Abstract
Since the discovery of mouse hybridoma technology by Kohler and Milstein in 1975, significant progress has been made in monoclonal antibody production. Advances in B cell immortalization and phage display technologies have generated a myriad of valuable monoclonal antibodies for diagnosis and treatment. Technological breakthroughs in various fields of ‘omics have shed crucial insights into cellular heterogeneity of a biological system in which the functional individuality of a single cell must be considered. Based on this important concept, remarkable discoveries in single-cell analysis have made in identifying and isolating functional B cells that produce beneficial therapeutic monoclonal antibodies. In this review, we will discuss three traditional methods of antibody discovery. Recent technological platforms for single-cell antibody discovery will be reviewed. We will discuss the application of the single-cell analysis in finding therapeutic antibodies for human immunodeficiency virus and emerging Zika arbovirus
1. Introduction
The humoral adaptive immunity elicits a protective immunological response against a pathogen. Unlike the innate response, humoral immunity is dependent on the extensive diversity of antigen recognition repertoires of the receptors expressed on B cells. Governed by allelic exclusion, each B cell should express solely one heavy chain and one light chain allele of immunoglobulin, therefore should produce an antibody that binds to one specific antigen. Some of these antigen-specific B cells differentiate into plasma cells to produce potent monoclonal antibodies (mAbs) and some develop into memory B cells to be reactivated for subsequent pathogen exposure. Plasma cells are short-lived, whereas memory B cells are rare and difficult to reactivate in culture. Major technological advances have been attempted to harness the therapeutic power of adaptive immunity, specifically the effector function of mAbs. Since the discovery of mouse hybridoma technology by Kohler and Milstein in 1975, where immortalized myeloma and spleen cells were fused to produce anti-sheep red blood cell antibodies1, the field of antibody discovery and application has evolved into a thriving industry comprised of basic research, diagnoses, and therapy. Global sale of monoclonal antibody products has increased dramatically from $39 billion in 2008 to $75 billion in 20132. Global sales revenue is expected to grow to $122.6 billion in 20193. The impetus for antibody development is the high specificity and affinity to an antigenic target that can either activate, inhibit, or block the target. Furthermore, the continued interest for antibody products is driven by the technological advancement of genomics, transcriptomics, proteomics, and metabolomics which identifies new targets of specific biological pathways that can be utilized to mitigate the disease process.
The Food and Drug Administration approved the first mouse mAb specific against CD3 (known as orthoclone OKT3; Ortho Biotech) for treatment of acute rejection of cadaveric renal transplantation. OKT3 was highly effective for acute renal-allograft rejection in a prospective randomized multicenter trial 4. However, antibodies of mouse origin have not been successful due to human anti-mouse immune response in patients. To circumvent these challenges, a number of engineering approaches have been undertaken. For example, chimeric antibodies with mouse variable domain regions fused to human constant regions were tested5,6. Another approach is antibody humanization in which by grafting mouse complementary determining regions (CDRs) that were evolved to bind to specific antigen into human immunoglobulin (Ig) backbone7. Other approaches have been attempted including human hybridoma technology and humanized transgenic animals in which the mouse Ig repertoires are replaced with human Ig repertoires. These technological variations have helped expand the therapeutic mAb product market in which 36 FDA-approved therapies constitute nearly 40% of the biologics market and 350 mAbs are currently in clinical trials 8-10.
Hybridoma technology and immortalization of antigen-specific B cells have been the traditional methods of mAb production. Sorting of desired B cell subset using fluorescence activated cell sorting (FACS), recombinant phage display technologies, and application of humanized transgenic mice have remarkably advanced the field. Some of these methods only capture average measurement from bulk or whole cell population undermines the heterogeneity or the autonomy of individual cells11,12. Recent developments in microfluidic chamber devices and microfabrication of nanowells designed to identify antigen-specific single cell have revolutionized the process of antibody discovery. In this review, we will discuss the traditional methods of monoclonal antibody production, specifically immortalization of antigen-specific human B cell by Epstein-Barr virus, hybridoma generation, and phage display. We will focus on current platforms for single-cell antibody discovery including fluorescence activated cell sorting, microfluidic devices, and single-cell antibody nanowells. Lastly, we will discuss the application of the single-cell analysis in finding therapeutic antibodies for human immunodeficiency virus and emerging Zika arbovirus.
2. Traditional methods of antibody discovery
2.1. Hybridoma technology and immortalization of antigen-specific human B cells
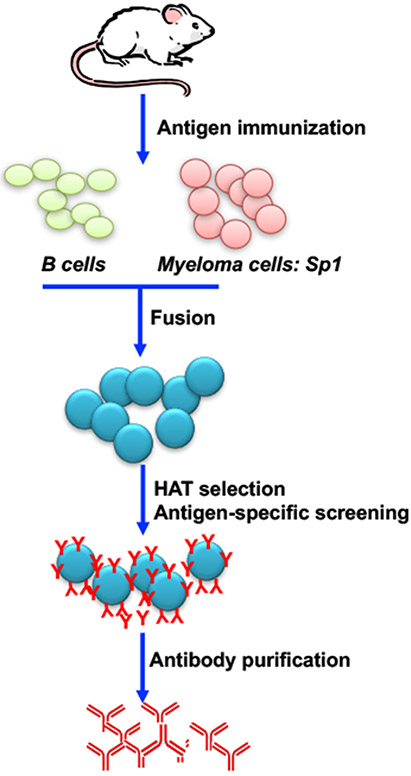
As mentioned, the hybridoma technique was first introduced to make mouse mAbs1. The technique, which has been refined over the years, took sheep red blood cell (SRBC) as immunogen and immunized BALB/c mice. The splenocytes of immunized mice were collected and fused with myeloma cells (Sp-1) to produce hybridoma cells. Immortalized hybridoma cells were selected in the presence of hypoxanthine-aminopterin-thymidine (HAT) selection medium. Unfused cells lack the hypoxanthine-guanine phosphoribosyltransferase (HGPRT) gene which makes them sensitive to the HAT selection. The aminopterin blocks the de novo DNA nucleotide synthesis pathway, therefore cells must alternatively utilize the salvage pathway to replicate in the presence of hypoxanthine and thymidine. However, the myelomas deficient in HGPRT are unable to replicate. As a result, only fused cells inherit a functional HGPRT gene from B cells can proliferate and produce antibodies. Antibody-producing B cells are further cloned and expanded by limited dilution using 96- or 384-well plates. The cloning is typically performed in multiple rounds to possibly obtain expanded clones from a single cell. Supernatants are screened by enzyme-linked immunosorbent assay (ELISA) to identify antigen-specific B cell clones (Figure 1). The process is efficient, but it can be time-consuming and labor intensive. Additionally, the resultant antibodies are of mouse origin, thereby preventing direct therapeutic translation to humans. To avoid some of these obstacles, the Epstein-Barr virus (EBV) has also been utilized to help immortalize B cells. The transformation is achieved by the activation of EBV-encoded nuclear proteins (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP) and the latent membrane proteins (LMP1, LMP2A, LMP2B) in latently infected B cells. These proteins have multiple functions, but mainly induction of survival, proliferation, and inhibition of apoptosis by upregulating expression of the anti-apoptotic proteins13. The advantages of EBV-transformed B cells are the more rapid and efficiency of screening for antigen-specific B cells in comparison to hybridoma method. Additionally, human B cells can be directly transformed to obtain antibodies, therefore there is little concern for anti-human antibody reaction. While the EBV-transformed B cells produce immunoglobulins, they yield lower quantities, which is sub-optimal for application purposes; these cells are notoriously difficult to clone and propagate14. The hybridoma technology and EBV transformation have shown promise and are methods that research and industry have adapted and improved, leading to several approved monoclonal antibodies15-17. Furthermore, significant advances in antibody engineering have been made to avoid adverse effects like acute anaphylaxis in patients, when treated with hybridoma derived mAbs18.
Figure 1. Schematic of monoclonal antibody production by hybridoma technology.
Laboratory mice are immunized with antigen of interest. Spleen cells are isolated and fused with immortalized myeloma cells such as SP1 cell line. Transformed fused cells are selected under HAT media. Antigen-specific B cells are screened using protein analytic methods (ELISA, Western blotting, flow cytometry). Once B cells of interest are identified, serial dilution will be performed to select for single cells which will be clonal expanded. Lastly, antigen-specific B cell clones will be cultured and antibodies will be purified.
2.2. Phage display
Phage display was first introduced in 198519, and has proven an effective method for mAb production, in addition to quicker production time when compared to hybridoma technology. The phage display technology is dependent on libraries naïve, immune, semi-synthetic, or synthetic of antibodies20-22, which represent non-infected, cleared infection or immunized, random sequences paired with those naturally occurring, and purely generated sequences, respectively. Since a naïve library has antibodies which have not undergone a maturation process, many will have poor binding affinity23, however high affinity mAbs have been generated24,25. In contrast, the immune library is taken from individuals that are immune to the disease, the library is inherently biased for antibodies that are mature and specific to the disease in question20,26; hence, a higher frequency of high affinity antibodies can be obtained20, but new immune libraries must be created for a specific infection or disease, resulting in a limited repertoire. Both semi-synthetic and synthetic libraries use synthetic oligonucleotides to generate diversity within the library, however may generate sequences that negative selection may have removed20,27. While semi-synthetic libraries use some natural sequences, both types of libraries are devoid of a natural maturation process22. These libraries can be utilized to design phages which present antibodies or fragments of antibodies as part of their protein coat. Diversity is key to the success of a library, however frame-shift mutations and transformation efficiency or the ability for cells to take up extracellular DNA and encode it can be a major concern20. Original cloning methods presented a multitude of technical difficulties, which resulted in several other innovations, including PCR-based assemblies28. Furthermore, in an effort to increase diversity, molecular methods, such as mutagenesis and sequence evolution29-33, have been utilized to help increase the diversity in libraries; this can be done at various intervals in the development process.
After diverse libraries are created, the phages expressing various fragmented or entire antibodies must be selected34 via a process called “panning”. The antigen-antibody complex is put through a series of parameters –e.g. toxicity to host35– then the phages are eluted, often via ELISA, amplified, and sequenced20,36. Panning can be performed against cells or nanoparticles (semi-automated), which ensures the reactivity/selectivity of the antibodies. However, both are labor intensive and the latter must be restricted to a lesser number of samples, to remain manageable20,37. Nevertheless, semi-automated is a robust, reproducible, and efficient method of panning38. Panning can be enhanced via the use of next generation sequencing (NGS), which can help eliminate unwanted clones, identify frequent sequences, and the reveal the evolution of the phages; this helps to reduce the number of rounds of panning39-41. However, binding affinities are not taken into account in this process. These methods can be used to select desirable phages from diverse libraries, producing sequences that can be tested in a much shorter timeframe than hybridomas, but this method is still tedious and laborious3.
3. New platforms for single-cell antibody discovery
3.1. Fluorescence activated cell sorting
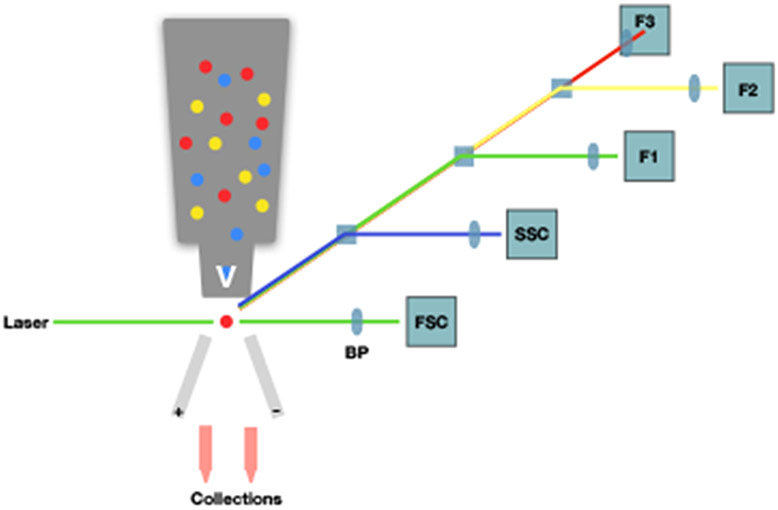
Fluorescence activated cell sorting (FACS) is an engineering adaption of flow cytometry, in which, cells are obtained or “sorted” based on fluorescent markers. The markers are commonly fluorescent-labeled antibodies against cell-specific proteins/receptors. While flow cytometry has only developed since the 1960s, when the first Coulter counter was produced, it has become a standard for identification of cell subsets. The original Coulter counter was based on the principle that the movement of a cell could be detected via changes in electrical signals as it passed through a microchannel. This has evolved over time, to the current flow cytometer which detects defraction of signal through a series of detectors when a fluorescently labeled cell passes through a microchannel42. The number of detectors available for a flow cytometer (and therefore the number of different fluorescent wavelengths it can distinguish) varies widely, with the high end being able to distinguish nearly 20 signals. As shown in Figure 2, as the cell passes through the microchannel, a laser excites the fluorescent molecule at its specific wavelength(s). The emitted signal passes through a series of bandpass filters (BP) and is able to be distinguished via the detectors. Sorting by FACS is initiated by applying a pulse of electricity to disrupt the droplet containing the cell, so that it is diverted into an appropriate receptacle. Sorting can be used restrict cell populations to those desired for subsequent experiments. Sorting for single cell is becoming a popular tool. For example, sorting of individual antigen-specific B cells were used to isolate HIV-specific antibodies. The high-throughput feature of FACS expedites the downstream applications, such as sequencing VDJ heavy and VJ light chains of single cell and, in this case, via transfection of these amplified DNA sequences into human kidney epithelial (HEK293) cells, produced monoclonal antibodies against an antigen43. This technique is heavily translational and can be used for a variety of diseases.
Figure 2. Principles of fluorescence activated cell sorting (FACS).
Fluorescence activated cell sorting (FACS) diagram. Yellow, blue, and red circles represent fluorescently-labelled cells in the flow cytometer, where, upon exiting, they are excited by a laser. Diffraction of the beam is measured through a series of detectors. Forward scatter (FSC) and side scatter (SSC) represent the size and complexity of the cell, respectively. Here, F1-F3 are the detectors and the tubes indicate user-defined positive and negative selection criteria.
3.2. Microfluidic devices
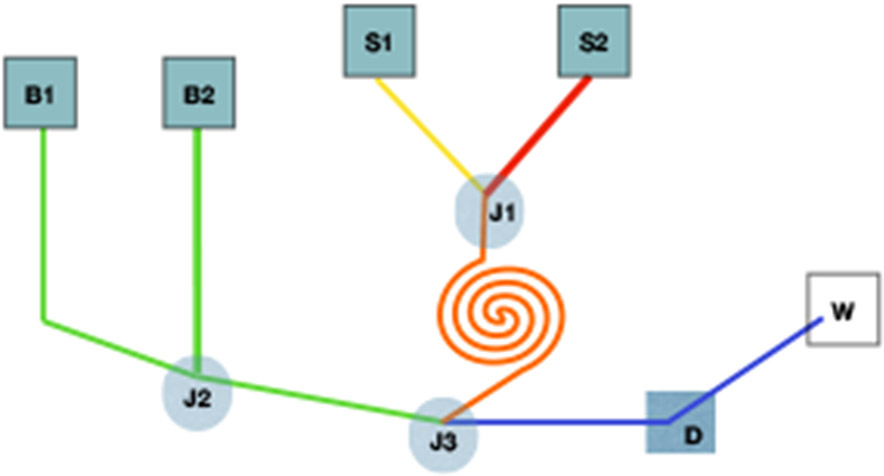
Microfluidic devices, or chips, started being used in biological applications in the late 1990s, where it was often used as a new immunoassay. The design of these early chips allowed for controlled release of specific reagents, in conjunction with the ability to mix reagents directly on the chip. Pore sizes/lengths and electrolytic buffers control flow rates of each of the components44,45 (Figure 3). For example, if two reagents need to be mixed on the chip at a disproportionate ratio, the pore size of the lesser component can be made smaller, physically limiting the amount that can be mixed at a time. This protocol is useful when performing whole cell ELISAs, especially when a cell population is limited. While with a conventional ELISA, a significantly larger number of cells and more reagents are necessary. As it is common to replicate samples in duplicate or triplicate, the amounts of reagents can become astronomically higher, making this device an optimal example of high content cell screening (HSC)46. With microfluidic devices, the human error is removed; there is no pipetting error nor inconsistencies in plated densities, since these are significantly smaller sample sizes.
Figure 3. Illustration of microfluidic device for antibody selection.
Basic schematic of a microfluidic device. “B” indicates a buffer, “S” a sample, “J” a junction, “W” is waste, and “D” is a detector. Here, two samples join at junction 1, they then continue through a coil to mix. After exiting the coil, the cells pass through a separation microchannel where cells are further diluted by the buffers (at junction 3). Here they form a single cell suspension for detection before proceeding to waste.
In the same fashion as the flow cytometer, this can be used to collect a pool of, or single cells; it may also be used to detect the response or endogenous state of individual cells, e.g. cytokines produced. It should be noted that microfluidic devices encompass a wide range of processes and many different chips can be used. While the chip above is a basic schematic, many variations on this can be used based on the application. In fact, many laboratories design their own. For example, circulating tumor cells (CTCs) are rare, difficult to detect cells which are hypothesized to be the cause of metastatic cancers. A chip was designed for this which is composed simply of a series of posts coated with antibody against epithelial cell adhesion molecule (EpCAM), a common cancer marker. This antibody then captures any cancer cells which come into contact with them on the chip47. The CTC chip can be used to enumerate and evaluate this specific cell type, enabling purification from whole blood in a single step. Notably, while this is a fairly simple design for a chip, there were still many calculations and experiments necessary to optimize how cells can be adhered on this chip. The most important factors here were layout of the posts (including diameter and distance apart), the flow rate, where too high a rate would result in loss of cells, and shear stress, in which the cells would be lysed rendering them useless. By using this same layout, antibodies could be used for a myriad of capture chips. Once a device is optimized, a simple, streamlined process for isolation and/or characterization of single cells has been achieved. This technique allows the user to save both time and money, as the volumes of reagents necessary are quite small. The malleability of this technique to a specific protocol makes it one of the most useful devices, however the time and manpower necessary to establish a single technique may not be practical for some labs.
3.3. Single-cell antibody nanowells (SCAN)
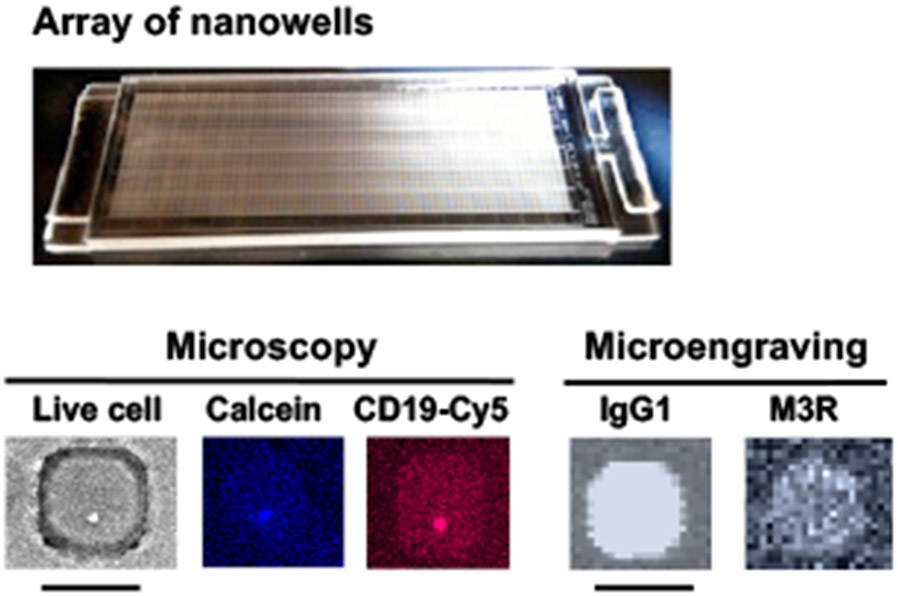
The technology was developed by Christopher Love and colleagues at MIT8. SCAN is a soft lithographic technique that uses a dense array of nanowells (50 x 50μm or 30 x 30μm, holding a volume of 0.1–1 nl each) fabricated of polydimethylsiloxane (PDMS) to isolate individual cells for printing of corresponding molecules secreted by each cell. The array of nanowells is fabricated on standard 1”x 3” glass slides containing 84,672 wells for 50μm size nanowells or 248,832 wells for 30μm size. A capture slide coated, for example, with immunoglobulins (Igs) can be hybridized by placement on the top of the nanochip to capture the antibodies being secreted by the corresponding individual live B cells that are seeded in the nanowells (Figure 4). Earlier works have shown that the nanowells with the rapid and high-throughput features were able to identify antigen-specific antibodies48,49. With the capability that the single ex-vivo cell can be cultured and confined in each nanowell for an extended amount of time, it facilitates the recovery and clonal expansion of cells with specific engraved phenotype50-52. Using the arrays of nanowells with multiplexing capability, the Love group was able to examine the isotypes of the secreted antibodies, the specificity and relative affinity for HIV antigens, identify the reactive subset of B cells (memory and plasma B cells), and sequence/identify the genes encoding the heavy and light chains53. Using this method, the group isolated HIV-specific neutralizing antibodies in colon biopsies53. This method is uniquely able to profile and isolate rare or low frequency B cells. The recent study by Tsioris et al. identified four novel West Nile virus (WNV) neutralizing antibodies in recently infected and post-convalescent subjects54. The most interesting aspect of the study was that given a low frequency of WNV-specific B cells (mean <24 events per 100,00 peripheral blood mononuclear cells), the group was able to identify some rare and potent neutralizing antibodies.
Figure 4. An application of SCAN to identify anti-muscarinic acetylcholine receptor type-3 (M3R) producing B cells.
Representative array of nanowells with microscopic micrographs showing a live cell in brightfield, calcein dye for live cell marker, and CD19 for B cell marker. Microengraving microarrays show the secretion of IgG1-isotypic anti-M3R antibody. Scale bar: 50 μm.
4. Application in infectious diseases
4.1. Human immunodeficiency virus (HIV)
According to UNAIDS / WHO, since the start of the HIV epidemic in the 1980s, worldwide 78 million people have become infected with HIV and 36 million people have died from HIV and AIDS-related diseases. As of 2016, 36.7 million people live with HIV. Combinations of highly active antiretroviral therapy (HAART) have been effective since their introduction in1996 and HIV-related mortality has been reduced since then, but remains above one million per year (1.1 in 2015 compared to 2 million in 2005), mainly due to insufficient access to screening and antiretroviral therapy in economically challenged countries which are often the most affected by the infection55. A recent meta-analysis shows that HIV-infected patients without access to HAART have a 2-year survival probability of progression from AIDS to AIDS-related death at 48% and the 6-year survival probability is 18%, whereas this life expectancy is 87% for the 2-year survival probability and 61% for 10-years survival probability for patients who received HAART56. An effective vaccine must achieve a production of protective antibodies against vaccine viral proteins. Due to an extensive genetic diversity of HIV, a prophylactic vaccine must provide global protection against all strains57,58. Currently there are three principal research directions on HIV treatment and vaccine development using neutralizing antibodies: 1) activation of B cells by sequential immunogens for expression neutralizing antibodies, 2) development of novel neutralizing antibodies due to passive administration, and 3) vector-mediated gene transfer using adeno-associated virus vectors for delivery of HIV broadly neutralizing antibodies (bNAbs) and antibody-like proteins59-61.
Human hybridoma, EBV transformation, FACS sorting of HIV-specific B cells, and combinatorial display technologies have been utilized in screening for single B cells that produce potent bNAbs. The interest in single cell antibody cloning has increased in the last few years due to advances in high-efficiency and throughput sequencing, which has reinvigorated studies on bNABs to obtain HIV-1 envelope-reactive antibodies58,62-64. Initially, cloning from single cells was introduced to examine the development and silencing of autoreactive B cells65. This method was performed for identification of single B cells expressing antibodies62,65,66 or to screen cultured B cells for the production of neutralizing activity58,67. Single cells from HIV infected patients are isolated by FACS, then sequences of immunoglobulin genes isolated from each cell are cloned into a vector for protein expression. Obtained bNAbs are analyzed to understand their specificity, protective capacity, binding conformation, and reactivity breadth and potency. Usually, screening of monoclonal antibodies is utilized to elicit a clonal assessment of specificities present in HIV infected patients68.
Passive administration of bNABs is advised for prevention and therapy of HIV infection. Studies on humans have proven safe and efficacious administration of monoclonal antibodies, yielding a promising approach of total control of HIV infection due to direct engagement in host immunity69. These bNABs must have high potency for HIV treatment with a capacity to reduce HIV viral load and minimize or prevent the risk of viral reactivation59. Pre-exposure prophylactic treatment has been studied in experiments with untreated non-human primate models infected with simian-human immunodeficiency virus (SHIV). Passive transfer or injection of HIV-1 bNABs protects host against viral infection70-74. A single bNABs infusion prevents virus acquisition with a single high dose72,75,76 or repeated low doses SHIV infection; this protection can be up to 23 weeks depending on antibody potency and half-life74. Furthermore, introduction of a mutation in the fragment cytallizable (Fc) domain extends the antibody half-life median protection74.
4.2. Emerging arboviruses: Zika
Zika virus (ZIKV) infections are an emerging health pandemic of significant medical importance. The current outbreak has garnered attention by exhibiting unique characteristics of devastating neurodevelopmental defects in newborns of infected pregnant women77,78. Over the past year, doctors in Brazil have documented over 4,000 cases of microcephaly in which infants are born with abnormally small heads79. Detection of ZIKV in fetal brain tissues and anti-ZIKV antibodies in these mothers and/or infants established a possible causal link between ZIKV infection and this birth defect80. Typical symptoms of ZIKV infection include joint pain, fever, and rash. In addition, there is emerging a potential link to the dramatic increase in the reported cases of Guillain-Barré syndrome, another rare disorder of the peripheral nervous system characterized by muscle weakness and paralysis81,82; in severe cases, Zika patients require life support. The spread of ZIKV has reached an alarming rate, particularly in the state of Florida. The influx of international travelers or tourists from ZIKV-infected areas, together with the warm tropical climate of the state, promotes the survival of the ZIKV-carrying mosquitoes, thus accelerating the spread of the virus. Responding to the Zika outbreak has been more than challenging. Unlike other well-known flaviviruses like Dengue, West Nile, Yellow Fever, and Japanese encephalitis viruses, there are no treatments or vaccinations, and diagnostic reagents are very limited. Although many investigations using immune-based therapies for arboviral infection have been pursued and have shown promise, there are no commercially available immune-based products for ZIKV. A better alternative would be to develop effective broadly neutralizing antibodies (bNAbs) as passive protection against ZIKV infection and more importantly prevent maternal-fetal transmission, reducing the likelihood of developing microcephaly in the newborns. As an emerging disease, there is a limited number of ZIKV monoclonal antibodies that are currently still at the testing phase (Table 2). Using EBV–immortalized memory B cells that were reactive to ZIKV NS1 or E proteins, Stettler et al. have identified 119 bNAbs capable of neutralizing ZIKV. The authors have shown that the most potent neutralizing antibodies were ZIKV-specific and targeted EDIII or quaternary epitopes83. Using tradition hybridoma technology in the mouse, Zhao et al. isolated six mAbs that recognized ZIKV evelope (E) protein after screening more than 2,000 hybridomas84. A recent study by Sapparapu et a. demonstrated that EBV-transformed ZIKV-specific B cells exhibited potent neutralizing capacity. Epitope mapping using X-ray crystallography indicated that the most effective bNAb recognized a unique quaternary epitope on the E protein dimer–dimer interface. Further studies showed the therapeutic efficacy in pregnant and non-pregnant mice in which mAb treatment markedly reduced tissue pathology, placental and fetal infection, and mortality in mice85. Future studies using single cell selection as proposed in Figure 5 will generate a complete repertoire of ZIKV-specific antibodies, develop better bNAbs and reveal essential epitopes for future structure-based vaccine design.
Table 2.
Neutralizing antibodies against Zika virus
| Neutralizing antibody | Viral unit | Epitope | Results |
|---|---|---|---|
| ZV-54 | Envelope subunit DIII | Lateral ridge | No cross-reactivity with DENV and/or JEV. Neutralization of 4 ZIKV strains in-vitro. Potency: 0.087-0.582 μg/mL |
| ZV-67 | Envelope subunit DIII | Lateral ridge | No cross-reactivity with DENV and/or JEV. Neutralization of 4 ZIKV strains in-vitro. Potency: 0.143-0.511 μl/mL |
| VH3-23/VK1-5 | Envelope subunit DIII | Lateral ridge | Recognition and neutralization of DENV-1 and ZIKV Potency: 0.7–4.6 ng/mL |
| ZV-64 | Envelope subunit DIII | C-C’ loop | No cross-reactivity with DENV and/or JEV. Reduced inhibitory activity in-vitro against African and American strains |
| zEDIII | Premembrane-envelope EDIII | Recognition and neutralization of ZIKV. No exacerbation of DENV infection | |
| c10 | Envelope subunits DI, DII (near from fusion loop), DIII | Intradimer | Recognition and neutralization of ZIKV in-vitro and in-vivo |
| ZV-2 | Envelope subunit DIII | ABDE sheet | No cross-reactivity with DENV and/or JEV. Neutralization of 4 ZIKV strains in-vitro |
| ZV-13 | Envelope subunit DI-II | Fusion loop | Cross-reactivity with DENV-1, 2, 3, 4, WNV, and JEV. No inhibitory activity in-vitro |
| ZIV-117 | Envelope subunit DII, | Dimer-dimer interface. Quaternary epitope | Broad neutralization of African, Asian, and American ZIKV strains. Potency: 5-25 ng/ml |
Potency is the measure by the IC50 (μg/mL) in a panel of 100–200 pseudoviruses. DENV: Dengue virus. JEV: Japanese encephalitis virus. ABDE :
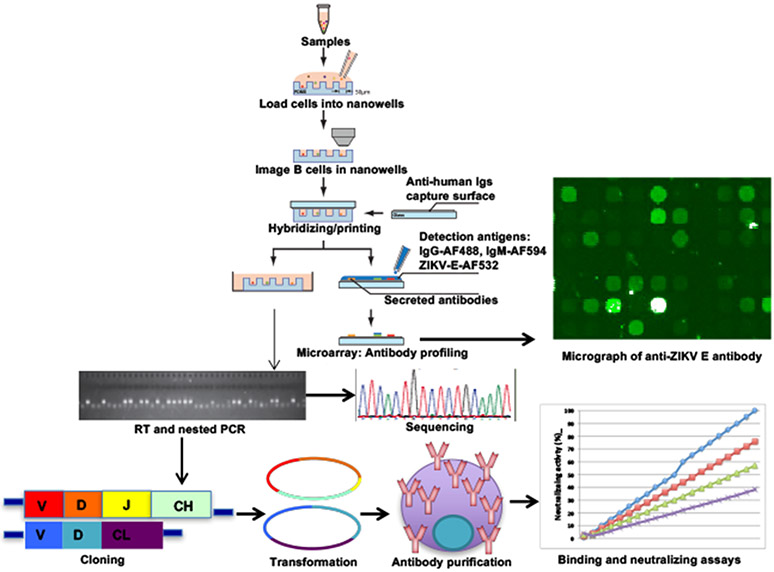
Figure 5. Screening for ZIka virus neutralizing antibodies using SCAN.
Peripheral blood cells of Zika infected patients will be isolated. Purified single-cell suspension will be labeled with anti-CD20-FITC and Calcein violet-405 for live cell and plated onto fabricated nanowells. Labeled cells in the nanowells will be imaged for surface markers and locations on the chips. Capture slide coated with anti-human immunoglobulins will be hybridized. Detection antibody mixture containing IgG-AF-488, IgM-AF594, and ZIKV E-AF532 will be added. Micrograph of anti-ZIKV E-secreting B cells will be generated. Individual ZIKV E-secreting B cells will be picked and performed RT/nested PCR for heavy/light chain sequences. Both chains will be cloned into an expression vector and expressed in 293T cell line. Secreted antibodies will be purified and screened for binding and neutralizing activity against ZIKV. AF: Alexa Fluor
5. Conclusion
Single-cell analysis is a powerful tool in examining a comprehensive repertoire of antigen-specific Abs from the most abundant to the least abundant B cells that are highly specific. Single-cell antibody discovery is critically important in selecting the few potent B cells with important capacity to produce the most competent therapeutic mAbs and broadly capable of neutralizing pathogens in infected individuals. Diseases in which vaccines are not readily available or effective, therapeutic mAbs can provide significant protection as passive immunity. The two quintessential examples are HIV and Zika as discussed. These technologies, while strong and important tools currently, have the potential to become widely utilized and even more powerful. They have the potential to be used in diagnostics and beyond that, these techniques are currently being used to develop treatments for other infectious diseases and cancer. In conjunction with shotgun mutagenesis and X-ray crystallography, antigenic epitopes can be mapped and the structural interactions between Abs and antigens can be examined. On a more fundamental level, single-cell analysis will be an essential player in creating immune-therapeutics and eventually vaccines.
Table 1.
Characterization of HIV broadly neutralizing antibodies
| Broadly neutralising antibody |
Envelope subunit |
Epitope | Breadth (%) | Potency (μg/mL) | Development stage |
|---|---|---|---|---|---|
| 4E10 | gp41 | Gp41 membrane-proximal external region | 13 (32) | 3.41 (32) | Phase I/II clinical trial (NCT00219986) |
| 2FS | gp41 | 19(33) | 2.30 (33) | Phase I/II clinical trial (NCT00219986) | |
| 10E8 | gp41 | 72 (35) | 0.35 (35) | In-vivo (rhesus macaque) (62) | |
| b12 | gp120 | CD4 binding site | 10 (32) | 2.82 (32) | In-vivo (rhesus macaque) (105) |
| VRC01 | gp120 | 74 (32) | 0.33 (32) | Phase II clinical trial (NCT02664415) | |
| VRC01-N | gp120 | ||||
| VRC07 | gp120 | 83 (41) | 0.11 (41) | Phase I clinical trial (NCT03015181) | |
| 3BNC117 | gp120 | 77 35) | 0.11 (35) | Phase II clinical trial (NCT02446847) | |
| NIH45-46 | gp120 | 76 (35) | 0.2 (35) | In-vivo (rhesus macaque) (106) | |
| N6 | gp120 | 96 (43) | 0.038 (43) | In vitro (43) | |
| ECD4-Ig | gp120 | 100 (100) | 0.05 (100) | In-vivo (rhesus macaque) (100) | |
| PG9 | gp120 | V1/V2 domain | 54 (32) | 0.23 (32) | Phase I clinical trial (NCT01937455) |
| PGDM1400 | gp120 | 83 (42) | 0.003 (42) | In-vitro (42) | |
| PGT145 | gp120 | 52 (32) | 0.2 (32) | In-vitro (32) | |
| PG16 | gp120 | 59 (35) | 0.15 (35) | In-vitro (35) | |
| 10-1074 | gp120 | V3 domain | 54 (66) | 0.4 (66) | Phase I clinical trial (NCT02824536) |
| PGT121 | gp120 | 57 (32) | 0.03 (32) | Phase I clinical trial (NCT02960581) | |
| PGT 128 | gp120 | 60 (32) | 0.02 (32) | In vivo (rhesus macaque) (107) |
Breadth is the percentage of viruses neutralized at IC50 > 1 μg/mL in a panel of 100–200 pseudoviruses. Potency is the measure by the IC50 (μg/mL) in a panel of 100–200 pseudoviruses.
6. Acknowledgements
This work was supported by the Florida Department of Health, Biomedical Research Program (JKY, CQN) and NIAID 1R21AI130561-01A1 (CQN).
References
- 1.Kohler G & Milstein C Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975). [DOI] [PubMed] [Google Scholar]
- 2.Ecker DM, Jones SD & Levine HL The therapeutic monoclonal antibody market. MAbs 7, 9–14, doi: 10.4161/19420862.2015.989042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald V & Leonard P Single cell screening approaches for antibody discovery. Methods 116, 34–42, doi: 10.1016/j.ymeth.2016.11.006 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Ortho Multicenter Transplant Study, G. A randomized clinical trial of OKT3 monoclonal antibody for acute rejection of cadaveric renal transplants. N Engl J Med 313, 337–342, doi: 10.1056/NEJM198508083130601 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Brekke OH & Sandlie I Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat Rev Drug Discov 2, 52–62, doi: 10.1038/nrd984 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Sun LK et al. Chimeric antibody with human constant regions and mouse variable regions directed against carcinoma-associated antigen 17-1A. Proc Natl Acad Sci U S A 84, 214–218 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PT, Dear PH, Foote J, Neuberger MS & Winter G Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 321, 522–525, doi: 10.1038/321522a0 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Love KR, Bagh S, Choi J & Love JC Microtools for single-cell analysis in biopharmaceutical development and manufacturing. Trends Biotechnol 31, 280–286, doi: 10.1016/j.tibtech.2013.03.001 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal SR What's fueling the biotech engine-2011 to 2012. Nat Biotechnol 30, 1191–1197, doi: 10.1038/nbt.2437 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Goodman M Market watch: Sales of biologics to show robust growth through to 2013. Nat Rev Drug Discov 8, 837, doi: 10.1038/nrd3040 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Urban PL et al. Carbon-13 labelling strategy for studying the ATP metabolism in individual yeast cells by micro-arrays for mass spectrometry. Molecular BioSystems 7, 2837–2840, doi: 10.1039/C1MB05248A (2011). [DOI] [PubMed] [Google Scholar]
- 12.Nemes P, Knolhoff AM, Rubakhin SS & Sweedler JV Metabolic Differentiation of Neuronal Phenotypes by Single-cell Capillary Electrophoresis–Electrospray Ionization-Mass Spectrometry. Analytical Chemistry 83, 6810–6817, doi: 10.1021/ac2015855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuppers R B cells under influence: transformation of B cells by Epstein-Barr virus. Nat Rev Immunol 3, 801–812, doi: 10.1038/nri1201 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wang S Advances in the Production of Human Monoclonal Antibodies. Antibody Technology Journal 2011, 4, doi: 10.2147/ANTI.S20195 (2011). [DOI] [Google Scholar]
- 15.Yamashita M, Katakura Y & Shirahata S Recent advances in the generation of human monoclonal antibody. Cytotechnology 55, 55–60, doi: 10.1007/s10616-007-9072-5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichert JM Monoclonal antibodies in the clinic. Nat Biotechnol 19, 819–822, doi: 10.1038/nbt0901-819 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Reichert JM, Rosensweig CJ, Faden LB & Dewitz MC Monoclonal antibody successes in the clinic. Nat Biotechnol 23, 1073–1078, doi: 10.1038/nbt0905-1073 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Hansel TT, Kropshofer H, Singer T, Mitchell JA & George AJ The safety and side effects of monoclonal antibodies. Nat Rev Drug Discov 9, 325–338, doi: 10.1038/nrd3003 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Smith GP Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985). [DOI] [PubMed] [Google Scholar]
- 20.Lim BN et al. Principles and application of antibody libraries for infectious diseases. Biotechnol Lett 36, 2381–2392, doi: 10.1007/s10529-014-1635-x (2014). [DOI] [PubMed] [Google Scholar]
- 21.Thie H, Meyer T, Schirrmann T, Hust M & Dubel S Phage display derived therapeutic antibodies. Curr Pharm Biotechnol 9, 439–446 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Miersch S & Sidhu SS Synthetic antibodies: concepts, potential and practical considerations. Methods 57, 486–498, doi: 10.1016/j.ymeth.2012.06.012 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Pansri P, Jaruseranee N, Rangnoi K, Kristensen P & Yamabhai M A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol 9, 6, doi: 10.1186/1472-6750-9-6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan TJ et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol 14, 309–314, doi: 10.1038/nbt0396-309 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Willats WG, Gilmartin PM, Mikkelsen JD & Knox JP Cell wall antibodies without immunization: generation and use of de-esterified homogalacturonan block-specific antibodies from a naive phage display library. Plant J 18, 57–65 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Ayat H et al. Isolation of scFv antibody fragments against HER2 and CEA tumor antigens from combinatorial antibody libraries derived from cancer patients. Biologicals 41, 345–354, doi: 10.1016/j.biologicals.2013.05.004 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Braunagel M & Little M Construction of a semisynthetic antibody library using trinucleotide oligos. Nucleic Acids Res 25, 4690–4691 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andris-Widhopf J, Rader C, Steinberger P, Fuller R & Barbas CF 3rd. Methods for the generation of chicken monoclonal antibody fragments by phage display. J Immunol Methods 242, 159–181 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Low NM, Holliger PH & Winter G Mimicking somatic hypermutation: affinity maturation of antibodies displayed on bacteriophage using a bacterial mutator strain. J Mol Biol 260, 359–368 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Sblattero D & Bradbury A Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol 18, 75–80, doi: 10.1038/71958 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Christensen PA et al. Modifying antibody specificity by chain shuffling of V / V between antibodies with related specificities. Scand J Immunol 69, 1–10, doi: 10.1111/j.1365-3083.2008.02164.x (2009). [DOI] [PubMed] [Google Scholar]
- 32.Fujii R, Kitaoka M & Hayashi K One-step random mutagenesis by error-prone rolling circle amplification. Nucleic Acids Res 32, e145, doi: 10.1093/nar/gnh147 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland EG et al. AXM mutagenesis: an efficient means for the production of libraries for directed evolution of proteins. J Immunol Methods 394, 55–61, doi: 10.1016/j.jim.2013.05.003 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Hammers CM & Stanley JR Antibody phage display: technique and applications. J Invest Dermatol 134, e17, doi: 10.1038/jid.2013.521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schirrmann T, Meyer T, Schutte M, Frenzel A & Hust M Phage display for the generation of antibodies for proteome research, diagnostics and therapy. Molecules 16, 412–426, doi: 10.3390/molecules16010412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukushi S et al. Antigen-capture ELISA for the detection of Rift Valley fever virus nucleoprotein using new monoclonal antibodies. J Virol Methods 180, 68–74, doi: 10.1016/j.jviromet.2011.12.013 (2012). [DOI] [PubMed] [Google Scholar]
- 37.de Kruif J, Terstappen L, Boel E & Logtenberg T Rapid selection of cell subpopulation-specific human monoclonal antibodies from a synthetic phage antibody library. Proc Natl Acad Sci U S A 92, 3938–3942 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turunen L, Takkinen K, Soderlund H & Pulli T Automated panning and screening procedure on microplates for antibody generation from phage display libraries. J Biomol Screen 14, 282–293, doi: 10.1177/1087057108330113 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Dias-Neto E et al. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One 4, e8338, doi: 10.1371/journal.pone.0008338 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravn U et al. By-passing in vitro screening--next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res 38, e193, doi: 10.1093/nar/gkq789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.t Hoen PA et al. Phage display screening without repetitious selection rounds. Anal Biochem 421, 622–631, doi: 10.1016/j.ab.2011.11.005 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Fulwyler MJ Electronic separation of biological cells by volume. Science 150, 910–911 (1965). [DOI] [PubMed] [Google Scholar]
- 43.Smith K et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4, 372–384, doi: 10.1038/nprot.2009.3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiem NH & Harrison DJ Microchip systems for immunoassay: an integrated immunoreactor with electrophoretic separation for serum theophylline determination. Clin Chem 44, 591–598 (1998). [PubMed] [Google Scholar]
- 45.Wheeler AR et al. Microfluidic device for single-cell analysis. Anal Chem 75, 3581–3586 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Cheong R, Paliwal S & Levchenko A High-content screening in microfluidic devices. Expert Opin Drug Discov 5, 715–720, doi: 10.1517/17460441.2010.495116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagrath S et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239, doi: 10.1038/nature06385 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogunniyi AO, Story CM, Papa E, Guillen E & Love JC Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat Protoc 4, 767–782, doi: 10.1038/nprot.2009.40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronan JL, Story CM, Papa E & Love JC Optimization of the surfaces used to capture antibodies from single hybridomas reduces the time required for microengraving. J Immunol Methods 340, 164–169, doi: 10.1016/j.jim.2008.10.018 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG & Ploegh HL A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol 24, 703–707, doi: 10.1038/nbt1210 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Esfandiary L et al. Single-cell antibody nanowells: a novel technology in detecting anti-SSA/Ro60- and anti-SSB/La autoantibody-producing cells in peripheral blood of rheumatic disease patients. Arthritis research & therapy 18, 107, doi: 10.1186/s13075-016-1010-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen CQ, Ogunniyi AO, Karabiyik A & Love JC Single-Cell Analysis Reveals Isotype-Specific Autoreactive B Cell Repertoires in Sjögren’s Syndrome. PLoS One 8, e58127, doi: 10.1371/journal.pone.0058127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogunniyi AO et al. Profiling human antibody responses by integrated single-cell analysis. Vaccine 32, 2866–2873, doi: 10.1016/j.vaccine.2014.02.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsioris K et al. Neutralizing antibodies against West Nile virus identified directly from human B cells by single-cell analysis and next generation sequencing. Integr Biol (Camb) 7, 1587–1597, doi: 10.1039/c5ib00169b (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahy M et al. Producing HIV estimates: from global advocacy to country planning and impact measurement. Glob Health Action 10, 1291169, doi: 10.1080/16549716.2017.1291169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poorolajal J, Hooshmand E, Mahjub H, Esmailnasab N & Jenabi E Survival rate of AIDS disease and mortality in HIV-infected patients: a meta-analysis. Public Health 139, 3–12, doi: 10.1016/j.puhe.2016.05.004 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Lelievre JD & Levy Y HIV-1 prophylactic vaccines: state of the art. J Virus Erad 2, 5–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein F et al. Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204, doi: 10.1126/science.1241144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmad M, Ahmed OM, Schnepp B & Johnson PR Engineered Expression of Broadly Neutralizing Antibodies Against Human Immunodeficiency Virus. Annu Rev Virol, doi: 10.1146/annurev-virology-101416-041929 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Brady JM, Baltimore D & Balazs AB Antibody gene transfer with adeno-associated viral vectors as a method for HIV prevention. Immunol Rev 275, 324–333, doi: 10.1111/imr.12478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deal CE & Balazs AB Vectored antibody gene delivery for the prevention or treatment of HIV infection. Curr Opin HIV AIDS 10, 190–197, doi: 10.1097/COH.0000000000000145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheid JF et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods 343, 65–67, doi: 10.1016/j.jim.2008.11.012 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiller T et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124, doi: 10.1016/j.jim.2007.09.017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wardemann H et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377, doi: 10.1126/science.1086907 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Scheid JF et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640, doi: 10.1038/nature07930 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Klein F et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med 209, 1469–1479, doi: 10.1084/jem.20120423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker LM et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470, doi: 10.1038/nature10373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho IY et al. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J Immunol Methods 438, 67–70, doi: 10.1016/j.jim.2016.09.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caskey M et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491, doi: 10.1038/nature14411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baba TW et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med 6, 200–206, doi: 10.1038/72309 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Hessell AJ et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 15, 951–954, doi: 10.1038/nm.1974 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moldt B et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A 109, 18921–18925, doi: 10.1073/pnas.1214785109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mascola JR et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol 73, 4009–4018 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gautam R et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature 533, 105–109, doi: 10.1038/nature17677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shingai M et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 211, 2061–2074, doi: 10.1084/jem.20132494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saunders KO et al. Sustained Delivery of a Broadly Neutralizing Antibody in Nonhuman Primates Confers Long-Term Protection against Simian/Human Immunodeficiency Virus Infection. J Virol 89, 5895–5903, doi: 10.1128/JVI.00210-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Araujo AQ, Silva MT & Araujo AP Zika virus-associated neurological disorders: a review. Brain 139, 2122–2130, doi: 10.1093/brain/aww158 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Baud D, Gubler DJ, Schaub B, Lanteri MC & Musso D An update on Zika virus infection. Lancet, doi: 10.1016/S0140-6736(17)31450-2 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Schuler-Faccini L et al. Possible Association Between Zika Virus Infection and Microcephaly - Brazil, 2015. MMWR Morb Mortal Wkly Rep 65, 59–62, doi: 10.15585/mmwr.mm6503e2 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Reagan-Steiner S et al. Evaluation of Placental and Fetal Tissue Specimens for Zika Virus Infection - 50 States and District of Columbia, January-December, 2016. MMWR Morb Mortal Wkly Rep 66, 636–643, doi: 10.15585/mmwr.mm6624a3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oehler E et al. Zika virus infection complicated by Guillain-Barre syndrome--case report, French Polynesia, December 2013. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 19 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Frontera JA & da Silva IR Zika Getting on Your Nerves? The Association with the Guillain-Barre Syndrome. N Engl J Med 375, 1581–1582, doi: 10.1056/NEJMe1611840 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Stettler K et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826, doi: 10.1126/science.aaf8505 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Zhao H et al. Structural Basis of Zika Virus-Specific Antibody Protection. Cell 166, 1016–1027, doi: 10.1016/j.cell.2016.07.020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sapparapu G et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature, doi: 10.1038/nature20564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861, doi: 10.1126/science.1187659 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheid JF et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637, doi: 10.1126/science.1207227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McLellan JS et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343, doi: 10.1038/nature10696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu X et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602, doi: 10.1126/science.1207532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonsignori M et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol 86, 4688–4692, doi: 10.1128/JVI.07163-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robbiani DF et al. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 169, 597–609 e511, doi: 10.1016/j.cell.2017.04.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang M, Dent M, Lai H, Sun H & Chen Q Immunization of Zika virus envelope protein domain III induces specific and neutralizing immune responses against Zika virus. Vaccine 35, 4287–4294, doi: 10.1016/j.vaccine.2017.04.052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang S et al. Neutralization mechanism of a highly potent antibody against Zika virus. Nature communications 7, 13679, doi: 10.1038/ncomms13679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai L et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 19, 696–704, doi: 10.1016/j.chom.2016.04.013 (2016). [DOI] [PubMed] [Google Scholar]