Summary
Background
SARS-CoV-2 vaccines currently authorized for emergency use have been highly successful in preventing infection and lessening disease severity. The vaccines maintain effectiveness against earlier SARS-CoV-2 Variants of Concern but the heavily mutated, highly transmissible Omicron variant presents an obstacle both to vaccine protection and monoclonal antibody therapies.
Methods
Pseudotyped lentiviruses were incubated with serum from vaccinated and boosted donors or therapeutic monoclonal antibody and then applied to target cells. After 2 days, luciferase activity was measured in a microplate luminometer. Resistance mutations of the Omicron spike were identified using point-mutated spike protein pseudotypes and mapped onto the three-dimensional spike protein structure.
Findings
Virus with the Omicron spike protein was 26-fold resistant to neutralization by recovered donor sera and 26-34-fold resistance to Pfizer BNT162b2 and Moderna vaccine-elicited antibodies following two immunizations. A booster immunization increased neutralizing titres against Omicron. Neutralizing titres against Omicron were increased in the sera with a history of prior SARS-CoV-2 infection. Analysis of the therapeutic monoclonal antibodies showed that the Regeneron and Eli Lilly monoclonal antibodies were ineffective against the Omicron pseudotype while Sotrovimab and Evusheld were partially effective.
Interpretation
The results highlight the benefit of a booster immunization to protect against the Omicron variant and demonstrate the challenge to monoclonal antibody therapy. The decrease in neutralizing titres against Omicron suggest that much of the vaccine efficacy may rely on T cells.
Funding
The work was funded by grants from the NIH to N.R.L. (DA046100, AI122390 and AI120898) and 55 to M.J.M. (UM1AI148574).
Keywords: SARS-CoV-2 variant, Omicron, Vaccine, Antibody neutralization, Monoclonal antibodies
Research in context.
Evidence before this study
Previous analyses of vaccine efficacy against SARS-CoV-2 variants showed relatively modest decreases in neutralizing antibody titres against the Alpha, Beta, Gamma and Delta variants. Booster immunization was known to increase antibody titres against the variants as did previous SARS-CoV-2 infection. Regeneron and Eli Lilly therapeutic monoclonal antibodies retained neutralizing activity against the variants.
Added value of this study
The study shows that the large number of mutations present in the Omicron spike protein cause a considerable decrease in the antibody titres that are elicited by vaccination with either the Pfizer/BioNtech mRNA vaccines. The decreased titre was significantly more pronounced than the decrease against the Delta variant. Omicron could not be neutralized by the current Regeneron or Eli Lilly monoclonal antibodies and was only partially neutralized by the newer Sotrovimab and Evusheld monoclonal antibodies. Mapping of the Omicron point mutations responsible for escape from the monoclonal antibodies showed that these lie directly at the interaction sites with the individual monoclonal antibodies, demonstrating that the escape is caused by direct alteration of the epitopes.
Implications of all the available evidence
The decreased antibody titres against Omicron suggest that full vaccination (two immunizations) will show decreased protection against infection and severe disease but that boosting with a third immunization will restore protection afforded by vaccination. The findings also suggest that the development of an Omicron vaccine is warranted and that booster immunization will be beneficial. The findings suggest that the Current Regeneron and Eli Lilly monoclonal antibody cocktails will not be effective for the treatment of COVID-19. The reduction in neutralizing titres by Evusheld and Sotrovimab monoclonal antibodies may result in a significantly loss of therapeutic efficacy but this will depend on their concentrations in relevant tissues in vivo.
Alt-text: Unlabelled box
Introduction
The vaccines that have been granted emergency use authorization (EUA) have proven highly protective against SARS-CoV-2, resulting in a major decrease in infection rates, hospitalization and deaths1; however, the appearance of recently evolved viral variants classified as variants of concern (VOC)2 with multiple mutations in the viral spike protein have raised concerns about potential decreases in vaccine effectiveness. These concerns have been assuaged by laboratory findings of modest 2-5-fold decreases in neutralizing antibody titre against the VOCs3, 4, 5, 6, 7, 8 and epidemiological evidence of continued vaccine protection.9,10 Vaccination has been found to provide 78% protection against infection by the Delta variant, 90% protection against hospitalization and 91% protection against death.11 Monoclonal antibody cocktails from Regeneron consisting of REGN10933 (Casirivamab) and REGN10987 (Imdevimab), and from Eli Lilly consisting of LY-CoV016 (Etesevimab) and LY-CoV555 (Bamlanivimab) have proven effective at decreasing the frequency of hospitalization of COVID-19 patients.12, 13, 14 More recently, the monoclonal antibody VIR-7183 (Sotrovimab) from GlaxoSmithKline (GSK)/Vir Biotechnology was found to decrease hospitalization and risk of death by 79% and was granted EUA authorization by the U.S. Food and Drug Agency.15 In addition, the monoclonal antibody cocktail (Evusheld) from AstraZeneca consisting of AZD8895 (Tixagevimab) and AZD1061 (Cilgavimab) was found to reduce the risk of developing COVID-19 by 77%16 and was approved for prophylactic use in immunocompromised individuals.
The recently emerged Omicron (B.1.1.529) variant was identified in Botswana in early November, 2021 where it rapidly rose to a prevalence of 71%. The variant was shortly thereafter identified in South Africa.17 The prevalence of the variant continued to increase rapidly as a result of increased transmissibility with Omicron having replaced Delta as the predominant variant in the U.S. with a current prevalence of 99.9% in metropolitan areas (as of Jan 29th, 2022). The large number of mutations in the Omicron spike protein raises the possibility of decreased antibody neutralizing titres by vaccine-elicited antibodies.
As compared to the previously designated VOC spike proteins that contain 9–11 missense mutations, the Omicron spike protein has 34, 20 of which have not been found in previous VOCs or variant of interests (VOIs). These include 15 mutations in the receptor binding domain (RBD), 8 of which lie in the receptor binding motif (RBM) that directly contacts the receptor. The N-terminal domain (NTD) has 8 mutations, 3 of which are deletions and one is a 3 amino acid insertion. The carboxy-terminal CTD has 10 mutations, 4 of which are close to the furin proteolytic processing site and three of which are close to the secondary processing site. The concomitant appearance of the multiple mutations in the Omicron virus suggests that some may have arisen from recombination with a related β-coronavirus or from extended replication in a chronically infected immunodeficient individual.18
The large number of mutations in the Omicron RBD and NTD, which are the primary sites targeted by neutralizing antibodies, raises the possibility that the variant may be resistant to neutralization by current EUA approved vaccine-elicited antibodies, resulting in decreased protection from infection. It also raises the possibility that individuals previously infected with an earlier version of the virus might not be protected against re-infection by the Omicron variant. In addition, it raises a concern that the heavily mutated Omicron RBD might cause the failure of therapeutic monoclonal antibodies currently in clinical use, decreasing the effectiveness of their use in the treatment of severe COVID-19.
In this study, we used spike protein-pseudotyped lentiviral particles to measure the sensitivity of the Omicron variant to neutralization by vaccine-elicited antibodies in the sera of both naïve and recovered individuals and analysed the neutralizing activity of the widely used therapeutic monoclonal antibodies. We found that Omicron spike protein was highly resistant to neutralization by the serum antibodies of individuals fully vaccinated with two immunizations of the Pfizer or Moderna mRNA vaccines. A homologous booster vaccination with an mRNA vaccine increased neutralizing antibody titres 6-8-fold to a level predicted to provide a high degree of protection. The monoclonal antibodies that constitute the Regeneron and Eli Lilly cocktails failed to neutralize virus with the Omicron spike protein while the potency of VIR-7183 (Sotrovimab) and the Evusheld monoclonal antibodies was significantly decreased.
Methods
Plasmids
Plasmid expression vectors used in the production of lentiviral pseudotypes pMDL, pcVSV.G, pRSV.Rev and the lentiviral dual reporter virus genome pLenti.GFP.nLuc have been previously described.19 The SARS-CoV-2 Omicron spike expression vector pc.Δ19.Omicron was synthesized in two fragments encoding the codon-optimized open reading frame overlapping by 50 bp. The full-length coding sequence was generated by overlap extension PCR with the two fragments amplified with external primers containing a Kpn-I and Xho-I sites and then cloned into pcDNA6. Expression vectors encoding spike proteins with the individual mutations of the Omicron spike protein were generated by overlap extension PCR mutagenesis using the D614G spike protein plasmid pcCOV2.Δ19. D614G as template.
Cells
293T (ATCC Cat# CRL-3216, RRID: CVCL_0063), ACE2.293T and Vero cells (CLS Cat# 605372/p622_VERO, RRID:CVCL_0059) were grown in Dulbecco's Modified Eagle's Medium/10% fetal bovine serum at 37 °C under 5% CO2. All cells were tested for mycoplasma negative.
Human sera and monoclonal antibodies
Sera from convalescent individuals were collected 32–57 days post-symptom onset (n = 10). Sera were collected prior to February, 2021, before the spread of the Delta variant which was first detected in May in the U.S. and began to spread in July in New York. The experience donors were presumably infected with either the D614G, Alpha or Iota variant. Sera from Pfizer BNT162b2-vaccinated (n = 9), Moderna mRNA-1273-vaccinated (n = 8) study participants which were shown in Figure 1b were collected 90 and 80 days mean post-second immunization, respectively. Serum samples from study participants previously infected and subsequently vaccinated with BNT162b2 mRNA vaccine (n = 12 and 7) which shown in Figure 1c and d were collected 1 month and 7, 8 months post-second immunization. Sera from study participants vaccinated with BNT162b2 mRNA boost vaccine were collected 1-month post-vaccination. Previous infection was documented by COVID-19 symptoms and a positive PCR test or serology.
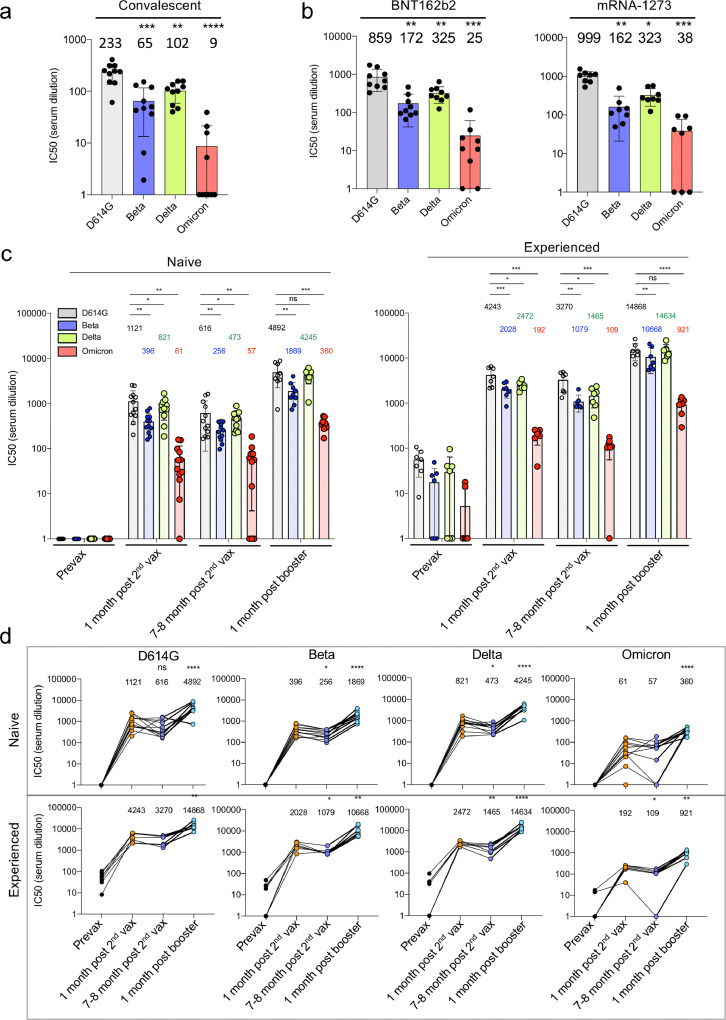
Figure 1.
Decreased neutralization of Omicron spike protein-pseudotyped viruses by convalescent sera, mRNA vaccine-elicited antibodies.
D614G, Beta, Delta and Omicron spike protein-pseudotyped viruses expressing dual GFP/nanoluciferase reporter genes with codon-optimized spike proteins deleted for the carboxy-terminal 19 amino acids were prepared as previously described.19 Equivalent amounts of virus were mixed with a 2-fold serial dilution of donor serum and then applied to ACE2.293T cells. Luciferase activity was measured two days post-infection. Each serum dilution was measured in triplicate and the experiment was done twice with similar results and IC50 was determined. Statistical significance was calculated by two-sided testing. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
a. Neutralizing antibody titres of sera from study participants who had recovered from infection prior to the appearance of the current VOCs was measured against viruses pseudotyped by current VOCs (n = 10). IC50 of each donor serum is shown with the Geometric mean titre (GMT) for each group shown above the bar.
b. Neutralization of variant spike protein pseudotyped viruses by the sera of study participants fully vaccinated (two immunizations) with Pfizer BNT162b2 (n = 9) and Moderna mRNA-1273 mRNA vaccines (n = 8).
c. Neutralizing antibody titres of study participants without or with a previous history of SARS-CoV-2 infection were measured on the pseudotyped viruses. Sera were collected from study participants pre-vaccination, 1-month post-second vaccination with Pfizer BNT162b2, 7, 8 months post-second vaccination, and 1-month post-boost. Study participants were without previous SARS-CoV-2 infection (naïve) (left) (n = 12) or previously infected (experienced) (right) (n = 7). COVID-19 history was determined by symptoms and a PCR+ test or serology. GMTs for each group are shown above the bar.
d. Sequential neutralizing antibody titres of sera from individual study participants without or with previous history of SARS-CoV-2 infection is shown for each of the study participants shown above in C. GMTs are shown above.
SARS-CoV-2 spike protein lentiviral pseudotypes
Spike protein pseudotyped lentiviruses were produced by cotransfection of 293T cells with pMDL Gag/Pol packaging vector, lentiviral vector plenti.GFP.nLuc and spike protein expression vectors encoding 19 amino acid cytoplasmic tail deletions, as previously reported.19 Transfected cell supernatants were harvested two days post-transfection and concentrated by ultracentrifugation. The viruses were normalized for reverse transcriptase (RT) activity and frozen in aliquots at -80 °C.
Antibody neutralization assay
Sera or monoclonal antibody was serially two-fold diluted and then incubated with an amount of virus corresponding to a volume that resulted in MOI=0.2 on ACE2.293T or Vero cells for pseudotyped virus. After 30 min incubation at room temperature, the virus was added to 1 × 104 target cells in a 96 well culture dish. The cells were cultured for 2 days after which the culture medium was removed and 50μl Nano-Glo luciferase substrate (Nanolight) was added. Luminescence was read in an Envision 2103 microplate luminometer.
Ethics
Human sera were collected at the NYU Vaccine Center with written consent of participants under IRB-approved protocols 18-02035 and 18-02037.
Statistics
All samples were tested in duplicate or triplicate. Neutralization assays were done by laboratory personnel blinded to the experimental groups. Power analysis for sample size were calculated assuming a power value (beta) of 0.95 to eliminate Type I error. A significance level of 0.05 was used for sample size calculations. The calculations showed that required sample sizes were 5, 8, 3 for Figure 1a and b, respectively. For Figure 1c and d, a power value (beta) of 0.9 was assumed and 10 and 5 samples were required, respectively. The sample numbers are within the sample sizes used in this study. Gaussian distribution was determined by Shapiro-Wilk and Kolmogorov-Smirnov test with GraphPad Prism 8 software which confirmed that all data sets passed the normality test (alpha=0.05). Additional statistical significance was determined by the two-tailed unpaired t-test or nonparametric ANOVA test. Significance was based on two-sided testing and attributed to p < 0.05. Confidence intervals are shown as the mean ± SD or SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Analyses of the structures of the SARS-CoV-2 spike protein with antibody Fabs was performed with the PyMOL Molecular Graphics System, v2.1.1 (Schrödinger, LLC).
Role of funding source
The funders of this study had no role in study design, sample collection, data collection, data analyses, interpretation, or writing of the report.
Results
Increased resistance of virus with the Omicron spike to serum antibodies elicited by natural infection and vaccination
To determine the effectiveness of antibodies induced by infection with earlier SARS-CoV-2 variants to protect from re-infection with the Omicron variant, we tested neutralizing antibody titres in the sera of unvaccinated participants involved in an ongoing clinical study that had been collected 32 to 57 days post-COVID-19 symptom onset. Neutralizing antibody titres were measured using lentiviral virions pseudotyped by the parental D614G, Alpha, Beta and Delta spike proteins, an assay that accurately reflects titres obtained in the plaque reduction neutralization test (PRNT).20 The results showed modest reductions in neutralizing titre against Beta and Delta as compared to the parental D614G but a more substantial average 26-fold reduction in titre against Omicron. Approximately 60% of the donor sera had titres below the IC50 of 20, the limit of detection in the assay (Figure 1a). To determine the effectiveness of antibodies elicited by vaccination, we tested sera collected 70 days post-immunization from study participants who had been fully vaccinated (two immunizations) with BNT162b2 or Moderna mRNA-1273 mRNA vaccines (Figure 1b). Notably, neutralizing antibody titres against the Omicron pseudotype was decreased 26-34-fold compared to D614G.
Previous infection has been shown to strengthen and broaden the neutralizing antibody response to SARS-CoV-2 variants upon vaccination. To determine whether previous infection would increase neutralizing antibody titres against the Omicron variant, we tested sera from study participants who were vaccinated with BNT162b2 and had, or had not, been previously infected with SARS-CoV-2 (Figure 1c). Sera from study participants without previous infection, collected one month post-second vaccination, had high titres of neutralizing antibody against D614G virus; titres against Beta compared to D614G were decreased 2.8-fold, against Delta 1.4-fold and against Omicron 18-fold. Titres had only slightly declined 7, 8 months post-vaccination. One-month post-boost, titres increased for all variants. Titres against Omicron remained 14-fold lower than against D614G. Notably, study participants who had poor neutralizing titres against Omicron after two immunizations had increased their titres following the boost (Figure 1d). Sera from previously infected study participants post-boost achieved high neutralizing titres against the Beta and Delta variants. While titres against Omicron also rose, they remained 16-fold lower on average than that of D614G virus (14,868 for D614G; 921 for Omicron).
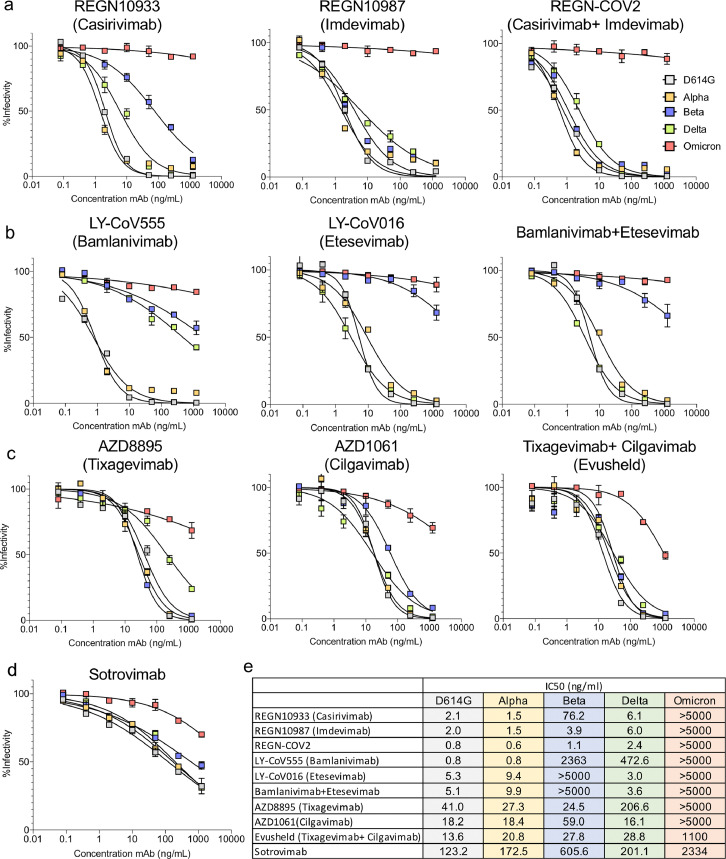
Virus with the Omicron spike protein is resistant to therapeutic monoclonal antibodies. To determine the sensitivity of the Omicron variant to the therapeutic monoclonal antibodies, we analysed the neutralizing titres of the Regeneron monoclonal antibodies REGN10933 (Casirivamab) and REGN10987 (Imdevimab), the Eli Lilly cocktail consisting of LY-CoV016 (Etesevimab) and LY-CoV555 (Bamlanivimab), GlaxoSmithKline/VIR Biotechnology VIR-7831 (Sotrovimab) and AstraZeneca AZD8895 and AZD1061 (Evusheld) against the D614G, Beta, Delta and Omicron spike protein-pseudotyped viruses. REGN10933 (Casirivamab) potently neutralized D614G and Delta, was less active against Beta but had no detectable activity against Omicron (Figure 2a). REGN10987 (Imdevimab) potently neutralized the earlier viruses but lacked activity against Omicron virus as did the REGN10933/REGN10987 cocktail. LY-CoV555 (Bamlanivimab) neutralized D614G and Alpha virus, had weak activity against Beta and Delta but was inactive against the Omicron virus (Figure 2b). LY-CoV016 (Etesevimab) potently neutralized the earlier viruses but lacked activity against Beta and Omicron virus as did the combined LY-CoV555/LY-CoV016 cocktail. Individually, the Evusheld monoclonal antibodies AZD8895 (Tixagevimab) and AZD1061 (Etesevimab) had modest activity against Omicron and synergized in combination yet the neutralizing titre of the cocktail remained 81-fold lower than its activity against D614G (Figure 2c). VIR-7831 (Sotrovimab) was active against Omicron but its IC50 was 19-fold higher than against D614G (Figure 2d) and yet less active compared to the other monoclonal antibodies against the D614G virus. IC50s calculated from the curves in Figure 2a–d are shown in Figure 2e.
Figure 2.
Therapeutic monoclonal antibodies have lost neutralizing activity against virus with the Omicron spike protein.
a. Neutralization of viruses with the VOC spike proteins by Regeneron REGN10933 (Casirivamab) and REGN10987 (Imdevimab) monoclonal antibodies and the REGN-CoV-2 cocktail was measured using spike variant spike protein-pseudotyped viruses.
b. Neutralization of viruses pseudotyped by the VOC spike proteins by LY-CoV555 (Bamlanivimab), LY-CoV016 (Etesevimab) monoclonal antibodies and cocktail were measured as in a above.
c. Neutralization of viruses, pseudotyped by the VOC spike proteins, by AZD8895 (Tixagevimab), AZD1061 (Cilgavimab) and combination Evusheld.
d. Neutralization of viruses pseudotyped by the VOC spike proteins by VIR-7831 (Sotrovimab) was measured as in a above.
e. The table shows the IC50s of the therapeutic monoclonal antibodies calculated using the data from the antibody neutralization curves shown in a, b and c. Larger numbers indicate decreased neutralization potency. Antibodies that did not reach >70% infectivity at the highest concentration tested are listed as IC50>5000; for antibodies that reached 51–70% infectivity at the highest concentration tested, the IC50 was extrapolated using GraphPad Prism 8 software.
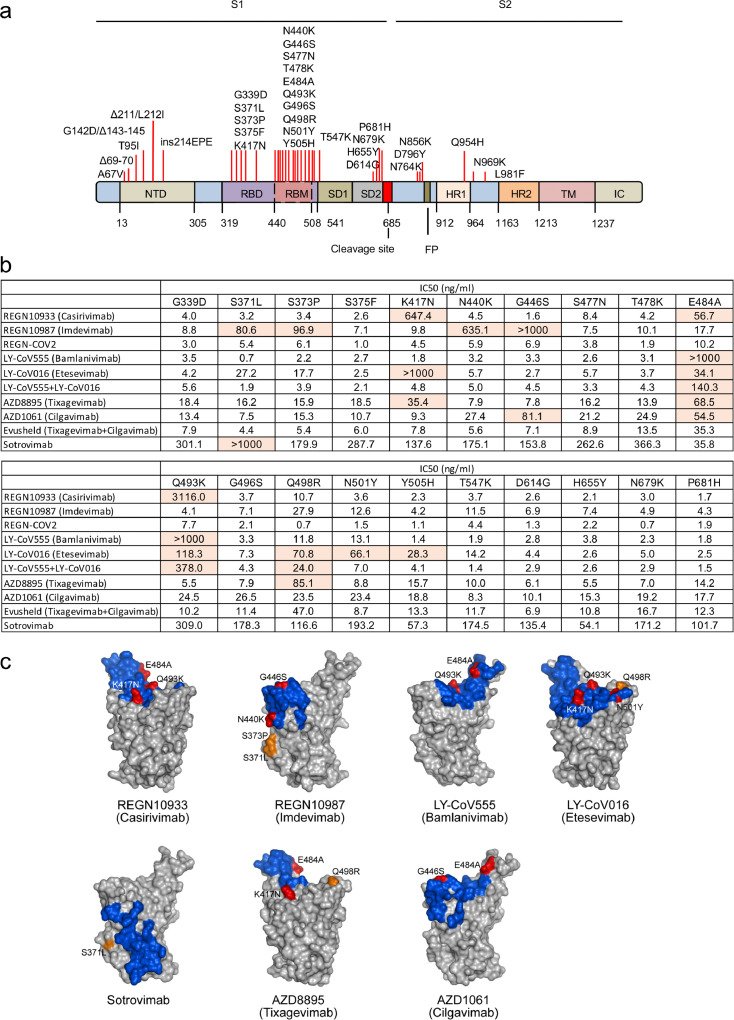
To identify the mutations in the Omicron spike protein that allowed the virus to escape neutralization we used a panel of viruses pseudotyped by spike proteins with the individual Omicron RBD mutations (Figure 3a). The results showed that REGN10933 (Casirivamab) was affected by K417N, E484A and Q493K (Figure 3b, c and Supplementary Fig. 1); REGN10987 (Imdevimab) was affected by S371L, S373P, N440K, G446S with minor effects of several other mutations. The REGN10933/REGN10987 cocktail maintained most of its neutralization potency against the single point mutated virus. LY-CoV016 (Etesevimab) neutralization of Omicron was largely affected by K417N, Q493K, Q498R and N501Y; LY-CoV555 (Bamlanivimab) was affected by E484A and Q493K with a lesser effect of several other mutations. AZD8895 (Tixagevimab) neutralization was affected by K417N, E484A and Q498R and AZD1061 was affected by G446S and E484A but the Evusheld cocktail retained most of its neutralization potency against the singly point-mutated viruses. VIR-7183 (Sotrovimab) was affected by the single mutation S371L (Figure 3b, c and Supplementary Fig. 1). Most of the mutations that affected neutralizing titre, with the exception of E484A, had modest effects on their own, suggesting that the loss of titre by each monoclonal antibody against Omicron results from the combinatorial effect of the mutations.
Figure 3.
Omicron spike protein mutations that cause escape from therapeutic monoclonal antibodies are located at the antibody interaction interface.
a. The location of Omicron mutations on the spike protein is diagrammed. The location of the S1 and S2 subunits of the processed spike protein, NTD, RBD, SD1, SD2, HR1, HR2, TM and IC domains are shown. Amino acid positions of the domains are labelled below. The furin cleavage site and hydrophobic fusion peptide (FP) are indicated.
b. The table shows the IC50s calculated from the neutralization curves shown in Supplementary Fig. 1. Mutations that caused >5-fold increase in IC50 are highlighted.
c. The footprints of neutralizing antibodies on the SARS-CoV-2 spike protein (residues within 5 Å of a Fab atom) are coloured blue. For each antibody, Omicron mutations that cause a 5-fold or greater decrease in binding are labelled. Mutated residues within the Fab footprint are coloured red, and mutated residues outside of the footprint are coloured orange. The PDB accession codes for the structures shown are 6XDG (Casirivimab and Imdevimab), 7KMG (Bamlanivimab), 7C01 (Etesevimab), 7R6W (Sotrovimab), and 7L7E (Tixagevimab and Cilgavimab).
The published crystal and cryo-electron microscopy structures of the monoclonal antibody Fabs bound to the spike protein provide insight into how mutations in the Omicron spike protein interfere with antibody binding (Figure 3c). The mutations K417N, E484A, and Q493K that affect REGN10933 (Casirivamab) are situated in the interface with the Fab heavy chain. K417N would cause a loss of hydrogen bonding with T28 and T102 of the immunoglobulin heavy chain and the loss of a favourable electrostatic interaction with D31. E484A would cause the loss of hydrogen bonding with heavy chain Y53 and S56, and Q493K would cause the loss of hydrogen bonding with heavy chain N74. REGN10987 (Imdevimab) is affected by mutations S371L, S373P, N440K, and G446S. N440K creates a steric clash between K440 and the antibody heavy and light chains and a charge repulsion with light chain K55. Mutation of G446 to a larger residue, as is the case for G446S, would cause a steric clash with heavy chain N57. S371 and S373, while not in direct contact with the Fab, could alter the stability of this loop segment, affecting the conformation of the nearby region (N440). LY-CoV555 (Bamlanivimab) was affected by E484A that would cause the loss of salt bridges with heavy chain R50 and light chain R96. Q493K would cause the loss of a hydrogen bond heavy chain R104 and the lysine is predicted to cause steric and electrostatic clashes with heavy chain R104. LY-CoV016 was affected by Omicron mutations K417N, E484A, Q493K, Q498R, N501Y and Y505H, all of which fall within the antibody interaction site. K417N causes the loss of a salt bridge with light chain D104; Q493K causes the loss of a hydrogen bond with heavy chain Y102; the arginine in Q498R is predicted to cause a charge repulsion with light chain R31; and N501Y destabilizes the local conformation of the spike protein causing a steric clash with light chain S28. Sotrovimab was affected by the single mutation S371L, which is not formally in the antibody footprint but may destabilize the nearby structure. AZD8895 was affected by mutations E484A, K417N and Q498R. E484A would result in the loss of an internal backbone hydrogen bond to spike protein F490 which may destabilize the proximal loop to which the light and heavy chains bind. The structural explanations for K417N and Q498R are not obvious although both involve a charge change. AZD1061 was affected by G446S and E484A. The G446S mutation would cause a steric clash with light chain Y55 and E484A would cause the loss of a hydrogen bond with light chain S32.
Discussion
The emergence of the Omicron variant represents an evolutionary leap by SARS-CoV-2 in which 15 mutations were introduced into the RBD along with mutations and deletions in the NTD and CTD. As a result, the Omicron variant has developed resistance to neutralization by the serum antibodies of recovered individuals who had been infected with earlier SARS-CoV-2 variants to a degree that is expected to increase the number of individuals who become re-infected. In addition, virus with the Omicron spike has a high degree of resistance to neutralization by vaccine-elicited antibodies. The resistance might be expected given that current EUA approved vaccines encode the earlier D614G spike protein. While Alpha, Beta, Gamma and Delta VOCs show about a 3-4-fold resistance to neutralization by vaccine-elicited antibodies,3, 4, 5, 6, 7, 8 virus with the Omicron spike protein has increased its resistance to neutralization by the serum antibodies of individuals fully vaccinated with BNT162b2 or Moderna-1273 by about 26-34-fold, resulting in titres that are predicted by mathematical modelling to cause an increased frequency of breakthrough infections.21,22 Several reports published during the preparation of this manuscript found levels of Omicron resistance to neutralization by vaccine-elicited antibodies, increase in neutralizing antibody titres following booster immunization and loss of activity of the therapeutic monoclonal antibodies similar to those found in our study.23, 24, 25, 26, 27, 28, 29, 30, 31
Homologous boosting of SARS-CoV-2-uninfected individuals by immunization with the Pfizer BNT162b2 vaccine increased neutralizing antibody titres against Omicron to levels that are predicted to be highly protective, although the titres remained about 10-fold below those against the other VOCs post-boost and the durability of the titres remain to be determined. Booster immunization of SARS-CoV-2 experienced individuals resulted in neutralizing antibody titres against Omicron approaching an IC50 of 1000, which as predicted by modelling will provide 90% protection against infection.
The loss of neutralizing titres of the therapeutic monoclonal antibodies suggest that they will be rendered ineffective for the treatment of COVID-19. REGN10933 (Casirivimab) and REGN10987 (Imdevimab) of the Regeneron cocktail32,33 and LY-CoV555 (Bamlanivimab)34,35 and LY-CoV016 (Etesevimab)36,37 of the Eli Lilly cocktail were ineffective against Omicron while Sotrovimab38 and Evusheld16 neutralizing titters against Omicron were significantly decreased compared to their titres against the other VOCs (19-fold for Sotrovimab and 80-fold for Evusheld as compared to D614G). Other groups have also found decreased Omicron neutralization by Sotrovimab and Evusheld monoclonal antibodies.27,39,29 Whether these monoclonal antibody therapies will remain efficacious in Omicron infected patients is unclear. Sotrovimab achieves a serum concentration of 24 g/ml following a 500 mg dose, a concentration that is well above its IC50 but the concentration of the antibodies in relevant tissues of the body and the extent of virus neutralization needed to provide clinical benefit are not known.
Mapping of the amino acid residues responsible for the escape from the monoclonal antibodies showed that most of the mutations had no effect but that several had partial effects. The only mutation that had a dramatic effect were E484A and Q493K which ablated neutralization by LY-CoV555 (Bamlanivimab). The other mutations that compromised antiviral activity had modest effects. Thus, it was the cumulative effect of several mutations that abrogated antiviral activity of the other monoclonal antibodies. REGN10933 (Casirivamab), the neutralizing activity of which has been previously found to be affected by E484K and K417N of the Beta spike protein,4,40,41 is decreased another 8-fold by E484A of Omicron. REGN10987 (Imdevimab), which is nearly impervious to mutations in the earlier VOCs, was compromised by four Omicron mutations (S371L, S373P, N440K, G446S). K417N had a major effect on the activity of LY-CoV016 (Etesevimab). Thus, the Regeneron and Eli Lilly cocktails are not likely to be effective for the treatment Omicron-infected patients.
Our findings suggest that the titres achieved by full vaccination followed by a booster immunization will protect most individuals from developing severe disease. Our findings provide further support for the benefits of booster immunization and point to the need to develop additional therapeutics. The emergence of the Omicron variant raises concern about the possibility of additional evolutionary leaps for the virus and the need to pre-empt variants before they emerge. While the current surge in Omicron infections has increased the number of COVID-19 hospitalizations and deaths, the resulting herd immunity provided by the combination of the antibody and T cell response is likely to protect against current and future variants, partially. Given the ability of the viral spike protein to accumulate mutations in the spike protein, the inclusion of additional antigens in future vaccines that broaden the T cell response may prove beneficial.
Study Limitations. This study involved a relatively small number of participants (n = 7–12), limiting the resolution of differences in antibody titres of the different experimental groups. Neutralizing antibody titres reported here were determined using lentiviral pseudotyped viruses rather than live virus which have been found to yield generally similar results, but could differ in some respects.20 In the comparison of sera from naïve and experienced study participants, we concluded that prior infection increased neutralizing antibody titres; The study participants in the two groups were of similar age and sex distribution but factors not evaluated such as body mass index (Data for BMI of participants was not available) could be a confounding covariate. Study participants were a random sampling of individuals in the New York area; and might not be representative of a wider population.
Contributors
T.T. and N.R.L. designed the experiments. T.T., H.Z., B.M.D. and V.C. carried out the experiments and analysed data. S.R.H. provided protein structural analyses. T.T., H.Z. and N.R.L. wrote the manuscript. M.I.S., R.H. and M.J.M supervised specimen selection and the collection of clinical information. T.T, H.Z., B.M.D. and V.C. have verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Raw data are available from the corresponding author upon request.
Declaration of interests
M.J.M. received research grants from Lilly, Pfizer, and Sanofi and serves on advisory boards for Pfizer, Merck, and Meissa Vaccines.
Acknowledgment
The work was funded by grants from the NIH to N.R.L. (DA046100, AI122390 and AI120898) and to M.J.M. (UM1AI148574).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103944.
Appendix. Supplementary materials
References
- 1.EUA. 2021. https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization.
- 2.WHO. 2021: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
- 3.Tada T., Dcosta B.M., Samanovic M.I., et al. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. mBio. 2021;12(3) doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 5.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 6.Wall E.C., Wu M., Harvey R., et al. AZD1222-induced neutralising antibody activity against SARS-CoV-2 Delta VOC. Lancet. 2021;398(10296):207–209. doi: 10.1016/S0140-6736(21)01462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis C., Logan N., Tyson G., et al. Reduced neutralisation of the delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021;17(12) doi: 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi A., Koch M., Wu K., et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. J Virol. 2021;95(23) doi: 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez Bernal J., Andrews N., Gower C., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas S.J., Moreira E.D., Kitchin N., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N Engl J Med. 2021;385(19):1761–1773. doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scobie H.M., Johnson A.G., Suthar A.B., et al. Monitoring incidence of COVID-19 cases, hospitalizations, and deaths, by vaccination status - 13 U.S. jurisdictions, April 4-July 17, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(37):1284–1290. doi: 10.15585/mmwr.mm7037e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razonable R.R., Pawlowski C., O'Horo J.C., et al. Casirivimab-Imdevimab treatment is associated with reduced rates of hospitalization among high-risk patients with mild to moderate coronavirus disease-19. EClinicalMedicine. 2021;40 doi: 10.1016/j.eclinm.2021.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ACTIV-3/TICO LY-CoV555 Study Group. Lundgren J.D., Grund B., et al. A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N Engl J Med. 2021;384(10):905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilly. New data show treatment with Lilly's neutralizing antibodies bamlanivimab (LY-CoV555) and etesevimab (LY-CoV016) together reduced risk of COVID-19 hospitalizations and death by 70 percent. 2021.
- 15.ACTIV-3/Therapeutics for Inpatients with COVID-19 (TICO) Study Group Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phase III double-blind, placebo-controlled study of AZD7442 for pre-exposure prophylaxis of COVID-19 in adult. (PROVENT). ClinicalTrialsgov.
- 17.NGS-SA. SARS-CoV-2 sequencing update. 2021.
- 18.Kannan S.R., Spratt A.N., Sharma K., Chand H.S., Byrareddy S.N., Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J Autoimmun. 2022;126 doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada T., Fan C., Chen J.S., et al. An ACE2 microbody containing a single immunoglobulin Fc domain is a potent inhibitor of SARS-CoV-2. Cell Rep. 2020;33(12) doi: 10.1016/j.celrep.2020.108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noval M.G., Kaczmarek M.E., Koide A., et al. Antibody isotype diversity against SARS-CoV-2 is associated with differential serum neutralization capacities. Sci Rep. 2021;11(1):5538. doi: 10.1038/s41598-021-84913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert P.B., Montefiori D.C., McDermott A.B., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2021:eab3435. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruell H., Vanshylla K., Tober-Lau P., et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med. 2022 doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng S.M.S., Mok C.K.P., Leung Y.W.Y., et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022 doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt F., Muecksch F., Weisblum Y., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2021 doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann M., Krüger N., Schulz S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2021 doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas D., Saunders N., Maes P., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 28.Cameroni E., Bowen J.E., Rosen L.E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y., Wang J., Jian F., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021 doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Iketani S., Guo Y., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 31.Cele S., Jackson L., Khoury D.S., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021 doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinreich D.M., Sivapalasingam S., Norton T., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum A., Ajithdoss D., Copin R., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370(6520):1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P., Nirula A., Heller B., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384(3):229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones BE, Brown-Augsburger PL, Corbett KS, et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv 2020.
- 36.Dougan M., Nirula A., Azizad M., et al. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N Engl J Med. 2021;385(15):1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi R., Shan C., Duan X., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 38.Gupta A., Gonzalez-Rojas Y., Juarez E., et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N Engl J Med. 2021;385(21):1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 39.VanBlargan L.A., Errico J.M., Halfmann P.J., et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022 doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen R.E., Winkler E.S., Case J.B., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596(7870):103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onodera T., Kita S., Adachi Y., et al. A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site. Immunity. 2021;54(10):2385–2398. doi: 10.1016/j.immuni.2021.08.025. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.