Abstract
Type 1 diabetes (T1D) is a T-cell mediated autoimmune disease in which the insulin-producing β-cells within the pancreas are destroyed. Identification of target antigens and epitopes of the β-cell-reactive T cells is important both for the understanding T1D pathogenesis and for the rational development of antigen-specific immunotherapies for the disease. Several studies suggest that proinsulin is an early and integral target autoantigen in T1D. However, proinsulin epitopes recognized by human CD4+ T cells have not been comprehensively characterized. Using a dye-dilution based T-cell cloning method, we generated and characterized 24 unique proinsulin-specific CD4+ T-cell clones from the peripheral blood of 17 individuals who carry the high-risk DR3-DQ2 and/or DR4-DQ8 HLA class II haplotypes. Some of the clones recognized previously reported DR4-restricted epitopes within the C-peptide (C25-35) or A-chain (A1-15) of proinsulin. However, we also characterized novel DR3-restricted epitopes within both the B-chain (B16-27 and B22-C3) and C-peptide (C25-35). Moreover, we identified novel DQ2-restricted epitopes within the B-chain and several DQ2- or DQ8-restricted epitopes within the C-terminal region of C-peptide that partially overlap with previously reported DQ-restricted epitopes. Two of the DQ2-restricted epitopes, B18-26 and C22-33, were shown to be naturally processed from whole human proinsulin. Finally, we observed a higher frequency of CDR3 sequences matching the TCR sequences of the proinsulin-specific T-cell clones in pancreatic lymph node samples compared to spleen samples. In conclusion, we confirmed several previously reported epitopes but also identified novel epitopes within proinsulin, which are presented by HLA class II molecules associated with T1D risk.
INTRODUCTION
Type 1 diabetes (T1D) is a T-cell mediated autoimmune disease characterized by the destruction of insulin-producing β-cells in the pancreas (1). Antigen-specific immunotherapies that target autoreactive T cells are an attractive approach for attenuating T1D-associated autoimmunity. However, currently no effective antigen-specific treatment exists for T1D (2). This results partially from the lack of fundamental knowledge on important target autoantigens and their T-cell epitopes. Better characterization of epitopes recognized by autoreactive T cells is also critical for the development of T-cell assays that can be used as surrogate biomarkers to monitor the disease process and the effect of immunotherapies on autoreactive T-cell responses (3).
Several lines of evidence suggest that proinsulin is likely the most important target for CD4+ T-cell autoimmunity during the development of T1D. In the NOD mouse model, CD4+ T-cell responses to proinsulin are a prerequisite for the development of autoimmune diabetes (4, 5). In humans, autoantibodies against insulin are commonly the first autoantibodies to appear in prediabetic children (6, 7). Finally, a genetic polymorphism in the insulin gene promoter region is associated with the risk of T1D. This polymorphism alters proinsulin expression in the thymus, which in turn, likely affects central tolerance to proinsulin (8–10). The HLA class II region has the strongest genetic association with risk of developing T1D. HLA-DR3-DQ2 and HLA-DR4-DQ8 are well-established risk haplotypes for T1D (11, 12) and around 90% of Caucasian patients with T1D carry one or both of these haplotypes. Taken together, there is a strong rationale that T cell responses against proinsulin epitopes, that are restricted by HLA class II molecules encoded by the high-risk HLA-DR3-DQ2 and HLA-DR4-DQ8 haplotypes, play a role in the pathogenesis of human T1D.
In the last few years, three separate studies have reported a total of 12 proinsulin-reactive CD4+ T-cell clones isolated directly from the pancreatic islets of organ donors with T1D (13–15). Interestingly, ten of these clones recognized epitopes within the C-peptide segment of human proinsulin. Most of these clones were restricted either with HLA-DQ8 or HLA-DQ8trans, a transdimer that is formed when the alpha-chain of HLA-DQ2 pairs with the β-chain of HLA-DQ8 in individuals heterozygous for DR3-DQ2/DR4-DQ8 (16).
In contrast to T cells isolated from pancreatic islets, most proinsulin-specific CD4+ T-cell clones isolated from peripheral blood recognize epitopes restricted by HLA-DR4 (10, 17–21). However, in a very recent study where C-peptide instead of whole proinsulin was used to stimulate T cells in vitro, the majority of reactive clones isolated from blood were DQ-restricted (19). Of note, the majority of previous studies addressing T-cell responses to proinsulin have focused heavily on T-cell responses restricted either by HLA-DR4 or HLA-DQ8 (13–15, 17–20, 22–28). To our knowledge, only single epitopes from proinsulin presented in the context of HLA-DQ2 (19) and HLA-DR3 (29) have been reported in the literature.
In the current study, we used an unbiased T-cell cloning approach to generate and characterize the largest collection of unique proinsulin-specific human CD4+ T-cell clones reported thus far, to our knowledge. We were able to confirm previously reported DR4-restricted proinsulin epitopes but also identified several novel DR3-, DQ2- and DQ8-restricted epitopes.
MATERIALS AND METHODS
Study subjects
Peripheral blood samples were collected from healthy islet autoantibody-negative children with HLA-conferred susceptibility to T1D (mean age 4.2 years ± SD 4.3, range 0.2-15.6 years). All study subjects were homozygous either for DR3-DQ2 or DR4-DQ8, or heterozygous for DR3-DQ2/DR4-DQ8 HLA class II haplotypes and participated in the Finnish Type 1 Diabetes Prediction and Prevention (DIPP) study (6).The study was approved by local ethics committees at the participating university hospitals. All families participating in the study have provided written informed consent.
Antigens
Peptides used in this study were synthesized, purified by HPLC to an average 90% (range 75-97%) purity and lyophilized by Mimotopes. Peptides were reconstituted in 40% acetonitrile at 10 mg/mL. The maximal acetonitrile concentration used in T-cell assays was 0.2%. Recombinant human proinsulin (Lilly) was reconstituted in sterile H2O at 2 mg/mL. Peptides and proteins were stored in aliquots at −80°C. The sequences of the peptides used are summarized in Supplemental Figure 1.
PBMC proliferation dye dilution assay
Fresh PBMCs were isolated from whole blood samples by Ficoll gradient centrifugation, labeled with CytoTell Green (AAT Bioquest) and cultured at 2x105 cells per well in 96-well U-bottomed plates in five replicate wells with a panel of 25 overlapping proinsulin peptides that covers the whole proinsulin sequence (5 μg/mL for each peptide, Supplemental Fig.1) in RPMI 1640 culture medium supplemented with 2 mM L-glutamine, 20 μM 2-mercaptoethanol, 1 mM sodium pyruvate, nonessential amino acids, 100 IU/mL penicillin, 100 μg/mL streptomycin, 10 mM HEPES (all from Lonza) and 5% inactivated human AB serum (Sigma-Aldrich). IL-7 at 0.1 ng/mL was added to the culture on day 0, and IL-2 at 10 IU/mL on day 2-4 (both from Miltenyi Biotec). On days 6 or 7, a portion of the cells from the replicate wells were pooled together and stained with anti-CD4-APC (clone RPA-T4, BioLegend) and proliferative responses were analyzed by the dilution of CytoTell Green with a FACSCanto II flow cytometer (BD Biosciences).
Generation and maintenance of proinsulin-specific CD4+ T-cell clones
T-cell cultures that displayed a positive proliferative response to proinsulin peptides were used for T-cell cloning experiments. Specifically, on days 7 or 8 after stimulation proliferating CD4+ T cells were single-cell sorted on a FACSAria III (BD Biosciences) to 96-well round bottom plates containing 5x104 γ-irradiated (3000 rad) allogeneic PBMCs and 5x103 γ-irradiated allogeneic EBV-transformed B cells. Clones were expanded in the presence of 1 μg/mL PHA (Oxoid) and 50 IU/mL IL-2 for two weeks. Thereafter, growing clones were expanded and maintained by stimulating every 2 to 3 weeks with anti-CD2/CD3/CD28 beads (T Cell Activation and Expansion kit, Miltenyi Biotec) at a 1:2 bead:cell ratio in the presence of IL-2.
Proliferation assays for T-cell clones
Proliferation assays for T-cell clones were set up in duplicates or triplicates in 96-well U-bottomed plates with 2.5x104 T cells and 2.5x104 γ-irradiated (3000 rad) HLA class II -matched EBV-transformed B cells, together with peptides (10 μg/mL, except 100 μg/mL for C-peptide). After 3 days, 0.5 μCi 3H-thymidine (Perkin Elmer) was added to the wells, and after an additional 16 hours, the cells were harvested onto glass fiber filters (Perkin Elmer). Radioactivity was measured by scintillation counting (Wallac Micro Beta 1540) and results were expressed as CPM. Stimulation indices (SI: CPM in the presence of antigen divided by CPM in the absence of antigen) and CPM differences (ΔCPM) between stimulated and unstimulated wells were determined. Effective concentration 50 (EC50) values, i.e. the peptide concentration needed to induce a half-maximal proliferative response, were determined from dose-response curves of individual T-cell clones stimulated with different concentrations (50 μg/mL to 0.001 μg/mL) of their cognate peptides. For the determination of reactivity to human proinsulin, monocyte-derived DCs (moDCs) were generated from HLA-matched PBMCs as previously described (30). The moDCs were pulsed with recombinant human proinsulin (200 μg/mL) for 4h and subsequently matured by stimulating them with 100 ng/mL LPS (Sigma) for two days before using them as APCs in proliferation assays.
HLA class II restriction of the T-cell clones
HLA restriction of the proinsulin-specific T-cell clones was determined by two separate assays. 1) HLA-matched EBV-transformed B cells were incubated at +37°C for 20 minutes with 10 μg/mL blocking monoclonal antibodies to HLA-DR (clone L243, Leinco), HLA-DP (clone B7/21, Leinco) or HLA-DQ (clone SPVL3, produced in-house), followed by addition of each individual T-cell clone and its cognate peptide. 2) EBV-transformed B cells homozygous for either DR3-DQ2 or DR4-DQ8, or heterozygous for DR3-DQ2/DR4-DQ8, or cell lines transfected with single HLA class II molecules were used as APCs in the proliferation assays. EBV-transformed B cells from a donor with bare lymphocyte syndrome (BLS) that had been stably transfected with different HLA genes were kindly provided by Dr. William W. Kwok (Benaroya Research Institute, Seattle, USA). RM3 cell lines transfected with different HLA genes were generated as previously described (31).
Determination of paired TCR αβ sequences of the T-cell clones and in silico analyses
The TCRα and TCRβ sequences of the clones were amplified by series of three nested PCRs, as previously reported (32). In brief, total RNA was isolated from 1x105 T cells using the RNeasy Mini Kit (Qiagen) and utilized for reverse transcription and preamplification using the One-Step RT PCR kit (Qiagen), followed by two nested PCR steps using Hot-Star Taq Polymerase (Qiagen) and specific forward and reverse primers. Finally, the generated amplified TCRα and TCRβ sequences were combined, purified and sequenced by next-generation sequencing (Illumina MiSeq). The TCR β-chain CDR3 sequences were queried in the Network for Pancreatic Organ donors with Diabetes (nPOD) datasets from FACS-isolated CD8+ T cells, CD4+CD25+CD127−/lo regulatory T cells (Treg), and CD4+CD127+ conventional T cell (Tconv) subsets isolated from pancreatic draining lymph nodes and spleen samples. For each query sequence, the presence of a sequence in the dataset with a maximum Hamming distance (HD) of 1 was considered a match for the specific query sequence. Identification of sequences with a maximum HD of 1 was determined by first filtering for sequences of the same length as the query sequence and then calculating Levensthein distance using the editdistance package (https://github.com/aflc/editdistance) in Python.
HLA class II peptide-binding assays
HLA-DR and HLA-DQ molecules were purified from homozygous EBV-transformed B cell lines by affinity chromatography using the monoclonal antibodies L243 and SPVL3, respectively (33, 34). Peptide binding to HLA-DR and -DQ molecules was assessed by competitive ELISA as previously reported (33, 34). In brief, the peptide concentration that prevented binding of 50% of the biotinylated peptide (IC50) was evaluated. Data were expressed as relative affinity (i.e., peptide IC50:reference peptide IC50), using a reference peptide with high binding affinity to the HLA-DR or HLA-DQ molecule analyzed.
Statistical analyses
Statistical analyses were conducted using Prism software (GraphPad) and R, as indicated. P < 0.05 was considered to indicate statistical significance.
RESULTS
Isolation of proinsulin-specific CD4+ T-cell clones from peripheral blood of individuals carrying HLA class II genotypes associated with a high risk of T1D
To characterize human CD4+ T-cell responses to proinsulin, we utilized blood samples from healthy children genetically at-risk for T1D (i.e., homozygous either for DR3-DQ2 or DR4-DQ8, or heterozygous for DR3-DQ2/DR4-DQ8 haplotypes) to ensure that the responding T cells would be restricted by HLA class II molecules encoded by the highest T1D-risk HLA haplotypes. Our experimental approach is outlined in Fig. 1. We stimulated PBMC samples with a panel of 25 overlapping peptides covering the entire sequence of human proinsulin (Supplemental Fig. 1) and analyzed the proliferation of CD4+ T cells with a dye dilution assay (Fig. 2A). However, no proliferative responses to proinsulin peptides were detected (Fig. 2B). We reasoned that autoreactive T-cell responses may arise from the naïve T-cell pool in healthy children, and this could result in an inefficient expansion of these T cells in vitro. To counter this, we added low concentrations of IL-2 and IL-7 to the cultures, and indeed T-cell responses to proinsulin peptides became detectable (Fig. 2B).
Figure 1.
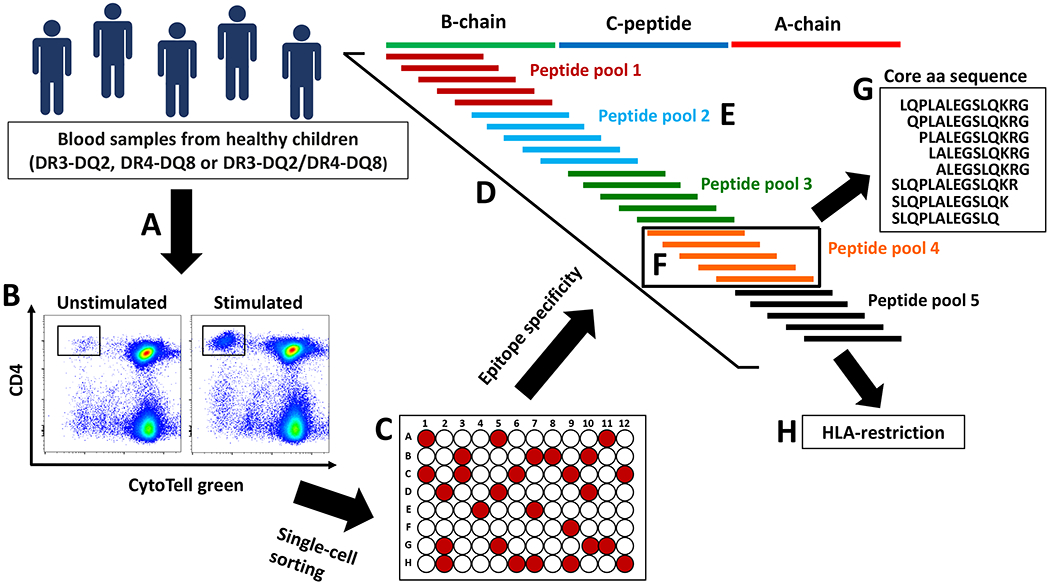
Generation of proinsulin-specific T-cell clones. PBMCs from healthy children homozygous for DR3-DQ2 or DR4-DQ8, or heterozygous for DR3-DQ2/DR4-DQ8 were stimulated with a panel of 25 overlapping peptides (15-mer with 11-13 amino acid overlap) covering the complete sequence of proinsulin (A). Proliferating cells were identified by a dye dilution assay (B), single cell-sorted and grown as T-cell clones (C). The epitope specificity of the growing clones was determined by stimulating the cells first with the full panel of proinsulin peptides (D), then with five peptide pools (E) and finally with individual peptides within the specific peptide pool (F). The core amino acid sequence of the clones was determined by a set of truncated peptides (G). HLA-restriction was determined by anti-HLA-DR, -DP and -DQ blocking antibodies, and by using HLA-matched EBV cell lines or cell lines transfected with single HLA class II molecules as APCs (H).
Figure 2.
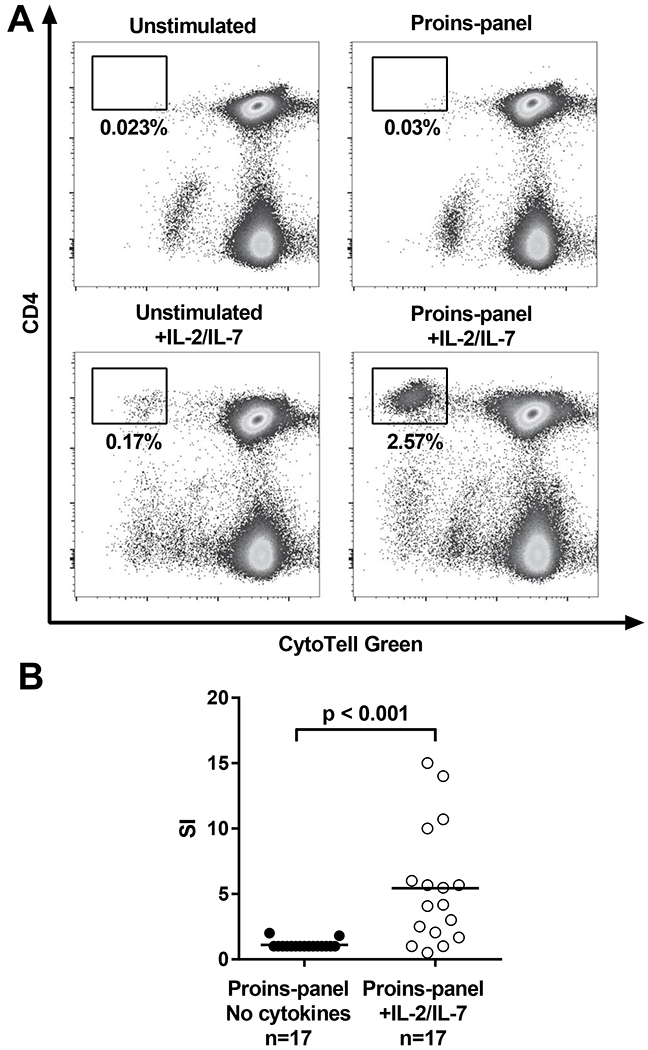
Detection of proinsulin-specific T cells in children carrying high-risk HLA class II haplotypes. PBMCs were stimulated with a panel of 25 overlapping peptides covering the sequence of proinsulin and proliferating CD4+ T cells were detected by flow cytometry. (A) Representative examples of proliferative responses to proinsulin peptides in the absence (top) or presence (bottom) of IL-2 and IL-7 in the culture. (B) Addition of IL-2 and IL-7 to the cell culture unmasked proliferative T-cell responses to proinsulin peptides. Results are expressed as stimulation indices (SI; percentage of divided cells in stimulated sample divided by the percentage in unstimulated sample). Mann-Whitney U-test was used in the statistical comparison.
To characterize the proinsulin-reactive CD4+ T cells in detail, we isolated proinsulin-specific T-cell clones by sorting single proliferating cells from proinsulin peptide-stimulated cultures (Fig. 1). We attempted to generate clones from 20 DR3-DQ2 homozygous, seven DR4-DQ8 homozygous and 53 DR3-DQ2/DR4-DQ8 heterozygous subjects. In total, 7,756 proliferating clones were screened for specificity. Excluding sister clones with similar TCRs, we established a total of 24 unique T-cell clones recognizing proinsulin peptides isolated from 17 different subjects.
Most proinsulin-specific T-cell clones recognize epitopes within the C-terminus of C-peptide
To determine the specificity of the T-cell clones, their proliferative response was tested first to the complete panel of proinsulin peptides, then to five proinsulin peptide pools and finally to individual peptides within the responsive pool (Fig. 1). The core amino acid sequences recognized by the T-cell clones were then determined using additional truncated versions of the proinsulin peptide recognized (Fig. 1). Reactivity to several separately synthesized peptide variants also excluded the possibility of the T-cell clones recognizing rare peptide synthesis contaminants (35, 36) instead of an epitope within the peptide sequence.
Thirteen of the 24 characterized clones (54%) recognized proinsulin epitopes located at the C-terminus of the C-peptide. Eleven of these responded to peptides spanning C21-35 (GSLQPLALEGSLQKR; clones 1-7, 11, 13, 15 and 22; Fig. 3, Supplemental Fig. 2 and Table I), with slightly differing core amino acid sequences. One clone (clone 23) recognized proinsulin peptides spanning C16-26 (GGPGAGSLQPL) and one clone (clone 20) peptides spanning C18-28 (PGAGSLQPLAL). All of the thirteen clones, excluding those that recognized a core amino acid sequence spanning also the two amino acids (C34-35) removed during C-peptide cleavage, responded to the complete cleaved proinsulin C-peptide fragment (C3-33; Fig. 3, Supplemental Fig. 2 and Table I).
Figure 3.

Epitope mapping results of representative proinsulin-specific T-cell clones. The clones were first stimulated with five proinsulin peptide pools (A), and then separately stimulated with the five peptides within the responsive pool (B). The core amino acid sequence recognized by the clones was determined by stimulating the clones with a panel of truncated versions of the responsive peptide (C). Reactivity of the clones to proinsulin protein was examined by using moDCs pulsed with recombinant human proinsulin as APCs. Results are expressed as mean CPM ± SEM of triplicate cultures, and are representative of two separate experiments (D). The HLA class II genotype of the subject from which the clone was isolated is indicated below the clone and subject numbers.
Table I.
HLA class II-restriction and core amino acid sequences recognized by the proinsulin-specific T-cell clones.
| Clone | Subject | HLA genotype | Core sequence | Position | Reactivity to | HLA-restriction | ||
|---|---|---|---|---|---|---|---|---|
| C-pep | B9-23 | Proinsulin | ||||||
| 1 | 001 | DR3/DR4 | PLALEGSLQK | C25-34 | No | * | No | DRB1*0404 |
| 2 | 001 | DR3/DR4 | PLALEGSLQ | C25-33 | Yes | * | No | DRB1*0404 |
| 3 | 001 | DR3/DR4 | PLALEGSLQK | C25-34 | Yes | * | No | DRB1*0404 |
| 4 | 002 | DR3/DR4 | GSLQPLALEGSL | C21-32 | Yes | * | No | DRB4*0101 |
| 5 | 003 | DR3/DR4 | PLALEGSLQ | C25-33 | Yes | * | No | DRB1*0404 |
| 6 | 003 | DR3/DR4 | PLALEGSLQKR | C25-35 | No | * | No | DRB1*0301 |
| 7 | 004 | DR3/DR4 | LQPLALEGSLQKR | C23-35 | No | * | No | DRB1*0401/0404 |
| 8 | 005 | DR3/DR3 | YLVCGERGFF | B16-25 | No | * | No | DR3 |
| 9 | 006 | DR3/DR3 | YLVCGERGFF | B16-25 | * | * | No | DR3 |
| 10 | 006 | DR3/DR3 | YLVCGERGFFYT | B16-27 | No | * | * | DR3 |
| 11 | 007 | DR3/DR3 | QPLALEGSLQKR | C24-35 | No | * | No | DRB1*0301 |
| 12 | 008 | DR3/DR4 | RGFFYTPKTRRE | B22-C3 | No | * | No | DRB1*0301 |
| 13 | 009 | DR3/DR4 | SLQPLALEGSL | C22-32 | Yes | * | No | DRB1*0401 |
| 14 | 010 | DR3/DR4 | YLVCGERGF | B16-24 | No | * | No | DR3 |
| 15 | 011 | DR3/DR4 | GSLQPLALEGSL | C21-32 | * | * | No | DR4 |
| 16 | 012 | DR3/DR4 | GIVEQCCTSICSLY | A1-14 | No | * | * | DR4 |
| 17 | 012 | DR3/DR4 | GIVEQCCTSICSLYQ | A1-15 | * | * | * | DR4 |
| 18 | 013 | DQ2/DQ8 | SHLVEALYLV | B9-18 | No | Yes | * | DQ2 |
| 19 | 014 | DQ2/DQ8 | VCGERGFFY | B18-26 | No | * | Yes | DQ2 |
| 20 | 005 | DQ2/DQ2 | PGAGSLQPLAL | C18-28 | Yes | * | * | DQ2 |
| 21 | 015 | DQ2/DQ2 | HLVEALYLVCG | B10-20 | * | Yes | * | DQ2 |
| 22 | 007 | DQ2/DQ2 | SLQPLALEGSLQ | C22-33 | Yes | * | Yes | DQ2 |
| 23 | 016 | DQ8/DQ8 | GGPGAGSLQPL | C16-26 | Yes | * | * | DQ8 |
| 24 | 017 | DQ2/DQ2 | ALYLVCGERG | B14-23 | No | Yes | * | DQ2 |
, not tested
Eight of the characterized clones recognized epitopes within the proinsulin B-chain. Six of these responded to peptides spanning B14-27 (ALYLVCGERGFFYT; clones 8-10, 14, 19 and 24; Fig. 3, Supplemental Fig. 2 and Table I) with slightly differing core amino acid sequences (Table I). Moreover, two clones responded to proinsulin peptides spanning B9-20 (SHLVEALYLVCG; clones 18 and 21; Fig. 3, Supplemental Fig. 2 and Table I). Of note, two of the three clones responding to proinsulin peptides spanning B9-23 also recognized the B9-23(B22E) variant peptide widely used in previous studies (25, 27, 28) (clones 18 and 21; Fig. 3, Supplemental Fig. 2 and Table I). In addition, we identified one clone that recognized an epitope located between the B-chain and C-peptide (B22-C3; RGFFYTPKTRRE; clone 12; Supplemental Fig. 2 and Table I).
Two of the isolated clones recognized an epitope located at the N-terminus of the A-chain. These clones responded to peptides spanning A1-15 (GIVEQCCTSICSLYQ; clones 16 and 17; Supplemental Fig. 2 and Table I).
Finally, the reactivity of the clones to whole proinsulin protein was tested using moDCs pulsed with recombinant proinsulin as APCs. Two clones that recognized peptide either in the B-chain (B18-26; clone 19) or C-peptide (C22-33; clone 22) responded to whole proinsulin (Figure 3D, Table I). Fourteen of the clones (clones 1-9, 11-15) did not respond to human proinsulin and the reactivity of eight clones could not be verified (Table I).
Proinsulin-specific T-cell clones are restricted by multiple HLA class II molecules encoded by the high-risk HLA haplotypes
Next, we characterized the HLA restriction of the proinsulin-specific T-cell clones (Fig. 1). The responses of 17 of the 24 clones (71%) were inhibited by anti-HLA-DR and the remaining seven (29%) by anti-HLA-DQ antibodies. Of the 17 HLA-DR-restricted clones, ten recognized epitopes within the C-terminal C-peptide epitope region (C21-35). Four of these clones were restricted by DRB1*04:04 (clones 1, 2, 3 and 5), one by DRB1*04:01 (clone 13), one by both DRB1*04:01 and 04:04 (clone 7), one by DRB4*01:01 (clone 4), and two by DRB1*03:01 (clones 6 and 11; Fig. 4, Table I and Supplemental Fig. 3). One clone reactive to C21-32 (clone 15) was restricted by HLA-DR4, although the exact restricting HLA class molecule (DRB1*04:04 or DRB4*01:01) could not be experimentally verified (Table I and Supplemental Fig. 3). Four clones recognizing epitopes in the B-chain (B16-27; clones 8-10 and 14) were restricted by HLA-DR3 (DRB1*03:01 or DRB3; Table I and Supplemental Fig. 3). One clone (clone 12) recognizing B22-C3 was restricted by DRB1*03:01 (Table I and Supplemental Fig. 3). The two clones responding to proinsulin peptides within the A-chain (A1-15; clones 16 and 17) were restricted by HLA-DR4 (Table I and Supplemental Fig. 3).
Figure 4.

HLA class II restriction results of four representative proinsulin-specific T-cell clones. The restriction of the clones was determined by inhibiting proliferative responses to cognate peptide by anti-HLA-DR, -DQ and -DP antibodies (A), and by using DR3-DQ2 and DR4-DQ8 homozygous EBV-transformed B cell lines (B) or BLS or RM3 cell lines transfected with single HLA class II molecules (C) as APCs. The HLA class II genotype of the subject from which the clone was isolated is indicated below the clone and subject numbers.
The DQ-restricted T-cell clones recognized epitopes either between B9-26 or C16-33. The four clones recognizing epitopes within B9-26 were all DQ2-restricted (clones 18, 19, 21 and 24; Fig. 4, Table I and Supplemental Fig. 3). Of the clones recognizing epitopes within C16-33, two were DQ2-restricted (clones 20 and 22) and one DQ8-restricted (clone 23; Fig. 4, Table I and Supplemental Fig. 3).
Next, the identified proinsulin T-cell epitopes were aligned with previously reported T-cell epitopes recognized by T cells isolated either from blood, pancreatic lymph nodes (pLN) or pancreatic islets of patients with T1D (Fig. 5). The DR4-restricted epitopes identified here appear to align with the previously identified C22-A3 and A1-15 regions (15, 17, 19, 21, 22, 24). However, based on our data there appear to be two distinct DR4-restricted minimal core sequences (C25-35 and C21-32) within the longer C22-A3 region. Moreover, based on our data, C25-35 appears to also contain a previously unidentified DR3-restricted epitope. The DR3-restricted B16-27 epitope identified here aligns with a previously reported epitope (B11-30) (29). The DQ2-restricted epitopes, as well as the DQ8-restricted epitope identified here appear to partially align with previously reported DQ2- and DQ8-restricted epitopes in the B-chain (26–28) and the C-terminus of the C-peptide (13, 14, 19).
Figure 5.
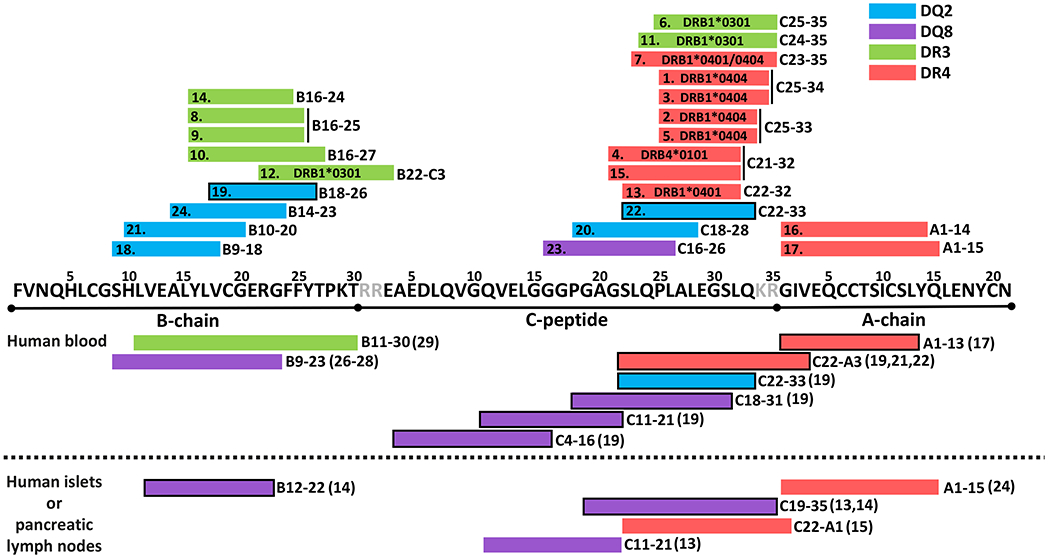
Summary of the epitope mapping results. HLA class II restriction and minimal core amino acids recognized by the clones analyzed in this study are aligned above the proinsulin sequence. Previously reported T-cell epitopes recognized by human T cells isolated from blood, pancreatic islets or pancreatic lymph nodes of patients with T1D are aligned below the proinsulin sequence. The original publications reporting these epitopes are indicated in parentheses after the minimal core sequence. Epitopes with a black frame have been demonstrated to be naturally processed from proinsulin protein.
Proinsulin C22-A1 binds to both DRB1*04:01 and DRB1*03:01 molecules whereas C19-33 binds to DRB4*01:01, DQ2 and DQ8 molecules
To verify our epitope mapping results, we measured HLA class II binding affinities of the proinsulin peptides that were recognized by the T-cell clones with a competitive HLA class II binding assay. In all, we identified two moderate- to high-binding peptides for T1D-associated HLA-DR molecules and five for HLA-DQ molecules (Table II).
Table II.
Relative binding affinity of the proinsulin peptides to HLA-DR and DQ molecules.
| Peptide | DR3 | DR4 | DRB4 | DQ2 | DQ8 |
|---|---|---|---|---|---|
| B8-22 | >1 250 | >3 333 | >1 000 | >50 | 1 |
| B11-25 | 1 250 | 333 | >1 000 | 15 | 1 |
| B14-28 | 1 000 | 2 667 | >1 000 | >50 | 61 |
| B17-C1 | >1 250 | >3 333 | >1 000 | >50 | >143 |
| B20-C4 | 6 | >3 333 | >1 000 | >50 | >143 |
| C10-24 | >1 250 | >3 333 | >1 000 | >50 | >143 |
| C13-27 | >1 250 | >3 333 | >1 000 | >50 | >143 |
| C16-30 | >1 250 | >3 333 | >1 000 | >50 | >143 |
| C19-33 | >1 250 | 1 764 | 141 | 15 | 49 |
| C22-A1 | 48 | 3 | >1 000 | >50 | >143 |
| C25-A4 | >1 250 | 153 | >1 000 | >50 | >143 |
| A1-15 | 106 | >3 333 | >1 000 | 5 | 9 |
Results are expressed as a relative binding ratio obtained by dividing the IC50 of the peptides by that of a reference peptide that binds strongly to the HLA molecule. Lower numbers correspond to a higher binding affinity. Bold: binding ratio below 100 (moderate binding) or below 10 (high binding). Each peptide-HLA combination was evaluated in two independent experiments. Peptides were tested at 10 different concentrations up to a maximal concentration of 100 000 nM. This maximal concentration was used to define the maximal relative binding ratio for each HLA molecule tested. The sequences and mean IC50 values of the reference peptides used were: MT2-16 (AKTIAYDEEARRGLE) for DRB1*03:01 (75 nM), HA306-318 (PKYVKQNTLKLAT) for DRB1*04:01 (30 nM), E2/E7 (AGDLLAIETDKATI) for DRB4*01:01 (63 nM), HCI46-63 (EPRAPWIEQEQGPEYWDQE) for DQ2 (DQA1*05:01/DQB1*02:01; 1400 nM) and DQB45-57 (ADVEVYRAVTPLGPPD) for DQ8 (DQA1*03:01/DQB1*03:02; 730 nM). DR3=DRB1*03:01, DR4=DRB1*04:01, DRB4=DRB4*01:01.
The peptide C22-A1 had a high binding affinity to DRB1*04:01 and a moderate binding affinity to DRB1*03:01, which correlated with the T-cell responses observed (clones 1-3, 5-7, 11 and 13; Table I and Fig. 5). Of note, this binding was abrogated in the peptide C19-33, implicating residues C34 and C35 as critical parts of the peptide-binding motif. In addition, B20-C4 had a high binding affinity to DRB1*03:01 (corresponding to clone 12). Low binding of C19-33 was observed to DRB4*01:01 (clones 4 and 15). Similarly, B11-25 and B14-28 exhibited a low binding affinity to DRB1*03:01 (clones 8-10 and 14), and A1-15 to DRB1*04:01 (clones 16 to 17)
A moderate binding affinity to DQ2 was observed for peptides B11-25 (clone 24) and C19-33 (clones 20 and 22). However, several peptides recognized by DQ-restricted T-cell clones did not show detectable binding affinity to DQ2 or DQ8. This was the case for peptides B8-22 (clones 18 and 21) as well as B14-28 and B17-C1 (clone 19) with DQ2-binding and peptides C13-27 and C16-30 (clone 23) with DQ8-binding. Of note, several peptides that exhibited a moderate to high binding affinity to DQ2 (peptide A1-15) and especially to DQ8 (B8-22, B11-25, B14-28, C19-33 and A1-15) were not recognized by the DQ2- or DQ8-restricted T-cell clones generated.
To analyze the functional TCR avidity of the T-cell clones, an EC50 value for each proinsulin-specific T-cell clone was determined by stimulating them with different concentrations of their cognate peptide. We observed that proinsulin-specific T-cell clones were in general of low functional avidity compared to two DRB1*0401-restricted reference clones recognizing influenza hemagglutinin HA306-318 (Fig. 6). Of note, this was observed also for DRB1*0401/0404-restricted clones recognizing peptide C22-A1, which displays an HLA-binding affinity comparable to HA306-318 (Table II). Proinsulin-specific T-cell clones restricted with HLA-DR molecules had a higher functional avidity compared to HLA-DQ-restricted clones (Fig. 6). Overall, however, no clear correlation with HLA-binding affinity and functional TCR avidity was observed, and several clones recognizing epitopes that bind poorly to HLA had comparable functional avidities to clones recognizing high-binding epitopes (Fig. 6).
Figure 6.
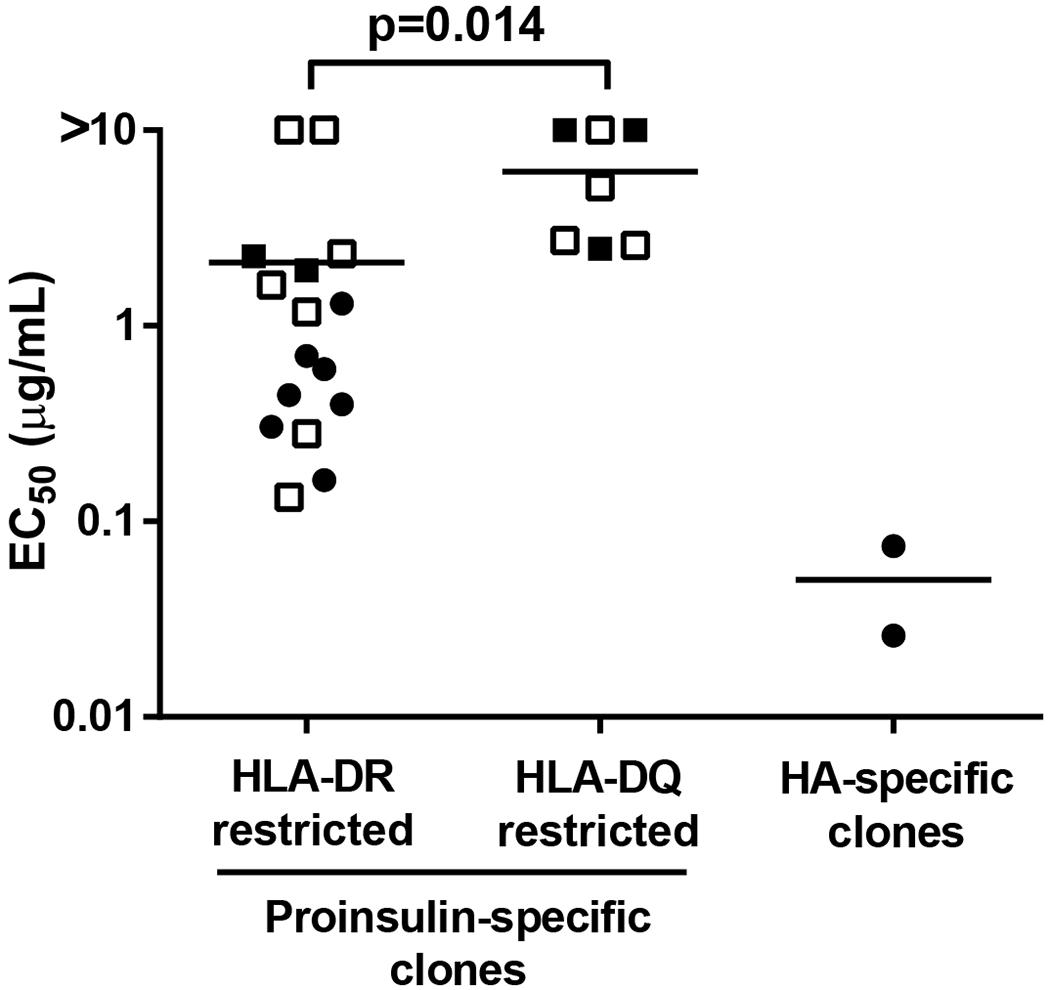
Functional avidities of the proinsulin-specific T-cell clones. EC50 values were determined from titrated dose-response curves for each T-cell clone. Two DRB1*0401-restricted clones generated from naïve CD4+ T cells of two different subjects recognizing the influenza hemagglutinin peptide HA306-318 were used as reference clones. T-cell clones recognizing peptides with a high, moderate or low HLA-binding affinity (Table II) are indicated as black circles, black squares or open squares, respectively. Mann-Whitney U-test was used in the statistical comparison.
TCR β-chain CDR3 sequences from proinsulin-specific T-cell clones are enriched in pLN compared to spleen
To determine the clonal diversity of the proinsulin-specific T-cell clones, their TCR genes were sequenced. The clones used a wide range of TRAV and TRBV genes, and most of the TRAV or TRBV genes were identified only in a single T-cell clone (Table III). Interestingly, the same TRBV gene, TRBV4-3, was used by four of the seven DQ-restricted clones (clones 18, 20, 22, 23). Moreover, three of the four T-cell clones recognizing the DR3-restricted epitope in B16-27 used the TRBV27 gene (clones 9, 10, 14). However, there were no similarities in TRAV gene usage or amino acid sequences of the TCR β-chain CDR3 among these clones. Finally, the TCR β-chain CDR3 amino acid sequences of the clones were compared in silico to polyclonal TCR repertoires compiled from CD4+ Tconv and CD8+ T cells as well as Treg cells isolated from pLN and spleen samples obtained from the nPOD (37). Interestingly, we observed a higher frequency of CDR3 sequences matching the proinsulin-specific TCR sequences in pLN than spleen samples from both controls and organ donors with T1D (Fig. 7A). When compared in enriched cell subsets, the clearest difference was observed in the sequences in the CD8+ T-cell compartment of patients with T1D (Fig. 7B).
Table III.
TCR sequences of the proinsulin-specific T-cell clones.
| Clone | TRBV | TRBJ | TRB CDR3 | TRB CDR3 length | TRAV | TRAJ | TRA CDR3 | TRA CDR3 length |
|---|---|---|---|---|---|---|---|---|
| 1 | TRBV14 | TRBJ2-7 | CASSQALAGVAEQYF | 15 | TRAV41 | TRAJ49 | CAASNTGNQFYF | 12 |
| 2 | TRBV6-1 | TRBJ1-2 | CASSMTQGLDYGYTF | 15 | TRAV13-1 | TRAJ17 | CAASSKAAGNKLTF | 14 |
| 3 | TRBV9 | TRBJ1-2 | CASRLSGTNIPPTYGYTF | 18 | TRAV19 | TRAJ49 | CALSDRTGNQFYF | 13 |
| 4 | TRBV4-1 | TRBJ2-7 | CASSRPPTSSSYEQYF | 16 | * | |||
| 5 | TRBV12-3/12-4 | TRBJ1-2 | CASSVQGLGYGYTF | 14 | * | |||
| 6 | TRBV4-1 | TRBJ2-7 | CASSPGQTSSYEQYF | 15 | * | |||
| 7 | TRBV6-2 | TRBJ1-6 | CASSFTQGSPLHF | 13 | TRAV20 | TRAJ37 | CAVAGSGNTGKLIF | 14 |
| 8 | TRBV6-5 | TRBJ1-2 | CASSYRGANGYTF | 13 | TRAV12-3 | TRAJ42 | WDYGGSQGNLIF | 12 |
| 9 | TRBV27 | TRBJ1-1 | CASSPRGYNTEAFF | 14 | TRAV26-2 | TRAJ54 | CILSLIIQGAQKLVF | 15 |
| 10 | TRBV27 | TRBJ1-5 | CASSFRGPASQPQHF | 15 | TRAV12-2 | TRAJ40 | CGTGSGTYKYIF | 12 |
| 11 | TRBV18 | TRBJ1-2 | CASSPDRQGVEYTF | 14 | TRAV13-1 | TRAJ47 | CAAMCKLVF | 9 |
| 12 | TRBV12-3 | TRBJ2-3 | CASSLGRDESTDTQYF | 16 | TRAV22 | TRAJ6 | CAVGGGGSYIPTF | 13 |
| 13 | TRBV12-3 | TRBJ2-1 | CASRQQGVGYNEQFF | 15 | TRAV17 | TRAJ11 | CATALISGYSTLTF | 14 |
| 14 | TRBV27 | TRBJ2-7 | CASRYRGAYEQYF | 13 | TRAV13-1 | TRAJ54 | CASLGIIQGAQKLVF | 15 |
| 15 | TRBV3-1 | TRBJ2-2 | CASSQLTGGVTGELFF | 16 | TRAV12-3 TRAV20 |
TRAJ40 TRAJ47 |
CATRTTSGTYKYIF CAVQVLMEYGNKLVF |
14 15 |
| 16 | TRBV20-1 | TRBJ2-2 | CSARDPLAGVRTGELFF | 17 | * | |||
| 17 | TRBV7-8 | TRBJ2-2 | CASSLYGGNTGELFF | 15 | TRAV8-2 | TRAJ4 | CVVSVAFSGGYNKLIF | 16 |
| 18 | TRBV4-3 | TRBJ1-6 | CASSQDQGGLGYNSPLHF | 18 | TRAV38-2 | TRAJ40 | CAYRFSGTYKYIF | 13 |
| 19 | TRBV7-2 | TRBJ1-1 | CASSFRRGSMNTEAFF | 16 | TRAV29 | TRAJ33 | CAASPSDSNYQLIW | 14 |
| 20 | TRBV4-3 | TRBJ2-7 | CASSQYYEQYF | 11 | * | |||
| 21 | TRBV20-1 | TRBJ2-2 | CSASRGDPTGELFF | 14 | TRAV4 | TRAJ18 | CLVPRGSTLGRLYF | 14 |
| 22 | TRBV4-3 | TRBJ2-3 | CASSQETGPTQYF | 13 | TRAV2 | TRAJ8 | CAVEDHGTGFQKLVF | 15 |
| 23 | TRBV4-3 | TRBJ2-3 | CASSKMGPRTDTQYF | 15 | TRAV12-3 | TRAJ33 | CAMSARSNYQLIW | 13 |
| 24 | TRBV5-8 | TRBJ2-5 | CASTEGIQETQYF | 13 | TRAV8-2 | TRAJ20 | CAVSEAYKLSF | 11 |
, sequence not detected
Figure 7.

In silico search for the proinsulin-specific TCR β-chain CDR3 sequences in the repertoire of CD8+, conventional CD4 + (Tconv) and regulatory CD4+ (Treg) T cells combined (A) and subsetted by cell type (B) obtained from nPOD pancreatic lymph node (pLN) and spleen samples from healthy controls and patients with T1D. Each CDR3 sequence was considered matched if the query sequence had a Hamming distance of 1 or less. Relative frequencies of each match were summed across all query proinsulin-specific sequences. Each point is one sample, and the x-axis is the source tissue in which the sequence was searched. The y-axis is the relative frequency of all matches for all proinsulin-specific sequences identified in the experiment. Mann-Whitney U-test together with Benjamini-Hochberg procedure to control for false discovery rate was used in the statistical comparison. *P<0.05, ***P<0.001
DISCUSSION
Despite the importance of proinsulin as an important target of T-cell autoimmunity in T1D, the T-cell epitopes recognized by autoreactive CD4+ T cells have remained only partially characterized. Here, we have addressed this question by analyzing the epitope-specificity and HLA restriction of a large collection of proinsulin-reactive T-cell clones from the peripheral blood of healthy, autoantibody-negative subjects carrying genetic risk for T1D.
Until recently (19), most proinsulin-specific human T-cell clones isolated from peripheral blood have mainly targeted DR4-restricted epitopes (10, 17–21). In line with these studies, ten of 24 clones characterized here also recognized DR4-restricted epitopes. Five of these clones responded to a DRB1*04:01/04-restricted epitope with a predicted binding motif between C26-34 (LALEGSLQK, IEDB analysis resource consensus tool (38, 39)), consistent with previous analyses (20–22). We also experimentally confirmed high binding to DRB1*04:01 with a peptide containing this motif (C22-A1). Importantly, DRB1*04:01-restricted T-cell responses against this epitope have consistently been observed against this epitope among T cells isolated from blood (20, 22) and pancreatic islets (15) of patients with T1D. Interestingly, three of the ten DR4-restricted clones characterized here recognized a slightly shifted minimal core, with a predicted binding motif for DRB1*04:01 and DRB4*01:01 between C23-31 (LQPLALEGS; IEDB). The corresponding peptide, C19-33, not containing the C26-34 motif, showed only weak binding to both DRB1*04:01 and DRB4*01:01. The final two DR4-restricted clones responded to an epitope within A1-15, which displayed low binding to DRB1*04:01. DRB1*04:01-restricted clones recognizing A1-15 have previously been isolated both from human blood (17) and pLN (24).
In contrast to DR4-restricted epitopes, DR3-restricted proinsulin epitopes have not been extensively characterized previously. To our knowledge, only one DR3-restricted clone recognizing epitope B11-30 has been reported earlier (29). Here, we identified and characterized seven unique T-cell clones recognizing DR3-restricted epitopes. Four of these clones recognized an epitope within B16-27, with a predicted DRB1*03:01 binding motif between B16-25 (YLVCGERGFF; IEDB). The corresponding peptide, B14-28 displayed low binding to DRB1*03:01, and the epitope matches the DRB1*03:01-restricted epitope previously reported (29). We also identified a novel DRB1*03:01-restricted epitope between B22-C3 (RGFFYTPKTRRE) that demonstrated high binding to DRB1*03:01. Finally, two clones recognized a DRB1*03:01-restricted epitope within C25-35, likely sharing the HLA-binding motif C26-34 (LALEGSLQK, IEDB) with DRB1*04:01/04.
We were able to establish one DQ8-restricted clone that recognized an epitope within C16-25. This epitope does not completely align with previously reported DQ8-restricted epitopes within C-peptide (13, 14, 19) and did not display detectable binding affinity to DQ8. Two of the DQ2-restricted clones characterized here recognized epitopes within C18-28 and C22-33. The latter epitope, which displayed moderate binding to DQ2 and was confirmed to be naturally processed, corresponds to the motif recognized by four distinct DQ2-restricted clones isolated by So et al. (19). Finally, we identified four DQ2-restricted clones recognizing slightly differing minimal cores between B9-26 with low to moderate binding to DQ2. Of these, the epitope within B18-26 was also confirmed to be naturally processed.
Isolation of DQ2- or DQ8-restricted proinsulin-specific T-cell clones has been more difficult than isolation of DR4-restricted clones from peripheral blood (10, 17, 18, 20, 21), despite the predominance of DQ8- (or DQ8trans-) restricted T-cell clones isolated from pancreatic islets (13, 14). Interestingly, most of the epitopes recognized by DQ-restricted T cells isolated from blood and pancreatic islets by us and others (13, 14, 19) do not display detectable binding to either DQ2 or DQ8. Consistent with this observation, several DQ-restricted clones isolated in the study by So et al. (19) appeared to prefer long flanking regions around the minimal binding motif, which may be necessary to stabilize the weak binding to DQ-molecules. In line with this, DQ8-restricted T-cell responses to B9-23 have also only been detected in peripheral blood by using a variant peptide, B9-23(R22E), in which one amino acid is artificially modified to improve binding to DQ8 (25, 27, 28). Together these observations support the idea that pathogenic DQ-restricted responses may be focused on epitopes with low HLA binding, which may have helped T cells recognizing these epitopes escape central tolerance. This notion is consistent with both murine (40, 41) and human studies (42), which suggest that low MHC-binding epitopes may be more important for T1D-autoimmunity than high-binding epitopes.
Identification of proinsulin epitopes is an essential prerequisite for the development of T-cell assays or immunotherapy for T1D. A proinsulin peptide C19-A3 from the C-terminal sequence of C-peptide has been successfully used both in ELISPOT assays (22, 43–45) as well as in one phase I immunotherapy trial (46). Based on our results, this long peptide contains epitopes recognized by different HLA class II molecules (DRB1*03:01, DRB1*04:01/04, DRB4*01:01, DQ2 and DQ8), each with slightly differing binding motifs. Hence, it can be argued that using two shorter peptides (C19-33 and C22-A1) instead of the long C19-A3 peptide could be beneficial for both immunoassays and peptide immunotherapy. The C19-33 peptide does not contain the dominant DRB1*03:01/*04:01 binding motif present in C22-A1, but instead contains DRB4*01:01, DQ2 and DQ8 binding motifs according to both our current data and the recent study by So et al (19). By using two distinct peptides, potential competition of these motifs with the high-binding DRB1*03:01/DRB1*04:01 binding motif would be avoided.
An obvious caveat of our study is that we isolated T-cell clones from healthy children genetically at-risk for T1D instead of patients with T1D. This was, however, necessary, since identifying a sufficient number of DR3-DQ2- or DR4-DQ8-homozygous, or DR3-DQ2/DR4-DQ8-DQ2-DQ8-heterozygous children with T1D within our cohort for cloning experiments would have been impossible. Hence, it is possible that the T-cell clones reported herein may have originated from the naïve T-cell pool and therefore, the potential relevance for these epitopes in T1D pathogenesis needs to be verified by additional studies. However, both murine (47) and human (48–50) studies suggest that T-cell precursor frequencies in the naïve repertoire largely dictate the hierarchy of memory T-cell responses, making it plausible that the epitopes identified here are significant contributors to the autoimmune response. In support of this, several of our epitopes match previously reported epitopes recognized by T cells isolated from blood, pLN or pancreatic islets of patients with T1D (Fig. 5) (10, 14, 15, 19–25). Another potential caveat of the current study is that majority of the T-cell clones analyzed here did not respond to stimulation with native proinsulin and we could experimentally verify only two DQ2-restricted epitopes to be naturally processed. However, many of the other epitopes characterized here have previously been shown to be naturally processed from native proinsulin (Fig. 5) (14, 19, 21, 22, 27). Moreover, whether natural processing is a strict requirement for the pathogenicity of proinsulin epitopes is currently unclear (51). For example, proinsulin B12-20 epitope, the main initiator of autoimmunity in the NOD mouse model, is not generated through natural antigen processing (52–54).
Finally, our in silico analysis of TCR sequences generated from nPOD tissue samples (37) suggested that TCR β-chain CDR3 sequences matching those expressed by our proinsulin T-cell clones were more readily observed in pLN samples compared to spleen samples, suggesting enrichment of T cells with autoreactive potential in the pLN samples. However, it is important to note that diversity as measured by Shannon evenness, a normalized diversity metric, was different in pLN and spleen datasets for all cell subsets (Supplemental Fig. 4), and due to limitations of the nPOD datasets only containing bulk β-chain sequencing data, this is all that we could query.
In summary, we were able to confirm previously reported DR4-restricted proinsulin epitopes and extend their characterization here. Importantly, we also identified novel DR3-, DQ2- and DQ8-restricted epitopes within proinsulin. Our current data have important implications for understanding human autoimmune T-cell responses to proinsulin, and can potentially be utilized in refining T-cell assays for immune monitoring as well as in the rational development of peptide-based immunotherapy for T1D.
Supplementary Material
KEY POINTS.
Proinsulin contains several T-cell epitopes recognized by human CD4+ T cells
HLA-DQ2-restricted proinsulin epitopes B18-26 and C22-33 are naturally processed
Proinsulin-reactive TCR sequences are enriched in pancreatic lymph node samples
Acknowledgements
The skillful technical assistance of Anne Suominen (University of Turku) as well as Virpi Fisk, Hanna Eskelinen and Mirja Saarelainen (University of Eastern Finland) is gratefully acknowledged
1) The study was supported by the Academy of Finland (Decision number 307320), the Sigrid Jusélius Foundation, State Research Funding (VTR) and the Finnish Diabetes Research Foundation. The DIPP study was supported by the Academy of Finland (Decision numbers 250114 and 286765), the Sigrid Jusélius Foundation and the JDRF. nPOD is supported by the JDRF, with cooperative mechanistic study support by The Helmsley Charitable Trust to T.M.B. The funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
2). Abbreviations used in the article:
- moDC
monocyte-derived dendritic cell
- nPOD
the Network for Pancreatic Organ donors with Diabetes
- pLN
pancreatic lymph nodes
- T1D
type 1 diabetes
REFERENCES
- 1.Bluestone JA, Herold K, and Eisenbarth G. 2010. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 464: 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnamurthy B, Selck C, Chee J, Jhala G, and Kay TWH. 2016. Analysis of antigen specific T cells in diabetes-Lessons from pre-clinical studies and early clinical trials. J. Autoimmun 71: 35–43. [DOI] [PubMed] [Google Scholar]
- 3.Roep BO, and Peakman M. 2010. Surrogate end points in the design of immunotherapy trials: Emerging lessons from type 1 diabetes. Nat. Rev. Immunol 10: 145–152. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliot JF, and Eisenbarth GS. 2005. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 435: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamurthy B, Dudek NL, McKenzie MD, Purcell AW, Brooks AG, Gellert S, Colman PG, Harrison LC, Lew AM, Thomas HE, and Kay TWH. 2006. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J. Clin. Invest 116: 3258–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilonen J, Hammais A, Laine AP, Lempainen J, Vaarala O, Veijola R, Simell O, and Knip M. 2013. Patterns of β-cell autoantibody appearance and genetic associations during the first years of life. Diabetes 62: 3636–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krischer JP, Lynch KF, Schatz DA, Ilonen J, Lernmark Å, Hagopian WA, Rewers MJ, She JX, Simell OG, Toppari J, Ziegler AG, Akolkar B, and Bonifacio E. 2015. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia 58: 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett ST, Lucassen A, Gough S, Powel E, Undlien D, Pritchard L, Merriman M, Kawaguchi Y, Dronsfield M, Pociot F, Nerup J, Bouzekri N, Cambon-Thomsen A, Ronningen K, Barnett A, Bain S, and Todd J. 1995. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. 9: 369–375. [DOI] [PubMed] [Google Scholar]
- 9.Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, and Polychronakos C. 1997. VNTR alleles at the IDDM2 locus. Nat. Genet 15: 289–292. [DOI] [PubMed] [Google Scholar]
- 10.Durinovic-Belló I, Wu RP, Gersuk VH, Sanda S, Shilling HG, and Nepom GT. 2010. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes Immun. 11: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermann R, Turpeinen H, Laine AP, Veijola R, Knip M, Simell O, Sipilä I, Åkerblom HK, and Ilonen J. 2003. HLA DR-DQ-encoded genetic determinants of childhood-onset type 1 diabetes in Finland: An analysis of 622 nuclear families. Tissue Antigens 62: 162–169. [DOI] [PubMed] [Google Scholar]
- 12.Erlich H, Valdes A, Noble J, Carlson J, Varney M, Concannon P, Mychaleckyj J, Todd J, Bonella P, Fear A, Lavant E, Louey A, and Moonsamy P. 2008. HLA DR-DQ Haplotypes and Genotypes and Type 1 Diabetes Risk. Diabetes 57: 1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathiraja V, Kuehlich JP, Campbell PD, Krishnamurthy B, Loudovaris T, Coates PTH, Brodnicki TC, O’Connell PJ, Kedzierska K, Rodda C, Bergman P, Hill E, Purcell AW, Dudek NL, Thomas HE, Kay TWH, and Mannering SI. 2015. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes 64: 172–182. [DOI] [PubMed] [Google Scholar]
- 14.Michels AW, Laundry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW, Jones KL, Gottlieb PA, Kappler JW, Tang Q, Roep BO, Atkinson MA, Mathews CE, and Nakayama M. 2017. Islet-Derived CD4 T Cells Targeting Proinsulin in Human Autoimmune Diabetes. Diabetes 66: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babon JAB, Denicola ME, Blodgett DM, Crèvecoeur I, Buttrick TS, Maehr R, Bottino R, Naji A, Kaddis J, Elyaman W, James EA, Haliyur R, Brissova M, Overbergh L, Mathieu C, Delong T, Haskins K, Pugliese A, Campbell-Thompson M, Mathews C, Atkinson MA, Powers AC, Harlan DM, and Kent SC. 2016. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med 22: 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nepom BS, Schwarz D, Palmer JP, and Nepom GT. 1987. Transcomplementation of HLA genes in IDDM. HLA-DQ α- and β-chains produce hybrid molecules in DR3/4 heterozygotes. Diabetes 36: 114–117. [DOI] [PubMed] [Google Scholar]
- 17.Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, Kay TWH, Rossjohn J, Falk BA, Nepom GT, and Purcell AW. 2005. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J. Exp. Med 202: 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannering SI, Pang SH, Williamson NA, Naselli G, Reynolds EC, O’Brien-Simpson NM, Purcell AW, and Harrison LC. 2009. The A-chain of insulin is a hot-spot for CD4+T cell epitopes in human type 1 diabetes. Clin. Exp. Immunol 156: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So M, Elso CM, Tresoldi E, Pakusch M, Pathiraja V, Wentworth JM, Harrison LC, Krishnamurthy B, Thomas HE, Rodda C, Cameron FJ, McMahon J, Kay TWH, and Mannering SI. 2018. Proinsulin C-peptide is an autoantigen in people with type 1 diabetes. Proc. Natl. Acad. Sci 115: 10732–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Danke N, Roti M, Huston L, Greenbaum C, Pihoker C, James E, and Kwok WW. 2008. CD4+ T cells from type 1 diabetic and healthy subjects exhibit different thresholds of activation to a naturally processed proinsulin epitope. J. Autoimmun 31: 30–41. [DOI] [PubMed] [Google Scholar]
- 21.Durinovic-Belló I, Rosinger S, Olson JA, Congia M, Ahmad RC, Rickert M, Hampl J, Kalbacher H, Drijfhout JW, Mellins ED, Al S, Kamradt T, Maeurer MJ, Nhan C, Roep BO, Boehm BO, Polychronakos C, Nepom GT, Karges W, Mcdevitt HO, and Sønderstrup G. 2006. DRB1 * 0401-restricted human T cell clone specific for the major proinsulin 73-90 epitope expresses a down-regulatory T helper 2 phenotype. Proc. Natl. Acad. Sci. U. S. A 103: 11683–11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, Roep BO, and Peakman M. 2004. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J. Clin. Invest 113: 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Congia M, Patel S, Cope AP, De Virgiliis S, and Sonderstrup G. 1998. T cell epitopes of insulin defined in HLA-DR4 transgenic mice are derived from preproinsulin and proinsulin. Proc. Natl. Acad. Sci 95: 3833–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, and Hafler DA. 2005. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435: 224–228. [DOI] [PubMed] [Google Scholar]
- 25.Spanier JA, Sahli NL, Wilson JC, Martinov T, Dileepan T, Burrack AL, Finger EB, Blazar BR, Michels AW, Moran A, Jenkins MK, and Fife BT. 2017. Increased effector memory insulin-specific CD4+ T cells correlate with insulin autoantibodies in patients with recent-onset type 1 diabetes. Diabetes 66: 3051–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alleva DG, Crowe PD, Jin L, Kwok WW, Ling N, Gottschalk M, Conlon PJ, Gottlieb PA, Putnam AL, and Gaur A. 2001. A disease-associated cellular immune response in type 1 diabetics to an immunodominant epitope of insulin. J. Clin. Invest 107: 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Chow I-T, Sosinowski T, Torres-Chinn N, Greenbaum CJ, James EA, Kappler JW, Davidson HW, and Kwok WW. 2014. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc. Natl. Acad. Sci 111: 14840–14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama M, McDaniel K, Fitzgerald-Miller L, Kiekhaefer C, Snell-Bergeon JK, Davidson HW, Rewers M, Yu L, Gottlieb P, Kappler JW, and Michels A. 2015. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc. Natl. Acad. Sci 112: 4429–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tree TIM, Lawson J, Edwards H, Skowera A, Arif S, Roep BO, and Peakman M. 2010. Naturally arising human CD4 T-cells that recognize islet autoantigens and secrete interleukin-10 regulate proinflammatory T-cell responses via linked suppression. Diabetes 59: 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parviainen S, Kinnunen T, Rytkönen-Nissinen M, Nieminen A, Liukko A, and Virtanen T. 2013. Mammal-derived respiratory lipocalin allergens do not exhibit dendritic cell-activating capacity. Scand. J. Immunol 77: 171–176. [DOI] [PubMed] [Google Scholar]
- 31.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CSL, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, Wood R, Alam R, Peters B, Sidney J, and Sette A. 2013. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics 65: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han A, Glanville J, Hansmann L, and Davis MM. 2014. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol 32: 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Texier C, Pouvelle S, Busson M, Herve M, Charron D, Menez A, and Maillere B. 2000. HLA-DR Restricted Peptide Candidates for Bee Venom Immunotherapy. J. Immunol 164: 3177–3184. [DOI] [PubMed] [Google Scholar]
- 34.Pancré V, Georges B, Angyalosi G, Castelli F, Delanoye A, Delacre M, Hachulla E, Maillere B, Bouzidi A, and Auriault C. 2002. Novel promiscuous HLA-DQ HIV Nef peptide that induces IFN-γ-producing memory CD4+ T cells. Clin. Exp. Immunol 129: 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannering SI, Purcell AW, Honeyman MC, McCluskey J, and Harrison LC. 2003. Human T-cells recognise N-terminally Fmoc-modified peptide. Vaccine 21: 3638–3646. [DOI] [PubMed] [Google Scholar]
- 36.Brezar V, Culina S, Østerbye T, Guillonneau F, Chiappetta G, Verdier Y, Vinh J, Wong FS, Buus S, and Mallone R. 2011. T cells recognizing a peptide contaminant undetectable by mass spectrometry. PLoS One 6: e28866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seay HR, Yusko E, Rothweiler SJ, Zhang L, Posgai AL, Campbell-Thompson M, Vignali M, Emerson RO, Kaddis JS, Ko D, Nakayama M, Smith MJ, Cambier JC, Pugliese A, Atkinson MA, Robins HS, and Brusko TM. 2016. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight 1: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P, Sidney J, Dow C, Mothé B, Sette A, and Peters B. 2008. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol 4: e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, and Peters B. 2010. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11: 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levisetti MG, Suri A, Petzold SJ, and Unanue ER. 2007. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J. Immunol 178: 6051–6057. [DOI] [PubMed] [Google Scholar]
- 41.Levisetti MG, Lewis DM, Suri A, and Unanue ER. 2008. Weak proinsulin peptide-major histocompatibility complexes are targeted in autoimmune diabetes in mice. Diabetes 57: 1852–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu JRF, Martina S, Verrijn Stuart AA, Fillié YE, Franken KLMC, Drijfhout JW, and Roep BO. 2012. CD8 T cell autoreactivity to preproinsulin epitopes with very low human leucocyte antigen class I binding affinity. Clin. Exp. Immunol 170: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arif S, Moore F, Marks K, Bouckenooghe T, Dayan CM, Planas R, Vives-Pi M, Powrie J, Tree T, Marchetti P, Huang GC, Gurzov EN, Pujol-Borrell R, Eizirik DL, and Peakman M. 2011. Peripheral and Islet Interleukin-17 Pathway Activation Characterizes Human Autoimmune Diabetes and Promotes Cytokine-Mediated β-Cell Death. Diabetes 60: 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arif S, Leete P, Nguyen V, Marks K, Nor NM, Estorninho M, Kronenberg-Versteeg D, Bingley PJ, Todd JA, Guy C, Dunger DB, Powrie J, Willcox A, Foulis AK, Richardson SJ, de Rinaldis E, Morgan NG, Lorenc A, and Peakman M. 2014. Blood and islet phenotypes indicate immunological heterogeneity in type 1 diabetes. Diabetes 63: 3835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arif S, Gibson VB, Nguyen V, Bingley PJ, Todd JA, Guy C, Dunger DB, Dayan CM, Powrie J, Lorenc A, and Peakman M. 2017. β-cell specific T-lymphocyte response has a distinct inflammatory phenotype in children with Type 1 diabetes compared with adults. Diabet. Med 34: 419–425. [DOI] [PubMed] [Google Scholar]
- 46.Ali MA, Liu Y-F, Arif S, Tatovic D, Shariff H, Gibson VB, Yusuf N, Baptista R, Richmann M, Petrov N, Heck S, Yang JHM, Tree TIM, Pujol-Autonell I, Yeo L, Baumard LR, Stenson R, Howell A, Clark A, Boult Z, Powrie J, Adams L, Wong FS, Luzio S, Dunseath G, Green K, ÓKeefe A, Bayly G, Thorogood N, Andrews R, Leech N, Joseph F, Nair S, Seal S, Cheung H, Beam C, Hills R, Peakman M, and Dayan CM. 2017. Metabolic and immune effects of immunotherapy with proinsulin peptide in human new-onset type 1 diabetes. Sci. Transl. Med 9: eaaf7779. [DOI] [PubMed] [Google Scholar]
- 47.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, and Jenkins MK. 2007. Naive CD4+ T Cell Frequency Varies for Different Epitopes and Predicts Repertoire Diversity and Response Magnitude. Immunity 27: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castelli FA, Szely N, Olivain A, Casartelli N, Grygar C, Schneider A, Besse A, Levy Y, Schwartz O, and Maillere B. 2013. Hierarchy of CD4 T Cell Epitopes of the ANRS Lipo5 Synthetic Vaccine Relies on the Frequencies of Pre-Existing Peptide-Specific T Cells in Healthy Donors. J. Immunol 190: 5757–5763. [DOI] [PubMed] [Google Scholar]
- 49.Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, Yang J, James EA, and DeLong JH. 2012. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J. Immunol 188: 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uchtenhagen H, Rims C, Blahnik G, Chow I-T, Kwok WW, Buckner JH, and James EA. 2016. Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat. Commun 7: 12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unanue ER, and Wan X. 2019. The immunoreactive platform of the pancreatic islets influences the development of autoreactivity. Diabetes 68: 1544–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohan JF, Levisetti MG, Calderon B, Herzog JW, Petzold SJ, and Unanue ER. 2010. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol 11: 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohan JF, Petzold SJ, and Unanue ER. 2011. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J. Exp. Med 208: 2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan X, Zinselmeyer BH, Zakharov PN, Vomund AN, Taniguchi R, Santambrogio L, Anderson MS, Lichti CF, and Unanue ER. 2018. Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature 560: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


