Funnel plots, Egger regression asymmetry tests, and Begg rank correlation tests were conducted to check for potential publication bias (33). Stata 16.0 software analysis was applied to all data in this meta-analysis.
Results
Literature Search
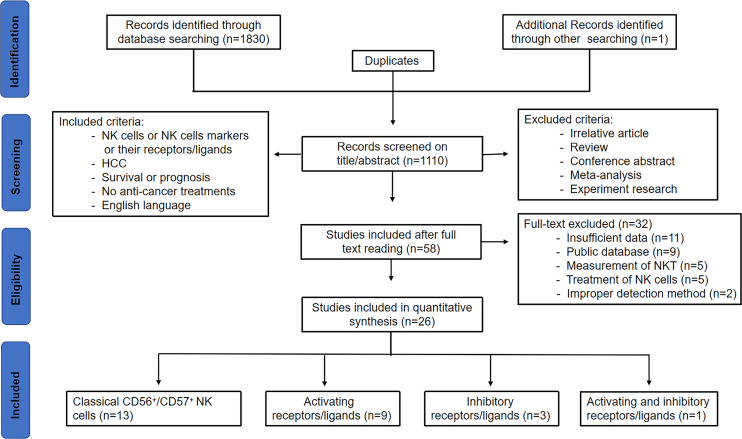
As shown in Figure 1 of the flowchart, a total of 1831 records were initially identified. After removing duplicate studies, 1110 studies were retained. After screening for titles and abstracts, 1052 studies were excluded. After reviewing the remaining 58 studies through full text, 32 studies were excluded due to insufficient data (11), public database (9), measurement of NKT (5), improper detection method including Ficoll separation or radiotaged NK-sensitive K-562 cell separation (2), and treatment of NK cells (5), including intravenous infusion of NK cells alone or combined with other modalities, such as radiofrequency ablation and irreversible electroporation. Finally, 26 studies were included in this analysis, including 13 on classical NK cells, 9 on activating receptors/ligands, 3 on inhibitory receptors/ligands, and 1 on both activating and inhibitory receptors/ligands.
Figure 1.
Study flow chart of the data extraction process and selection of studies for meta-analysis.
The Basic Characteristics of Included Studies About NK Cells
Table 1 summarized the basic characteristics of the included studies about NK cells. Sample sizes of the eligible studies ranged from 36 to 258, for a total of 1711, and these studies were conducted primarily in two countries: twelve in China and one in Italy. Seven studies reported on CD56+ NK cells, 4 on CD57+ NK cells, and 2 on NK cells. Among these studies, 5 studies detected NK cells in peripheral blood, and 8 detected intratumor NK cells. In total, 12 studies mentioned the correlation between NK cell levels and OS (20–22, 25–27, 34–37, 39, 40). 9 studies mentioned the correlation between NK cells levels and DFS/RFS/TTR/PFS (20, 25–27, 35, 36, 38–40). NOS score > 6 was defined as high quality, and ≤ 6 was defined as low quality ( Supplementary Table 1 ).
Table 1.
The characteristics of all included eligible studies about NK cells.
| Author | Year | Country | Sample size | Male/Female | Measurement | Marker | Treatment | Source | Tumor stage | VS | Numberof VS | Divide | Outcome | Follow-up times | Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhuang et al. (34) | 2019 | China | 78 | 64/14 | NA | CD56 | mixed+SBRT | Peripheral blood | NA | High/Low | NA | cutoff value | OS;PFS | median:32 (4.1-80) month |
7 |
| Hu et al. (25) | 2021 | China | 182 | NA | Immuno histochemistry |
CD57 | resection | Intratumor | I-IV | High/Low | NA | NA | OS;TTR | until 30/06/2016 | 7 |
| Lin et al. (35) | 2013 | China | 132 | NA | Immuno histochemistry |
CD56 | resection | Intratumor | I-III | High/Low | NA | cutoff value | OS;DFS | total:72 month | 8 |
| Wu et al. (20) | 2013 | China | 256 | 115/15 | Immuno histochemistry |
CD57 | resection/RFA | Intratumor | I-IV | High/Low | 126/130 | median | OS;DFS | NA | 9 |
| Tao et al. (26) | 2020 | China | 258 | NA | Immuno histochemistry |
CD56 | resection | Intratumor | I-III | High/Low | 129/129 | median | OS;TTR | NA | 9 |
| Chew et al. (22) | 2012 | China | 36 | NA | Immuno histochemistry |
CD56 | resection | Intratumor | I-IV | High/Low | NA | median | OS | median:3.94 (0.9-5.5) year |
6 |
| Zhao et al. (21) | 2014 | China | 163 | 131/32 | Immuno histochemistry |
CD57 | resection | Intratumor | NA | High/Low | 82/81 | median | OS | total:>60 month | 9 |
| Gao et al. (36) | 2012 | China | 206 | NA | Immuno histochemistry |
CD57 | liver transplantation | Intratumor | I-III | High/Low | NA | median | CSS;RFS | median:48.1 (3.4- 111.9) month |
8 |
| Cariani et al. (23) | 2016 | Italy | 70 | 41/29 | Flow cytometry | NK cells | resection/RFA | Peripheral blood | NA | High/Low | NA | median | OS;TTR | median OS:64 month; median TTR:16.5 month |
7 |
| Pan et al. (37) | 2014 | China | 121 | NA | Flow cytometry | CD56 | resection+CIK | Peripheral blood | NA | High/Low | 60/61 | median | OS | until 31/12/2012 | 9 |
| Liu et al. (38) | 2021 | China | 100 | NA | Immuno histochemistry |
CD56 | resection | Intratumor | NA | Positive/ Negative |
31/68 | score | RFS | until:20/06/2020 | 6 |
| Pan et al. (27) | 2020 | China | 48 | 39/9 | Flow cytometry | CD56 | resection+CIK | Peripheral blood | NA | High/Low | 24/24 | median | OS;RFS | total:>60 month | 7 |
| Che et al. (39) | 2014 | China | 61 | NA | Flow cytometry | NK cells | resection | Peripheral blood | NA | High/Low | 33/28 | median | OS;PFS | total:36 month | 6 |
SBRT, Stereotactic body radiation therapy; RFA, Radiofrequency ablation; CIK, Cytokine-induced killer; Mixed, TACE or RFA or PEI or surgery or no treatment; TAE, Transcatheter arterial embolization; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; TTR, Time-to recurrence; PFS, Progression-free survival; CSS, Cancer-specific survival.
NA, Not available.
Prognostic Value of NK Cells in Patients With HCC
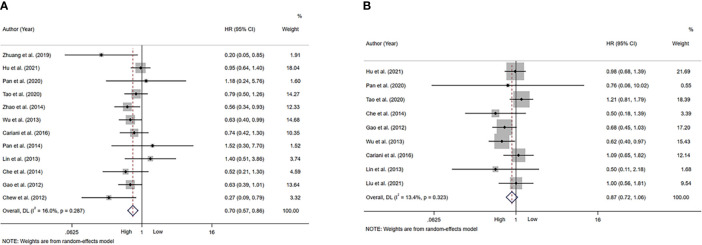
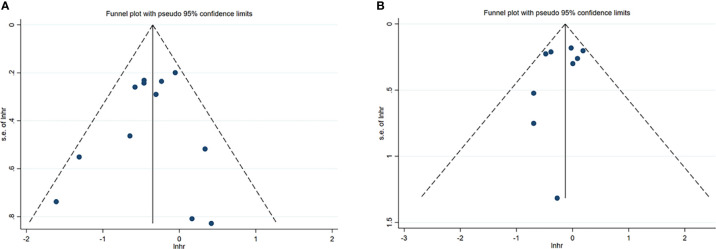
A total of 12 studies, involving 1611 patients, investigated the prognostic value of NK cells for OS. The pooled results from the 12 comparative studies were significant (HR=0.70, 95%CI 0.57-0.86, p=0.001), and the data were not heterogeneous (I2 = 16.0%, P=0.287; Figure 2A ). No bias was observed in the funnel plot ( Figure 3A ). In order to better understand the prognostic value of NK cells, we further performed subgroup analysis according to the marker and source of NK cells ( Table 2 ). In subgroup analysis of CD57+ NK cells, the pooled results from 4 comparative studies were significant (HR=0.70, 0.55-0.89, P=0.004), while it was insignificant in CD56+ NK cells (HR=0.69, 95%CI 0.38-1.25, P=0.224; Supplementary Figure 1 ). Compared to peripheral NK cells (HR=0.66, 95%CI 0.41-1.06, P=0.088), the level of intratumor NK cells had better prognostic value (HR=0.71, 95%CI 0.55-0.90, P=0.005) for HCC patients ( Supplementary Figure 2 ).
Figure 2.
Forest plot of NK cells in HCC. (A) Forest plot of NK cells and OS in HCC. (B) Forest plot of NK cells and DFS/RFS/TTR/PFS in HCC. CI, Confidence interval; HR, Hazard ratio; HCC, Hepatocellular carcinoma; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; TTR, Time-to recurrence; PFS, Progression-free survival.
Figure 3.
Funnel plots of NK cells in HCC. (A) Funnel plots of HR for OS of NK cells. (B) Funnel plots of HR for DFS/RFS/TTR/PFS of NK cells. CI, Confidence interval; HR, Hazard ratio; HCC, Hepatocellular carcinoma; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; TTR, Time-to recurrence; PFS, Progression-free survival.
Table 2.
Subgroup meta-analysis of the prognostic role of NK cells in HCC.
| Factor | No. of study | No. of patients | HR (95%CI) | P-value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I2(%) | P-value | |||||
| OS | ||||||
| Total | 12 | 1611 | 0.70 (0.57-0.86) | 0.001 | 16.0 | 0.287 |
| Marker | ||||||
| CD56 | 6 | 673 | 0.69 (0.38-1.25) | 0.224 | 45.6 | 0.102 |
| CD57 | 4 | 807 | 0.70 (0.55-0.89) | 0.004 | 12.8 | 0.328 |
| NK cells | 2 | 131 | 0.67 (0.41-1.08) | 0.103 | 0.0 | 0.532 |
| Source | ||||||
| Peripheral blood | 5 | 378 | 0.66 (0.41-1.06) | 0.088 | 12.0 | 0.337 |
| Intratumor | 7 | 1233 | 0.71 (0.55-0.90) | 0.005 | 29.0 | 0.207 |
| DFS/RFS/TTR/PFS | ||||||
| Total | 9 | 1313 | 0.87 (0.72-1.06) | 0.164 | 13.4 | 0.323 |
| Marker | ||||||
| CD56 | 4 | 538 | 1.09 (0.79-1.50) | 0.602 | 0.0 | 0.689 |
| CD57 | 3 | 644 | 0.76 (0.57-1.01) | 0.059 | 32.3 | 0.228 |
| NK cells | 2 | 131 | 0.84 (0.41-1.73) | 0.641 | 43.8 | 0.182 |
| Source | ||||||
| Peripheral blood | 3 | 179 | 0.93 (0.59-1.46) | 0.746 | 0.0 | 0.406 |
| Intratumor | 6 | 1134 | 0.86 (0.68-1.09) | 0.213 | 32.1 | 0.195 |
| Outcome | ||||||
| DFS | 2 | 388 | 0.61 (0.40-0.93) | 0.022 | 0.0 | 0.784 |
| RFS | 3 | 354 | 0.77 (0.55-1.08) | 0.134 | 0.0 | 0.568 |
| TTR | 3 | 510 | 1.08 (0.85-1.36) | 0.543 | 0.0 | 0.737 |
| PFS | 1 | 61 | 0.50 (0.18-1.39) | 0.185 | / | / |
HCC, Hepatocellular carcinoma; NK, Natural killer; CI, Confidence interval; HR, Hazard ratio; OS, Overall survival; DFS, Disease-free survival; TTR, Time-to recurrence; RFS, Recurrence-free survival; PFS, Progression-free survival.
A total of 9 studies, involving 1313 patients, investigated the prognostic value of NK cells for DFS/RFS/TTR/PFS. The pooled results from the 9 comparative studies were not significant (HR=0.87, 95%CI 0.72-1.06, P=0.164), and the data were not heterogeneous (I2 = 13.4%, P=0.323; Figure 2B ). No bias was observed in the funnel plot ( Figure 3B ). We also performed subgroup analysis to better understand prognostic value of NK cells ( Table 2 ). The pooled HR (95%CI) for CD57+ NK cells and CD56+ NK cells was 0.76 (0.57-1.01, P=0.059) and 1.09 (0.79-1.50, P=0.602), respectively ( Supplementary Figure 3 ). For NK cells derived from peripheral blood and intratumor, the pooled HR (95%CI) was 0.93 (0.59-1.46, P=0.746) and 0.86 (0.68-1.09, P=0.213), respectively ( Supplementary Figure 4 ). In addition, in subgroup analysis of outcome, we found that high NK cells levels could be a good predictor for DFS (HR=0.61, 95%CI 0.40-0.93, P=0.022), but not for RFS, TTR and PFS ( Supplementary Figure 5 ).
Prognostic Value of Activating Receptors/Ligands on NK Cells
Table 3 summarized the outcomes of 3 studies that reported on NKp30+ NK cells. One study mentioned that high NKp30+ NK cell level was associated with better survival (41), while another study reported no effect of NKp30+ NK cell level on patient outcome (43). Other study suggested that high NKp30+ NK cell level was associated with good PFS, but not with OS (42). In addition, one study investigated the prognostic role of NKG2D. They concluded that low frequency of circulating NKG2D+CD56dim NK cells one month after hepatectomy may predict a poor prognosis for patients with HBV-related HCC (23).
Table 3.
The characteristics of included studies about the NK cells activating receptors and their ligands.
| Marker | Author | Year | Country | Sample size | Measure ment | Treatment | Source | Tumor stage | VS | Number of VS | Divide | Outcome | P-value | Follow-up times | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activating receptors of the NK cells | NKp30 | Chew et al. (41) | 2010 | Singapore | 61 | Immuno histochemistry | resection | Intratumor | I-III | High/Low | NA | median | OS | HR (95%CI) 0.34 (0.13,0.85) P=0.0144 | median: 2.56 (0.02-9.11) year |
| NKp30 | Li et al. (42) | 2021 | China | 25 | Flow cytometry | untreated | Peripheral blood | NA | High/Low | 16/9 | cutoff value | OS;PFS | Log-rank test P=0.279; Log-rank test P=0.016 | NA | |
| NKG2D | Gao et al. (23) | 2016 | China | 20 | Flow cytometry | resection | Peripheral blood | I-III | High/Low | 10/10 | median | OS;RFS | Log-rank test P=0.014; Log-rank test P=0.010 | until:2014.11 | |
| NKp30 | Rochigneux et al. (43) | 2019 | France | 57 | Flow cytometry | RFA | Peripheral blood | NA | High/Low | 28/29 | median | PFS | HR (95%CI) 0.61 (0.29,1.29) P=0.20 | until:12/2016 | |
| Ligands of the NK cells activating receptors | soluble MICA | Li et al. (44) | 2013 | China | 60 | ELISA | TACE | serum | III/IV | High/Low | 28/32 | median | OS | HR (95%CI) 1.47 (1.01,1.95) P<0.001 | until:31/08/2010 |
| B7-H6 | Qiu et al. (45) | 2021 | China | 90 | Immuno histochemistry | resection | Intratumor | I/II | High/Low | 33/57 | mean-H score | OS;DFS | HR (95%CI) 0.47 (0.24,0.93) P=0.029; HR (95%CI) 0.72 (0.36,1.43) P=0.1013 | total:>60 month | |
| MICA | Zhang et al. (28) | 2014 | China | 143 | Immuno histochemistry | resection | Intratumor | I-IV | High/Low | NA | NA | OS;RFS | HR (95%CI) 0.91 (0.49,1.69) P=0.774; HR (95%CI) 1.43 (0.90,2.27) P=0.135 | until:08/2013 | |
| MICA/B | Fang et al. (46) | 2014 | China | 96 | Immuno histochemistry | resection | Intratumor | I-IV | High/Low | 75/21 | MICA/B expression score | OS | HR (95%CI) 0.32 (0.11,0.92) P<0.001 | until:08/2012 | |
| ULBP1 | Kamimura et al. (47) | 2012 | Japan | 54 | Immuno histochemistry | untreated/ resection | Intratumor | NA | Positive/Negative | 25/47 | expression | OS;RFS | HR (95%CI) 0.72 (0.09,5.70) P=0.120; HR (95%CI) 0.2 (0.06,0.65) P=0.006 | NA | |
| ULBP1 | Easom et al. (48) | 2020 | England | 72 | ELISA | untreated | serum | NA | High/Low | NA | NA | OS | HR (95%CI) 2.11 (1.02,4.02) P=0.0029 | NA |
NK, Natural killer; CI, Confidence interval; HR, Hazard ratio; RFA, Radiofrequency ablation; ELISA, Enzyme-linked immunosorbent assay; TACE, Transcatheter arterial chemoembolization; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; PFS, Progression-free survival; NA, Not available.
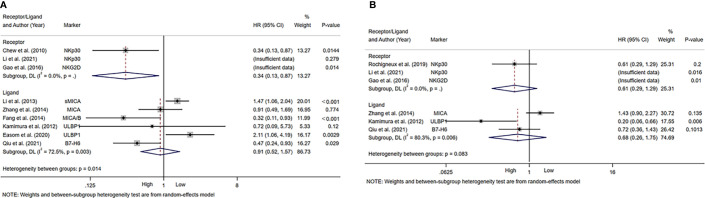
A total of 6 studies reported on ligands of NK cell activating receptors, including MICA, MICB, soluble MICA (sMICA), ULBP1 and B7-H6 (28, 44–48). The pooled HR (95%CI) for OS and DFS/RFS/PFS was 0.91 (0.52-1.57, P=0.726; Figure 4A ) and 0.68 (0.26-1.75, P=0.422; Figure 4B ), respectively.
Figure 4.
Forest plot of NK cells activating receptors/ligands in HCC. (A) Forest plot of NK cells activating receptors/ligands and OS in HCC. (B) Forest plot of NK cells activating receptors/ligands and DFS/RFS/PFS in HCC. CI, Confidence interval; HR, Hazard ratio; HCC, Hepatocellular carcinoma; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; PFS, Progression-free survival.
Prognostic Value of Inhibitory Receptors/Ligands on NK Cells
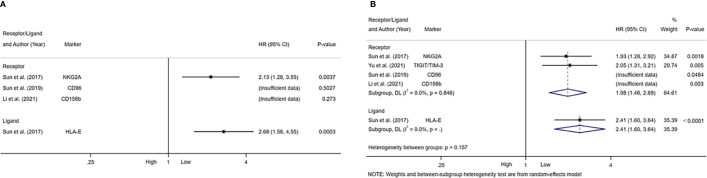
Table 4 summarized the outcomes of 4 studies that mentioned NK cell inhibitory receptors, including NKG2A, CD96, CD158b, TIGIT and TIM-3 (24, 42, 49, 50). They all concluded that high level of NK cell inhibitory receptors predicted increased recurrence of HCC patients. Sun and Li et al. suggested that the level of NK cell inhibitory receptors was not associated with survival of HCC patients, while other studies revealed that high NK cell inhibitory receptors level predicted poor survival of HCC patients. One study found that intratumor level of HLA-E was increased, and high HLA-E level was correlated with poor prognosis of HCC patients ( Figures 5A, B ).
Table 4.
The characteristics of included studies about the NK cells inhibitory receptors and their ligands.
| Marker | Author | Year | Country | Sample size | Measurement | Treatment | Source | Tumor stage | VS | Number of VS | Divide | Outcome | P-value | Follow-up times | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibitory receptors of the NK cells | NKG2A | Sun et al. (24) | 2017 | China | 177 | Immune histochemistry | resection | Intratumor | NA | High/Low | 68/109 | cutoff value | OS;DFS | HR (95%CI) 2.13 (1.28,3.56) P=0.0037; HR (95%CI) 1.93 (1.28,2.93) P=0.0018 |
median OS:1299.7 ± 1974.2 day; median DFS:980.0 ± 1796.1 day |
| CD96 | Sun et al. (49) | 2019 | China | 236 | Flow cytometry | resection | Intratumor | NA | High/Low | NA | cutoff value | OS;DFS | Log-rank test P=0.5027; Log-rank test P=0.0484 | NA | |
| CD158b | Li et al. (43) | 2021 | China | 13 | Flow cytometry | SBRT | Peripheral blood | NA | High/Low | 5/8 | cutoff value | OS;PFS | Log-rank test P=0.273; Log-rank test P=0.003 | NA | |
| TIGIT TIM-3 |
Yu et al. (50) | 2021 | China | 133 | Flow cytometry | palliative/minimally invasive/resection | Peripheral blood | NA | High/Low | 65/68 | cutoff value | PFS | HR (95%CI) 2.05 (1.24,3.04) P=0.005 | NA | |
| Ligands of the NK cells inhibitory receptors | HLA-E | Sun et al. (49) | 2017 | China | 177 | Immune histochemistry | resection | Intratumor | NA | High/Low | 79/98 | cutoff value | OS;DFS | HR (95%CI) 2.68 (1.58,4.56) P=0.0003; HR (95%CI) 2.41 (1.60,3.64) P<0.0001 |
median OS:1299.7 ± 1974.2 day; median DFS:980.0 ± 1796.1 day |
NK, Natural killer; CI, Confidence interval; HR, Hazard ratio; SBRT: Stereotactic body radiation therapy; OS, Overall survival; DFS, Disease-free survival; PFS, Progression-free survival; NA, Not available.
Figure 5.
Forest plot of NK cells inhibitory receptors/ligands in HCC. (A) Forest plot of NK cells inhibitory receptors/ligands and OS in HCC. (B) Forest plot of NK cells inhibitory receptors/ligands and DFS/PFS in HCC. CI, Confidence interval; HR, Hazard ratio; HCC, Hepatocellular carcinoma; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; PFS, Progression-free survival.
Assessment of Sensitivity Analysis and Publication Bias
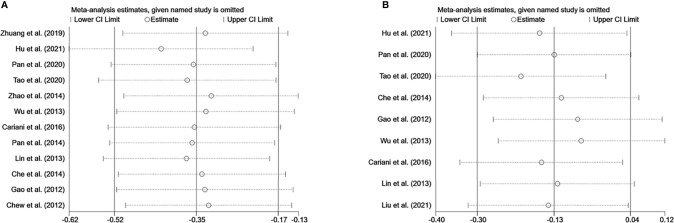
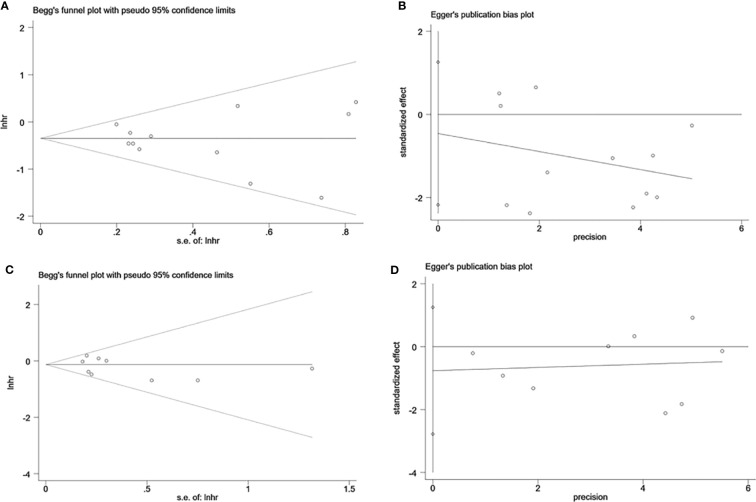
Sensitivity analysis was performed to evaluate the stability of NK cells for predicting survival and recurrence of HCC patients. After removing any of the studies, the results did not exceed the 95% CI range of the pooled results ( Figures 6A, B ). Begg’s test and Egger’s linear regression test were used to assess whether there was potential publication bias in this meta-analysis. The results showed that no apparent publication bias for the analysis was found between NK cells and OS (Begg’s test: P=0.732, Figure 7A ; Egger’s test: P=0.564, Figure 7B ). Similarly, no significant publication bias was found for DFS/RFS/TTR/PFS analysis (Begg’s test: P=0.602, Figure 7C ; Egger’s test: P=0.401, Figure 7D ).
Figure 6.
Sensitivity analysis of NK cells. (A) Sensitivity analysis for OS of NK cells. (B) Sensitivity analysis for DFS/RFS/TTR/PFS of NK cells. CI, Confidence interval; HR, Hazard ratio; NK, Natural killer; OS, Overall survival; DFS, Disease-free survival; RFS, Recurrence-free survival; TTR, Time-to recurrence; PFS, Progression-free survival.
Figure 7.
Evaluation of publication bias of NK cells using Begg’s test and Egger’s test. (A) Begg’s test for OS of NK cells, P=0.732. (B) Egger’s test for OS of NK cells, P=0.564. (C) Begg’s test for DFS/RFS/TTR/PFS of NK cells, P=0.602. (D) Egger’s test for DFS/RFS/TTR/PFS of NK cells, P=0.401. lnhr, the ln of HR; s.e., standard error; NK, Natural killer; OS, Overall survival, DFS, Disease-free survival; RFS, Recurrence-free survival; TTR, Time-to recurrence; PFS, Progression-free survival.
Discussion
NK cells are innate lymphocytes that can kill virus-infected or cancer cells, and have a vital role in early hepatocarcinogenesis (51, 52). Different to T cells which require somatic gene rearrangement to produce highly antigen-specific receptors (53), NK cells are innately equipped with germline-encoded activating and inhibitory receptors that can directly determine whether NK cells are activated or inhibited (12, 54). NK cells can deliver cytotoxic granules, secrete effector cytokines, and are involved in death receptor induced apoptosis (55). NK cells can also rapidly produce cytokines with anti-tumor effects, such as IFN-γ, to exert their killing effects in the early stage of disease (56). In addition, NK cells can bind to target cells through surface CD16 and kill them through exerting antibody-dependent cell-mediated cytotoxicity (ADCC) (57). These results imply that NK cells play an essential role in the body’s immune process in the defense against HCC.
In this study, we found that high NK cell level could predict better survival for patients with HCC. Similar results were previously reported in a meta-analysis of solid tumors (58). In subgroup analysis, CD57+ NK cells had better prognostic value over CD56+ NK cells. On the one hand, it may be that CD56+ NK cells, accounting for the majority of circulating NK cells, also expressed inhibitory molecules, which may strive for a dynamic balance between activating and inhibitory molecules. Moreover, lower IFN-γ production was also described in HCC, in accordance with the decreased cytotoxicity of NK cells (59). On the other hand, acquisition of CD57 represents a shift toward a higher cytotoxic capacity, greater responsiveness to signaling via CD16 and natural NCRs (60). The same result was also observed in Hu et al.’s study (61). In addition, compared to peripheral NK cells, NK cells from intratumor had better prognostic value for prognosis of HCC patients, possibly because NK cells are abundant in human liver.
NK cells express activating and inhibitory receptors in order to perceive signals and display their activity. Depending on the received signal, NK cells can be activated or restricted (62). In this study, activating receptors/ligands mainly contain NKG2D, NKp30, and their ligands. NKG2D ligands mainly consist of ULBPs and MICA/B. MICA/B molecules expressed on HCC cells are recognized by NKG2D to induce ubiquitination-mediated endocytosis of the NKG2D-DAP10 complex, thereby activating NK cells to kill HCC (46, 57). NKp30 contains two ligands. One is BAT3, a nuclear protein that induces apoptosis in target cells by interacting with P53. The other is B7-H6, a newly discovered member of the B7 family that is expressed on the surface of tumor cells (45). B7-H6/NKp30 pathway is involved in the NK cell-mediated immune responses, and NK cells can recognize and eliminate B7-H6-expressing tumors, including HCC. However, tumors can also impair NK cell function by shedding B7-H6 membranes or decreasing NKp30 expression, leading to tumor immune escape and tumor progression (63). In addition, hypoxia, some soluble forms of NCRs ligands, or soluble factors produced by tumor/tumor-associated cells, can induce a decrease in both NCR expression and function (10, 64, 65), and protect tumor from NK cell-mediated cytotoxicity (44, 66). Until now, the prognostic value of activating receptors remained inconsistent from different studies and deserved further investigation.
Inhibitory receptors/ligands mainly contain NKG2A, CD96, CD158b, TIGIT and TIM-3, and HLA-E. The NKG2A is expressed approximately in half of the peripheral blood NK cells, and is also expressed on CD8+ T cells (67). The inhibitory signals induced by NKG2A engagement can result in decreased capacity of NK cells and CD8+ T cells to lyse target cells (68), and enhanced expression of the HLA-E on tumor may result in resistance and immune escape by binding to NKG2A (68). Therefore, blocking the interaction of NKG2A with HLA-E has shown promising therapeutic effects in animal study (69). In addition, there is increasing evidence that PD-1 is also expressed on the surface of NK cells and exerts a suppressive function on T cell responses (16). In this study, though we found that inhibitory receptors of NK cells may be a good predictor for recurrence of HCC, however, their prognostic value in predicting survival was unclear. Therefore, more high-quality prospective studies are needed to explore the prognostic value of NK cells and their receptors/ligands for HCC.
The strength of this study is that it explored for the first time the prognostic value of NK cells and their receptors/ligands in HCC. Almost all relevant articles that could be collected were included and a comprehensive analysis was provided. However, the following limitations should also be considered. First, some of the data were not obtained directly from the included studies. HRs and 95% CIs were calculated using survival curves or 95% CIs were calculated from known P values and HRs, which may result in data inaccuracy to some extent. Second, although most studies used median as the cut-off value for NK cells level, these values were complex and related to the clinicopathological characteristics of the HCC patients. Third, although DFS/RFS/TTR/PFS of HCC patients are considered as composite outcome indicators, there are still slightly difference between them. Fourth, NK cells receptors/ligands are diverse. The pooled results may exist in bias to some extent. Fifth, this was an aggregate data rather than an individual data meta-analysis, and the data were various between studies, which somehow diminished the significance of the study. Finally, some surface markers were expressed not only in NK cells, but also in other immune cells, such as T cells, Dendritic cells (DCs), etc. Moreover, liver NK cells are composed of several subsets including conventional, type 1 innate lymphoid cells (ILC1) like, and liver-resident NK cells. All of them can express similar molecules and potentially be responsible for the clinical benefit observed (70).
Conclusions
In summary, we concluded that NK cells could be a good predictor for survival of HCC. More importantly, CD57+ NK cells may have better prognostic value over CD56+ NK cells, and intratumor NK cells have better prognostic value over peripheral NK cells. Inhibitory receptors of NK cell may be a good predictor for recurrence of HCC, but the value of activating and inhibitory receptors in predicting the survival of HCC was unclear. More high-quality prospective studies are essential to evaluate the prognostic value of NK cells and their receptors/ligands for HCC.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author Contributions
J-SX and TL were responsible for designing the study. J-SX, Z-ND, and G-XM conducted the systematic search and performed the screening. J-SX, Z-ND, G-XM, L-JY, HL, H-CL, S-YY, B-WT, J-GH, Z-RD, Z-QC, and D-XW primarily performed the quality assessment as well as supervision. J-SX analyzed, interpreted the data, and drafted the manuscript. TL revised the manuscript. All data and material analyzed during this study were included in this article. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the grants from the Taishan Scholars Program for Young Expert of Shandong Province (Grant No. tsqn20161064), National Natural Science Foundation of China (Grant No. 82073200 & 81874178), funds for Independent Cultivation of Innovative Team from Universities in Jinan (Grant No. 2020GXRC023), and Major basic research of Shandong Provincial Natural Science Foundation (Grant No. ZR202105070027).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
| NK | Natural killer |
| HCC | Hepatocellular carcinoma |
| HRs | Hazard ratios |
| CI | Confidence interval |
| NKG2D | NK group 2 member D |
| NCRs | Natural cytotoxic receptors |
| IL | Interleukin |
| IFNR | Interferon receptor |
| TGF-βR | Tumor growth factor-β receptor |
| BAT3 | HLA-B-associated transcript 3 |
| BAG6 | Bcl-2-associated athanogene 6 |
| MLL5 | Mixed lineage leukemia 5 |
| PCNA | Proliferating cell nuclear antigen |
| B7-H6 | B7 homolog 6 |
| MIC | Major histocompatibility complex class I chain-related protein |
| ULBPs | UL16-binding proteins |
| KIRs | Killer immunoglobulin-like receptors |
| TIM-3 | T cell immunoglobulin domain and mucin domain-3 |
| TIGIT | T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domain |
| PD-1 | Programmed cell death-1 |
| PD-L1 | Programmed cell death ligand-1 |
| LAG3 | Lymphocyte activation gene-3 |
| LAIRs | Leukocyte-associated immunoglobulin-like receptors |
| A2AR | Adenosine 2A receptor |
| ILTs | Immunoglobulin-like transcripts |
| HLA-E | Histocompatibility leucocyte antigen E |
| MHC-I | Major histocompatibility complex class I |
| PRISMA | Preferred Reporting Items for Systematic reviews and Meta-Analyses |
| OS | Overall survival |
| CSS | Cancer-specific survival |
| DFS | Disease-free survival |
| RFS | Recurrence-free survival |
| TTR | Time-to recurrence |
| PFS | Progression-free survival |
| NOS | Newcastle–Ottawa Scale |
| sMICA | Soluble MICA |
| ADCC | Antibody-dependent cell-mediated cytotoxicity |
| DCs | Dendritic cells |
| ILC1 | Type 1 innate lymphoid cells |
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.872353/full#supplementary-material
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of Natural Killer Cells. Nat Immunol (2008) 9(5):503–10. doi: 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- 4. Mikulak J, Bruni E, Oriolo F, Di Vito C, Mavilio D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front Immunol (2019) 10:946. doi: 10.3389/fimmu.2019.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caligiuri MA. Human Natural Killer Cells. Blood (2008) 112(3):461–9. doi: 10.1182/blood-2007-09-077438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell KS, Hasegawa J. Natural Killer Cell Biology: An Update and Future Directions. J Allergy Clin Immunol (2013) 132(3):536–44. doi: 10.1016/j.jaci.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solana R, Tarazona R, Gayoso I, Lesur O, Dupuis G, Fulop T. Innate Immunosenescence: Effect of Aging on Cells and Receptors of the Innate Immune System in Humans. Semin Immunol (2012) 24(5):331–41. doi: 10.1016/j.smim.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 8. Huntington ND, Cursons J, Rautela J. The Cancer-Natural Killer Cell Immunity Cycle. Nat Rev Cancer (2020) 20(8):437–54. doi: 10.1038/s41568-020-0272-z [DOI] [PubMed] [Google Scholar]
- 9. Hammer Q, Rückert T, Romagnani C. Natural Killer Cell Specificity for Viral Infections. Nat Immunol (2018) 19(8):800–8. doi: 10.1038/s41590-018-0163-6 [DOI] [PubMed] [Google Scholar]
- 10. Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human Nk Cells: Surface Receptors, Inhibitory Checkpoints, and Translational Applications. Cell Mol Immunol (2019) 16(5):430–41. doi: 10.1038/s41423-019-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H, Wang S, Xin J, Wang J, Yao C, Zhang Z. Role of Nkg2d and Its Ligands in Cancer Immunotherapy. Am J Cancer Res (2019) 9(10):2064–78. [PMC free article] [PubMed] [Google Scholar]
- 12. Martinet L, Smyth MJ. Balancing Natural Killer Cell Activation Through Paired Receptors. Nat Rev Immunol (2015) 15(4):243–54. doi: 10.1038/nri3799 [DOI] [PubMed] [Google Scholar]
- 13. Li J, Tao L, Wang X. Cytotoxic Immune Cell-Based Immunotherapy for Hepatocellular Carcinoma. Hepatoma Res (2020) 6:15. doi: 10.20517/2394-5079.2019.34 [DOI] [Google Scholar]
- 14. Kalathil SG, Thanavala Y. Natural Killer Cells and T Cells in Hepatocellular Carcinoma and Viral Hepatitis: Current Status and Perspectives for Future Immunotherapeutic Approaches. Cells (2021) 10(6):1332. doi: 10.3390/cells10061332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and Inhibitory Receptors of Natural Killer Cells. Immunol Cell Biol (2011) 89(2):216–24. doi: 10.1038/icb.2010.78 [DOI] [PubMed] [Google Scholar]
- 16. Quatrini L, Mariotti FR, Munari E, Tumino N, Vacca P, Moretta L. The Immune Checkpoint Pd-1 in Natural Killer Cells: Expression, Function and Targeting in Tumour Immunotherapy. Cancers (2020) 12(11):3285. doi: 10.3390/cancers12113285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eugène J, Jouand N, Ducoin K, Dansette D, Oger R, Deleine C, et al. The Inhibitory Receptor Cd94/Nkg2a on Cd8(+) Tumor-Infiltrating Lymphocytes in Colorectal Cancer: A Promising New Druggable Immune Checkpoint in the Context of Hlae/B2m Overexpression. Modern Pathol (2020) 33(3):468–82. doi: 10.1038/s41379-019-0322-9 [DOI] [PubMed] [Google Scholar]
- 18. Hiraoka N, Ino Y, Hori S, Yamazaki-Itoh R, Naito C, Shimasaki M, et al. Expression of Classical Human Leukocyte Antigen Class I Antigens, Hla-E and Hla-G, Is Adversely Prognostic in Pancreatic Cancer Patients. Cancer Sci (2020) 111(8):3057–70. doi: 10.1111/cas.14514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Hla-E and Hla-G Predicts Prognosis in Colon Cancer Patients. Br J Cancer (2014) 110(2):459–68. doi: 10.1038/bjc.2013.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, et al. Monocyte/Macrophage-Elicited Natural Killer Cell Dysfunction in Hepatocellular Carcinoma Is Mediated by Cd48/2b4 Interactions. Hepatol (Baltimore Md) (2013) 57(3):1107–16. doi: 10.1002/hep.26192 [DOI] [PubMed] [Google Scholar]
- 21. Zhao J-J, Pan Q-Z, Pan K, Weng D-S, Wang Q-J, Li J-J, et al. Interleukin-37 Mediates the Antitumor Activity in Hepatocellular Carcinoma: Role for Cd57+Nk Cells. Sci Rep (2014) 4:5177. doi: 10.1038/srep05177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chew V, Chen J, Lee D, Loh E, Lee J, Lim KH, et al. Chemokine-Driven Lymphocyte Infiltration: An Early Intratumoural Event Determining Long-Term Survival in Resectable Hepatocellular Carcinoma. Gut (2012) 61(3):427–38. doi: 10.1136/gutjnl-2011-300509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao J, Duan Z, Zhang L, Huang X, Long L, Tu J, et al. Failure Recovery of Circulating Nkg2d(+)Cd56(Dim)Nk Cells in Hbv-Associated Hepatocellular Carcinoma After Hepatectomy Predicts Early Recurrence. Oncoimmunology (2016) 5(1):e1048061. doi: 10.1080/2162402x.2015.1048061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun C, Xu J, Huang Q, Huang M, Wen H, Zhang C, et al. High Nkg2a Expression Contributes to Nk Cell Exhaustion and Predicts a Poor Prognosis of Patients With Liver Cancer. Oncoimmunology (2017) 6(1):e1264562. doi: 10.1080/2162402x.2016.1264562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu ZQ, Xin HY, Luo CB, Li J, Zhou ZJ, Zou JX, et al. Associations Among the Mutational Landscape, Immune Microenvironment, and Prognosis in Chinese Patients With Hepatocellular Carcinoma. Cancer Immunol Immunother: CII (2021) 70(2):377–89. doi: 10.1007/s00262-020-02685-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tao P, Hong L, Tang W, Lu Q, Zhao Y, Zhang S, et al. Comprehensive Characterization of Immunological Profiles and Clinical Significance in Hepatocellular Carcinoma. Front Oncol (2020) 10:574778. doi: 10.3389/fonc.2020.574778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan QZ, Liu Q, Zhou YQ, Zhao JJ, Wang QJ, Li YQ, et al. Cik Cell Cytotoxicity Is a Predictive Biomarker for Cik Cell Immunotherapy in Postoperative Patients With Hepatocellular Carcinoma. Cancer Immunol Immunother: CII (2020) 69(5):825–34. doi: 10.1007/s00262-020-02486-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Xu Z, Zhou X, Zhang H, Yang N, Wu Y, et al. Loss of Expression of Mhc Class I-Related Chain a (Mica) Is a Frequent Event and Predicts Poor Survival in Patients With Hepatocellular Carcinoma. Int J Clin Exp Pathol (2014) 7(6):3123–31. [PMC free article] [PubMed] [Google Scholar]
- 29. Shamseer L, Moher D, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (Prisma-P) 2015: Elaboration and Explanation. BMJ (Cli Res ed) (2015) 350:g7647. doi: 10.1136/bmj.g7647 [DOI] [PubMed] [Google Scholar]
- 30. Stang A. Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur J Epidemiol (2010) 25(9):603–5. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 31. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical Methods for Incorporating Summary Time-To-Event Data Into Meta-Analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altman DG, Bland JM. How to Obtain the Confidence Interval From a P Value. BMJ (Cli Res ed) (2011) 343:d2090. doi: 10.1136/bmj.d2090 [DOI] [PubMed] [Google Scholar]
- 33. Egger M, Davey Smith G, Schneider M, Minder C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ (Cli Res ed) (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhuang Y, Yuan BY, Chen GW, Zhao XM, Hu Y, Zhu WC, et al. Association Between Circulating Lymphocyte Populations and Outcome After Stereotactic Body Radiation Therapy in Patients With Hepatocellular Carcinoma. Front Oncol (2019) 9:896. doi: 10.3389/fonc.2019.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin SZ, Chen KJ, Xu ZY, Chen H, Zhou L, Xie HY, et al. Prediction of Recurrence and Survival in Hepatocellular Carcinoma Based on Two Cox Models Mainly Determined by Foxp3+ Regulatory T Cells. Cancer Prev Res (Philadelphia Pa) (2013) 6(6):594–602. doi: 10.1158/1940-6207.Capr-12-0379 [DOI] [PubMed] [Google Scholar]
- 36. Gao Q, Zhou J, Wang XY, Qiu SJ, Song K, Huang XW, et al. Infiltrating Memory/Senescent T Cell Ratio Predicts Extrahepatic Metastasis of Hepatocellular Carcinoma. Ann Surg Oncol (2012) 19(2):455–66. doi: 10.1245/s10434-011-1864-3 [DOI] [PubMed] [Google Scholar]
- 37. Pan K, Wang QJ, Liu Q, Zheng HX, Li YQ, Weng DS, et al. The Phenotype of Ex Vivo Generated Cytokine-Induced Killer Cells Is Associated With Overall Survival in Patients With Cancer. Tumour Biol (2014) 35(1):701–7. doi: 10.1007/s13277-013-1096-1 [DOI] [PubMed] [Google Scholar]
- 38. Liu J, Kuang S, Zheng Y, Liu M, Wang L. Prognostic and Predictive Significance of the Tumor Microenvironment in Hepatocellular Carcinoma. Cancer Biomarkers: Section A Dis Markers (2021) 32(1):99–110. doi: 10.3233/cbm-203003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Che Y-Q, Feng L, Rong W-Q, Shen D, Wang Q, Yang L, et al. Correlation Analysis of Peripheral Blood T Cell Subgroups, Immunoglobulin and Prognosis of Early Hepatocellular Carcinoma After Hepatectomy. Int J Clin Exp Med (2014) 7(11):4282–90. [PMC free article] [PubMed] [Google Scholar]
- 40. Cariani E, Pilli M, Barili V, Porro E, Biasini E, Olivani A, et al. Natural Killer Cells Phenotypic Characterization as an Outcome Predictor of Hcv-Linked Hcc After Curative Treatments. Oncoimmunology (2016) 5(8):e1154249. doi: 10.1080/2162402x.2016.1154249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chew V, Tow C, Teo M, Wong HL, Chan J, Gehring A, et al. Inflammatory Tumour Microenvironment Is Associated With Superior Survival in Hepatocellular Carcinoma Patients. J Hepatol (2010) 52(3):370–9. doi: 10.1016/j.jhep.2009.07.013 [DOI] [PubMed] [Google Scholar]
- 42. Li TT, Sun J, Wang Q, Li WG, He WP, Yang RC, et al. The Effects of Stereotactic Body Radiotherapy on Peripheral Natural Killer and Cd3(+)Cd56(+) Nkt-Like Cells in Patients With Hepatocellular Carcinoma. Hepatobil Pancreatic Dis Int: HBPD Int (2021) 20(3):240–50. doi: 10.1016/j.hbpd.2020.12.015 [DOI] [PubMed] [Google Scholar]
- 43. Rochigneux P, Nault JC, Mallet F, Chretien AS, Barget N, Garcia AJ, et al. Dynamic of Systemic Immunity and Its Impact on Tumor Recurrence After Radiofrequency Ablation of Hepatocellular Carcinoma. Oncoimmunology (2019) 8(8):1615818. doi: 10.1080/2162402X.2019.1615818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li JJ, Pan K, Gu MF, Chen MS, Zhao JJ, Wang H, et al. Prognostic Value of Soluble Mica Levels in the Serum of Patients With Advanced Hepatocellular Carcinoma. Chin J Cancer (2013) 32(3):141–8. doi: 10.5732/cjc.012.10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qiu H, Gao S, Sun Z, Wang J. Dual Role of B7-H6 as a Novel Prognostic Marker in Hepatocellular Carcinoma. APMIS: Acta Pathol Microbiol Immunol Scand (2021) 129(3):105–17. doi: 10.1111/apm.13099 [DOI] [PubMed] [Google Scholar]
- 46. Fang L, Gong J, Wang Y, Liu R, Li Z, Wang Z, et al. Mica/B Expression Is Inhibited by Unfolded Protein Response and Associated With Poor Prognosis in Human Hepatocellular Carcinoma. J Exp Clin Cancer Res: CR (2014) 33(1):76. doi: 10.1186/s13046-014-0076-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kamimura H, Yamagiwa S, Tsuchiya A, Takamura M, Matsuda Y, Ohkoshi S, et al. Reduced Nkg2d Ligand Expression in Hepatocellular Carcinoma Correlates With Early Recurrence. J Hepatol (2012) 56(2):381–8. doi: 10.1016/j.jhep.2011.06.017 [DOI] [PubMed] [Google Scholar]
- 48. Easom NJW, Marks M, Jobe D, Gillmore R, Meyer T, Maini MK, et al. Ulbp1 Is Elevated in Human Hepatocellular Carcinoma and Predicts Outcome. Front Oncol (2020) 10:971. doi: 10.3389/fonc.2020.00971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun H, Huang Q, Huang M, Wen H, Lin R, Zheng M, et al. Human Cd96 Correlates to Natural Killer Cell Exhaustion and Predicts the Prognosis of Human Hepatocellular Carcinoma. Hepatol (Baltimore Md) (2019) 70(1):168–83. doi: 10.1002/hep.30347 [DOI] [PubMed] [Google Scholar]
- 50. Yu L, Liu X, Wang X, Yan F, Wang P, Jiang Y, et al. Tigit(+) Tim-3(+) Nk Cells Are Correlated With Nk Cell Exhaustion and Disease Progression in Patients With Hepatitis B Virus−Related Hepatocellular Carcinoma. Oncoimmunology (2021) 10(1):1942673. doi: 10.1080/2162402x.2021.1942673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu S, Galat V, Galat Y, Lee YKA, Wainwright D, Wu J. Nk Cell-Based Cancer Immunotherapy: From Basic Biology to Clinical Development. J Hematol Oncol (2021) 14(1):7. doi: 10.1186/s13045-020-01014-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lim O, Jung MY, Hwang YK, Shin EC. Present and Future of Allogeneic Natural Killer Cell Therapy. Front Immunol (2015) 6:286. doi: 10.3389/fimmu.2015.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reindl LM, Albinger N, Bexte T, Müller S, Hartmann J, Ullrich E. Immunotherapy With Nk Cells: Recent Developments in Gene Modification Open Up New Avenues. Oncoimmunology (2020) 9(1):1777651. doi: 10.1080/2162402x.2020.1777651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Podhorzer A, Machicote A, Belén S, Lauferman L, Imventarza O, Montal S, et al. Intrahepatic and Peripheral Blood Phenotypes of Natural Killer and T Cells: Differential Surface Expression of Killer Cell Immunoglobulin-Like Receptors. Immunology (2018) 154(2):261–73. doi: 10.1111/imm.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: Nk Cell Therapies at the Forefront of Cancer Control. J Clin Invest (2019) 129(9):3499–510. doi: 10.1172/jci129338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang S, Saha B, Kodys K, Szabo G. Ifn-Γ Production by Human Natural Killer Cells in Response to Hcv-Infected Hepatoma Cells Is Dependent on Accessory Cells. J Hepatol (2013) 59(3):442–9. doi: 10.1016/j.jhep.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mantovani S, Oliviero B, Varchetta S, Mele D, Mondelli MU. Natural Killer Cell Responses in Hepatocellular Carcinoma: Implications for Novel Immunotherapeutic Approaches. Cancers (2020) 12(4):926. doi: 10.3390/cancers12040926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang S, Liu W, Hu B, Wang P, Lv X, Chen S, et al. Prognostic Significance of Tumor-Infiltrating Natural Killer Cells in Solid Tumors: A Systematic Review and Meta-Analysis. Front Immunol (2020) 11:1242. doi: 10.3389/fimmu.2020.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang QF, Yin WW, Xia Y, Yi YY, He QF, Wang X, et al. Liver-Infiltrating Cd11b(-)Cd27(-) Nk Subsets Account for Nk-Cell Dysfunction in Patients With Hepatocellular Carcinoma and Are Associated With Tumor Progression. Cell Mol Immunol (2017) 14(10):819–29. doi: 10.1038/cmi.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nielsen CM, White MJ, Goodier MR, Riley EM. Functional Significance of Cd57 Expression on Human Nk Cells and Relevance to Disease. Front Immunol (2013) 4:422. doi: 10.3389/fimmu.2013.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu G, Wang S. Prognostic Role of Tumor-Infiltrating Cd57-Positive Lymphocytes in Solid Tumors: A Meta-Analysis. Oncotarget (2018) 9(8):8111–9. doi: 10.18632/oncotarget.23621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roma S, Carpen L, Raveane A, Bertolini F. The Dual Role of Innate Lymphoid and Natural Killer Cells in Cancer. From Phenotype to Single-Cell Transcriptomics, Functions and Clinical Uses. Cancers (2021) 13(20):5042. doi: 10.3390/cancers13205042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Semeraro M, Rusakiewicz S, Minard-Colin V, Delahaye NF, Enot D, Vély F, et al. Clinical Impact of the Nkp30/B7-H6 Axis in High-Risk Neuroblastoma Patients. Sci Trans Med (2015) 7(283):283ra55. doi: 10.1126/scitranslmed.aaa2327 [DOI] [PubMed] [Google Scholar]
- 64. Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, et al. Soluble Ligands for Nk Cell Receptors Promote Evasion of Chronic Lymphocytic Leukemia Cells From Nk Cell Anti-Tumor Activity. Blood (2013) 121(18):3658–65. doi: 10.1182/blood-2013-01-476606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schlecker E, Fiegler N, Arnold A, Altevogt P, Rose-John S, Moldenhauer G, et al. Metalloprotease-Mediated Tumor Cell Shedding of B7-H6, the Ligand of the Natural Killer Cell-Activating Receptor Nkp30. Cancer Res (2014) 74(13):3429–40. doi: 10.1158/0008-5472.Can-13-3017 [DOI] [PubMed] [Google Scholar]
- 66. Luo Q, Luo W, Zhu Q, Huang H, Peng H, Liu R, et al. Tumor-Derived Soluble Mica Obstructs the Nkg2d Pathway to Restrain Nk Cytotoxicity. Aging Dis (2020) 11(1):118–28. doi: 10.14336/ad.2019.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van Montfoort N, Borst L, Korrer MJ, Sluijter M, Marijt KA, Santegoets SJ, et al. Nkg2a Blockade Potentiates Cd8 T Cell Immunity Induced by Cancer Vaccines. Cell (2018) 175(7):1744–55.e15. doi: 10.1016/j.cell.2018.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Borst L, van der Burg SH, van Hall T. The Nkg2a-Hla-E Axis as a Novel Checkpoint in the Tumor Microenvironment. Clin Cancer Res (2020) 26(21):5549–56. doi: 10.1158/1078-0432.Ccr-19-2095 [DOI] [PubMed] [Google Scholar]
- 69. Ruggeri L, Urbani E, André P, Mancusi A, Tosti A, Topini F, et al. Effects of Anti-Nkg2a Antibody Administration on Leukemia and Normal Hematopoietic Cells. Haematologica (2016) 101(5):626–33. doi: 10.3324/haematol.2015.135301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jacquelot N, Seillet C, Souza-Fonseca-Guimaraes F, Sacher AG, Belz GT, Ohashi PS. Natural Killer Cells and Type 1 Innate Lymphoid Cells in Hepatocellular Carcinoma: Current Knowledge and Future Perspectives. Int J Mol Sci (2021) 22(16):9044. doi: 10.3390/ijms22169044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.