Abstract
The gene numbers and evolutionary rates of birds were assumed to be much lower than those of mammals, which is in sharp contrast to the huge species number and morphological diversity of birds. It is, therefore, necessary to construct a complete avian genome and analyze its evolution. We constructed a chicken pan-genome from 20 de novo assembled genomes with high sequencing depth, and identified 1,335 protein-coding genes and 3,011 long noncoding RNAs not found in GRCg6a. The majority of these novel genes were detected across most individuals of the examined transcriptomes but were seldomly measured in each of the DNA sequencing data regardless of Illumina or PacBio technology. Furthermore, different from previous pan-genome models, most of these novel genes were overrepresented on chromosomal subtelomeric regions and microchromosomes, surrounded by extremely high proportions of tandem repeats, which strongly blocks DNA sequencing. These hidden genes were proved to be shared by all chicken genomes, included many housekeeping genes, and enriched in immune pathways. Comparative genomics revealed the novel genes had 3-fold elevated substitution rates than known ones, updating the knowledge about evolutionary rates in birds. Our study provides a framework for constructing a better chicken genome, which will contribute toward the understanding of avian evolution and the improvement of poultry breeding.
Keywords: chicken, pan-genome, missing genes, noncanonical DNA secondary structure, avian evolution
Introduction
The ∼10,770 species of birds described (Gill et al. 2020) show complex and diverse morphology and behavior; however, the currently available avian genomes present a reduced rate of evolution and much lower gene numbers than those of all other tetrapods (Zhang et al. 2014). The apparent discordance remained a major evolutionary conundrum. Some studies have shown that birds tend to have fewer genes than other tetrapods due to the large segmental deletions found in their genomes (Lovell et al. 2014; Zhang et al. 2014), whereas other researchers suggested that these missing genes may not have been sequenced (Bornelöv et al. 2017; Botero-Castro et al. 2017; Yin et al. 2019; Zhu et al. 2021). Using more advanced sequencing technologies and methodologies, the Vertebrate Genomes Project (VGP) found many genes were missing in previous genome assemblies, and this was clearly not a biological difference as some of the previous and VGP assemblies were from the same individuals. The missing genes were biased toward GC-rich and repeat-rich regions that they proposed were hard to sequence using prior technologies (Kim et al. 2021; Rhie et al. 2021). It still remains unclear how many genes are within single bird species, and the reasons why some genes are missing in the currently available genomes need to be further explored.
Comprehensive analyses indicate multiple high-quality de novo genome assemblies possess more power to capture the complete set of genes, which leads to the appearance and prevalence of “pan-genome” in various species (Wong et al. 2018, 2020; Duan et al. 2019; Tian et al. 2020). The pan-genome of mammals is typically of the “closed” pattern with a limited number of variable genes (Duan et al. 2019; Li et al. 2019; Tian et al. 2020), which means the number of genes in mammalian species is relatively conserved. Whereas bacteria, fungi, and plants exhibit the characteristic of an “open” pattern, where the proportion of core genes size is <80% in many species (Golicz et al. 2019). Recent research using population resequencing data found that the core genome of chickens is only 76% of the genome (Wang et al. 2021), which puzzles us because it seems to be inconsistent with the status of chickens in evolution. As the most abundant class of tetrapod vertebrates, birds have not yet had a de novo pan-genome established, which is essential to solve many biological questions.
Chicken (Gallus gallus) as one of the most important farm animals plays a major role in human food production and has been widely used as a model organism in studies of developmental biology, virology, oncogenesis, and immunology (Cooper et al. 1966; Stehelin et al. 1976; Brown et al. 2003; Vogt 2011). In this study, we utilized 20 new high-quality assemblies of diverse chicken breeds to generate the first de novo assembled-based chicken pan-genome. As many as 1,335 genes missing in previous genome assemblies were identified, verified, and localized. Importantly, most of the novel genes actually exist in all of the chicken genomes but were prone to be missing in the DNA sequencing leaded by high proportions of tandem repeats (TRs) and secondary structures. Hence, unwinding complex DNA structures should be one of the most important advances to improve the sequencing quality for the assembly of complete avian genomes. Our study revealed that the numbers of chicken genes are comparable to those of other tetrapod vertebrates and a new pan-genome pattern of birds.
Results
Identification and Validation of Nonredundant Novel Sequences
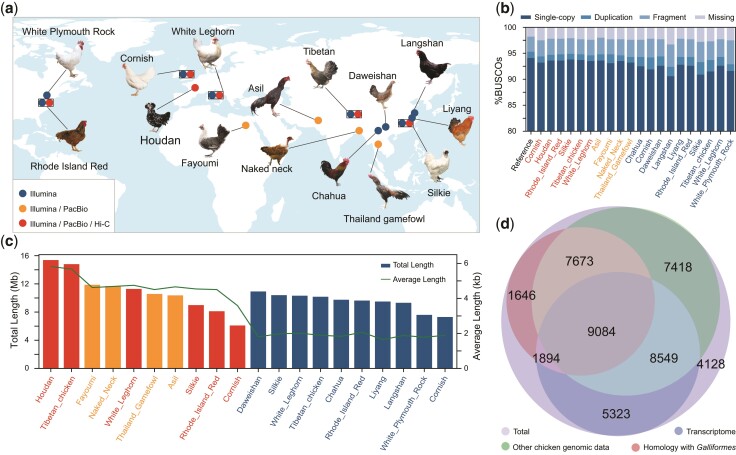
Twenty chickens from four continents representing widespread indigenous chicken breeds, commercial broilers, and layer lines were sampled for de novo genome assembly (fig. 1a, supplementary table S1, Supplementary Material online). Ten assemblies were constructed by integrating both PacBio (53–95×) and Illumina data (45–70×), resulting in a contig N50 size ranging from 5.89 to 16.72 Mb (supplementary table S2, Supplementary Material online). Six of them were further clustered at the chromosome level by using high-throughput chromatin conformation capture (Hi-C) (112–125×) data (see Methods, supplementary figs. S1–S4 and tables S2 and S3, Supplementary Material online). The remaining ten samples were assembled based on Illumina reads from a combination of libraries with multiple insert sizes, ranging from 500 bp to 5 Kb (with a depth of ∼134× per genome, supplementary table S2, Supplementary Material online). These ten samples showed a contig N50 size ranging from 80.30 to 137.59 Kb (supplementary table S2, Supplementary Material online), in accordance with high-quality Illumina genomes (Schatz et al. 2010). The completeness of the 20 assemblies was evaluated through the Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis. Most (from 92.4% to 95.3%) of the 4,915 core genes in the Aves dataset were identified in the 20 assemblies, which is comparable to the percentage in the reference chicken genome (GRCg6a: 95.4%) and thus supports a high-quality genome assembly (fig. 1b, supplementary fig. S5 and table S4, Supplementary Material online).
Fig. 1.
Chicken novel nr sequences identified by 20 de novo assemblies. (a) Geographic locations of the original chicken breeds used for de novo assembly and their sequencing platforms. The rectangle indicates this breed has two individuals. (b) Genome assembly completeness assessed by BUSCO. (c) Length of novel sequences initially obtained from 20 de novo assemblies. The polygonal line represents the average length and the column represents the total length. (d) The number of novel sequences validated by other chicken genomes, homology with Galliformes, and transcriptome. The colors of the breed name in (b) and (c) are consistent with (a).
To identify novel sequences, all 20 de novo assemblies were aligned against GRCg6a (see Methods, supplementary fig. S6, Supplementary Material online). For stability, we used GRCg6a from a same red junglefowl as the reference genome in the past two decades, not the newly unpublished GRCg7b from a broiler. The genome length of GRCg6a (1.06 Gb) and GRCg7b (1.05 Gb) are almost the same. Unaligned sequences or sequences with <90% identity and >500 bp in length compared with GRCg6a were retained and potentially contaminating non-Chordata sequences were removed. After these screening, each assembly left 6.10–15.40 Mb of novel sequences (fig. 1c). We merged the novel sequences from all 20 assemblies and built a pan-genome of chicken. The set of the pan-genome contained GRCg6a and 158.98 Mb of nonredundant (nr) novel sequences which were obtained from 45,715 contigs with an average length of 3,478 bp (table 1, supplementary fig. S7 and table S5, Supplementary Material online). The chicken pan-genome expanded the size of GRCg6a by 14.92% which is the highest percentage among the published vertebrate pan-genomes.
Table 1.
The Characteristics of Novel Sequences in this Study.
| Characteristic | |
|---|---|
| Total novel sequence length (bp) | 158,981,245 |
| Total gap length (bp) | 1,405,623 |
| Number of novel sequences | 45,715 |
| Novel sequence N50 (bp) | 6,784 |
| Mean novel sequence length (bp) | 3,478 |
| GC ratio | 57.20% |
| G4 motif content | 37.08% |
| Tandem repeat content | 79.13% |
We next validated the reproduced 158.98 Mb novel sequences. 71.58% novel sequence can be detected in other individual genomes, including the other 19 de novo assemblies from this study or 922 resequenced chicken genomes from previous studies. 44.40% are orthologs found in 14 other publicly available Galliformes genomes. 54.36% are detected from transcriptomes 263 transcriptomes from multiple tissues from 54 chickens, including 46 transcriptomes obtained from 11 tissues/organs from six individuals in our study and 217 publicly available chicken RNA sequencing (RNA-Seq) datasets from 48 individuals (Cardoso-Moreira et al. 2019) (see fig. 1d, supplementary figs. S8–S10, tables S5–S8, Note, and Dataset, Supplementary Material online). In total, 90.97% of the novel sequences were verified in at least one of the above data sources.
Distribution of Cryptic Novel Sequences Across Chicken Individuals
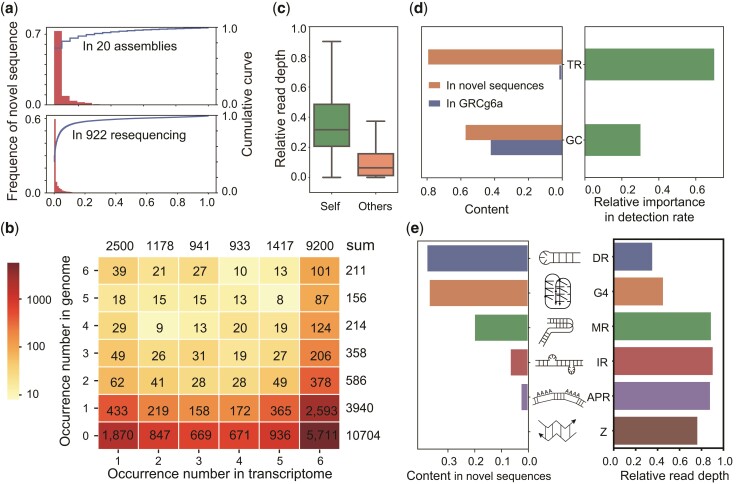
We found that the distribution of the novel sequences is obviously inconsistent across different verified sources. The detection rate of novel sequences in one genome is extremely low, the median is only 0.43% among the 922 resequenced data, and 5% among the 20 assemblies (fig. 2a). Among all 159 Mb novel sequences, the ten Illumina assemblies independently detected about 60 Mb, containing only 3.44 Mb intersection with PacBio assemblies. Due to the higher detection rate of RNA-Seq, we picked up the transcribed novel sequences according to the 263 transcriptomes for further validation. RNA-Seq confirmed that 60.51% of the transcribed regions of the cryptic novel sequences were shared among more than half of the chicken genomes (supplementary fig. S11, Supplementary Material online). In the six individuals with both PacBio genome assembly and transcriptome data, the transcriptomes of the six individuals supported a total number of 16,169 novel sequences, 9,200 (56.90%) of which were detected in all the transcriptomes of six individuals. However, 5,711 (62.08%) of the 9,200 novel sequences were completely absent in the PacBio assemblies of the six individuals (fig. 2b). By mapping the PacBio reads to the novel sequences, 76.35% and 52.81% of the novel sequences were covered by at least one read across more than half or all PacBio-sequenced individuals, respectively. Moreover, although the GRCg6a assembly did not contain our novel sequences, 6.30% (2,879) of the sequences were covered by the Illumina sequencing reads of the GRCg6a individual with at least 7× coverage (corresponding to 25% of the genome-wide depth) (supplementary fig. S12 and Dataset, Supplementary Material online). To explain the prevalence of ubiquitously transcribed yet missing novel sequences in the assemblies, we compared the median sequencing depth of the novel sequences with the whole-genome depth in the individuals. We found that the median sequencing depth of the novel sequences was only one-third of the whole-genome depth in the individuals in which the novel sequences were successfully assembled. Furthermore, in the individuals in which a given novel sequence was missing from the assembly, the median sequencing depth of the novel sequences was only one-twentieth of the whole-genome depth, which is insufficient for successful assembly (fig. 2c). Collectively, the results indicated that the novel sequences were most likely present in most or all the chicken genomes but were prone to be missing in the assemblies due to their extremely low DNA sequencing depth.
Fig. 2.
Characterization of novel sequences. (a) The distribution and cumulative curve of observed frequencies of novel sequences in 20 assemblies and 922 resequenced individuals. (b) The observed frequency of the expressed novel sequences in the transcriptomes of six chickens (column) and their corresponding genomes (row). (c) Relative read depth of novel sequences in the specific assembly in which the novel sequence was present (green) and absent (orange). The whole-genome read depth was set to one. (d) Left: TR and GC content of the GRCg6a and novel sequences, respectively; right: the feature importance of TR and GC for the detection rate of novel sequences. (e) Left: the content of noncanonical DNA structures in the novel sequences; middle: the putative structures of noncanonical DNA; right: the read depth ratio of novel sequences with or without noncanonical DNA structures. TR, tandem repeat; DR, direct repeat; G4, G-quadruplexes; MR, mirror repeat; IR, inverted repeat; APR, A-phased repeat; Z, Z-DNA.
Cryptic Novel Sequences have a High Content of TRs
We observed a higher GC content in the novel sequences than in the reference genome (57.2% vs. 42.30%). Notably, we found the content of TRs in the novel sequences was 79.13%, which is extremely high and significantly higher than in GRCg6a (2.2%; χ2 test, P-value = 0) (fig. 2d and table 1). Other interspersed repeats such as LTR and LINE were low (0.09% in novel sequence vs. 9.6% in GRCg6a, supplementary fig. S13, Supplementary Material online). We predicted the relative importance of TR and GC content in detection rate in assembly using random forest classifier and found the TR content had a greater influence than GC (fig. 2d, supplementary fig. S14, Supplementary Material online). The TR can form noncanonical DNA structures, such as G-quadruplexes (four-stranded noncanonical DNA/RNA topologies, hereafter referred to as G4 motifs), Z-DNA, A-phased repeats, and inverted repeats, which can form cruciforms, triplexes, and slipped structures, leading to genomic instability (Zhao et al. 2010) and incapable DNA sequencing (Guiblet et al. 2018). We found these noncanonical structures are highly intersected with TR regions (supplementary fig. S15, Supplementary Material online). Among these structures, the content of direct repeats (DRs) (37.96%) and G4 motifs (37.08%), are the highest in novel sequences, whereas their occurrence in GRCg6a is only 1.47% and 0.77%. DR and G4 also showed the largest negative correlation with read depth, the novel sequence with DR and G4 motif had only 1/3 and 1/2 read depth of all novel sequences (fig. 2e). It is worth noting that as particularly stable noncanonical DNA structures, G4 motifs typically form in guanine-rich regions of genomes, which may be one of the reasons why GC-rich sequences are difficult to sequence. We also found that the transcribed regions of novel sequences showed a lower TR content (supplementary fig. S16, Supplementary Material online), which might be the reason why RNA-Seq resulted in a higher observed frequency than DNA sequencing.
Abundantly Expressed Genes are Embedded in Novel Sequences
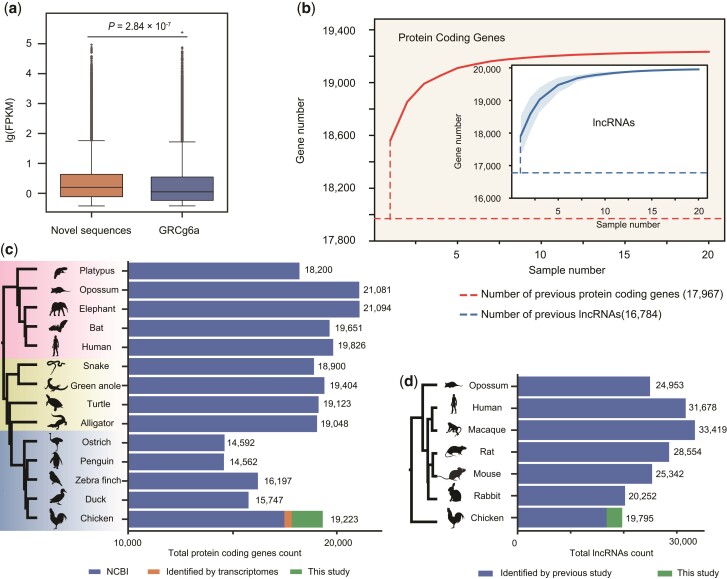
Within the novel sequences, the expressed sequences are the most interesting for potentially discovering novel candidate genes. To identify novel chicken genes, we performed gene annotation for all 20 assemblies by de novo and reference-guided methods using the multi-tissue transcriptomes (see Methods, supplementary table S8, Supplementary Material online). The median expression level of these putative novel genes was significantly higher than the median expression of GRCg6a-annotated genes (P-value = 2.84 × 10−7) (fig. 3a and supplementary figs. S17 and S18, Supplementary Material online). Furthermore, the orthologs of the novel genes showed expression levels that were higher than the median levels observed in other species, such as human and mouse (supplementary fig. S19, Supplementary Material online), suggesting plausible functions and active expression of these genes.
Fig. 3.
Abundantly expressed genes are embedded in novel sequences. (a) Relative expression of transcripts of reference and novel sequences, respectively. (b) The identification of protein-coding genes/lncRNAs increased with sample numbers. The shaded area indicates the 95% confidence interval. (c) The number of protein-coding genes in representative species including mammals, reptiles, and birds. Blue, orange, and green columns refer to protein-coding genes identified by NCBI, Yin et al (2019), and our study, respectively. (d) Total lncRNAs numbers of mammalian representative species and chicken. The blue and green columns refer to lncRNAs identified by Sarropoulos et al (2019) and this study, respectively.
We identified 1,335 novel coding genes with fragments per kilobase per million mapped reads (FPKM) > 1, and completely missing from GRCg6a (see Methods, supplementary fig. S20 and table S9, Supplementary Material online). The novel coding genes were distributed across 1,100 novel sequences, with an average length of 1,047 bp. By searching against the nr protein database of NCBI (E-value ≤ 1 × 10−5), 969 of the novel coding genes were found to show Chordata protein orthologs, 738 of which belonged to Aves (supplementary table S9, Supplementary Material online). In addition to novel coding genes, we also identified 3,874 confident transcripts which complemented 1,336 partially missing coding genes in GRCg6a (supplementary fig. S21 and table S10, Supplementary Material online).
To validate the novel coding genes, proteomic analysis of multiple tissues (hypothalamus, spleen, and cecal tonsil) was performed via an LC–mass spectrometry (MS)/MS strategy (supplementary table S8, Supplementary Material online). A total of 255 (19.10%) novel coding genes were confirmed by the existence of corresponding proteins (supplementary table S9, Supplementary Material online), compared with 6,201 (35.48%) of all the coding genes present in the reference genome. The lower detection rate of novel genes in proteomics may be affected by the differences in protein length and the quality of the protein database used for searching. Notably, after removing novel coding genes <1 Kb in length, the proteomic verification ratio of the remaining novel coding genes increased to 29.11%.
We found that most of the novel coding genes were present and expressed in most chicken breeds. According to the DNA data, 92.47% of the novel sequences containing novel coding genes were supported by at least one PacBio read in each sample (supplementary fig. S22, Supplementary Material online). According to the comparison of multi-tissue transcriptomes of six individuals, 55.13% and 80.97% of the novel coding genes were detected in all six or at least three individuals, respectively (supplementary table S9, Supplementary Material online). Based on our sequencing platform, assembly strategy, and annotation pipeline, the modeling of the saturation curve by iteratively randomly sampling individuals suggested that the number of novel genes detected by genome assembly did not significantly increase beyond a sample size of ten (fig. 3b). We also checked if the novel genes were successfully assembled in the recently VGP chicken genome assembly, GRCg7b. We found that 331 novel coding genes were also partially assembled in GRCg7b, including 52 of them were assembled with more than 90% coverage (supplementary table S9, Supplementary Material online). A previous study (Yin et al. 2019) based on the de novo assembly of massive chicken transcriptomes increased the number of known chicken coding genes from 17,477 to 17,967 (fig. 3c, supplementary fig. S23, Supplementary Material online). According to our chicken pan-genome, we found that the total number of chicken coding genes reached at least 19,223 (fig. 3c, supplementary fig. S23 and table S11, Supplementary Material online).
In addition to coding genes, we identified 3,011 long noncoding RNAs (lncRNAs) (see Methods, supplementary table S12, Supplementary Material online). Among these novel lncRNAs, 87.85% were supported by at least one PacBio read in each sample (supplementary fig. S22, Supplementary Material online). In our multi-tissue transcriptomes of six individuals, 47.72% and 75.09% of novel lncRNA genes were detected in all six or at least three individuals, respectively (supplementary table S12, Supplementary Material online). The increasing saturation curve of the observed novel lncRNA genes was similar to that of novel coding genes (fig. 3b). And there are 371 novel lncRNA genes that were partially assembled in GRCg7b, including 136 of them were assembled with more than 90% coverage (supplementary table S12, Supplementary Material online). Using the same pipeline as in a previous study (Sarropoulos et al. 2019), we showed that the total number of chicken lncRNAs was at least 19,795 (fig. 3d). Therefore, our study revealed that the numbers of both the protein-coding and lncRNA genes of chicken are comparable to those of other tetrapod vertebrates (fig. 3c and d).
Novel Sequences and Genes are Concentrated in Microchromosomes and Subtelomeric Regions with Elevated Substitution Rates
We anchored the novel sequences to GRCg6a based on flanking sequence alignment and chromosome interaction mapping (see Methods). A total of 27,966 (61.17%) novel sequences containing 1,043 novel coding genes and 1,567 novel lncRNAs were anchored to GRCg6a by at least one end (supplementary tables S5, S9, and S12, Supplementary Material online). Among these sequences, 6,735 novel sequences containing 388 coding genes were fully anchored by both ends. The fully anchored novel sequences were further classified as insertions, alternate alleles, or multiple alternative alleles (supplementary figs. S24b–d, Supplementary Material online) and were dispersed on every chromosome of GRCg6a, filling 72 of 946 gaps in GRCg6a (supplementary fig. S24a and e, Supplementary Material online).
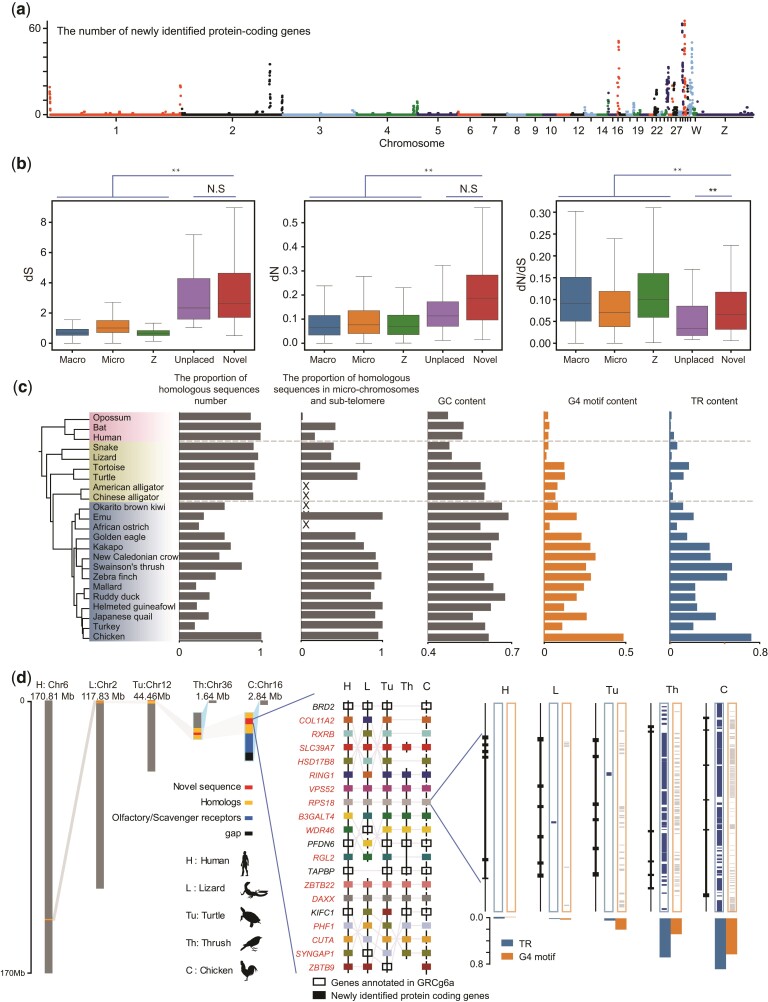
The fully anchored novel sequences and genes were overrepresented on microchromosomes (GGA11–38) (<10 Mb) or the terminal 5 Mb ends of macrochromosomes (fig. 4a, supplementary fig. S25, Supplementary Material online), which are termed as subtelomeric regions. By comparison with the random distribution, we estimated 2.5- (P-value < 1 × 10−6, permutation) and 5-fold (P-value < 1 × 10−6, permutation) increases in fully anchored novel sequences and gene density within subtelomeric regions, respectively (supplementary fig. S26, Supplementary Material online). The novel sequences nearly doubled the length of the microchromosomes such as chromosomes 16, 25, 30, 31, 32, and 33, adding a total of 421 coding genes (supplementary fig. S24f and Supplementary Note, Supplementary Material online). Notably, ten microchromosomes of the newly VGP zebra finch genome were also greatly expanded by ∼36–97% (Kim et al. 2021).
Fig. 4.
The novel coding genes clustered in the microchromosomes and subtelomere of chromosomes. (a) The location of the novel coding gene clusters on chromosomes. (b) Box plot for dS, dN, dN/dS values of genes on macrochromosomes, microchromosomes, the Z chromosome, unplaced scaffold of chicken reference genome, and the novel sequences. (c) The proportion of orthologous sequences detected number, homologous sequences detected in microchromosomes and subtelomere, and contents of GC, G4 motif, and TR of homologous novel coding genes clusters in chicken and 22 other species. The region containing more than three genes are considered as clusters, and genes located within 5-MB of the end of chromosomes are considered as subtelomeric regions. (d) A detailed synteny conservation of novel coding genes on chromosome 16 of chicken with mammal (human), reptilia (lizard, turtle), and aves (thrush, chicken), respectively. Hollow rectangles represent annotated genes in the genome, and other color rectangles, with gene name in red, represent novel coding genes in chickens.
It is widely accepted that subtelomeric regions of the chromosomes and microchromosomes of birds exhibit high rates of recombination and mutation (International Chicken Genome Sequencing Consortium 2004; Burt 2005; Linardopoulou et al. 2005; Bell et al. 2020). We investigated the evolutionary rates of 160 high-quality orthologs of the novel coding genes by comparing chicken genes with those of human and mouse. The synonymous substitution rate (dS) and nonsynonymous substitution rate (dN) of these novel genes were 3.3- and 2.5-fold higher than that of anchored GRCg6a genes, respectively. And the dN/dS ratio of these novel genes was lower than that of the reference genes. Interestingly, the unlocalized genes of GRCg6a, which may also be located in microchromosomes or subtelomeric regions, showed a similar mutation pattern as the novel genes (fig. 4b). This suggested that the novel coding genes in microchromosomes and subtelomeric regions showed a higher mutation rate.
We next identified novel gene clusters to investigate collinearity. Screening according to the existence of more than three novel coding genes within 1 Mb bin across the genome revealed 19 regions containing 201 of 388 fully anchored genes. The 19 gene clusters were all located in microchromosomes or subtelomeric regions (fig. 4a, supplementary fig. S25, Supplementary Material online). By checking the orthologous sequences detected in each lineage, we found that almost all 201 novel coding genes had homologs in mammalian and reptile genomes and showed good collinearity (fig. 4d, supplementary fig. S25 and table S13, Supplementary Material online). Some novel gene clusters likely existed in the microchromosomes or subtelomeric regions before the divergence of testudines and avian. However, the significant increase of the TR clusters with high content of noncanonical DNA structures only happened on the bird lineage (fig. 4c). Unlike the previous notion that large segmental deletions occurred in the evolution process (Lovell et al. 2014; Zhang et al. 2014), our results provided a large number of confident new gene clusters in microchromosomes and subtelomeric regions, filling gaps in which genes were often missing due to insufficient sequencing.
Functional Assignment of Novel Regions and Genes
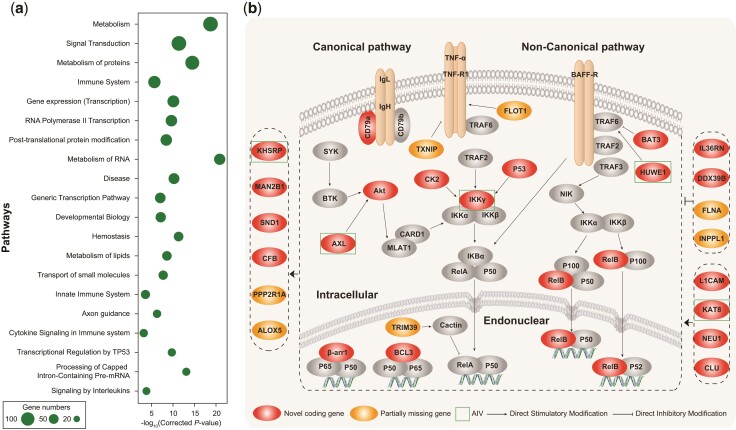
Among the novel coding genes that we identified, 176 of them were identified as housekeeping genes in human and mouse (Hounkpe et al. 2021) (supplementary table S9, Supplementary Material online). Through the annotation and enrichment analyses, we also found that a large number of them were involved in essential biological reactions and pathways, such as metabolism, signal transduction, basic biological functions, the immune system, and disease (fig. 5a, supplementary tables S14–S16, Supplementary Material online).
Fig. 5.
Function enrichment of novel coding genes and the case of NF-κB pathway-related novel coding genes. (a) The top 20 significant Reactome pathway with the largest number of novel coding genes. (b) Novel coding genes (red) and partially missing gene (yellow) are related to NF-κB signaling pathway. The green boxes represent differentially expressed genes (DEGs) in avian influenza virus.
In the novel regions, we dissected chromosome 16 and the subtelomeric part of chromosome 1 as two examples to reveal their plausible gene arrangement and functions. Chromosome 16 is a microchromosome that contains many immune system-related genes (fig. 4d) and spans only 2.84 Mb of GRCg6a. We assembled 3.76 Mb of novel sequences and identified 61 novel coding genes and 80 lncRNA genes on chromosome 16. The novel gene clusters showed good syntenic relationships with other tetrapods (fig. 4d). One of the novel gene clusters showed that birds had experienced regional complications in the cluster and lacked a large number of coding genes (fig. 4d). One novel coding gene, the complement factor B (CFB) gene, which is an important immune gene involved in the alternative complement pathway of the immune system (supplementary fig. S27, Supplementary Material online) and is regulated by the nuclear factor kappa B (NF-κB) pathway, was de novo identified on chromosome 16. This gene is highly and uniquely expressed in the liver of chickens and confirmed based on our MS/MS data (supplementary fig. S27, Supplementary Material online). In addition, we identified two novel ribosomal genes, mitochondrial ribosomal protein S18B (MRPS18B), and ribosomal protein S18 (RPS18) on chromosome 16 (fig. 4d, supplementary table S13, Supplementary Material online).
Another novel gene cluster, including the leptin gene, is located on chromosome 1 (supplementary fig. S25, Supplementary Material online). Based on RNA-Seq, previous research has shown that the leptin gene does exist in the chicken genome, yet it was absent from the chicken reference genome (Seroussi et al. 2017). Interestingly, we found that two divergent haplotypes of the leptin gene were assembled from two individuals. The entire gene region and its flanking regions had extremely high TR and G4 motif contents (supplementary fig. S28, Supplementary Material online). Based on chromosome interaction data, leptin was assigned to the distal tip of chromosome 1p, showing collinearity with SND1 and LRRC4 (supplementary figs. S25 and S28, Supplementary Material online). We found that leptin exon 2 was conserved, whereas exon 1 was variable in chicken. The length of its intron also varied among different chicken individuals (supplementary fig. S28, Supplementary Material online). Neither of the two exons showed good homology with other species. In this region, we found another novel gene, ovocleidin-17 (OC-17), which plays a key role in avian eggshell biomineralization and is not contained in the reference genome (supplementary table S9, Supplementary Material online).
Application of the Chicken Pan-Genome in Avian Influenza
The chicken pan-genome identified novel genes related to avian diseases resistance that had not been discovered previously. Chickens are susceptible to several diseases that have far-reaching effects on human society, such as avian influenza. Here, we reanalyzed the transcriptome data (Smith et al. 2015) of chicken lung and ileum samples after infection with low pathogenic (H5N2) and highly pathogenic (H5N1) avian influenza virus. Compared with the expression levels observed in the control group, 30 novel coding genes, 65 novel lncRNAs, and 79 partially missing genes showed differential expression in these samples (false discovery rate [FDR] < 0.05) (supplementary tables S9, S10, and S12, Supplementary Material online). B-cell-related genes (CD22, CD79A, PRMT1, and SND1), T cell-related genes (CD2BP2), immunoglobulin genes (IGLL5), and ribosome genes (RPS18) were screened among these differential expression genes (supplementary tables S9 and S17, Supplementary Material online). Notably, several significantly differentially expressed genes (AXL, HUWE1, IKKγ, KAT8, and KHSRP) belonged to or were regulated by the NF-κB signaling pathway, which is the master regulator of the immune response to infection due to its role in regulating cytokine and antimicrobial peptide expression (fig. 5b). RELB, a subunit of NF-κB, associated with the immune responses to influenza A (Rückle et al. 2012) and severe acute respiratory syndrome-associated coronavirus (Chen et al. 2006), was identified, anchored, and validated in our study (supplementary fig. S29, Supplementary Material online). Another novel gene, IKKγ (supplementary table S9, Supplementary Material online), a subunit of the IκB kinase complex, was essential for the activation of NF-κB transcriptional activity. Besides, the newly identified genes AXL (Schmid et al. 2016), CSNK2B (Marjuki et al. 2008), DDX39B (Wisskirchen et al. 2011), KHSRP (Liu et al. 2015), and TP53 (Wang et al. 2018) have also been reported to play a role in the immune response to influenza A. In total, there were 21 novel coding genes and 7 partially missing coding genes that belonged to or were regulated by the NF-κB signaling pathway (fig. 5b). The NF-κB pathway is essential in defense against viral infections, such as those caused by influenza viruses.
Discussion
The chicken is the modern descendant of the dinosaurs being the first fully sequenced genome among nonmammalian amniotes (International Chicken Genome Sequencing Consortium 2004). Despite several major updates, the completeness of the chicken genome still needs to be improved and the number of genes in the chicken genome still underestimated. Our study suggests that the chicken pan-genome exhibits a more complex mammalian-like “closed” genome pattern. More specifically, we identified 1,335 and 3,011 novel coding genes and novel long noncoding genes, respectively, containing mostly core genes, which appear different from previous mammalian pan-genome studies that reported fewer novel genes (Golicz et al. 2019; Sherman et al. 2019; Sherman and Salzberg 2020; Tian et al. 2020). The highly complex noncanonical DNA structure across the novel genes might be the main reason to prevent the efficient genome assembly of identified novel genes in lots of individuals in the past. The seldomly detection of DNA sequencing in the regions of novel sequences due to the secondary DNA structure might be the reason why there are still so many genes missing in the recent high-quality VGP avian assemblies, which may suggest there are still more challenges to complete the avian reference genome. Nevertheless, we increased the number of protein-coding genes in chicken to 19,223 and denied the gene loss hypothesis during avian evolution. Furthermore, some genes may still hide in some more complex genomic regions and waiting to be discovered. Our study not only revealed the gene number in birds is comparable to that found in other tetrapods but also presented a novel closed pattern of avian pan-genome. The complete avian genomes will greatly contribute to studies on comparative genomics and functional genomics research in birds.
It has been believed that both the evolutionary substitution rate and the rate of chromosomal rearrangement in the avian lineage are lower compared with mammals (Burt et al. 1999; Zhang et al. 2014). However, we found a large number of novel genes that have three times the substitution rate than the known ones, which can greatly increase the average substitution rate of the chicken genome. We find that the novel sequences and genes were concentrated in the microchromosomes and subtelomeric regions of the chromosomes, in which the recombination rates tend to be higher (Linardopoulou et al. 2005; Bell et al. 2020). This may drive the base composition evolution via biased gene conversion (Marais 2003) and cause repeat expansions or contractions (Richard and Paques 2000; Polleys et al. 2017), and might be the critical factor driving the development of the special characteristics in microchromosomes and subtelomeric regions. These genes may have a pivotal role on the formation and development of some unique phenotypes of the dinosaurs-avian branch. For instance, some differentially expressed novel genes were associated with immune response, which may be an ingenious design of the bird immune system to resist viruses with high mutation rates. With the high recombination rate, the novel sequences may represent a large unexplored part of the chicken genetic map, which will contribute to the comprehensive understanding of the genetic variation and pinpoint the causal variations of important traits and thus promote the development of chicken breeding.
In conclusion, our chicken pan-genome provides a comprehensive resource and a great platform for the research of avian evolution, functional genomics, and chicken breeding. These results highlight the complexity of species genomes and suggest that many functionally important regions may be cryptic in reference genomes across the tree of life.
Materials and Methods
Sample Collection
A total of 20 chicken individuals were collected from all around the world for genomic sequencing. Transcriptome sequencing was also performed in 11 tissues of 6 individuals, including breast muscle, bursa of Fabriclus, cecal tonsil, Harderian gland, hypophysis, hypothalamus, liver, ovary, spleen, testis, and thymus tissues. Moreover, tandem MS/MS data were generated from three tissues (hypothalamus, spleen, and cecal tonsil) from four of the six individuals by RNA-Seq. The tissue sources and the institutes in charge of the collection are listed in supplementary table S1, Supplementary Material online. All animal specimens were collected legally in accordance with the policies for Animal Care and Use Ethics of each institution, making all efforts to minimize invasiveness.
Library Construction and Genome Sequencing
For PacBio continuous long reads sequencing, genomic DNA was extracted from chicken liver using a QIAamp DNA Mini Kit (QIAGEN). The integrity of the DNA was determined with an Agilent 4200 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Eight micrograms of genomic DNA were sheared using g-Tubes (Covaris), and concentrated with AMPure PB magnetic beads. Each SMRT bell library was constructed using the Pacific Biosciences SMRTbell template prep kit 2.1. The constructed libraries were size-selected on a BluePippin system for molecules ≥20 kb, followed by primer annealing and the binding of SMRT bell templates to polymerases with the DNA/Polymerase Binding Kit. Sequencing was carried out using P6-C4 chemistry on the Pacific Bioscience Sequel II platform by Annoroad Gene Technology Company.
For short-read DNA sequencing, the genomic DNA of ten samples used for next-generation sequencing (NGS) assembly was extracted from ethylenediaminetetraacetic acid-anticoagulated blood randomly fragmented. Two paired-end libraries and two mate-pair libraries with insert sizes of 500 bp, 800 bp, 3 Kb, and 5 Kb were constructed. All libraries were sequenced on the Illumina HiSeq 2000 platform according to the manufacturer’s protocol. After filtering out adapter sequences and low-quality reads, a total of 1.61 Tb (average 134× coverage of chicken genome) of data were retained for assembly. In addition, the libraries of ten samples used for PacBio sequencing were also constructed using an amplification-free method with an insert size of 350 bp and sequenced on the Illumina XTen platform with paired-end 150 bp sequence reads.
Whole-Transcriptome Sequencing
For transcriptome analysis, total RNA was extracted using TRIzol extraction reagent (Thermo Fisher). The RNA quality analysis method was the same as DNA quality analysis method described above. Libraries with 250–350 bp insert sizes were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, USA). To obtain transcriptome profiles, all libraries were sequenced on Illumina XTen system platform using the manufacturer’s protocol.
Hi-C Sequencing
Hi-C experiments were performed according to a previously published protocol (Lieberman-Aiden et al. 2009). Hi-C libraries were created from the breast muscle samples of six of the above individuals. All libraries were sequenced on an Illumina HiSeq X Ten sequencer (paired-end sequencing with a 150 bp read length). On average, 127 Gb of data with ∼120-fold genomic coverage and 271,268,477 read pairs could be uniquely aligned to GRCg6a (supplementary tables S2 and S3, Supplementary Material online).
Tandem Mass Spectrometry Analysis
The samples were ground into a cell powder in liquid nitrogen and then sonicated in lysis buffer (8 M urea, 1% protease inhibitor cocktail) three times on ice using a high-intensity ultrasonic processor (Scientz). The remaining debris was removed by centrifugation at 12,000 × g at 4 °C for 10 min. Thereafter, the supernatant was collected, and the protein concentration was determined with a BCA kit according to the manufacturer’s instructions. Then, the protein solution was subjected to trypsin digestion. Next, the tryptic peptides were fractionated by high-pH reverse-phase HPLC using a Thermo Betasil C18 column (5 μm particles, 10 mm ID, and 250 mm length).
The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and directly loaded onto a homemade reversed-phase analytical column (15-cm length, 75 μm i.d.). The gradient consisted of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, an increase from 23% to 35% over 8 min and then to 80% over 3 min, withholding at 80% for the last 3 min, all at a constant flow rate of 400 nl/min in an EASY-nLC 1000 UPLC system. The peptides were introduced to a nanospray ionization source, followed by MS/MS in a Q ExactiveTM Plus system (Thermo) coupled online to the UPLC system.
The MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8) (Cox and Mann 2008). Tandem mass spectra were searched against the human UniProt database concatenated with the reverse decoy database. Trypsin/P was specified as the cleavage enzyme, allowing up to four missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification and acetylation modifications and oxidation on Met were specified as variable modifications. The FDR was adjusted to <1%, and the minimum score for modified peptides was set as >40. For protein identification, peptides containing a minimum of seven amino acids and at least one unique peptide were required. Only proteins with at least two peptides and at least one unique peptide were considered to have been identified and used for further data analysis.
De novo Genome Assembly, Evaluation, and Repeat Annotation
Assembly Based on PacBio SMRT Sequencing Platform
The raw PacBio SMRT reads were corrected by itself with Canu v1.7 (Koren et al. 2017), and assembled with WTDBG v2.2 (Ruan and Li 2019) to generate the contig layout and edge sequences, and WTPOA-CNS v1.2 was used to obtain the initial consensus in FASTA format. Then, we used minimap2 v2.14-r883 (Li 2018) to map the corrected reads to the consensus, and they were subsequently polished by using WTPOA-CNS v1.2. This process was repeated three times. Next, the consensus sequence obtained in the previous step was mapped by using the NGS reads from the same individual with BWA-MEM v0.7.17-r1188 (Li and Durbin 2010) and then polished with Pilon v1.22 (Walker et al. 2014). This process was repeated three times to obtain the final contigs.
We performed further scaffolding based on the results for six individuals with Hi-C data. Using the final contigs as a reference, we mapped the Hi-C data to the final contigs using Juicer v1.5 (Durand et al. 2016) to obtain the interaction matrix. Finally, 3d-dna v180419 (Dudchenko et al. 2017) was used for scaffolding contigs.
Assembly Based on NGS Platform
The genomes sequenced on the NGS platform were de novo assembled into contigs by using a pipeline that combined the Fermi package (Li 2012) and Phusion assembler (Mullikin and Ning 2003) for 500/800 bp paired-end libraries. For the 3/5 Kb mate-pair libraries, we used SOAPdenovo (Li et al. 2010) with 77 kmers to build contigs. Furthermore, SSPACE (Boetzer et al. 2011) was used to build scaffolds, and the contigs assembled by Fermi and Phusion were used for the substitution of sequences and bases and for further rectifying to rectify the local assembly error. After the inspection of the initial scaffolds, gaps were closed using Gap5 (Bonfield and Whitwham 2010) software.
Genome Evaluation and Annotation
BUSCO v3.0.2 (Simão et al. 2015) was used to assess assembly completeness by estimating the percentage of expected single-copy conserved orthologs captured in our assemblies and the reference genome, referring to the lineage dataset aves_odb9 (Creation date: 2016-02-13, number of species: 40, number of BUSCOs: 4,915). Repeat sequences were annotated using RepeatMasker v4.0.8 (with the parameters: -engine ncbi -species “Gallus gallus” -s -no_is -cutoff 255 -frag 20000). Subsequently, TRs were further annotated using Tandem Repeats Finder v4.07b (Benson 1999) (with the settings 2 7 7 80 10 50 2000 -d -h). In addition, Quadron software (Sahakyan et al. 2017) was used to predict G4 motifs, and only nonoverlapping hits with a score >19 were used for subsequent analysis.
Chicken Pan-Genome Construction
The de novo assemblies were aligned to the chicken reference genome (GRCg6a; GCF_000002315.6) using minimap2 (Li 2018) (-cx asm10). Based on the pairwise alignment, unaligned or low-identity sequences (showing more than 10% sequence divergence relative to GRCg6a) were extracted. Then, the adjacent sequences within 200 bp were merged. BLASTN 2.6.0+ (Camacho et al. 2009) (with the parameters -word_size 20 -max_hsps 1 -max_target_seqs 1 -dust no -soft_masking false -evalue 0.00001) was further used to align the unaligned sequences from the previous step to GRCg6a, and the sequences showing identity >90% to GRCg6a sequences were removed. The remaining sequences were merged according to the adjacent regions within 200 bp, and sequences of <500 bp in length were removed. Subsequently, the unaligned and low-identity sequences obtained from all of the assemblies were combined, redundancies were removed with CD-HIT v4.7 (Fu et al. 2012) (parameter: -c 0.9 -aS 0.8 -d 0 -sf 1), and the longest sequence in the cluster was selected as the representative sequence. To further exclude potential contaminants in the dataset, we used BLASTN to compare the nr set with the nr database of NCBI (v20181220). The sequences that were aligned to non-Chordata species were removed from the final novel sequence set (supplementary table S5, Supplementary Material online).
Observed Present or Absent Analysis of Novel Sequences in Resequenced Individuals
The whole-genome resequencing data of 922 chickens (Li et al. 2017; Wang et al. 2020) (supplementary table S6, Supplementary Material online) were downloaded for the present or absent analysis of novel sequences. To explore whether the different sequencing platforms affected the results, the Illumina sequencing reads of the GRCg6a individual (SRR3954707 [Warren et al. 2017], which were previously used for single-base error correction) were also included in this analysis. The presence and absence of each novel sequence were then determined according to the sequence coverage and depth. First, to obtain high-quality reads and minimize false genotyping results due to low-quality reads supplied by Illumina, we implemented the following quality control procedures to filter the reads before read mapping using Trimmomatic v0.36 (Bolger et al. 2014), and leading or trailing stretches of Ns and bases with a quality score below 3 were trimmed. Then, the reads were scanned using a 4-base wide sliding window and clipped when the average quality per base was below 15, and only reads of 40 nucleotides or longer were finally retained. Second, high-quality paired reads were aligned to GRCg6a using BWA-MEM v0.7.17 (Li and Durbin 2010) with the default parameters, except that “-M” was enabled. The BWA-aligned BAM files were then processed using Picard v2.1 (http://broadinstitute.github.io/picard/), including reads sorted and merged read groups belonging to the same sample and marked duplicates at the sample level. Finally, we estimated the coverage distribution at each called site for each sample using QualiMap v2.2 (Okonechnikov et al. 2016).
Poorly aligned or unaligned reads were extracted as follows: Samblaster v0.1.24 (Faust and Hall 2014) was used to extract clipped reads and unaligned reads, whereas sambamba v0.6.8 (Tarasov et al. 2015) and SAMTools v1.9 (Li et al. 2009) were used to collect other poorly aligned reads. The paired reads with unaligned/poorly aligned read pairs were extracted using seqtk v1.3-r106 (https://github.com/lh3/seqtk) and were then aligned to the novel sequence set using a previously described process. Novel sequences with a coverage above 0.8 and a depth greater than one-quartered of the whole-genome depth were identified as present.
Feature Importance Analysis
To estimate the influence of GC, G4 motif, and TR contents on the observed frequency of novel sequences, 9,200 novel sequences shared by all individuals were used to construct a random forest model. The sklearn package in Python was used to build the final model and perform classification.
Transcribed Region Annotation and Coding Potential Assessment
The raw RNA-Seq reads were processed to remove adapters, low-quality sequences, and sequences with poly A/T tails using Trimmomatic v0.36 (Bolger et al. 2014). The cleaned reads were de novo assembled using SPAdes v3.14.1 (Bushmanova et al. 2019). The expression levels of the de novo assembled transcripts were quantified by using Kallisto v0.46.2 (Bray et al. 2016). Additionally, the cleaned reads were assembled using a reference-guided method by alignment to the de novo genome assemblies using HISAT2 v2.0.3-beta (Kim et al. 2019) with the default parameters, except that “–dta” was enabled. Transcripts including novel splice variants were assembled using StringTie v1.2.2 (Pertea et al. 2015) with the default parameters. Then, StringTie (–merge) was used to merge all the transcript GTFs obtained from the samples mapped to this assembly to obtain a reference annotation. Finally, all samples were reassembled and quantified using StringTie with the reference annotation to obtain the expression level of each transcript. Notably, the transcripts with FPKM ≥ 1 were considered robustly expressed.
Redundancy among genes that were annotated based on the de novo and reference-guided methods and intersected with novel sequences was removed with CD-HIT (parameter: -c 0.9 -aS 0.8 -d 0 -sf 1). Then, the remaining genes were searched against the nr database and the genes of GRCg6a using BLASTN 2.6.0+. Genes with no hits to either non-Chordata species or GRCg6a were retained as “novel genes” that were completely absent in the chicken reference genome. Genes showing hits to GRCg6a genes with more than 95% identity were classified as partially missing in the chicken reference genome.
Next, the coding potential of these novel genes was assessed by using CPAT v1.2.3 (Wang et al. 2013) with the default parameters. CPAT uses an alignment-independent logistic regression model to detect coding potential based on sequence features. To select a cut-off for classification, we built hexamer tables and logit models for chicken using chicken CDSs and ncRNA sequences downloaded from Ensembl (release 98) as training data. Then, a two-graph receiver operating characteristic curve was used to determine the optimum cut-off value through ten random sample validations (supplementary fig. S20, Supplementary Material online). A cut-off of 0.69 was selected to classify the novel genes as potential protein-coding or noncoding genes. Then, the ORFs were searched by using TransDecoder v5.5.0 (http://transdecoder.github.io) and ORFfinder v0.4.3 (https://www.ncbi.nlm.nih.gov/orffinder/) with the default parameters. Genes showing values above the cut-off of the CPAT with a minimum ORF of at least 100 amino acids were classified as novel coding genes. For the remaining novel genes, RNAcode (Washietl et al. 2011) was used to further estimate the coding potential. To prevent the divergent homologous haplotypes that can caused false gene duplications (Ko et al. 2021), we merged novel coding genes that have high similarity (identify ≥95%) with each other or can be annotated to the same gene, and then performed the manual check. We generated customized whole-genome alignments for each de novo assembly against Japanese quail (GCF_001577835.1), turkey (GCF_000146605.3), and helmeted guineafowl (GCF_002078875.1), which we used to estimate coding potential. We used BLASTX 2.6.0+ (with the parameters “-evalue 0.00001”) to translate each novel genes from all six possible reading frames, and the results were compared with known proteins in the nr database. Genes with an E-value ≤ 10−5, alignment length of ≥10 amino acids, and identity ≥95% were removed from the final potential long noncoding gene set. Only multiple exon genes with more than 200 nucleotides and without any detectable protein-coding potential were classified as novel long noncoding genes.
We compared the DNA sequence of the novel coding genes with the genome of GRCg7b (GCF_016699485.2) by BLASTN with an E-value ≤ 10−5, identity ≥95%, and coverage ≥10% to check whether GRCg7b assembled the gene.
Protein-Coding Gene Annotation
Using the human (Homo sapiens) dataset as the background, the novel coding genes were annotated with the annotate module of online KOBAS 3.0 (Xie et al. 2011) (http://kobas.cbi.pku.edu.cn/). The Gene Ontology terms, KEGG pathways, and Reactome pathways of these genes were characterized by using the enrichment module of online KOBAS 3.0. P < 0.05 was set as the cut-off threshold.
InterProScan v5.36-75.0 (Jones et al. 2014) (parameter: -f tsv -dp) was used to classify the protein-coding gene fragments within the novel sequences and the protein-coding genes influenced by the location of novel sequences into protein families. The analysis results of Pfam 32.0 (http://pfam.xfam.org/) were selected to determine the families to which the proteins belonged.
Differential Expression Analysis
The expression levels of each gene obtained from the previous step were used for differential expression analysis. The R language was used to identify differentially expressed genes with the edgeR package (v3.28.1) (Robinson et al. 2010). The fold changes between the two groups were calculated as logFC = log2 (experimental/control group). Benjamini–Hochberg correction was used to correct for multiple comparisons (with a false discovery cut-off of < 0.05). Genes in the two groups with |logFC| > 2 and q-value < 0.05 were defined as differentially expressed genes.
Anchoring Novel Sequences onto the Reference Genome
Flanking Sequences
The novel sequences were anchored to GRCg6a based on alignment information between all de novo assemblies and GRCg6a. First, the scaffolds of the de novo assemblies that contained novel sequences were extracted and anchored on the chromosome/scaffold of GRCg6a which showed the most alignment hits with them. Then, the adjacent flanking sequences (more than 100 bp) of the novel sequences aligned to the same chromosome/scaffold were retained for further positioning. If the flanking sequences were perfectly aligned to GRCg6a with no gaps, an identity ≥90%, and a breakpoint shift of ≤5 bp, we recorded the sequences as “placed.” The other alignments were recorded as “ambiguously placed.” The novel sequences with two placed flanking sequences were reported as “localized.” The novel sequences with one or two ambiguously placed flanking sequences were reported as “unlocalized.” The final remaining sequences were reported as “unplaced.” Based on the genome placement information, the localized sequences could be further classified as insertions, alternate alleles, or ambiguous sequences. The insertions introduced only one sequence fragment to the reference genome and were no more than 10 bp in length. For alternate alleles, the novel sequences had to share <90% (or 0%) identity with their counterparts in the reference. Furthermore, the novel sequences and their counterparts had to have comparable lengths, with a length ratio between 1/3 and 3. The remaining sequences that did not meet the above criteria for insertions and alternate alleles were classified as ambiguous sequences.
Chromosome Interaction Mapping
The preprocessing of paired-end sequencing data, mapping of reads, and filtering of mapped di-tags were performed using the Juicer pipeline (version 1.5) (Durand et al. 2016). Briefly, short reads were mapped to the chicken pan-genome using BWA-MEM (version 0.7.17-r1188) (Li and Durbin 2010). Reads with low mapping quality were filtered using Juicer with the default parameters, discarding invalid self-ligated and unligated fragments as well as PCR artifacts. Filtered di-tags were further processed with Juicer command line tools to bin ditags (10 kb bins) and to normalize matrices with KR normalization (Knight and Ruiz 2013). We normalized all Hi-C matrices on the same scale by KR normalization, ensuring that any differences between Hi-C data were not attributable to variation in sequence length. The maximum 100-kb bin of each novel sequence interaction (interaction intensity ≥5) was collected as a potential location of novel sequences. Novel sequences that were validated in at least two individuals with Hi-C data and anchored to the same location were kept for further analysis.
Gene Orthology and dN/dS Analysis
The integrated toolkit TBtools v1.0 (Chen et al. 2020) was used for collinearity analysis between species. First, the protein sequence of each gene was obtained, and pairwise sequence similarities were calculated using BLASTP with a cut-off of E-value ≤ 10−10. Then, syntenic blocks were detected using MCScanX v1.0 (Wang et al. 2012) with the default parameters. OrthoFinder v2.4.0 (Emms and Kelly 2019) was used to identify orthologous genes with the default parameters. Among these genes, 1:1 orthologous genes between different species were used for downstream analysis. Using 1:1 orthologous genes as the input, Codeml in PAML version 4.9d (Yang 2007) was used for dN/dS analysis with the default parameters. The genome assemblies and corresponding annotations used in this analysis were: gray short-tailed opossum (GCF_000002295.2), greater horseshoe bat (GCF_004115265.1), human (GCF_000001405.39), western terrestrial garter snake (GCF_009769535.1), common lizard (GCF_011800845.1), Red-eared slider turtle (GCF_013100865.1), Goodes thornscrub tortoise (GCF_007399415.2), green sea turtle (GCF_015237465.1), American alligator (GCF_000281125.3), Chinese alligator (GCF_000455745.1), Australian saltwater crocodile (GCF_001723895.1), Okarito brown kiwi (GCF_003343035.1), African ostrich (GCF_000698965.1), emu (GCA_016128335.1), golden eagle (GCF_900496995.1), kakapo (GCF_004027225.2), New Caledonian crow (GCF_009650955.1), Swainson’s thrush (GCF_009819885.1), zebra finch (GCF_008822105.2), mallard (GCF_015476345.1), helmeted guineafowl (GCF_002078875.1), turkey (GCF_000146605.3), Japanese quail (GCF_001577835.2), and chicken (GCF_000002315.6).
Supplementary Material
Acknowledgments
We thank High-Performance Computing (HPC) of Northwest A&F University (NWAFU) for providing computing resources. We thank Margarida Cardoso-Moreira for kindly providing the individual source information of the 217 chicken transcriptomes. This project was supported by grant from the National Natural Science Foundation of China (31822052, 31572381) and the National Thousand Youth Talents Plan to Y.J., National Natural Science Foundation of China (No. 31930105) and China Agriculture Research System of MOF and MARA (CARS-40) to N.Y., National Natural Science Foundation of China (31761143014) to Q.N., The National Key Research and Development Program of China (2021YFD1300600) to C.S., and the Unit of Excellence 2021, University of Phayao, Thailand (UoE64003) to C.S.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
N.Y., Y.J., and X.H. conceived the project and designed the research. M.L., X.T., P.B., and N.X. performed the majority of the analysis with contributions from Y.W., X.D., R.L., Y.G., F.W., X.W., P.Y., S.Z., D.H., Z.L., W.F., C.S., C.W., F.L., X.L., A.S., and C.S. prepared the DNA samples. X.J. and Q.N. provided the genome of Fayoumi. M.L., X.W., and N.X. drafted the manuscripts with input from all authors and Y.J., C.S., Y.W., R.H., M.W., and X.Z. revised the manuscript.
Data Availability
All the data of our study are publicly available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession code BioProject: PRJNA573584 and PRJNA777393. The multiple genome alignment and novel sequences data are available at Zenodo (https://doi.org/10.5281/zenodo.5881830) and http://animal.nwsuaf.edu.cn/code/index.php/panChicken.
References
- Bell AD, Mello CJ, Nemesh J, Brumbaugh SA, Wysoker A, McCarroll SA. 2020. Insights into variation in meiosis from 31,228 human sperm genomes. Nature 583:259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield JK, Whitwham A. 2010. Gap5—editing the billion fragment sequence assembly. Bioinformatics 26:1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv S, Seroussi E, Yosefi S, Pendavis K, Burgess SC, Grabherr M, Friedman-Einat M, Andersson L. 2017. Correspondence on Lovell et al.: identification of chicken genes previously assumed to be evolutionarily lost. Genome Biol. 18:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero-Castro F, Figuet E, Tilak MK, Nabholz B, Galtier N. 2017. Avian genomes revisited: hidden genes uncovered and the rates versus traits paradox in birds. Mol Biol Evol. 34:3123–3131. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34:525–527. [DOI] [PubMed] [Google Scholar]
- Brown WRA, Hubbard SJ, Tickle C, Wilson SA. 2003. The chicken as a model for large-scale analysis of vertebrate gene function. Nat Rev Genet. 4:87–98. [DOI] [PubMed] [Google Scholar]
- Burt DW. 2005. Chicken genome: current status and future opportunities. Genome Res. 15:1692–1698. [DOI] [PubMed] [Google Scholar]
- Burt DW, Bruley C, Dunn IC, Jones CT, Ramage A, Law AS, Morrice DR, Paton IR, Smith J, Windsor D, et al. . 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402:411–413. [DOI] [PubMed] [Google Scholar]
- Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 8:giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinform 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, Liechti A, Ascenção K, Rummel C, Ovchinnikova S, et al. . 2019. Gene expression across mammalian organ development. Nature 571:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen W-J, Yang J-Y, Lin J-H, Fann CSJ, Osyetrov V, King C-C, Chen Y-MA, Chang H-L, Kuo H-W, Liao F, et al. . 2006. Nasopharyngeal shedding of severe acute respiratory syndrome-associated coronavirus is associated with genetic polymorphisms. Clin Infect Dis. 42:1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MD, Peterson RDA., South MA, Good RA. 1966. The functions of the thymus system and the bursa system in the chicken. J Exp Med. 123:75–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p. p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Duan Z, Qiao Y, Lu J, Lu H, Zhang W, Yan F, Sun C, Hu Z, Zhang Z, Li G, et al. . 2019. HUPAN: a pan-genome analysis pipeline for human genomes. Genome Biol. 20:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. . 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust GG, Hall IM. 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30:2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill F, Donsker D, Rasmussen P, editors. 2020. IOC World Bird List (v10.1). doi: 10.14344/ioc.Ml.10.1 [DOI]
- Golicz AA, Bayer PE, Bhalla PL, Batley J, Edwards D. 2019. Pangenomics comes of age: from bacteria to plant and animal applications. Trends Genet. 36:132–145. [DOI] [PubMed] [Google Scholar]
- Guiblet WM, Cremona MA, Cechova M, Harris RS, Kejnovská I, Kejnovsky E, Eckert K, Chiaromonte F, Makova KD. 2018. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res. 28:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hounkpe BW, Chenou F, de Lima F, De Paula EV. 2021. HRT Atlas v1.0 database: redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res. 49:D947–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium . 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. . 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee C, Ko BJ, Yoo D, Won S, Phillippy A, Fedrigo O, Zhang G, Howe K, Wood J, et al. . 2021. False gene and chromosome losses affected by assembly and sequence errors. bioRxiv. doi: 10.1101/2021.04.09.438906 [DOI] [Google Scholar]
- Knight PA, Ruiz D. 2013. A fast algorithm for matrix balancing. IMA J Numer Anal. 33:1029–1047. [Google Scholar]
- Ko BJ, Lee C, Kim J, Rhie A, Yoo D, Howe K, Wood J, Cho S, Brown S, Formenti G, et al. . 2021. Widespread false gene gains caused by duplication errors in genome assemblies. bioRxiv. doi: 10.1101/2021.04.09.438957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Che T, Chen B, Tian S, Zhou X, Zhang G, Li M, Gaur U, Li Y, Luo M, et al. . 2017. Genomic data for 78 chickens from 14 populations. Gigascience 6:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2012. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28:1838–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fu W, Su R, Tian X, Du D, Zhao Y, Zheng Z, Chen Q, Gao S, Cai Y, et al. . 2019. Towards the complete goat pan-genome by recovering missing genomic segments from the reference genome. Front Genet. 10:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. . 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. . 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A-L, Li Y-F, Qi W, Ma X-L, Yu K-X, Huang B, Liao M, Li F, Pan J, Song M-X. 2015. Comparative analysis of selected innate immune-related genes following infection of immortal DF-1 cells with highly pathogenic (H5N1) and low pathogenic (H9N2) avian influenza viruses. Virus Genes 50:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Wirthlin M, Wilhelm L, Minx P, Lazar NH, Carbone L, Warren WC, Mello CV. 2014. Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol. 15:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19:330–338. [DOI] [PubMed] [Google Scholar]
- Marjuki H, Scholtissek C, Yen H-L, Webster RG. 2008. CK2beta gene silencing increases cell susceptibility to influenza A virus infection resulting in accelerated virus entry and higher viral protein content. J Mol Signal. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z. 2003. The phusion assembler. Genome Res. 13:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F. 2016. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleys EJ, House NCM, Freudenreich CH. 2017. Role of recombination and replication fork restart in repeat instability. DNA Repair (Amst). 56:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. . 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Paques F. 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H. 2019. Fast and accurate long-read assembly with wtdbg2. Nat Methods 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. 2012. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-κB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J Virol. 86:10211–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan AB, Chambers VS, Marsico G, Santner T, Di Antonio M, Balasubramanian S. 2017. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci Rep. 7:14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. 2019. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL. 2010. Assembly of large genomes using second-generation sequencing. Genome Res. 20:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid ET, Pang IK, Carrera Silva EA, Bosurgi L, Miner JJ, Diamond MS, Iwasaki A, Rothlin CV. 2016. AXL receptor tyrosine kinase is required for T cell priming and antiviral immunity. Elife 5:e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroussi E, Pitel F, Leroux S, Morisson M, Bornelöv S, Miyara S, Yosefi S, Cogburn LA, Burt DW, Anderson L, et al. . 2017. Mapping of leptin and its syntenic genes to chicken chromosome 1p. BMC Genet. 18:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman RM, Forman J, Antonescu V, Puiu D, Daya M, Rafaels N, Boorgula MP, Chavan S, Vergara C, Ortega VE, et al. . 2019. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet. 51:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman RM, Salzberg SL. 2020. Pan-genomics in the human genome era. Nat Rev Genet. 21:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith J, Smith N, Yu L, Paton IR, Gutowska MW, Forrest HL, Danner AF, Seiler JP, Digard P, Webster RG, et al. . 2015. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 16:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehelin D, Varmus HE, Bishop JM, Vogt PK. 1976. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260:170–173. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Li R, Fu W, Li Y, Wang X, Li M, Du D, Tang Q, Cai Y, Long Y, et al. . 2020. Building a sequence map of the pig pan-genome from multiple de novo assemblies and Hi-C data. Sci China Life Sci. 63:750–763. [DOI] [PubMed] [Google Scholar]
- Vogt PK. 2011. Historical introduction to the general properties of retroviruses. In: Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. . 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lam TH, Soh MK, Ye Z, Chen J, Ren EC. 2018. Influenza A virus facilitates its infectivity by activating p53 to inhibit the expression of interferon-induced transmembrane proteins. Front Immunol. 9:1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Hu H, Tian Y, Li J, Scheben A, Zhang C, Li Y, Wu J, Yang L, Fan X, et al. . 2021. The chicken pan-genome reveals gene content variation and a promoter region deletion in IGF2BP1 affecting body size. Mol Biol Evol. 38:5066–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W. 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M-S, Thakur M, Peng M-S, Jiang Y, Frantz LAF, Li M, Zhang J-J, Wang S, Peters J, Otecko NO, et al. . 2020. 863 genomes reveal the origin and domestication of chicken. Cell Res. 30:693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, et al. . 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Tomlinson C, Minx P, Kremitzki M, Graves T, Markovic C, Bouk N, Pruitt KD, Thibaud-Nissen F, et al. . 2017. A new chicken genome assembly provides insight into avian genome structure. G3 (Bethesda) 7:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Findeiß S, Muller SA, Kalkhof S, von Bergen M, Hofacker IL, Stadler PF, Goldman N. 2011. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA 17:578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisskirchen C, Ludersdorfer TH, Müller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J Virol. 85:8646–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KHY, Levy-Sakin M, Kwok P-Y. 2018. De novo human genome assemblies reveal spectrum of alternative haplotypes in diverse populations. Nat Commun. 9:3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KHY, Ma W, Wei C-Y, Yeh E-C, Lin W-J, Wang EHF, Su J-P, Hsieh F-J, Kao H-J, Chen H-H, et al. . 2020. Towards a reference genome that captures global genetic diversity. Nat Commun. 11:5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39:W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin Z-T, Zhu F, Lin F-B, Jia T, Wang Z, Sun D-T, Li G-S, Zhang C-L, Smith J, Yang N, et al. . 2019. Revisiting avian ‘missing’ genes from de novo assembled transcripts. BMC Genom. 20:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, et al. . 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bacolla A, Wang G, Vasquez KM. 2010. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 67:43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Yin Z-T, Wang Z, Smith J, Zhang F, Martin F, Ogeh D, Hincke M, Lin F-B, Burt DW, et al. . 2021. Three chromosome-level duck genome assemblies provide insights into genomic variation during domestication. Nat Commun. 12:5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data of our study are publicly available at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession code BioProject: PRJNA573584 and PRJNA777393. The multiple genome alignment and novel sequences data are available at Zenodo (https://doi.org/10.5281/zenodo.5881830) and http://animal.nwsuaf.edu.cn/code/index.php/panChicken.