Abstract
The D-type cyclin (CYCD) gene, as the rate-limiting enzyme in the G1 phase of cell cycle, plays a vital role in the process of plant growth and development. Early studies on plant cyclin mostly focused on herbs, such as Arabidopsis thaliana. The sustainable growth ability of woody plants is a unique characteristic in the study of plant cyclin. Here, the promoter of PsnCYCD1;1 was cloned from poplar by PCR and genetically transformed into tobacco. A strong GUS activity was observed in the areas with vigorous cell division, such as stem tips, lateral buds, and young leaves. The PsnCYCD1;1-GFP fusion expression vector was transformed into tobacco, and the green fluorescence signal was observed in the nucleus. Compared with the control plant, the transgenic tobacco showed significant changes in the flower organs, such as enlargement of sepals, petals, and fruits. Furthermore, the stems of transgenic plants were slightly curved at each stem node, the leaves were curled on the adaxial side, and the fruits were seriously aborted after artificial pollination. Microscopic observation showed that the epidermal cells of petals, leaves, and seed coats of transgenic plants became smaller. The transcriptional levels of endogenous genes, such as NtCYCDs, NtSTM, NtKNAT1, and NtASs, were upregulated by PsnCYCD1;1. Therefore, PsnCYCD1;1 gene played an important role in the regulation of flower organ and stem development, providing new understanding for the functional characterization of CYCD gene and new resources for improving the ornamental value of horticultural plants.
Keywords: Populus, PsnCYCD1;1, cyclin, small cell, enlarged floral organs
Introduction
Plant development is accompanied by cell division, which is regulated by cell cycle activity (Inzé and De Veylder, 2006). Plant D-type cyclins control the process of cell cycle and play an important role in cell division and proliferation. D-type cyclins are preferentially induced by mitogen stimulants in the G1 phase to accumulate developmental signals (Oakenfull et al., 2002; Richard et al., 2002) and control cells to re-enter the cell cycle (Soni et al., 1995; Meijer and Murray, 2001). D-type cyclins (CYCDs) are called G phase-specific cyclins due to their important role in the transition from G1 to S phase (De Veylder et al., 1999).
Ten genes encoding CYCDs have been identified in Arabidopsis. They are divided into seven subfamilies (CYCD1-CYCD7), of which CYCD3 subfamily has three members, CYCD4 subfamily has two members, and the other five CYCD subfamilies are encoded by only one gene (Vandepoele et al., 2002). CYCD1;1, CYCD2;1, and CYCD3;1 are originally screened from Arabidopsis, which can restore the phenotype of yeast G1 cyclin mutant (Soni et al., 1995). Cyclins, as a regulatory subunit of protein kinase, closely bind to CDK, forming an active kinase complex, and directionally regulate plant growth and development (Morgan, 1995). Most CYCD proteins can interact with cyclin-dependent kinase A;1 (CDKA;1), and the overexpression of some CYCD genes promote the cells to enter the S phase (Masubelele et al., 2005; Menges et al., 2006; Mendez et al., 2020), indicating that the CDKA-CYCD complex regulates the conversion of G1/S phase.
Many studies have shown that overexpression of some CYCDs in plants can change plant growth and development. The constitutive expression of CYCD enables transgenic Arabidopsis leaves to undergo cell division without adding exogenous cytokinin (Riou-Khamlichi et al., 2000). The Populus trichocarpa PtCYCB1 promoter is used to drive the expression of the CYCD gene in tobacco, which improves the cell division of vascular cambium and increases the differentiation of secondary xylem in tobacco (Fujii et al., 2012). The ectopic expression of AtCYCD2 accelerates the growth and aboveground biomass accumulation rate of transgenic tobacco from the seedling stage to maturity (Boucheron et al., 2005; Kopertekh and Reichardt, 2021). Moreover, overexpressing the CYCD3 gene into tobacco also accelerates the growth rate of leaves and changes the structure of the shoot apical meristem (Boucheron et al., 2005). The CYCD3;1 transgenic Arabidopsis develops slowly, but the flowering time does not change. Meanwhile, overexpression of CYCD3;1 also causes changes in leaf morphology and structure, particularly, increased number of leaf epidermal cells and curled leaves toward the medial axis (Dewitte and Murray, 2003). Soybean D-type cyclin (GmCYCD6;1-6) forms a feedforward loop which is the key mechanism to regulate the initial division of soybean nodule primordium (Wang et al., 2022). The seed formation is controlled by the activity of CYCD7 in the central cell and in the developing endosperm (Sornay et al., 2015).
Previous reports on CYCDs were not comprehensive and mostly focused on genes, such as CYCD2, CYCD3, and CYCD7, from model plants and herbs. There are few reports on the function of cyclin genes in woody plants. We cloned a CYCD1;1 from poplar and performed a preliminary analysis in Arabidopsis (Zheng et al., 2021). Significant phenotypic changes were observed in transgenic tobacco by constructing plant expression vector, and these phenotypes were not found in previous transgenic Arabidopsis. Therefore, this study not only improves the function of PsnCYCD1;1, but also enriches the new understanding of plant cyclin.
Materials and Methods
Plant Material and Growth Conditions
The Populus simonii × P. nigra cross was obtained from the campus of Northeast Forestry University in Heilongjiang Province, China. The young leaves were sampled, immediately frozen in liquid nitrogen, and stored at −80°C before the extraction of genomic DNA. Nicotiana tabacum L. was planted in a pot containing cultivation substrate (turf peat and pearlite, 2:1 v/v) under controlled conditions with 16 h of light/8 h of darkness, 22 ± 2°C temperature, 40–50% humidity, and 200 μmol m–2 s–1 light intensity.
Cloning of PsnCYCD1;1 Gene
To verify the stable subcellular localization of PsnCYCD1;1 gene, PCR cloning was carried out with the specific primers (Supplementary Table 1) by removing the stop codon and using pUC18-PsnCYCD1;1 plasmid as a template (Zheng et al., 2021). The reaction system consisted of 2.5 μl 10 × Ex PCR buffer, 2.0 μl dNTP (10 mmol/L), and 1.0 μl upstream and downstream primers, 0.25 μl ExTaq (5 U/μl), 0.1 μl plasmid template (10 ng/μl), and added ddH2O to make final volume of 25 μl. The amplification procedure was as follows: 35 cycles at 94°C for 4 min, 94°C for 30 s, 56°C for 30 s, and 72°C for 1.5 min, and 72°C for 7 min. After the reaction, 5 μl of PCR products were detected in 1% agarose gel. Then, the PCR product gel was recovered, connected to the pEasy-T1 vector, transformed into Escherichia coli Trans1-T1 competent cells, and screened for positive clones.
Construction of Plant Expression Vector
Restriction endonucleases XbaI and KpnI were used to digest pEasy-PsnCYCD1;1 and pROKII-GFP vectors, respectively. The enzyme reaction system consisted of 1.0 μl XbaI, 1.0 μl KpnI, 2.0 μl 10× K buffer, 1 μg plasmid, and ddH2O to make the final volume of 20 μl. After the target fragment was recovered by gel and linked by T4 DNA ligase, the recombinants were transformed into E. coli Trans1-T1 competent cells and positive colonies were screened for sequencing verification. Finally, the pROKII-PsnCYCD1;1-GFP plasmid was transferred into Agrobacterium GV3101 competent cells using liquid nitrogen freezing and thawing method (Chen et al., 1994).
Cloning of PsnCYCD1;1 Promoter and Construction of Plant Expression Vector
The DNA of young leaves of P. simonii × P. nigra was isolated by hexadecyltrimethylammonium bromide (CTAB) method (Doyle, 1987), and was used as a template to amplify promoter fragment by specific primers that were designed based on the upstream 1,800 bp sequences of PsnCYCD1;1. To verify spatiotemporal expression pattern of PsnCYCD1;1 promoter, after cloning and purification of PsnCYCD1;1 promoter fragment, the target fragment, and pBI121-GUS were digested by HindIII and KpnI and linked by T4 DNA ligase to construct the target vector pBI121-PsnCYCD1;1pro-GUS. After verification by sequencing, the recombinant plasmid was transferred into Agrobacterium GV3101 competent cells using the liquid nitrogen freezing and thawing method.
Genetic Transformation
Recombinants (pROKII-PsnCYCD1;1-GFP and pBI121-PsnCYCD1;1pro-GUS) were transferred into wild-type tobacco by leaf disc method (Zheng et al., 2014). The final resistant plants were obtained through differentiation on a selective medium containing 50 mg/L kanamycin. After stem growth and rooting culture, the positive plantlets were used to extract DNA from the leaves by the CTAB method. PCR was performed using vector primers, and the products were evaluated by 1% agarose gel electrophoresis.
Quantitative Real-Time PCR Analysis
To validate the transcriptional level of the PsnCYCD1;1 gene, the total RNA of young leaves of transgenic plants was extracted using a MiniBEST Plant RNA Extraction Kit (TaKaRa, Beijing, China). Total RNA was reverse transcribed into cDNA and diluted 10 times with ddH2O as a template. The reaction system was 20 μl, containing 10 μl SYBR Green, 0.4 μl Rox dye II, 2 μl cDNA template, and 0.8 μl forward and reverse primers, respectively (Supplementary Table 1). The reaction procedure was as follows: 40 cycles at 95°C for 30 s; 95°C for 5 s, 60°C for 35 s; 95°C 15 s, 60°C 1 min, and 95°C 15 s. The Ntactin gene was used as an internal reference (Zheng et al., 2014). Three biological and technical repetitions were performed, and the relative expression of the target gene was calculated with the 2-delt cycle threshold (CT) method (Livak and Schmittgen, 2001).
To detect the transcriptional changes of endogenous gene-related stem and leaf development, the total RNA of young stems and leaves of 3-week-old control and transgenic tobacco was isolated. Fourteen gene-specific primers were designed using Integrated DNA Technologies (IDT) online tools1 (Supplementary Table 1). Housekeeping gene and reaction system were the same as above.
Subcellular Localization
The root tips of 2-week-old transgenic tobacco (pROKII-PsnCYCD1;1-GFP) seedlings were collected and flatten on a glass slide for microscopic observation. The wild-type and transgenic tobacco with a pROKII-GFP vector was used as control. The green fluorescent signals in cells were observed and photographed by Leica TCS SP8 laser confocal microscope (Leica, Wetzlar, Germany).
GUS Staining
Transgenic tobacco (pBI121-PsnCYCD1;1pro-GUS) seedlings and different tissues in different growth stages were immersed in β-glucuronidase (GUS) staining solution, and the interstitial gas of tissues were discharged with a vacuum pump to maintain the air pressure at 0.01 Pa for about 30 min. Subsequently, the tissues were dyed in a 37°C incubator for 5–7 h and fixed overnight with Carnot fixative, soaked in 75% alcohol for 2–3 days until completely faded, and photographed with Leica ED4 stereomicroscope (Leica, Germany).
Phenotypic Observation of Transgenic Tobacco
Wild-type and three transgenic lines (pROKII-PsnCYCD1;1-GFP) with high expression levels were planted in pots in a greenhouse with controlled environmental conditions. Leaf development and stem growth were recorded every 2 days. Five newly blooming flowers were randomly selected from each plant, and the width of the corolla, the length of petal, calyx, fruit, and seed were measured and statistically analyzed.
To observe the seed development, the transgenic and control plants planted under the same growth conditions were selected. After 3 days, when the flowers withered and the fruit was developing, the peel was peeled off and the developing seeds were observed and photographed under the microscope. One week later, the abortion of developing seeds was statistically analyzed.
Environmental Scanning Electron Microscope Observation
Dry seeds were sprayed with gold powder before scanning and observation. Fresh samples of petals and leaves were cut into sheets less than 1 cm wide and 1.5 cm long before being directly placed on the objective table of environmental scanning electron microscope (ESEM) for observation and photography with Philips Quanta 200 (FEI, Eindhoven, Netherlands). The environmental vacuum mode was selected for the electron microscope sample chamber, and the gas secondary electronic signal imaging was adopted. The working conditions were as follows: high voltage at 12.5 KV, pressure >7.5e−3 Torr, filament current of 2.34 A, and emission current at 97 M a.
The relative size of epidermal cell and seed between transgenic tobacco and controls were measured and statistically analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, United States).
Results
Cloning and Genetic Transformation of PsnCYCD1;1 Gene and Promoter in Tobacco
A specific band of about 1,000 bp was amplified from pUC18-PsnCYCD1;1 (Supplementary Figure 1), and the PsnCYCD1;1 gene was confirmed by sequencing. Then, the recombinant pROKII-PsnCYCD1;1-GFP was obtained by connecting PsnCYCD1;1 and pROKII-GFP by double enzyme digestion. A 1,941 bp nucleotide sequence of the upstream of PsnCYCD1;1 gene was cloned using leaf DNA from P. simonii × P. nigra as template (Supplementary Figure 2), while the recombinant pBI121-PsnCYCD1;1pro-GUS was obtained by inserting into pBI121-GUS with restriction endonuclease. Sequencing results showed that the target fragment was an upstream sequence of the PsnCYCD1;1 gene and the cis-elements of PsnCYCD1;1 promoter were analyzed using PlantCARE (Supplementary Figure 3).
The genetic transformation was carried out by leaf disc method with Agrobacterium GV3101 containing pROKII-PsnCYCD1;1-GFP and pBI121-PsnCYCD1;1pro-GUS vectors, respectively. Then, 19 independent 35S:PsnCYCD1;1-GFP transgenic tobacco and 16 independent PsnCYCD1;1pro-GUS transgenic tobacco T1 lines were obtained by DNA PCR, respectively. The positive plants were further detected by quantitative real-time PCR (qRT-PCR) to detect the relative expression level of PsnCYCD1;1 (Supplementary Figure 4). Finally, nine independent lines in the homologous T2 generation were screened, and four lines (OE1, OE2, OE6, and OE14) were further chosen to analyze the phenotypic changes.
GFP Observation of Transgenic Tobacco Root Tips
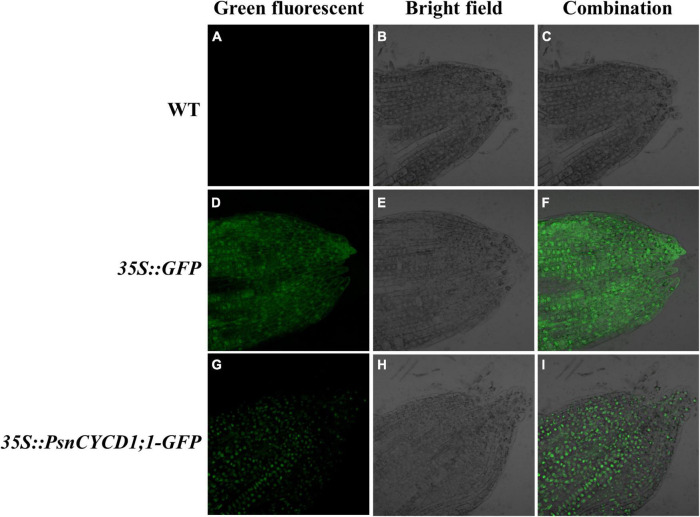
To validate the subcellular localization of PsnCYCD1;1 in plants, the root tips of T2 generation seedling of transgenic (35S:PsnCYCD1;1-GFP), empty vector (35:GFP), and wild-type tobacco were compressed and observed under laser confocal microscope. The results showed that in the root tips of wild-type tobacco, there was no fluorescent signal. In empty vector control (CK) and tobacco root tip cells, the green fluorescence signal was detected in the whole cell, while in 35S:PsnCYCD1;1-GFP transgenic tobacco root tip cells, the green fluorescence signal was only detected in the nucleus, indicating that PsnCYCD1;1 performs biological functions in the nucleus (Figure 1).
FIGURE 1.
Subcellular location of green fluorescent protein (GFP) and PsnCYCD1;1-GFP protein in tobacco root tip. Green fluorescent (A,D,G): green fluorescence signal; bright field (B,E,H): white light; and combination (C,F,I): combined signals of different fluorescence. WT, wild-type; 35::GFP, pROKII-GFP, empty vector control; 35S::PsnCYCD1;1-GFP, fusion vector of pROKII-PsnCYCD1;1-GFP.
GUS Activity of PsnCYCD1;1pro-GUS Transgenic Tobacco
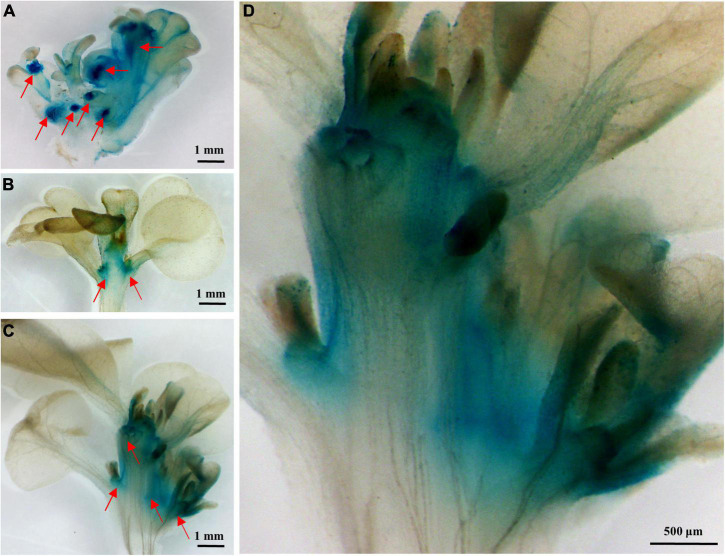
To study the function of the PsnCYCD1;1 promoter, the homozygous T2 generation was screened and observed by histochemical staining. The results showed deep GUS staining at the growth point, terminal bud, and leaf bud. The coloring was very light and almost invisible in other parts, such as in leaves or stem segments, suggesting that the strong expression of PsnCYCD1;1 in the meristem (Figure 2).
FIGURE 2.
Histochemical analysis of β-glucuronidase (GUS) activity of PsnCYCD1;1pro-GUS transgenic tobacco. (A) Adventitious buds; (B) stem tip; (C) axillary bud development of stem tip; and (D) partial enlargement of (C).
Morphological Phenotype of Tobacco Overexpressing PsnCYCD1;1
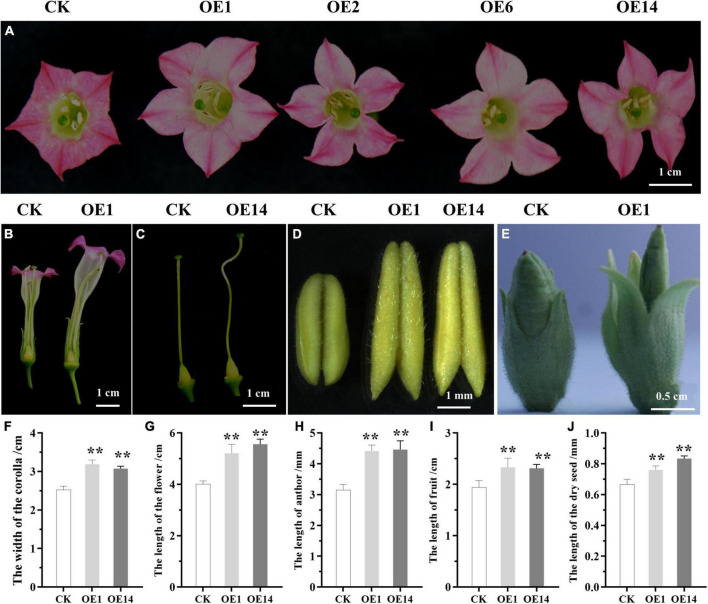
In the vegetative growth stage, the leaves of transgenic tobacco were hypertrophic and could not be flattened normally compared with the control lines (Supplementary Figure 5 and Figure 3). Meanwhile, the stem of transgenic tobacco was not straight, showing irregular zigzag between two leaf nodes (Figure 3). After entering the reproductive growth stage, compared with the wild-type and CK lines, the transgenic lines had larger corolla, larger petals and sepals, and longer fruits and seeds. In addition, the length of stigma increased and showed a twisted phenotype (Figure 4). Three transgenic lines (OE1, OE2, and OE14) and three CK lines were selected, and five flowers were randomly taken from the top of each plant. The width of corolla and the length of sepal, petal, fruit, and seed were measured and analyzed by using SPSS 19.0 software and the q test. The results showed that compared with CK plants, the corolla of transgenic plants was significantly wider, the petals were significantly larger, and the sepals, fruits, and seeds were significantly longer (Figure 4), suggesting that ectopic expression of PsnCYCD1;1 in tobacco affects the development of flower organs and fruits. Under normal conditions, the tobacco used in this study gradually withers after flowering and fruiting, and the whole growth cycle usually spans 3–5 months. However, some transgenic lines maintained long-term growth, and OE14 maintained flowering for more than 20 months, thus becoming a perennial plant with new individuals sprouting and growing from the axillary buds (Supplementary Figures 6, 7).
FIGURE 3.
Phenotype observation of empty vector control (CK) and 35S:PsnCYCD1;1-GFP tobacco. (A) Three-month old CK tobacco; (B) 3-month old 35S:PsnCYCD1;1-GFP transgenic tobacco; (C) stem morphology of transgenic, wild-type, and CK tobacco; and (D) comparison of leaf morphology between transgenic and CK tobacco; WT, wild-type; CK, empty vector control; OE1/2, two individual transgenic lines.
FIGURE 4.
The differences in reproductive organs between CK and transgenic tobacco. (A) Comparison of the morphology of corollas; (B) longitudinal cutting of floral organs flowers; (C) comparison of the morphology of pistils; (D) comparison of the size of anthers; (E) comparison of the morphology of fruits; (F–J) statistical analysis of the phenotype of corolla, flower, anther, fruit, and dry seed. CK, empty vector control; OE1/2/6/14, four individual transgenic lines. Asterisks on the column diagram showed the significant difference between different samples (p ≤ 0.01).
Comparative Analysis of Fertility of Transgenic Tobacco
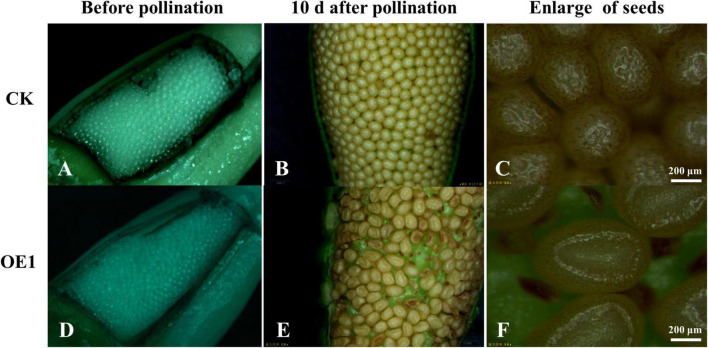
Although transgenic tobacco produces a large number of flowers, 100% of them fall off early and cannot develop into mature fruits. Through artificial pollination, few fruits can continue to develop, but the seed yield is very low (Supplementary Figure 8). Three transgenic lines and control lines were selected to observe seed development before pollination and 10 days after artificial pollination. After peeling, the unpollinated ovules and seeds were observed under the microscope and the abortion rate per unit area was calculated. The results of statistical analysis showed that the ovule development of control and transgenic plants was basically the same before pollination. After pollination with wild-type pollens, there were significant differences in fruit development between the CK and the transgenic line (PsnCYCD1;1), to which the abortion rates per unit area were 3.2 and 46.1%, respectively (Figure 5 and Supplementary Figures 8, 9), suggesting that the ectopic expression of PsnCYCD1;1 can cause severe male sterility and slight female sterility in transgenic tobacco.
FIGURE 5.
Ectopic expression of PsnCYCD1;1 caused seed fertility in transgenic tobacco. (A,D) Observation of ovules before pollination between CK and transgenic (OE1) line; (B,E) microscopic observation of seeds in the ovary 10 days after pollination with wild-type pollens between CK and transgenic (OE1) line; and (C,F) close-up of the image (B,E). CK, control plant (A–C); OE1, transgenic plant (D–F).
Comparative Analysis of Cell Size of Transgenic Tobacco
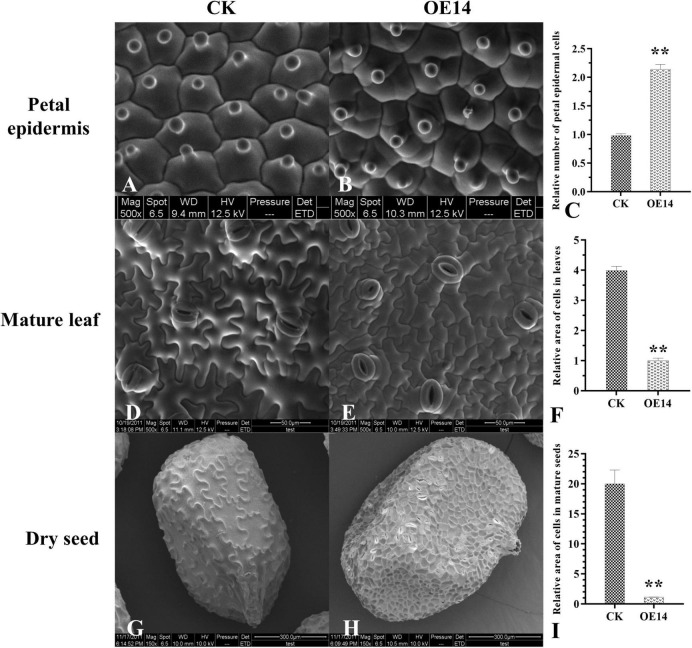
To reveal the reasons for the enlargement of flower organs, the cell size of different tissues of transgenic plants was observed. Three fully bloomed petals from OE14 and CK tobacco were observed by ESEM. The results indicated that the normal papillary cells existed in the adaxial petal epidermis of CK tobacco (Figures 6A–C). Interestingly, there were many small cells formed by subsequent division of the perianth cells, and there were no papillary apexes on these small cells, suggesting that the ectopic expression of PsnCYCD1;1 caused the morphological changes of perianth cells in transgenic tobacco.
FIGURE 6.
Comparative analysis of cell size between CK and transgenic (OE14) tobacco by environmental scanning electron microscope (ESEM). (A,B) ESEM observation of the petal epidermis between CK and transgenic (OE14) line. (D,E) ESEM observation of the abaxial epidermis of the leaves between CK and transgenic (OE14) line. (G,H) Microscopic observation of dry seeds between CK and transgenic (OE14) line. (C,F,I) Statistical analysis of the petal epidermis, mature leaf, and seed coat cells between CK and transgenic (OE14) lines. Asterisks on the column diagram showed the significant difference between different samples (p < 0.01).
The mature leaves of OE14 and CK tobacco with the same growth rates were selected, and the abaxial epidermis of the leaves was observed and photographed by ESEM. The results showed that the cell size of the abaxial epidermis cells of the OE14 was significantly different from that of the CK (Figures 6D,E). The relative cell size of the OE14 was only 25% of CK cells (Figure 6F).
To avoid the difference of growth period, the mature seeds were further selected for ESEM. The results showed that the surface of the seed coat of CK tobacco had reticular decoration (degenerated integument epidermis), in which the mesh outline was mostly irregular and in the form of round polygons with different sizes (Figure 6G). However, the net ridge of the reticular pattern on the seed coat surface of the OE14 seeds was thin, and the mesh outline was shaped like a regular polygon and looked like a honeycomb (Figure 6H). Statistical analysis of the size of a single cell on the seed epidermis showed that there was a great difference between the OE14 and the CK cells, and the relative sizes of OE14 cells were only about 1/20 of the CK cells (Figure 6I).
Detection of Endogenous Related Genes in Transgenic Tobacco
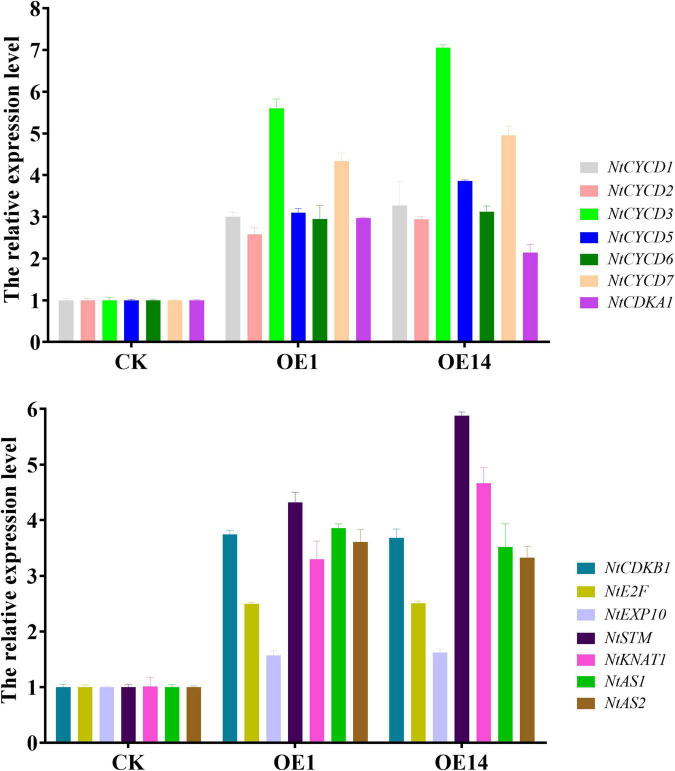
To detect the transcriptional changes of other D type cyclins, cyclin kinases, downstream division genes, and stem development-related genes in 35S:PsnCYCD1;1-GFP transgenic tobacco, the total RNA of young stems and young leaves of 3-week-old CK and transgenic (OE1 and OE14) seedlings was extracted, and the transcription levels of fourteen genes were detected by qRT-PCR. Compared with the CK, the endogenous NtCYCD genes were upregulated for more than two times in the transgenic plants. In particular, the expression of NtCYCD3 gene was upregulated for about seven times. Cyclin kinases, NtCDKA1 and NtCDKB1, and downstream division-related gene NtE2F were also upregulated more than two times in transgenic plants. In addition, stem development-related genes, NtSTM and NtKNAT1, and leaf polarity-related genes, NtAS1 and NtAS2, were upregulated more than two times in transgenic plants (Figure 7).
FIGURE 7.
Quantitative real-time PCR analysis of endogenous related genes in CK and transgenic tobacco. CK, empty vector control tobacco; OE1/14, two individual transgenic lines.
Discussion
The organs of higher plants continuously develop in the whole life cycle, which depends on the flexible control of cell division and cell proliferation. D-type cyclins are rate-limiting enzymes in the G1 phase, which determines the key period of cell differentiation. In Arabidopsis, ten genes encoding CYCDs have been identified and they are divided into seven subfamilies (CYCD1-CYCD7) (Vandepoele et al., 2002). In woody plant Populus tomentosa, 22 PtrCYCD genes are identified and divided into six subfamilies, including five CYCD1 members, two CYCD2/CYCD4 members, six CYCD3 members, three CYCD5 members, five CYCD6 members, and one CYCD7 gene (Dong et al., 2011). In view of the genome and secondary growth differences between woody plants and herbs, the number of cyclins in poplar is more than twice as compared to Arabidopsis, and some genes may have functional redundancy or special functions.
The subcellular localization results of PsnCYCD1;1 showed its location in the nucleus (Figure 1), which is consistent with the homologous genes located in the nucleus of Arabidopsis, such as AtCYCD 1;1, AtCYCD2;1, and AtCYCD3;1 (Planchais et al., 2004; Sanz et al., 2011). In transgenic tobacco, strong fluorescence signals were observed in root apical meristem (RAM) and lower epidermis of leaves (Supplementary Figure 10), suggesting that PsnCYCD1;1 is also a relatively stable protein, possibly due to the ectopic expression of PsnCYCD1;1 gene in tobacco. Strong GUS activity was detected in the growth points, axillary buds, and young leaves, similar to the qRT-PCR results and confirming the high PsnCYCD1;1 expression in the stem, flower, root, and young leaves (Zheng et al., 2021). The cis-element in PsnCYCD1;1 promoter was predicted, and it was found that the PsnCYCD1;1 promoter contained many tissue-specific expression elements (CAT-box, Skn-1_motif, and CCGTCC-box) and a variety of hormone response elements (ABRE and TGA elements), suggesting an important role of PsnCYCD1;1 in poplar stem growth and lateral bud formation through hormone signaling pathways. In chrysanthemum, the expression level of CYCD was upregulated by cytokinin, auxin, and sugar together, causing a short internodal distance (Sun et al., 2021).
Plant growth and development are controlled in RAM and stem apical meristem (SAM). Meristem is the most active region of cell division. Therefore, the change in cell cycle may affect the downstream development process. Overexpression of CYCDs can change plant growth and development. Due to the reduction of G1 phase, the development process of CYCD2 transgenic tobaccos is accelerated at all stages, from seed to adult plant growth, but the size of cells and meristem is normal (Cockcroft et al., 2000). However, overexpression of CYCD2 in Arabidopsis showed no significant phenotypic changes (Cockcroft et al., 2000). In contrast, overexpression of CYCD3 in Arabidopsis caused morphological changes, such as delayed SAM and leaf senescence and leaf explant tissues, proliferated independent of cytokinin (Riou-Khamlichi et al., 1999). Overexpression of PtoCYCD3;3 in poplar promoted the development of cambium and vascular bundle (Guan et al., 2021). Targeted expression of AtCYCD7;1 to the central cells and early endosperm reduced the seed number in each silique and increased seed size (Sornay et al., 2015, 2016). There are few reports on the overexpression of CYCD1 gene in Arabidopsis. Ectopic expression of PsnCYCD1;1 gene from poplar promoted cell division and produced curved rosette leaves in Arabidopsis (Zheng et al., 2021). Here, a new phenotype was found after the transfer of PsnCYCD1;1 gene into tobacco. Compared with the control plant, the flower organs of transgenic tobacco overexpressing PsnCYCD1;1 showed a significant increase in the width of corolla and the length of petals and sepals. At the same time, the seeds of transgenic tobacco had a high abortion. In addition, the cells of petals, lower epidermis of leaves, and seeds of transgenic plants became smaller, which was similar to the result of overexpression of AtCYCD2;1 in Arabidopsis, causing significantly smaller cells in the root tip division area (Qi and John, 2007). Flower bud differentiation is a physiological and morphological sign of transformation from vegetative to reproductive growth. The differentiation order of each round of flower organs on the flower primordium is generally a centripetal differentiation. In this study, the calyx, petal, anther, and pistil were significantly larger, suggesting an important role of PsnCYCD1;1 in heterologous regulation of cell cycle activities and plant reproductive development. Tobacco petal epidermis cells usually have papilla cells. We found comparatively smaller cells in the petal epidermis of transgenic plants, and some small cells did not have papillae. It is suggested that the overexpression of PsnCYCD1;1 not only accelerates cell division, but also affects cell differentiation. Overexpression of CYCD3;1 gene in Arabidopsis rendered the cells of transgenic plants smaller and less differentiated than wild-type cells. Meanwhile, there were no normal palisade and spongy tissue as these were replaced by a large number of smaller cells (Dewitte and Murray, 2003). Altogether, PsnCYCD1;1 accelerates the process of mitosis in transgenic plants, resulting in a large number of cells. This may be due to an increased need for energy and nutrients for cytoplasmic growth in transgenic plants.
To investigate the reason for formation of small cells in transgenic tobacco, genes related to cell division and stem growth were identified, including six NtCYCDs, two NtCDKs, one NtE2F, one NtEXP10, one NtSTM, one NtKNATs, and two NtASs. The result of qRT-PCR showed that two D-type cyclins (NtCYCD3 and NtCYCD7) were significantly upregulated (Figure 7), which is similar to the report of homologous genes. For example, AtCYCD3;3 is involved in the lateral bud and root tip differentiation of Arabidopsis (Gaudin et al., 2000), while AtCYCD7 regulates the development of embryo and root tip (Sornay et al., 2015). The CDKB is specific in plants (Boudolf et al., 2001), and CDKB2-GUS fusion protein showed intermittent expression patterns in Arabidopsis meristem (Adachi et al., 2006). The result of qRT-PCR showed that NtCDKA1 and NtCDKB1 were significantly upregulated, which is consistent with the report that CYCD and CDKA can form complexes (Mendez et al., 2020). The change of E2F expression can cause cell division and elongation of RAM and change the expression of some cell cycle regulatory factors, including E2Fa, E2Fb, and E2Fe/DEL1 (Sozzani et al., 2009). The result of qRT-PCR showed that the expression of downstream division gene NtE2F was significantly upregulated, suggesting that the interaction among E2Fs may control the cell proliferation during organ formation and tissue development. Stem and leaf development-related genes (NtSTM, NtKNAT1, NtAS1, and NtAS2) were also upregulated in transgenic lines. AS gene regulates the paraxial and abaxial polarity development in leaves (Rast and Simon, 2012). The loss of the ANT gene function leads to the formation of small organs, while its overexpression increases the size of organs (Mizukami and Fischer, 2000). KNAT, mainly expressed in apical meristem, is inhibited by auxin and AS gene, and it participates in meristem regulation by cytokinin and CYCD3 together with STM gene (Scofield et al., 2013).
In this study, the heterologous expression in tobacco may not always reflect the endogenous biological function in native species. Obtaining transgenic poplars to verify the phenotypic changes and evaluating the role of cis-elements in the regulation of flower organ and stem development is recommended to be the main objective in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZZ, TZ, and GQ conceived and drafted the manuscript. ZZ and TZ performed the experiment. ZZ, TZ, LD, YL, and SL analyzed the data. GQ provided plant resources. TZ and GQ contributed to the conception of the study and finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Sagheer Ahmad in Fujian Agriculture and Forestry University (Fuzhou, China) for the assistance in English editing.
Footnotes
Funding
This research was supported by the Heilongjiang Province Key R&D Program of China (GA21B010), Fundamental Research Funds for the Central Universities (2572020DY24), and Innovation Project of State Key Laboratory of Tree Genetics and Breeding of Northeast Forestry University (2015A01).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.868731/full#supplementary-material
References
- Adachi S., Uchimiya H., Umeda M. (2006). Expression of B2-type cyclin-dependent kinase is controlled by protein degradation in Arabidopsis thaliana. Plant Cell Physiol. 47, 1683–1686. [DOI] [PubMed] [Google Scholar]
- Boucheron E., Healy J. H. S., Bajon C., Sauvanet A., Rembur J., Noin M., et al. (2005). Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J. Exp. Bot. 56 123–134. 10.1093/jxb/eri001 [DOI] [PubMed] [Google Scholar]
- Boudolf V., Rombauts S., Naudts M., Inzeì D., De Veylder L. (2001). Identification of novel cyclin-dependent kinases interacting with the CKS1 protein of Arabidopsis. J. Exp. Bot. 52 1381–1382. 10.1093/jexbot/52.359.1381 [DOI] [PubMed] [Google Scholar]
- Chen H., Nelson R. S., Sherwood J. L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16 664–668, 670. [PubMed] [Google Scholar]
- Cockcroft C. E., den Boer B. G., Healy J. M., Murray J. A. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. 10.1038/35014621 [DOI] [PubMed] [Google Scholar]
- De Veylder L., de Almeida Engler J., Burssens S., Manevski A., Lescure B., Van Montagu M., et al. (1999). A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta 208 453–462. 10.1007/s004250050582 [DOI] [PubMed] [Google Scholar]
- Dewitte W., Murray J. A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54 235–264. 10.1146/annurev.arplant.54.031902.134836 [DOI] [PubMed] [Google Scholar]
- Dong Q., Zhao Y., Jiang H., He H., Zhu S., Cheng B., et al. (2011). Genome-wide identification and characterization of the cyclin gene family in Populus trichocarpa. Plant Cell Tissue Organ Cult. 107 55–67. 10.1007/s11240-011-9957-z [DOI] [Google Scholar]
- Doyle J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19 11–15. [Google Scholar]
- Fujii T., Sato K., Matsui N., Furuichi T., Takenouchi S., Nishikubo N., et al. (2012). Enhancement of secondary xylem cell proliferation by Arabidopsis cyclin D overexpression in tobacco plants. Plant Cell Rep. 31 1573–1580. 10.1007/s00299-012-1271-7 [DOI] [PubMed] [Google Scholar]
- Gaudin V., Lunness P. A., Fobert P. R., Towers M., Riou-Khamlichi C., Murray J. A., et al. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122 1137–1148. 10.1104/pp.122.4.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Xue Y., Jiang P., He C., Zhuge X., Lan T., et al. (2021). Overexpression of PtoCYCD3;3 promotes growth and causes leaf wrinkle and branch appearance in Populus. Int. J. Mol. Sci. 22:1288. 10.3390/ijms22031288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40 77–105. 10.1146/annurev.genet.40.110405.090431 [DOI] [PubMed] [Google Scholar]
- Kopertekh L., Reichardt S. (2021). At-CycD2 enhances accumulation of Above-Ground biomass and recombinant proteins in transgenic Nicotiana benthamiana plants. Front. Plant Sci. 12:712438. 10.3389/fpls.2021.712438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Masubelele N. H., Dewitte W., Menges M., Maughan S., Collins C., Huntley R. P., et al. (2005). D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102 15694–15699. 10.1073/pnas.0507581102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Murray J. A. (2001). Cell cycle controls and the development of plant form. Curr. Opin. Plant Biol. 4 44–49. 10.1016/s1369-5266(00)00134-5 [DOI] [PubMed] [Google Scholar]
- Mendez A., Pena L. B., Curto L. M., Sciorra M. D., Ulloa R. M., Garza Aguilar S. M., et al. (2020). Optimization of recombinant maize CDKA;1 and CycD6;1 production in Escherichia coli by response surface methodology. Protein Expr. Purif. 165:105483. 10.1016/j.pep.2019.105483 [DOI] [PubMed] [Google Scholar]
- Menges M., Samland A. K., Planchais S., James A. H. M. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-Phase transition in Arabidopsis. Plant Cell 18 893–906. 10.1105/tpc.105.039636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97 942–947. 10.1073/pnas.97.2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O. (1995). Principles of CDK regulation. Nature (London) 374 131–134. 10.1038/374131a0 [DOI] [PubMed] [Google Scholar]
- Oakenfull E. A., Riou-Khamlichi C., Murray J. A. H., Chua N. H. (2002). Plant D–type cyclins and the control of G1 progression. Philos. Trans. Biol. Sci. 357 749–760. 10.1098/rstb.2002.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S., Samland A. K., Murray J. A. H. (2004). Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 38 616–625. 10.1111/j.0960-7412.2004.02071.x [DOI] [PubMed] [Google Scholar]
- Qi R., John P. C. L. (2007). Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: implications for the control of plant growth. Plant Physiol. 144 1587–1597. 10.1104/pp.107.096834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast M. I., Simon R. (2012). Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24 2917–2933. 10.1105/tpc.112.099978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C., Lescot M., Inzé D., De Veylder L. (2002). Effect of auxin, cytokinin, and sucrose on cell cycle gene expression in Arabidopsis thaliana cell suspension cultures. Plant Cell Tissue Organ Cult. 69 167–176. 10.1023/A:1015241709145 [DOI] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A., James A. H. M. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544. 10.1126/science.283.5407.1541 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Menges M., Healy J. M., James A. H. M. (2000). Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. 10.1128/MCB.20.13.4513-4521.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Dewitte W., Forzani C., Patell F., Nieuwland J., Wen B., et al. (2011). The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23 641–660. 10.1105/tpc.110.080002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Nieuwland J., Murray J. A. H. (2013). The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 75 53–66. 10.1111/tpj.12198 [DOI] [PubMed] [Google Scholar]
- Soni R. U. O. C., Carmichael J. P., Shah Z. H., Murray J. A. H. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103. 10.1105/tpc.7.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornay E., Dewitte W., Murray J. A. H. (2016). Seed size plasticity in response to embryonic lethality conferred by ectopic CYCD activation is dependent on plant architecture. Plant Signal. Behav. 11:e1192741. 10.1080/15592324.2016.1192741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornay E., Forzani C., Forero Vargas M., Dewitte W., Murray J. A. H. (2015). Activation of CYCD7;1 in the central cell and early endosperm overcomes cell-cycle arrest in the Arabidopsis female gametophyte, and promotes early endosperm and embryo development. Plant J. 84 41–55. 10.1111/tpj.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R., Maggio C., Giordo R., Umana E., Ascencio-Ibañez J. T., Hanley-Bowdoin L., et al. (2009). The E2FD/DEL2 factor is a component of a regulatory network controlling cell proliferation and development in Arabidopsis. Plant Mol. Biol. 72 381–395. 10.1007/s11103-009-9577-8 [DOI] [PubMed] [Google Scholar]
- Sun D., Zhang L., Yu Q., Zhang J., Li P., Zhang Y., et al. (2021). Integrated signals of jasmonates, sugars, cytokinins and auxin influence the initial growth of the second buds of chrysanthemum after decapitation. Biology 10:440. 10.3390/biology10050440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., Veylder L. D., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. 10.1105/tpc.010445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li M., Zhao Y., Liang N., Li H., Li P., et al. (2022). SHORT-ROOT paralogs mediate feedforward regulation of D-type cyclin to promote nodule formation in soybean. Proc. Natl. Acad. Sci. U.S.A. 119:e2108641119. 10.1073/pnas.2108641119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Dai L., Liu Y., Li S., Zheng M., Zhao Z., et al. (2021). Overexpression Populus D-type cyclin gene PsnCYCD1;1 influences cell division and produces curved leaf in Arabidopsis thaliana. Int. J. Mol. Sci. 22:5837. 10.3390/ijms22115837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Li S., Zang L., Dai L., Yang C., Qu G.-Z., et al. (2014). Overexpression of two PsnAP1 genes from Populus simonii x P. Nigra causes early flowering in transgenic tobacco and Arabidopsis. PLoS One 9:e111725. 10.1371/journal.pone.0111725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.