Abstract
Objective:
This study was undertaken to identify shared functional network characteristics among focal epilepsies of different etiologies, to distinguish epilepsy patients from controls, and to lateralize seizure focus using functional connectivity (FC) measures derived from resting state functional magnetic resonance imaging (MRI).
Methods:
Data were taken from 103 adult and 65 pediatric focal epilepsy patients (with or without lesion on MRI) and 109 controls across four epilepsy centers. We used three whole-brain FC measures: parcelwise connectivity matrix, mean FC, and degree of FC. We trained support vector machine models with fivefold cross-validation (1) to distinguish patients from controls and (2) to lateralize the hemisphere of seizure onset in patients. We reported the regions and connections with the highest importance from each model as the common FC differences between the compared groups.
Results:
FC measures related to the default mode and limbic networks had higher importance relative to other networks for distinguishing epilepsy patients from controls. In lateralization models, regions related to somatosensory, visual, default mode, and basal ganglia showed higher importance. The epilepsy versus control classification model trained using a 400-parcel connectivity matrix achieved a median testing accuracy of 75.6% (median area under the curve [AUC] = .83) in repeated independent testing. Lateralization accuracy using the 400-parcel connectivity matrix reached a median accuracy of 64.0% (median AUC = .69).
Significance:
Machine learning models revealed common FC alterations in a heterogeneous group of patients with focal epilepsies. The distribution of the most altered regions supports the hypothesis that shared functional alteration exists beyond the seizure onset zone and its epileptic network. We showed that FC measures can distinguish patients from controls, and further lateralize focal epilepsies. Future studies are needed to confirm these findings by using larger numbers of epilepsy patients.
Keywords: functional MRI, lateralization, support vector machine, temporal lobe epilepsy
1 |. INTRODUCTION
Pharmacoresistant epilepsy results in the highest mortality, morbidity, and health care resource utilization in people with epilepsy.1,2 Intracranial electroencephalography (EEG)3 and combined EEG–functional magnetic resonance imaging (fMRI)4 studies provide evidence for extensive distribution of functional networks from the regions associated with seizure generation. Focal epilepsies are hence increasingly recognized as network disorders, and imaging and neurophysiologic methods to investigate large-scale network properties, paired with advanced analytic methods, are employed to study epilepsy.3,5,6,7 The role of fMRI in evaluation for surgical treatment of epilepsy by investigating epileptic network properties is not established. This role is relevant to the surgical workup for both lesional and nonlesional focal epilepsies, where an epileptic lesion is not visualized on standard clinical imaging.
Functional connectivity (FC) analysis provides a reliable method for identifying brain networks based on the correlation of low-frequency fMRI signal fluctuation between different brain regions.6 The epileptic networks associated with both generalized8 and focal9– 11 epilepsies can be characterized using FC. Focal epilepsies demonstrate FC alterations beyond apparent structural findings such as in the default mode network (DMN)12,13 and thalami,14,15 among other regions. Although group analysis of homogeneous focal epilepsy patients supports the use of FC for seizure focus lateralization,16 patient-to-patient variability limits the utility of FC analysis for this purpose at the individual level. Machine learning approaches are increasingly used to find subtle alterations in brain networks and to overcome the statistical limitations of conventional FC analysis.7,17
In this study, we first employed a machine learning approach based on individual FC characteristics to identify the most important features that distinguish epilepsy patients from healthy controls. We also trained machine learning models to identify features that distinguish patients with left versus right hemisphere of seizure onset (i.e., lateralization). We used a multicenter pediatric and adult dataset including lesional and nonlesional focal epilepsy patients as well as controls. We based our approach on the hypotheses that shared FC changes exist across focal epilepsies, and that patients with the same hemisphere of seizure onset share lateralizing FC features. The novelty of our approach lies in its inclusion of heterogeneous data to identify common FC attributes among different epilepsy localizations and pathologies.
2 |. MATERIALS AND METHODS
2.1 |. Participants and datasets
We included clinical and fMRI data from four epilepsy databases in a retrospective, sequential, and nonrandomized manner (Table S1). We included all available resting state fMRI studies from adult and pediatric patients with or without identified lesions obtained in 3-T MRI. An unmatched control group included healthy volunteers with no neurological or psychiatric history, and a confirmed normal MRI, previously collected as part of other fMRI studies. The diagnosis of pharmacoresistant focal epilepsy and the laterality of seizure onset were determined by each center based on their multidisciplinary clinical epilepsy surgery evaluation. Demographic, clinical, and imaging characteristics including EEG, epilepsy protocol structural MRI, and if performed, invasive EEG studies were documented in databases by the treating team at each center. We did not limit the study to those who subsequently underwent surgery; however, patients with a history of prior intracranial resective or ablative surgery were excluded. An epileptologist (T.G.) reviewed the clinical information in databases to confirm the eligibility. Using these criteria, 168 epilepsy patients (103 adults, >18 years old at the time of fMRI scan) were included. Of those, 110 patients had an identifiable lesion on their clinical MRI; 109 controls (92 adults) were included in the primary analysis; 58 patients and 107 control participants contributed to the dataset with two runs of resting state fMRI (Table 1). Those labeled as unclear laterality or bilateral foci by the treating physicians were excluded from the second part of the analysis. Imaging databases and current anonymous analysis of subject data were separately approved by institutional review boards at each institution. Anonymized preprocessed data will be shared upon request from any qualified investigator.
TABLE 1.
Demographic and clinical characteristics of participants
| Patients | Adult patients | Pediatric patients |
|---|---|---|
| n (female:male) | 103 (45:58) | 65 (34:31) |
| Median age, years (range) | 39 (19– 68) | 13 (5– 18) |
| Seizure focus laterality (left:right) | 49:53a | 41:20a |
| Seizure focus lobe, temporal/frontal/other | 87/8/8 | 32/11/22 |
| Abnormal clinical MRI, n (%)b | 56 (54.4%) | 54 (83.1%) |
| MTS/MTS+ | 42 | 12 |
| MCD/FCD | 5 | 18 |
| Neoplasm and cystic lesions | 5 | 11 |
| Vascular/encephalomalacia | 4 | 13 |
| Controls | Adult controls | Pediatric controls |
| n (female:male) | 92 (44:48) | 17 (7:10) |
| Mean age, years (range) | 37.5 (19– 71) | 11 (5– 18) |
Note: A total of 277 participants contributed to 442 acquisition runs.
Abbreviations: FCD, focal cortical dysplasia; MCD, malformations of cortical development; MRI, magnetic resonance imaging; MTS, mesial temporal sclerosis.
One adult patient and four pediatric patients had uncertain laterality and were excluded from secondary analysis.
MTS/MTS+: MTS and MTS plus another relevant lesion, such as FCD; MCD/FCD: MCD including mostly FCD but also polymicrogyria, nodular heterotopias, tuberous sclerosis, and hemimegalencephaly. Neoplasm and cystic lesions: low-g rade tumors, cystic calcified or noncalcified lesions such as infection sequelae deemed relevant to the patient’s diagnosis of epilepsy. Vascular/encephalomalacia: stroke and perinatal ischemic lesions, cavernous malformations, arteriovenous malformations deemed relevant to patient’s diagnosis of epilepsy.
2.2 |. Image processing
Resting state fMRI sequences were acquired with site-specific parameters on clinical 3-T MRI scanners (Table S1). An isometric T1-weighted anatomical MRI obtained on the same session was used for preprocessing of images using fMRIPrep18 (v20.2), a pipeline that uses a combination of tools from validated software packages to provide the best software implementation for each stage of preprocessing for high reproducibility. The anatomical preprocessing included intensity correction, brain extraction, segmentation, and volume-based spatial normalization to a standard brain image (ICBM152 Nonlinear Asymmetrical template v2009c, labeled as MNI152NLin2009cAsym)19 through nonlinear registration. Functional images were coregistered to the anatomical image with nine degrees of freedom, followed by head motion parameters estimation, slice-timing correction, and resampling into the standard space. The average values for root mean square frame-wise displacement (RMS) as the head motion estimate were compared, and were not different between the two groups when averaged across epilepsy patients (.26 mm, SD = .22) and controls (.22 mm, SD = .26, t- test p > .05). Similarly, RMS difference did not reach significant levels in left (.24 mm, SD = .25) and right (.28 mm, SD = .19, t- test p > .05) hemisphere onset patients. Please refer to the workflow section in fMRIPrep documentation for more details on the preprocessing pipeline.18
2.3 |. FC analysis
We used xcpEngine (v1.2.1)20 to denoise and extract residualized whole brain FC time series of cortical parcels from the preprocessed images. Cortical parcels were defined according to the validated functional parcellation atlases by Schaefer et al.21 dividing cerebral cortex into 400 parcels. Three types of features were calculated from the denoised time series for each subject in MATLAB (R2020a). Connectivity matrix is an adjacency matrix of Fisher–z-transformed Pearson correlation between each pair of 400-parcel fMRI time series. An individualized threshold to select the top 10% correlation values was applied to the matrix to increase the reliability of FC within and between subjects.22 Mean FC for each parcel, defined as the average of its nonzero correlation values, and degree of FC for each parcel were also calculated. The degree of FC is a graph theory measure defined as the number of connections (i.e., nonzero value in the top 10% matrix) divided by the total number of possible connections for each parcel.23 Cortical parcels in the Schaefer atlases are affiliated with one of the seven large- scale functional brain networks: visual, somatomotor, dorsal attention, ventral attention (salience), limbic, frontoparietal control, and DMN.21,24 Subcortical structures are not part of this parcellation scheme. To have full brain coverage, we added data from 14 subcortical regions with mean FC or degree of FC calculated in a similar fashion in the Harvard–Oxford atlas25: bilateral hippocampus, amygdala, thalamus, caudate, putamen, pallidum, and nucleus accumbens. This yielded 414 features for training models with mean FC or degree of FC measures. The connectivity matrix used cortical networks and for interpretation purposes, did not include the subcortical regions.
2.4 |. Classification models
Before building the classification model, we used neuroCombat,26 a data harmonization method, to adjust for site- or scanner-related biases, and to control for participant age and sex. To avoid bias in our independent testing process, harmonization was applied separately for each training and testing subset. We used linear support vector machine (SVM) binary classifier models from the scikit-learn and LIBSVM Python libraries.27 Three separate SVM models were trained using the three selected features: (1) mean FC, (2) degree of FC, and (3) connectivity matrix. We optimized the SVMs with a fivefold cross-validated grid search for the best C hyperparameter during the training step for all analyses.
Each model’s performance was reported in terms of its accuracy and the area under the curve (AUC) of the receiver operating characteristic curve analysis on the unseen testing data. We also explored the distribution of regions with the highest contribution to the classification model by ranking the features based on their weight (derived from the absolute value of their classification coefficients in each model), as well as their affiliation to each of the seven networks. For the mean and degree of FC models, we explored the network affiliation of 20 features with the highest importance. For the connectivity matrix SVM, the weight of each connection was tabulated, and each of the 400 connections with the highest weight features (near .5% of the 400 × 400 matrix connections) was reported along with the network affiliation of the pair of parcels it connects. Connections were considered “within network” if they connected two parcels affiliated with the same network (for example different parts of DMN) or were otherwise considered “between networks.”
In primary analysis (epilepsy vs. control), the participant list was divided equally in a stratified fashion into training and testing subsets; data from each participant contributed only to either the training or testing subset: 84 patients and 55 controls in training, 84 patients and 54 controls in testing. Next, to assess generalizability of the best performing model, we repeated the analysis 50 times by reiterating the stratified splitting of the dataset into 50% training and 50% testing followed by harmonization, and fivefold cross-validation with grid search optimization for training in each reiteration. We reported the median and range of accuracy and AUC in repeated independent testing.
Given the relatively high number of temporal lobe epilepsy (TLE) patients, we compared the likelihood of being misclassified as a control between TLE and non-TLE patients using chi-squared test. We repeated the epilepsy versus control analysis within a number of subgroups of patients. We used the values from the most accurate models for this purpose and reported the testing AUC as the model performance measure, which is a better measure than accuracy when there is imbalance in patient to control proportions.28 We repeated the primary analysis as TLE versus control after exclusion of non-TLE patients. In a similar approach, a subanalysis was performed limiting the participants to adult patients and controls (age 18 years or older at the time of the scan), and lesional and nonlesional patients.
In secondary analysis (i.e., lateralization), to identify the common important features leading to laterality classification, we included all patients in the training step, without a separate testing subset. The features were ranked based on their importance and reported similarly to the primary analysis. Next, we assessed the generalizability of the best performing model similarly to the primary analysis, by splitting the patient data into half, using fivefold cross-validation for training, and reporting the median and range of accuracy and AUC.
3 |. RESULTS
3.1 |. Primary analysis: Epilepsy versus control classification
From the mean FC model, parcels affiliated with DMN followed by dorsal attention and frontoparietal control networks were the most represented among the 20 most important features, along with hippocampus and thalamus from the subcortical regions. From the degree of FC model, the limbic network followed by DMN had the most representation in the top 20 features (Figure 1A,B and Table 2). All three SVM models achieved high classification accuracies during training. The mean FC and the degree of FC models achieved classification accuracies of 68% and 75%, respectively, and AUC of .74 and .80 (Figure 2A,B).
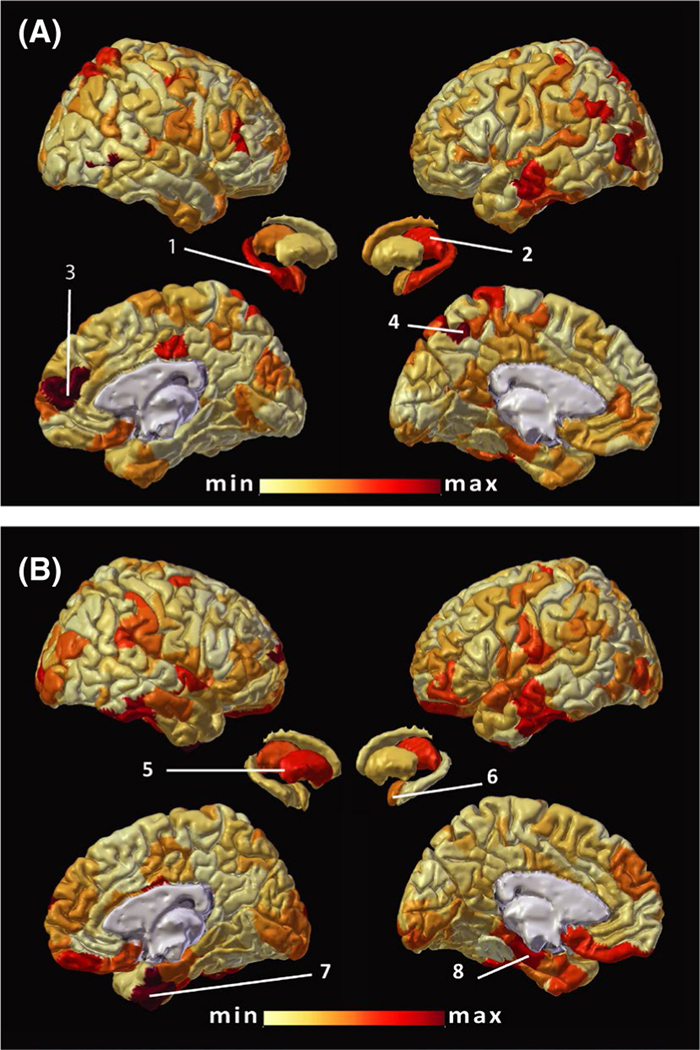
FIGURE 1.
Distribution of features with the highest importance for epilepsy versus control analysis. The absolute classification weight values for (A) mean functional connectivity (FC) and (B) degree of FC models are illustrated on a scale that ranges from the minimum (min; lightest) to maximum (max; darkest) values projected to the standard brain space using MMVT tools.56 A number of hemispheric parcels and subcortical regions are highlighted based on the Schaefer 400-parcel cortical atlas21 and Harvard–Oxford25 subcortical region naming for reference: right hippocampus (1), left thalamus (2), members of the default mode network in prefrontal region (3, RH_Default_PFCdPFCm_4), control network in precuneus (4, LH_Cont_pCun_2), right putamen (5), and left amygdala (6), as well as members of the limbic networks in both hemispheres (7, RH_Limbic_TempPole_1; 8, LH_Limbic_TempPole_8)
TABLE 2.
Network affiliations of the features with the highest importance
| Mean FC |
Degree of FC |
||||
|---|---|---|---|---|---|
| Network name | Count | Average weight | Network name | Count | Average weight |
| Default mode | 5 | 1.10 | Limbic | 9 | .11 |
| Dorsal attention | 4 | 1.15 | Default mode | 3 | .10 |
| Somatomotor | 3 | 1.18 | Somatomotor | 3 | .08 |
| Frontoparietal control | 3 | 1.05 | Frontoparietal control | 2 | .10 |
| Subcortical (hippocampus, thalamus) | 2 | 1.07 | Visual | 1 | .10 |
| Visual | 2 | .97 | Subcortical (putamen) | 1 | .09 |
| Limbic | 1 | .95 | Salience/ventral attention | 1 | .08 |
Note: The network distribution of the 20 features with the highest importance for epilepsy versus control classification for mean FC and degree of FC models. Count and average weight of the selected features per seven-network affiliation24 or subcortical parts are listed. Model classification weight is measured by an arbitrary unit to rank within each model’s results, but cannot be compared between models and datasets. Seven brain network affiliation is based on the Schaefer 400-parcel atlas,21 with the addition of subcortical regions from the Harvard–Oxford atlas.25
Abbreviation: FC, functional connectivity.
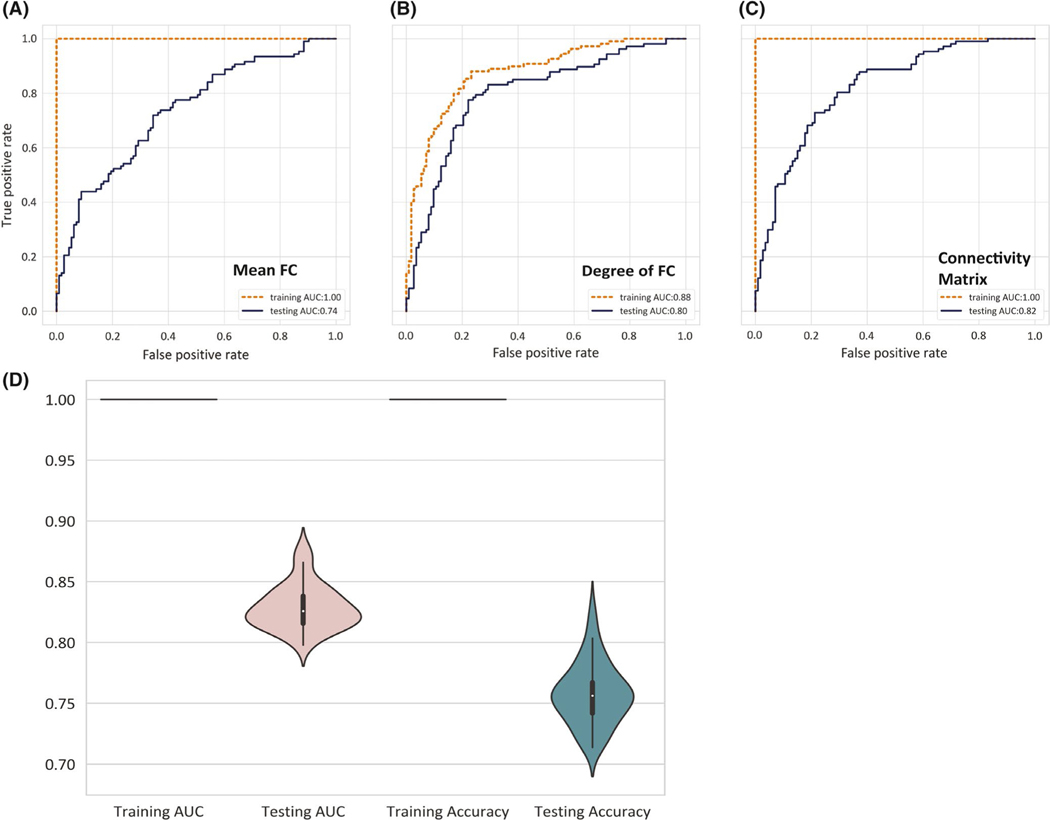
FIGURE 2.
Classification performance for epilepsy versus control analysis. Receiver operating characteristic curve analysis for epilepsy versus control classification compares (A) mean functional connectivity (FC), (B) degree of FC, and (C) connectivity matrix. Comparison of training and testing sets are illustrated by color lines (Orange: testing, Navy blue: training). Models in A and B include features from the Schaefer 400-parcel cortical atlas and 14 subcortical regions from the Harvard–Oxford atlas,25 whereas C uses only the connectivity matrix between Schaefer cortical atlases. (D) Violin plot represents the distribution of training and testing accuracies and area under the curve in 50 reiterations
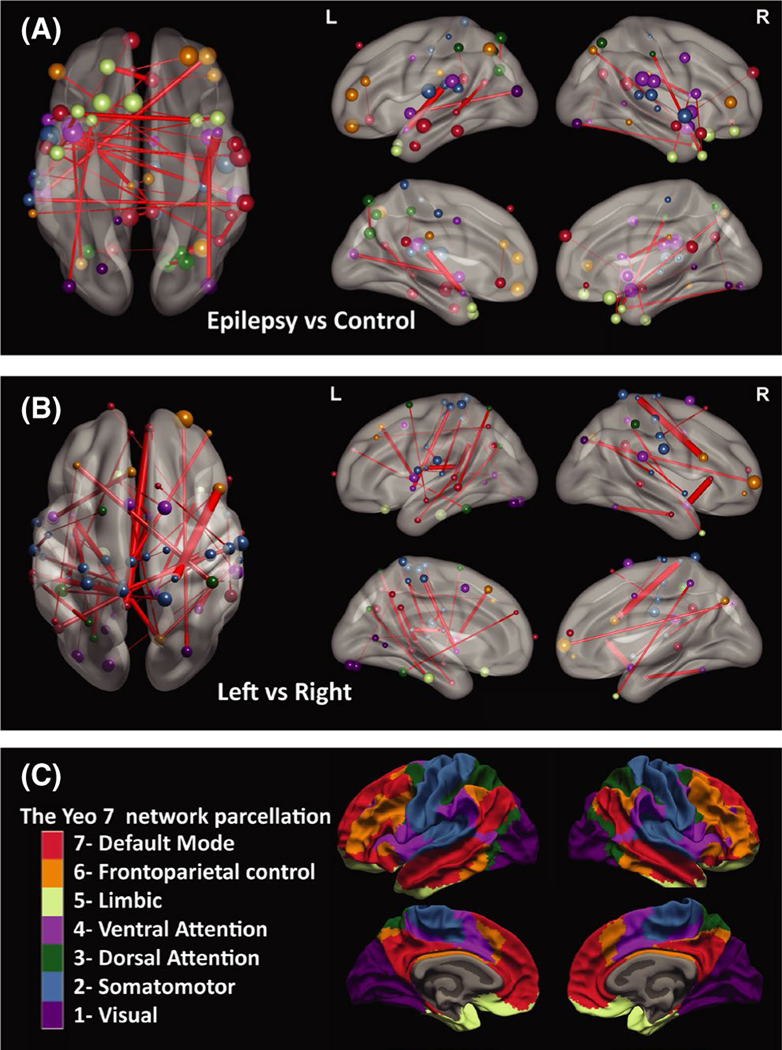
Using models trained on connectivity matrix for epilepsy versus control classification, we observed higher importance for within-network changes; 30% of the 400 most important connections were within-network, despite making up only 16% of all connections. Among the 400 most important features, 52 were within-network DMN connections (i.e., DMN–DMN), followed by higher representation from between-networks frontoparietal–DMN, within-network somatosensory, and within-network salience network connections. Both interhemispheric and intrahemispheric connections were among the most important features (Figure 3A).
FIGURE 3.
Distribution of connectivity matrix features with the highest classification importance. The connections with the highest importance (absolute classification weight) are illustrated for models using the connectivity matrix for (A) epilepsy patients versus controls (primary analysis) and (B) left versus right (lateralization, secondary analysis). Only 40 connections with the highest absolute classification weight out of 79 800 potential connections are illustrated. Edge thickness represents the weight value for that connection. Node size represents the degree of functional connectivity for the involved parcels. (C) Node colors represent each parcel’s affiliation with one of the seven Yeo networks.24 Images are illustrated using BrainNet Viewer
Patients with TLE were more likely to be misclassified in the testing set. For the epilepsy versus patient model using a 400-parcel connectivity matrix, TLE was misclassified as control in 25 of 84 cases (29.8%), of which 14 had lesions on MRI and 11 were considered nonlesional. This was compared to five of 29 (17.2%) misclassifications in non-TLE patients (χ2 = 8.30, p < .01). The same model misclassified 26 of 107 (24.3%) control participants as patients.
In reiteration of the training process using 400-parcel connectivity matrix measures, accuracies reached 100% with AUC = 1.0 during fivefold cross-validation training. The median testing accuracy was 75.6% (range = 71.4%–82.6%), and the median AUC was .83 (range = .80–. 88; Figure 2C,D).
3.2 |. Subset analyses results
Given the higher performance on the whole dataset, subset analyses were done using 400-parcel connectivity matrix measures (AUC = .82 when including all patients). In comparison, AUC = .79 was achieved when trained on only TLE patients against the control group (Figure S2). The subset analysis using only adult participants achieved an AUC of .88, and subset analysis using only lesional patients resulted in AUC = .87. Last, limiting the patient group to the nonlesional patients resulted in an AUC of .77.
3.3 |. Secondary analysis: Epilepsy lateralization
Using mean FC, the somatosensory and visual parcels followed by subcortical regions (pallidum, putamen, and amygdala) showed higher representation among the 20 most important features (Figure 4A). Mean FC model achieved lateralization accuracy of 78% (AUC = .87). The degree of FC model revealed regions in the default mode and visual networks, and subcortical regions (putamen, hippocampus, and caudate) as the 20 most important features (Figure 4B) and reached an accuracy of 94% (AUC = .98).
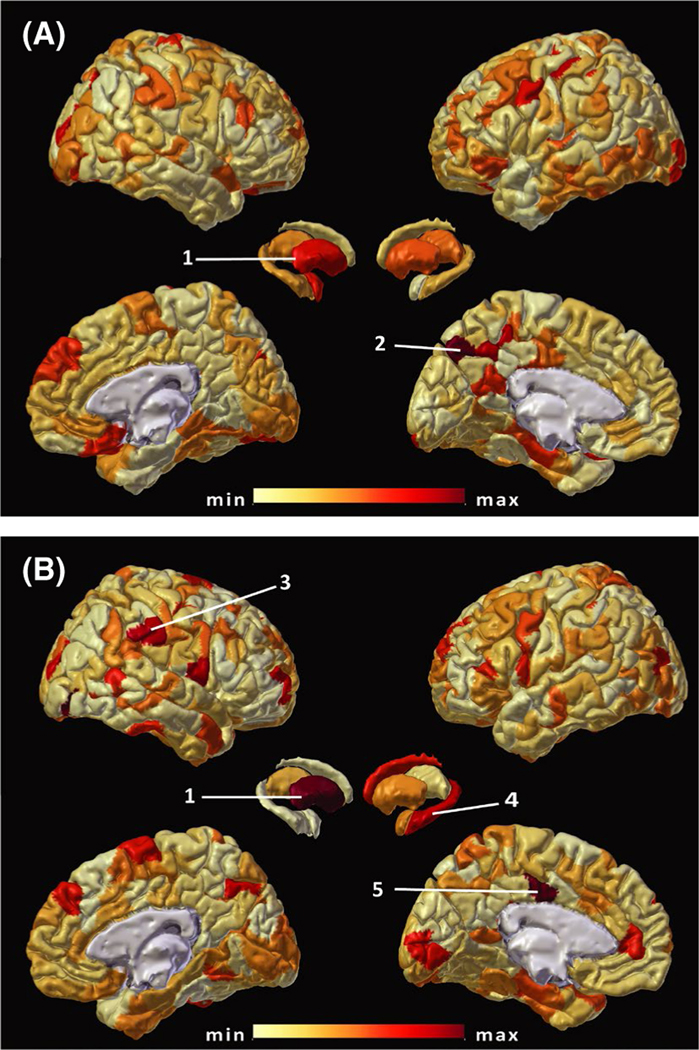
FIGURE 4.
Distribution of features with the highest importance for lateralization analysis. Similar to Figure 1, absolute feature weights from models trained with (A) mean functional connectivity (FC) and (B) degree of FC are illustrated. The color scale ranges from the minimum (min; lightest) to maximum (max; darkest) feature weight values. A number of hemispheric parcels and subcortical regions are labeled based on the Schaefer atlas21 and Harvard– Oxford25 atlas terminology for reference: right putamen (1) and left hippocampus (4), as well as members of the default mode network in precuneus and posterior cingulate anatomical regions (2, LH_Default_pCunPCC_10; 5, RH_Default_ pCunPCC_9), and ventral attention network (3, RH_SalVentAttn_TempOccPar_6)
Models trained on the connectivity matrix resulted in the best lateralization accuracies (accuracy = 94%, AUC = 1.0). We ranked connections based on their importance in this model. Compared to the primary analysis, there was a more distributed representation for each of the seven networks, and less within-network connections. Only 17.5% of the most important features from secondary analysis were within-network connections, compared to 30% in the primary analysis. Between-networks connections involving somatosensory, dorsal attention, default mode, salience, and visual networks had higher representation among the 400 most important features (Figure 3B).
In assessment of the generalizability of the findings, reiteration with training on 50% of the data resulted in 100% training accuracy. The median accuracy for the connectivity matrix using a 400-parcel atlas was 64.0% (range = 55.6%–7 1.9%), and the median AUC was .69 (range = .62–.74; Figure S1).
4 |. DISCUSSION
Our results demonstrate that a supervised machine learning method can identify common FC changes in focal epilepsies compared to controls, distinguish patients from controls, and potentially lateralize the seizure focus using features derived from resting state fMRI. Our approach is relevant to the day-to-day evaluation of patients with epilepsy, as this was achieved by using data from a heterogeneous group of adult and pediatric patients with epilepsy, with and without identifiable lesions. Our study is based on the assumption that there are common identifiable functional network attributes among several types of focal epilepsies. Although the roles of individual features used in machine learning methods are complex, we used a linear model to identify the relative importance of regions and connections in classification models. The distribution of the most prominent classification features supports the hypothesis that functional changes in focal epilepsy occur beyond the seizure onset zone and the epileptic network, in line with prior studies.7,29 Compared to prior fMRI studies, we used a larger, harmonized multisite dataset, and we optimized the machine learning approach in several ways. In addition to the inclusion of heterogeneous data, we used functional parcellation atlases to boost stability of FC signals and to increase the interpretability of findings. Functional parcellation allowed us to identify the most important features in each model in the context of their local and global functional significance and affiliation with each of the seven large-scale functional networks.21,24
4.1 |. Distinguishing FC features for epilepsy
In reviewing the features with the most importance in our primary analysis, parcels affiliated with DMN showed higher representation for both mean and degree of FC models. Differences in DMN connections between patients and controls were the strongest group of features driving the model using the connectivity matrix, as illustrated in Figure 3A and Figure S2C. Given that a majority of our patients had TLE, this is in line with previous studies showing significant alterations of DMN connections in TLE.30– 32 We attribute this to the rich connections of DMN to other brain networks, and its stability and symmetry in healthy individuals, which can produce a larger effect size when compared to patients. Our observations add to prior studies reporting changes in the DMN connectivity in epilepsy,4,12,13,33 with an overall decrease in DMN FC measures in patients (Figure S2A,B; see DMN parcels). Higher mean FC represents stronger correlations, and can be prominent in regions with strong within-network connectivity such as DMN or the primary somatosensory cortex.24 The degree of FC, however, is more representative of each node’s role in the higher level FC topology of the brain.34,35 The limbic network exhibits alteration in FC and reorganization of connections to DMN and other brain hubs, particularly in TLE.4,36 In our results, limbic parcels had higher weights in the models based on the degree of FC, and also showed overall higher values in patients compared to controls (Figure S2B; see limbic parcels). This is also in line with prior studies looking at graph theory measures in TLE.30,37,38
We also looked at the possible role of TLE overrepresentation in model training and found limited differences in the distribution of FC measure values between TLE and non-TLE groups (Figure S2), and repeated the analysis with only TLE patients. We concluded that our models are not biased toward TLE characteristics. Interestingly, the trained connectivity SVM was more likely to misclassify TLE patients as controls compared to non-TLE. This may support the notion that TLE is not a homogeneous group of epilepsy patients despite clinical similarities.39
4.2 |. Lateralizing FC features
In our secondary analysis, the features with the highest importance were distributed across different networks. Features from parcels with higher intersubject stability, such as somatosensory and visual networks, and the pallidum and putamen rose to higher ranks. The presence of an epileptic network can affect/disturb the normal FC networks5,16,38,40 and thalamocortical connections,15 and can in turn be used to predict neurocognitive41 and seizure outcomes.42,43 We postulate that our models are sensitive to changes in robust connections induced by hemispheric epilepsy networks. We postulate that compared to disease-specific FC changes in the limbic network, alterations in networks with higher intersubject stability are easier to detect.
4.3 |. Machine learning for disease identification and lateralization
FC analysis, particularly using graph theory measures, is increasingly being applied to epilepsy research.43,44 Several studies demonstrate promising results for disease classification or lateralization/localization of the seizure focus using FC.14,45 Machine learning has been used for disease classification in neurological disorders using a wide range of features, including those derived from structural MRI,7,46 fluorodeoxyglucose positron emission tomography uptake,47 and fMRI.48 In epilepsy, machine learning with features derived from volumetric46,49 and diffusion imaging7,50 shows promising results for classification and outcome prediction.
In our results, high classification performances were achieved during cross-validation training for both the primary and secondary analyses, which may represent an “overfitting” phenomenon.28 In both primary and secondary analyses, we demonstrated the generalizability of each model by applying it to an independent, equal size “unseen” testing set and repeating the training using fivefold cross-validation with grid search to achieve a range for model performance. We obtained high classification scores (near 100% accuracy) during training, but the testing accuracies were lower and within a wide range, suggesting an overfitting problem despite employing a cross-validation method. The results are, however, consistent with previous machine learning approaches using even larger structural datasets and more homogeneous patient populations.7 Although the accuracy on the testing data was lower compared to our primary analysis, rather than concluding that those FC features do not help with lateralization, we postulate that our secondary analysis was limited by a smaller sample size, or a less robust effect size and/or more noise (i.e., being underpowered). This view is based on the observed reproducibility of the primary analysis and the range of performance in certain iterations, particularly using the connectivity matrix features.
There is limited experience in analysis of fMRI data using machine learning approaches in epilepsy; however, the generalizability of those methods to larger groups of patients is not established.51– 53 An analysis of the Epilepsy Connectome Project data examined a more homogeneous sample, restricted to 60 TLE patients and 59 controls using three classification methods, including SVM with feature selection. The training achieved an average accuracy of 83% with FC derived from a slow frequency band; there was no separate testing set to test generalizability.54 In another disease classification study with 100 patients with focal and generalized epilepsies and 80 healthy controls, an SVM trained with local asymmetry and global characteristics of the DMN reached an average accuracy of 77.6% in the testing set.51 A study using independent component analysis-derived features to train an SVM for classifying TLE patients (n = 42) from controls (n = 90) reported 97.5% accuracy during training.55 In terms of efforts for developing lateralization models, one study using SVM trained on selected fMRI features from resting state in 12 patients resulted in 83% training accuracy.53 Compared to those studies, we have achieved similar or better training accuracies. We also examined the generalizability of our method in a separate testing dataset. Our results were achieved on a larger, multicenter, heterogeneous dataset and using harmonization methods. The linear SVM algorithm we used in this study finds and optimizes a decision boundary to separate the two groups using multiple features and is a more suitable method for common clinical settings with a limited number of subjects for training compared to other simple classifiers or deep learning methods.17
4.4 |. Limitations
Despite providing a statistical advantage to the analysis, machine learning methods have some limitations. Models are built on a finite number of data points, and the training dataset and features used essentially determine the generalizability to other populations. We addressed this limitation by pooling data from different sites to increase the number of training samples and by employing a harmonization method to account for sex, age, and site-specific differences. Despite harmonization, the difference between the adult and child population is significant and remains a limitation of this project. Limiting the analysis to adult participants showed comparable results to the whole dataset in terms of model classification performance. Evaluation of feature importance can provide a window into the physiologic basis of the classifications; however, this limited us to using linear SVMs. Future studies will focus on using nonlinear as well as data-driven methods to answer localization questions.
It is plausible that differences between epilepsy and controls derive from medication effects, motion-related artifacts, and other residual uncorrected or unbalanced conditions between the two groups. Effect of antiseizure medications is a relevant factor and an inherent limitation to our study, and to many studies comparing patients and controls. Medications may affect both epileptic and resting state networks in patient groups and affect the classification performance or the most important features. Our secondary analysis included only patients, and regions identified in our primary analysis are concordant with abnormalities reported by previous studies. Another limitation is related to imbalance in clinical characteristics of our patients. Comparing the distribution of the most important features and reviewing the misclassified patients suggest that TLE and non-TLE subjects share attributes in their FC alteration. As a real-world study, the imbalance in clinical characteristics might also be reflective of fMRI studies conducted at epilepsy centers for surgical patients. Last, we did not limit the study to those with good postsurgical outcomes or intracranial recording. We have adequate confidence in multidisciplinary evaluation of patients at our centers for lateralization of seizure onset to consider there to be a low risk for mislabeled side of seizure onset. With larger datasets, limiting analysis to patients with intracranial evaluation or with favorable surgical outcomes will be feasible.
5 |. CONCLUSIONS AND FUTURE DIRECTIONS
We demonstrate that our machine learning approach can identify alterations in FC characteristics and use them for classification despite real-world heterogeneity in data acquisition and clinical characteristics of patients. Our results using machine learning approaches need to be confirmed in larger datasets before serving as a biomarker discovery tool. This study was a first and necessary step in developing clinically relevant models for classification purposes. Future studies should assess whether training at one site can be validated at other sites for classification. Due to challenges in generating harmonized and comparable testing and training groups per site, we did not assess this across-site generalizability. The clinical implementations of this approach include distinguishing epilepsy patients from those with nonepileptic events, as well as lateralization of the seizure focus to guide invasive studies. The next goals are to achieve greater localization precision of the seizure focus for clinical use, and the development of predictors of surgical or pharmacological treatment outcomes using larger, multicenter datasets.
Supplementary Material
Key Points.
We used a heterogeneous, multicenter adult and pediatric epilepsy dataset including patients with and without MRI lesions to identify common functional connectivity alterations across focal epilepsies
The common functional connectivity alterations identified by our machine learning models for epilepsy versus control classification included the within-network connections of the default mode network, and the degree of functional connectivity in the limbic network
Functional connectivity measures in somatosensory and visual networks, basal ganglia, and the default mode network had higher importance in machine learning lateralization of focal epilepsy, suggesting common distinguishing characteristics within left and right onset focal epilepsies
We showed that supervised machine learning approaches can use resting state functional connectivity MRI measures to distinguish patients from controls and potentially lateralize the seizure focus
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (UL1TR001876/KL2TR001877 to T.G.), NIH National Institute of Neurological Disorders and Stroke (NS075270, NS110130, NS108445 to V.L.M.), and the NIH National Institute of Child Health and Human Development (Intellectual and Developmental Disabilities Research Center 1U54HD090257-01). We thank Emily Matuska for her help in manuscript preparation.
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: 1U54HD090257-01; National Center for Advancing Translational Sciences, Grant/Award Number: UL1TR001876/ KL2TR001877; National Institute of Neurological Disorders and Stroke, Grant/Award Number: NS075270, NS108445 and NS110130
Footnotes
CONFLICT OF INTEREST
None of the authors has any conflict of interest to disclose. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- 1.DeGiorgio CM, Markovic D, Mazumder R, Moseley BD. Ranking the leading risk factors for sudden unexpected death in epilepsy. Front Neurol. 2017;8:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342(5):314–9. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei F, Lagarde S, Wendling F, McGonigal A, Jirsa V, Guye M, et al. Defining epileptogenic networks: contribution of SEEG and signal analysis. Epilepsia. 2017;58(7):1131–47. [DOI] [PubMed] [Google Scholar]
- 4.Fahoum F, Lopes R, Pittau F, Dubeau F, Gotman J. Widespread epileptic networks in focal epilepsies: EEG-fMRI study. Epilepsia. 2012;53(9):1618–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil F, Padilla N, Soria-Pastor S, Setoain X, Boget T, Rumiá J, et al. Beyond the epileptic focus: functional epileptic networks in focal epilepsy. Cereb Cortex. 2020;30(4):2338–57. [DOI] [PubMed] [Google Scholar]
- 6.Pittau F, Vulliemoz S. Functional brain networks in epilepsy: recent advances in noninvasive mapping. Curr Opin Neurol. 2015;28(4):338–43. [DOI] [PubMed] [Google Scholar]
- 7.Gleichgerrcht E, Munsell BC, Alhusaini S, Alvim MKM, Bargalló N, Bender B, et al. Artificial intelligence for classification of temporal lobe epilepsy with ROI-level MRI data: a worldwide ENIGMA-Epilepsy study. Neuroimage Clin. 2021;31:102765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeller F, Maneshi M, Pittau F, Gholipour T, Bellec P, Dubeau F, et al. Functional connectivity in patients with idiopathic generalized epilepsy. Epilepsia. 2011;52(3):515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes MJ, Yang X, Landman BA, Ding Z, Kang H, Abou-Khalil B, et al. Functional networks in temporal-lobe epilepsy: a voxel-wise study of resting-state functional connectivity and gray-matter concentration. Brain Connect. 2013;3(1):22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nedic S, Stufflebeam SM, Rondinoni C, Velasco TR, dos Santos AC, Leite JP, et al. Using network dynamic fMRI for detection of epileptogenic foci. BMC Neurol. 2015;15(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HW, Arora J, Papademetris X, Tokoglu F, Negishi M, Scheinost D, et al. Altered functional connectivity in seizure onset zones revealed by fMRI intrinsic connectivity. Neurology. 2014;83(24):2269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang CH, Lu Y, Brinkmann B, Welker K, Worrell G, He B. Lateralization and localization of epilepsy related hemodynamic foci using presurgical fMRI. Clin Neurophysiol. 2015;126(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, Zhang Z, Chen H, Meng Y, Li J, Li Z, et al. Temporal variability profiling of the default mode across epilepsy subtypes. Epilepsia. 2021;62(1):61–73. [DOI] [PubMed] [Google Scholar]
- 14.Morgan VL, Sonmezturk HH, Gore JC, Abou-Khalil B. Lateralization of temporal lobe epilepsy using resting functional magnetic resonance imaging connectivity of hippocampal networks. Epilepsia. 2012;53(9):1628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, Doucet GE, Sperling M, Sharan A, Tracy JI. Reduced thalamocortical functional connectivity in temporal lobe epilepsy. Epilepsia. 2015;56(10):1571–9 . [DOI] [PubMed] [Google Scholar]
- 16.Dansereau CL, Bellec P, Lee K, Pittau F, Gotman J, Grova C. Detection of abnormal resting-state networks in individual patients suffering from focal epilepsy: an initial step toward individual connectivity assessment. Front Neurosci. 2014;8:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbasi B, Goldenholz DM. Machine learning applications in epilepsy. Epilepsia. 2019;60(10):2037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods. 2019;16(1):111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonov V, Evans A, McKinstry R, Almli C, Collins D. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage. 2009;47:S102. [Google Scholar]
- 20.Ciric R, Adebimpe A, Cieslak M, Rosen A, Satterthwaite TD, Tooley U, et al. PennBBL/xcpEngine: compatible with fmriprep outputs of version 20.07. Zenodo; 2020. [Google Scholar]
- 21.Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo X-N, Holmes AJ, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28(9):3095–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Heuvel MP, de Lange SC, Zalesky A, Seguin C, Yeo BTT, Schmidt R. Proportional thresholding in resting-state fMRI functional connectivity networks and consequences for patient-control connectome studies: issues and recommendations. NeuroImage. 2017;152:437–49. [DOI] [PubMed] [Google Scholar]
- 23.Farahani FV, Karwowski W, Lighthall NR. Application of graph theory for identifying connectivity patterns in human brain networks: a systematic review. Front Neurosci. 2019;13:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14(1):11–22. [DOI] [PubMed] [Google Scholar]
- 26.Fortin JP, Cullen N, Sheline YI, Taylor WD, Aselcioglu I, Cook PA et al. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage. 2018;167:104–120. 10.1016/j.neuroimage.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C-C, Lin C-J LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):1–27. 10.1145/1961189.1961199 [DOI] [Google Scholar]
- 28.Poldrack RA, Huckins G, Varoquaux G. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry. 2020;77(5):534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haneef Z, Lenartowicz A, Yeh HJ, Levin HS, Engel J Jr, Stern JM. Functional connectivity of hippocampal networks in temporal lobe epilepsy. Epilepsia. 2014;55(1):137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pittau F, Grova C, Moeller F, Dubeau F, Gotman J. Patterns of altered functional connectivity in mesial temporal lobe epilepsy. Epilepsia. 2012;53(6):1013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, et al. Default mode network abnormalities in mesial temporal lobe epilepsy: a study combining fMRI and DTI. Hum Brain Mapp. 2011;32(6):883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Campos BM, Coan AC, Lin Yasuda C, Casseb RF, Cendes F. Large-scale brain networks are distinctly affected in right and left mesial temporal lobe epilepsy. Hum Brain Mapp. 2016;37(9):3137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, He Z, Luo C, Qiu X, He S, Peng A, et al. Altered spontaneous brain activity in MRI-negative refractory temporal lobe epilepsy patients with major depressive disorder: a resting-state fMRI study. J Neurol Sci. 2018;386:29–35. [DOI] [PubMed] [Google Scholar]
- 34.Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017;20(3):353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang S, Stern JM, Engel J Jr, Levin HS, Haneef Z. Differences in graph theory functional connectivity in left and right temporal lobe epilepsy. Epilepsy Res. 2014;108(10):1770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maneshi M, Vahdat S, Fahoum F, Grova C, Gotman J. Specific resting-state brain networks in mesial temporal lobe epilepsy. Front Neurol. 2014;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tracy JI, Osipowicz K, Spechler P, Sharan A, Skidmore C, Doucet G, et al. Functional connectivity evidence of cortico-cortico inhibition in temporal lobe epilepsy. Hum Brain Mapp. 2014;35(1):353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughan DN, Rayner G, Tailby C, Jackson GD MRI-negative temporal lobe epilepsy. Neurology. 2016;87(18):1934–42. 10.1212/wnl.0000000000003289 [DOI] [PubMed] [Google Scholar]
- 40.Englot DJ, Konrad PE, Morgan VL. Regional and global connectivity disturbances in focal epilepsy, related neurocognitive sequelae, and potential mechanistic underpinnings. Epilepsia. 2016;57(10):1546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doucet GE, Rider R, Taylor N, Skidmore C, Sharan A, Sperling M, et al. Presurgery resting-state local graph-theory measures predict neurocognitive outcomes after brain surgery in temporal lobe epilepsy. Epilepsia. 2015;56(4):517–26. [DOI] [PubMed] [Google Scholar]
- 42.Larivière S, Weng Y, Vos de Wael R, Royer J, Frauscher B, Wang Z, et al. Functional connectome contractions in temporal lobe epilepsy: microstructural underpinnings and predictors of surgical outcome. Epilepsia. 2020;61(6):1221–33. [DOI] [PubMed] [Google Scholar]
- 43.He X, Doucet GE, Pustina D, Sperling MR, Sharan AD, Tracy JI Presurgical thalamic “hubness” predicts surgical outcome in temporal lobe epilepsy. Neurology. 2017;88(24):2285–93. 10.1212/wnl.0000000000004035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughan DN, Rayner G, Tailby C, Jackson GD. MRI-negative temporal lobe epilepsy: a network disorder of neocortical connectivity. Neurology. 2016;87(18):1934–42. [DOI] [PubMed] [Google Scholar]
- 45.Fallahi A, Pooyan M, Lotfi N, Baniasad F, Tapak L, Mohammadi-Mobarakeh N, et al. Dynamic functional connectivity in temporal lobe epilepsy: a graph theoretical and machine learning approach. Neurol Sci. 2021;42(6):2379–90. [DOI] [PubMed] [Google Scholar]
- 46.Bennett OF, Kanber B, Hoskote C, Cardoso MJ, Ourselin S, Duncan JS, et al. Learning to see the invisible: a data-driven approach to finding the underlying patterns of abnormality in visually normal brain magnetic resonance images in patients with temporal lobe epilepsy. Epilepsia. 2019;60(12):2499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerr WT, Nguyen ST, Cho AY, Lau EP, Silverman DH, Douglas PK, et al. Computer-aided diagnosis and localization of lateralized temporal lobe epilepsy using interictal FDG-PET. Front Neurol. 2013;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Y, Fu Z, Calhoun VD. Classification and prediction of brain disorders using functional connectivity: promising but challenging. Front Neurosci. 2018;12:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudie JD, Colby JB, Salamon N. Machine learning classification of mesial temporal sclerosis in epilepsy patients. Epilepsy Res. 2015;117:63–9. [DOI] [PubMed] [Google Scholar]
- 50.Gleichgerrcht E, Keller SS, Drane DL, Munsell BC, Davis KA, Kaestner E, et al. Temporal lobe epilepsy surgical outcomes can be inferred based on structural connectome hubs: a machine learning study. Ann Neurol. 2020;88(5):970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Cheng W, Wang Z, Zhang Z, Lu W, Lu G, et al. Pattern classification of large-scale functional brain networks: identification of informative neuroimaging markers for epilepsy. PLoS One. 2012;7(5):e36733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hekmati R, Azencott R, Zhang W, Chu ZD, Paldino MJ. Localization of epileptic seizure focus by computerized analysis of fMRI recordings. Brain Inform. 2020;7(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z, Choupan J, Reutens D, Hocking J. Lateralization of temporal lobe epilepsy based on resting-state functional magnetic resonance imaging and machine learning. Front Neurol. 2015;6:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang G, Nair VA, Mathis J, Cook CJ, Mohanty R, Zhao G, et al. Using low-frequency oscillations to detect temporal lobe epilepsy with machine learning. Brain Connect. 2019;9(2):184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bharath RD, Panda R, Raj J, Bhardwaj S, Sinha S, Chaitanya G, et al. Machine learning identifies “rsfMRI epilepsy networks” in temporal lobe epilepsy. Eur Radiol. 2019;29(7):3496–505. [DOI] [PubMed] [Google Scholar]
- 56.Felsenstein O, Peled N, Hahn L, Rockhill AP, Folsom L, Gholipour T, et al. Multi-modal neuroimaging analysis and visualization tool (MMVT). 2019;1(617):1–29. https://arxiv.org/abs/1912.10079. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.