Abstract
STUDY QUESTION
Do women with endometriosis have a different endometrial gene expression profile at the time of embryo implantation than women without endometriosis?
SUMMARY ANSWER
The endometrial gene expression profile of women with endometriosis differs from that of women without endometriosis at the mid-secretory phase, although the differences are small.
WHAT IS KNOWN ALREADY
About 50% of women with endometriosis suffer infertility. Several molecular studies have suggested impaired endometrial receptivity in women with endometriosis, while others have detected no dysregulation of endometrial receptivity. Nevertheless, the previous endometrial transcriptome studies comparing women with and without endometriosis have been performed in small sample size with limited statistical power. We set out to systematically search and compile data of endometrial gene expression signatures at the receptive phase in women with endometriosis versus control women. Based on the obtained data, we conducted a meta-analysis of differentially expressed genes in order to raise the power of the analysis for identifying the molecular profiles of receptive phase endometria in endometriosis.
STUDY DESIGN, SIZE, DURATION
A systematic literature search was conducted up to February 2022 following PRISMA criteria and included PubMed, Cochrane and Web of Science databases. For the systematic search, the term ‘endometriosis’ was paired with the terms ‘transcriptomics’, ‘transcriptome’, ‘gene expression’, ‘RNA-seq’, ‘sequencing’ and ‘array’, by using the Boolean operator ‘AND’ to connect them. Articles written in English were screened and interrogated for data extraction.
PARTICIPANTS/MATERIALS, SETTING, METHODS
A meta-analysis was performed on the selected studies to extract the differentially expressed genes described at the mid-secretory phase in women with endometriosis versus women without endometriosis in natural cycles, using the robust rank aggregation method. In total, transcriptome data of 125 women (78 patients and 47 controls) were meta-analysed, with a special focus on endometrial receptivity-specific genes based on commercial endometrial receptivity tests.
MAIN RESULTS AND THE ROLE OF CHANCE
In total, 8 studies were eligible for the quantitative meta-analysis, gathering transcriptome data from the mid-secretory phase endometria of 125 women. A total of 7779 differentially expressed transcripts between the study groups were retrieved (3496 up-regulated and 4283 down-regulated) and were meta-analysed. After stringent multiple correction, there was no differential expression of any single molecule in the endometrium of women with endometriosis versus controls, while enrichment analysis detected that the pathways of chemotaxis and locomotion are dysregulated in endometriosis. Further analysis of endometrial receptivity-specific genes highlighted dysregulation of C4BPA, MAOA and PAEP and enrichment of immune and defence pathways in women with endometriosis.
LIMITATIONS, REASONS FOR CAUTION
Most of the studies included into the meta-analysis were relatively small and had different study designs, which might have contributed to a bias.
WIDER IMPLICATIONS OF THE FINDINGS
The current meta-analysis supports the hypothesis that endometrial receptivity is altered in women with endometriosis, although the changes are small. The molecules and pathways identified could serve as future biomarkers and therapeutical targets in detecting and treating endometriosis-associated infertility.
STUDY FUNDING/COMPETING INTEREST(S)
The authors declare no competing interests. This work was supported by the Spanish Ministry of Education, Culture and Sport [grant FPU15/01193] and the Margarita Salas program for the Requalification of the Spanish University system [grant UJAR01MS]; Spanish Ministry of Economy, Industry and Competitiveness (MINECO) and European Regional Development Fund (FEDER): grants RYC-2016-21199 and ENDORE SAF2017-87526-R; Programa Operativo FEDER Andalucía (B-CTS-500-UGR18; A-CTS-614-UGR20); the Junta de Andalucía [BIO-302; and PAIDI P20_00158]; the University of Jaén [PAIUJA-EI_CTS02_2017]; the University of Granada, Plan Propio de Investigación 2016, Excellence actions: Units of Excellence; Unit of Excellence on Exercise and Health (UCEES), and by the Junta de Andalucía, Consejería de Conocimiento, Investigación y Universidades and European Regional Development Fund (ERDF), ref. SOMM17/6107/UGR; the Estonian Research Council (grant PRG1076); Horizon 2020 innovation (ERIN, grant no. EU952516) of the European Commission and Enterprise Estonia (grant EU48695).
TRIAL REGISTRATION NUMBER
The systematic review was registered at PROSPERO (identifier: CRD42020122054).
Keywords: endometriosis, endometrium, infertility, meta-analysis, transcriptomics
WHAT DOES THIS MEAN FOR PATIENTS?
This study investigates whether the gene expression profile of the endometrium (the inner lining of the uterus) of women with endometriosis is different from that of control women during the phase of the menstrual cycle when the uterus is receptive for embryo to implantation. Our extensive systematic review and meta-analysis identified endometrial receptivity-associated genes and molecular pathways that seem to be altered in women with endometriosis. The study findings could help to explain endometriosis-associated infertility in women suffering from this common gynaecological disease and could lead to the development of molecular biomarkers for detecting and treating infertility in endometriosis.
Introduction
Endometriosis is a debilitating gynaecological condition that affects ∼10% of women of reproductive age and is characterized by the implantation of endometrial tissue in ectopic locations (Zondervan et al., 2018). Among the main symptoms of endometriosis, pelvic pain, dysmenorrhoea and infertility are the most prevalent, with evidence of impaired fertility in up to 50% of affected women (Giudice and Kao, 2004; Practice Committee of the American Society for Reproductive Medicine, 2012). One of the suggested reasons for endometriosis-associated infertility is diminished endometrial receptivity and defective embryo implantation (Brosens et al., 2012; Altmäe and Aghajanova, 2015; Lessey and Kim, 2017; Horton et al., 2019).
Accumulating evidence suggests that endometriosis has a detrimental reproductive impact in both natural as well as assisted reproduction, where the potentially dysfunctional endometrium, aberrant uterine contractility and affected endometrium–myometrium interface could hinder embryo implantation (Horton et al., 2019). In addition, different pathological processes involving inflammation, immune modulation, aberrant angiogenesis, oxidative stress, extracellular matrix remodelling, genetic and epigenetic changes could impact endometrial receptivity and implantation process in women with endometriosis (Gupta et al., 2008; Kokcu, 2013; Vigano et al., 2015). However, there is no consensus about whether endometrial receptivity is dysregulated in endometriosis and which molecular mechanisms are involved.
Several studies focusing on single molecules have observed alterations in the expression levels of different genes and proteins in the endometria of women with endometriosis (Giudice et al., 2002; Wei et al., 2009; May et al., 2011). Nevertheless, the results obtained are controversial and lack confirmation and validation. With the advancement of ‘-omics’ technologies, several studies have investigated the gene expression profile of the whole genome (i.e. transcriptomics) in eutopic endometria from women with endometriosis (Giudice, 2003; Fassbender et al., 2012; Altmäe et al., 2014; Miravet-Valenciano et al., 2017; Saare et al., 2017; McKinnon et al., 2018; Poli-Neto et al., 2020). Regardless of the long lists of differentially regulated genes identified in the whole genome expression analyses, the studies are heterogeneous, performed on limited sample size, and lack power, consensus and validation. Therefore, whether endometrial transcriptome is dysregulated in the endometrium in the receptive phase in endometriosis remains an open debate.
We set out to perform a systematic literature search followed by a meta-analysis of the endometrial transcriptome at the mid-secretory phase eutopic endometria in women with endometriosis in comparison with endometria from women without the disease in order to raise the power in identifying endometrial transcriptome profiles.
Materials and methods
Search of the literature and data extraction
A systematic search of the literature was performed using PubMed, Cochrane and Web of Science databases up to February 2022 by two researchers (S.A. and E.V.) independently and in agreement with the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) (Page et al., 2021). For the systematic search, the term ‘endometriosis’ was paired with the terms ‘transcriptomics’, ‘transcriptome’, ‘gene expression’, ‘RNA-seq’, ‘sequencing’ and ‘array’, by using the Boolean operator ‘AND’ to connect them.
Titles of the papers were read to extract information of the potentially eligible abstracts, which were then carefully evaluated for further full-text evaluation. The reference lists of review articles and relevant studies were explored manually to identify other potentially eligible studies. No restrictions were applied. A detailed protocol for this systematic review was registered in PROSPERO under the title ‘Endometrial transcriptome in women with endometriosis compared with women without endometriosis during the window of implantation: systematic review and meta-analysis’ and can be accessed at: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020122054.
Abstracts of all the retrieved articles were read for the selection of eligible studies, and the full text of each potentially suitable article was evaluated. In the final step, we restricted the inclusion criteria only to original experimental studies concerning the endometrial transcriptome (cDNA microarray or RNA-sequencing techniques) in women with endometriosis in the mid-secretory phase of the menstrual cycle compared with control women without endometriosis from the same phase of the cycle. Articles focusing on the study of other phases of the menstrual cycle or including pathological conditions other than endometriosis were not included in the final list of articles.
For all eligible studies, the lists of differentially expressed genes in the mid-secretory phase in eutopic endometrial tissue of women with endometriosis versus normal endometrium from control women were extracted directly from the publication. When the gene lists were not available, the authors were contacted. In preparation for the subsequent analyses, all the gene lists were standardized to a common nomenclature system using the official gene symbols. Therefore, all lists that had other gene identifiers (GenBank IDs or Affymetrix array probes IDs, mainly) were converted into the official gene symbols by using the g:Convert tool in g:Profiler database (https://biit.cs.ut.ee/gprofiler/convert; Raudvere et al., 2019). All the array probes unable to be converted to official gene symbol were removed from the subsequent analyses. Finally, the lists of dysregulated genes were ranked by the absolute value of their fold changes (FCs). For the genes with duplicated values of FC, only the highest absolute value of the FC remained.
Meta-analysis
The robust rank aggregation (RRA) method (RRA package v.1.1) implemented in R (version 3.5.1) was used for the meta-analysis of the ranked gene lists (Kolde et al., 2012). The total number of official gene symbols ranked by their FCs in each study was used as an input. The RRA method assigns a significance ρ (rho) score for each transcript, which is used to order the genes based on that value (Kolde et al., 2012), resulting in a list of prioritized genes in accordance to the representation each gene has in each list of differentially expressed genes. This parameter is not itself a p-value and therefore has to be corrected. Hence, to adjust for multiple testing, a false discovery rate (FDR) correction was calculated following the Benjamini–Hochberg procedure using the p.adjust function included in R package stats. All the lists involved in the analysis reflect expression in the receptive phase of the menstrual cycle in the endometriosis group compared with the control group.
Next, we focused on the known endometrial receptivity genes by extracting gene lists from three commercially available endometrial receptivity tests: (i) the endometrial receptivity array (ERA; Igenomix, Spain) which includes 238 genes (Díaz-Gimeno et al., 2011); (ii) the ER Map®/ER Grade® test (iGLS, Spain) with focus on the 25 receptivity-sensitive genes (Enciso et al., 2018); and (iii) the beREADY® test (Competence Centre on Health Technologies, Estonia), integrated by a gene list of 57 genes (based on meta-analysis by Altmäe et al. (2017)). Every dataset included in our meta-analysis was assessed for any differentially expressed endometrial receptivity-specific transcript, and independently across the intersection between each receptivity test and the meta-analysed studies (ERA test versus studies; ER Map®/ER Grade® test versus studies; and beREADY® test versus studies). In preparation for the RRA method, the FC of the differentially expressed transcripts belonging to the receptivity-specific genes were extracted, and gene symbols were ranked according to that value.
Enrichment analyses
The functional enrichment analysis for Gene Ontology (GO) terms and biological pathways was performed using gprofiler2 package implemented in R (Raudvere et al., 2019) with Benjamini–Hochberg FDR multiple testing correction method applying a significance threshold at 0.05. The ‘ordered query’ option was applied. As data sources, the overrepresented signalling pathways were obtained by using GO for discovery of Biological Processes (BP), Molecular Functions (MF) and Cellular Components (CC), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome (REAC) databases. Functional enrichment analysis was performed for the ranked list of genes obtained after the application of the RRA method, and for the genes in the meta-analysed dataset specifically involved in endometrial receptivity.
Data mining of candidate genes
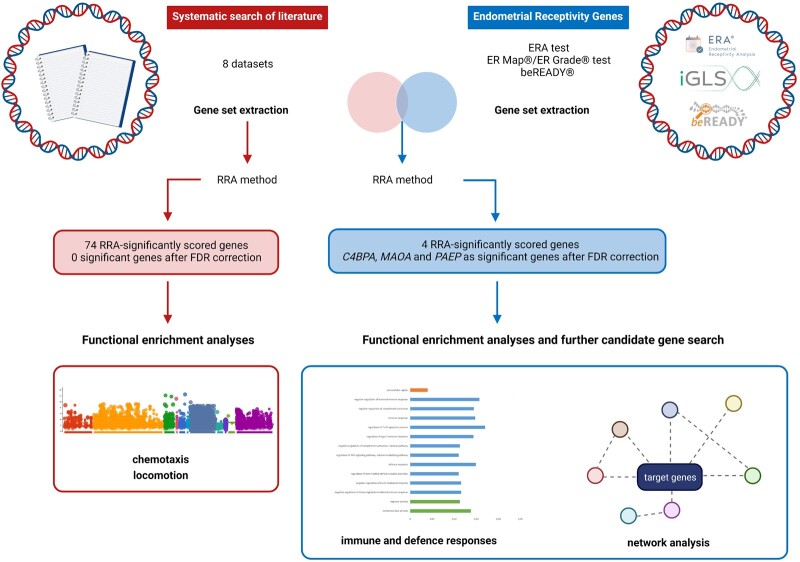
High-throughput experiments usually generate large sets of potential candidates among which only a few could be interpreted as truly relevant to the phenotype of study (Tranchevent et al., 2011). In our study, both web and desktop tools were applied to further investigate the main characteristics of the proposed candidates. DisGeNET, a discovery platform of genetic associations for human diseases (Bauer-Mehren et al., 2010; Piñero et al., 2020) was employed to interrogate for gene variants of endometriosis and also to check the diseases that have been reported in association with the candidate genes which arose from our study using the gene–disease associations and variant–disease associations tools, respectively (Bauer-Mehren et al., 2011; Piñero et al., 2020). Finally, and following the analyses based on network approaches, GeneMANIA plugin implemented in Cytoscape software was used to characterize and predict the interactions among the gene sets of interest (Warde-Farley et al., 2010). The complete pipeline of analysis we followed in this study is summarized in Fig. 1.
Figure 1.
Study design and the main results obtained. The initial systematic literature search resulted in eight studies suitable for the data extraction. In parallel, data from commercial endometrial receptivity tests were extracted and used to generate a dataset of endometrial receptivity-specific genes. Both independent datasets and their intersection were meta-analysed using the robust rank aggregation (RRA) method. Obtained results were utilized to perform functional enrichment analyses and further candidate gene search. FDR, false discovery rate. Figure was created using BioRender.
Results
Systematic literature search, meta-data creation and meta-analysis
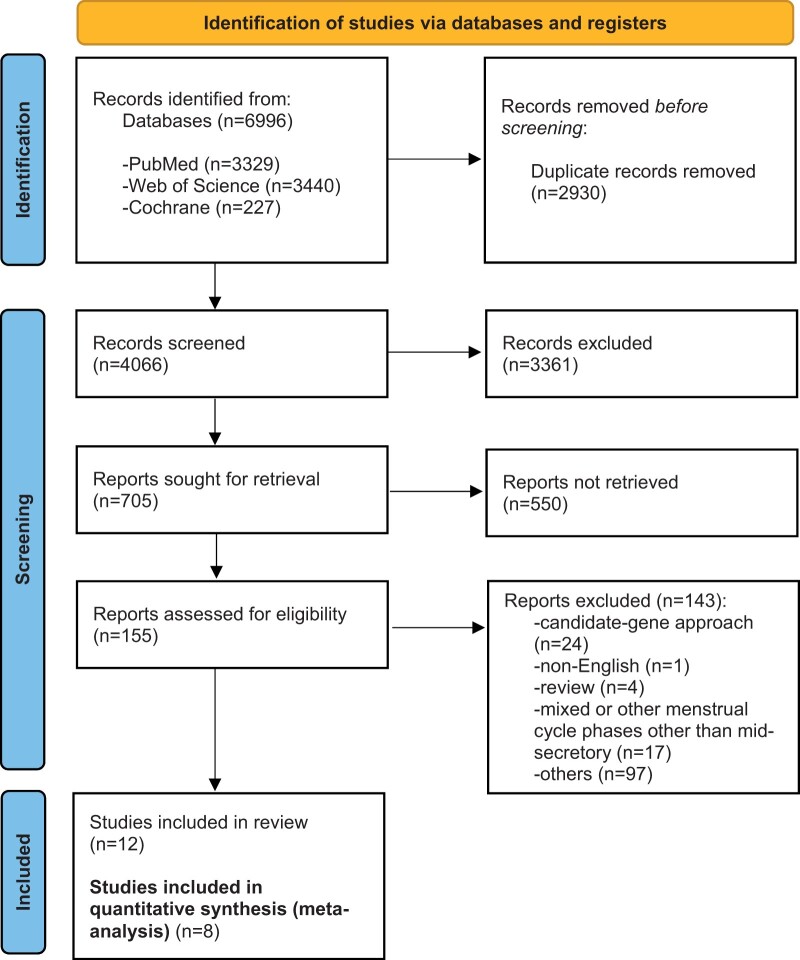
Out of the 155 total eligible studies obtained from the systematic literature search, 8 studies focusing on the analysis of the gene expression profile of the endometrial tissue of women with endometriosis at the receptive phase remained suitable for quantitative analysis (Fig. 2). The pooled dataset of differentially expressed genes composed in total of 125 samples, 78 eutopic endometrial samples from women with endometriosis and 47 endometrial samples from women without endometriosis, which served as the control group (Table I). After removal of the duplicated genes within studies, a total number of 7779 genes, 3496 up-regulated and 4283 down-regulated genes, were obtained for further analysis.
Figure 2.
Flow chart of the systematic literature search. Chart depicting the flow of information throughout the phases of the systematic review conducted following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement. After exclusion of non-eligible studies, a total number of 12 studies were included in the final list of articles suitable for meta-analysis. Of those, in 4 studies the data were not available, and 8 studies were finally subjected to meta-analysis.
Table I.
Studies selected for data extraction for meta-analysis after the systematic literature search.
| Authors | Controls/patients with endometriosis (sample size) | Array/sequencing platform | Cycle phase | Signif. threshold | FC threshold | Up-regulated transcripts | Down- regulated transcripts |
|---|---|---|---|---|---|---|---|
| Ahn et al. (2016) | Controls (n = 8): Healthy women undergoing tubal ligation. No hormonal therapy for 3 months before sampling. | nCounter human Immunology v2 panel | Sec.* | P ≤ 0.05 | >1.5 | 60 | 31 |
| Patients (n = 8): Stage III–IV endometriosis with infertility and/or pelvic pain. No hormonal therapy for 3 months before sampling. | |||||||
| Burney et al. (2007) | Controls (n = 8): Normally cycling women, volunteers to donate sample, no endometrial inflammation. No hormonal treatment for 3 months before sampling. | Affymetrix Human U133-Plus 2.0 | MS | FDR <0.05 | >1.5 | 428 | 293 |
| Patients (n = 9): Surgically documented and histologically validated moderate/severe-stage endometriosis. No hormonal treatment in preceeding 3 months. | |||||||
| Kao et al. (2003) | Controls (n = 7): Normally cycling women. Surgically confirmed without endometriosis. No medication. | Affymetrix Genechip Hu95A | MS | P < 0.05 | >2 | 63 | 86 |
| Patients (n = 8): Surgically documented mild/moderate endometriosis. No medication. | |||||||
| Matsuzaki et al. (2005) | Controls (n = 3): Fertile women with regular cycles undergoing tubal ligation or sterilization. No hormonal treatment in preceeding 6 months. | Clontech Atlashuman 1.2 cDNA expression array | MS | P < 0.05 | >3 | 6** | 20** |
| Patients (n = 3): Surgically confirmed deep endometriosis. No hormonal treatment for 6 months before surgery. | |||||||
| Tamaresis et al. (2014) | Controls (n = 8): No uterine/pelvic pathology, healthy volunteers for tubal ligation or endometrial biopsy. No hormonal treatment in preceeding 3 months. | Affymetrix Human U133-Plus 2.0 | MS | No threshold | >1.5 | 4020 | 6438 |
| Patients (n = 28): Laparoscopically confirmed endometriosis and pelvic pain and/or infertility. No hormonal treatment in preceeding 3 months. | |||||||
| Zhao et al. (2017) | Controls (n = 5): Laparoscopic surgery for examination or hydrotubation. No hormonal therapy. | Illumina HiSeq4000 system | Sec.* | FDR ≤0.05 | ≥2 (up) ≤0.5 (down) | 66 | 6 |
| Patients (n = 8): Endometriosis confirmed by histology, stage III–IV. No hormonal therapy. | |||||||
| Da Broi et al. (2019) | Controls (n = 5): Fertile healthy women with regular cycles, undergoing tubal ligation. No medication. | Illumina HiSeq2500 System | MS | adj P < 0.05 | log2 > 1 log2 < 1 | 0 | 0 |
| Patients (n = 6): Infertility exclusively associated to endometriosis (diagnosed by laparoscopy). No medication. | |||||||
| Joshi et al. (2021) | Controls (n = 3): Healthy endometriosis-free women. Regular cycles. No hormonal therapy for 3 months before sampling. | Affymetrix Human Gene 1.0 ST Arrays | MS | FDR ≤0.05 | >1.5 | 0 | 0 |
| Patients (n = 8): Endometriosis confirmed with laparoscopy and pathology. No hormonal therapy in preceding 3 months. | |||||||
| Total number of samples: | Total number of differentially expressed genes (with duplicates) | 4643 | 6874 | ||||
| Controls n = 47; Endometriosis n = 78 | |||||||
| Total differentially expressed genes (with real symbol and FC) | 4286 | 6465 | |||||
| Total number of unique differentially expressed genes | 3496 | 4283 | |||||
adj P: adjusted P-value; FC, fold change; FDR, false discovery rate; MS, mid-secretory phase; Sec.: secretory phase.
Secretory phase not specified but the focus on the study was on receptive phase.
Gene lists from epithelial and stromal cells were merged.
The integration of the lists of dysregulated genes using the RRA method led us to detect 74 genes from the 7779 meta-analysed genes showing significant RRA score (<0.05) (Table II). However, none of these genes remained under the significance threshold when the FDR multiple correction was applied. The complete results of the meta-analysis including RRA score and FDR values for each gene are presented in Supplementary Table SI.
Table II.
List of meta-analysed dysregulated genes after application of Robust Rank Aggregation (RRA) method.
| Gene symbol | Gene name | RRA score | FDR |
|---|---|---|---|
| FOSB ↑ | FosB Proto-Oncogene, AP-1 Transcription Factor Subunit | 4.12E−05 | 0.304 |
| S100A8 ↑ | S100 Calcium Binding Protein A8 | 0.00013341 | 0.492 |
| FOS ↑ | Fos Proto-Oncogene, AP-1 Transcription Factor Subunit | 0.00086691 | 1 |
| GUCY1B2 ↑↓ | Guanylate Cyclase 1 Soluble Subunit Beta 2 (Pseudogene) | 0.00157031 | 1 |
| CFB ↑ | Complement Factor B | 0.00211388 | 1 |
| IL32 ↑ | Interleukin 32 | 0.00247818 | 1 |
| SELL ↑↓ | Selectin L | 0.00329218 | 1 |
| CD4 ↑↓ | CD4 Molecule | 0.00421945 | 1 |
| EGR1 ↑ | Early Growth Response 1 | 0.00455357 | 1 |
| CYR61 ↑ | Cysteine-rich angiogenic inducer 61 | 0.00472531 | 1 |
| PPBP ↑ | Pro-Platelet Basic Protein | 0.0048764 | 1 |
| CASP5 ↑ | Caspase 5 | 0.0048764 | 1 |
| WT1 ↓ | WT1 Transcription Factor | 0.0048764 | 1 |
| RP11-319E12.2 ↑ | Clone-based (Vega) gene | 0.0048764 | 1 |
| SOCS1 ↑ | Suppressor of cytokine signalling 1 | 0.00724011 | 1 |
| IL17A ↑ | Interleukin 17A | 0.00974949 | 1 |
| LTB4R2 ↑ | Leukotriene B4 Receptor 2 | 0.00974949 | 1 |
| htMART ↑ | Putative mono-ADP-ribosyltransferase | 0.00974949 | 1 |
| ATP7B ↓ | ATPase Copper Transporting Beta | 0.00974949 | 1 |
| IGFBP1 ↑ | Insulin Like Growth Factor Binding Protein 1 | 0.00974949 | 1 |
| NR4A1 ↑ | Nuclear Receptor Subfamily 4 Group A Member 1 | 0.01027371 | 1 |
| CTNNB1 ↑↓ | Catenin Beta 1 | 0.01029457 | 1 |
| C4BPA ↑↓ | Complement Component 4 Binding Protein Alpha | 0.01211943 | 1 |
| HES1 ↑ | Hes Family BHLH Transcription Factor 1 | 0.01440884 | 1 |
| CXCL10 ↑ | C-X-C Motif Chemokine Ligand 10 | 0.01461928 | 1 |
| PTAFR ↑ | Platelet Activating Factor Receptor | 0.01461928 | 1 |
| NAB50 ↑ | RNA-binding protein CUG-BP/hNab50 | 0.01461928 | 1 |
| SLC1A1 ↓ | Solute Carrier Family 1 Member 1 | 0.01461928 | 1 |
| LRRC26 ↓ | Leucine Rich Repeat Containing 26 | 0.01461928 | 1 |
| LTBR ↑↓ | Lymphotoxin Beta Receptor | 0.01819934 | 1 |
| IL7R ↑ | Interleukin 7 Receptor | 0.01948577 | 1 |
| VSTM2L ↑ | V-Set and Transmembrane Domain Containing 2 Like | 0.01948577 | 1 |
| BSEP ↑ | Bile Salt Export Pump | 0.01948577 | 1 |
| SOS1 ↑↓ | SOS Ras/Rac Guanine Nucleotide Exchange Factor 1 | 0.01948577 | 1 |
| C1orf63 ↓ | Arginine And Serine Rich Protein 1 | 0.01948577 | 1 |
| SERPINB2 ↑ | Serpin Family B Member 2 | 0.01948577 | 1 |
| CD19 ↑ | CD19 Molecule | 0.02434897 | 1 |
| ASB2 ↑ | Ankyrin Repeat and SOCS Box Containing 2 | 0.02434897 | 1 |
| VDAC1P1 ↑ | Voltage Dependent Anion Channel 1 Pseudogene 1 | 0.02434897 | 1 |
| SMG1 ↓ | SMG1 Nonsense Mediated MRNA Decay Associated PI3K Related Kinase | 0.02434897 | 1 |
| CFH ↑ | Complement Factor H | 0.02920887 | 1 |
| GZMA ↑ | Granzyme A | 0.02920887 | 1 |
| NEAT1 ↑↓ | Nuclear Paraspeckle Assembly Transcript 1 | 0.02920887 | 1 |
| ZIC2 ↑ | Zic Family Member 2 | 0.02920887 | 1 |
| SLC6A7 ↓ | Solute Carrier Family 6 Member 7 | 0.02920887 | 1 |
| EREG ↑ | Epiregulin | 0.02920887 | 1 |
| MPPED2 ↑↓ | Metallophosphoesterase Domain Containing 2 | 0.02928315 | 1 |
| ANK3 ↑↓ | Ankyrin 3 | 0.03015014 | 1 |
| IER3 ↑↓ | Immediate Early Response 3 | 0.03035717 | 1 |
| CXCR2 ↑ | C-X-C Motif Chemokine Receptor 2 | 0.03406547 | 1 |
| LOC389332 ↑ | Small Integral Membrane Protein 32 | 0.03406547 | 1 |
| TRPM6 ↓ | Transient Receptor Potential Cation Channel Subfamily M Member 6 | 0.03406547 | 1 |
| CA1 ↑ | Carbonic Anhydrase 1 | 0.03406547 | 1 |
| ITGAL ↑ | Integrin Subunit Alpha L | 0.03406547 | 1 |
| MMP27 ↑ | Matrix Metallopeptidase 27 | 0.03406547 | 1 |
| HTRA3 ↑ | HtrA Serine Peptidase 3 | 0.03521064 | 1 |
| WAS ↑↓ | WASP Actin Nucleation Promoting Factor | 0.0375287 | 1 |
| CXCL9 ↑ | C-X-C Motif Chemokine Ligand 9 | 0.03891879 | 1 |
| SAP30L ↑↓ | SAP30 Like | 0.03891879 | 1 |
| PMS7 ↑ | HPMS7 protein | 0.03891879 | 1 |
| PTPN11 ↓ | Protein Tyrosine Phosphatase Non-Receptor Type 11 | 0.03891879 | 1 |
| ABP1 ↑ | Actin-binding protein | 0.03891879 | 1 |
| S100A7 ↓ | S100 Calcium Binding protein A7 | 0.03891879 | 1 |
| SAMD14 ↑ | Sterile Alpha Motif Domain Containing 14 | 0.04376881 | 1 |
| PRIM2 ↑ | DNA primase large subunit | 0.04376881 | 1 |
| DYNLL1 ↑ | Dynein Light Chain LC8-Type 1 | 0.04376881 | 1 |
| CTSW ↑ | Cathepsin W | 0.04376881 | 1 |
| MMP10 ↑ | Matrix Metallopeptidase 10 | 0.04376881 | 1 |
| KLRG2 ↑ | Killer Cell Lectin Like Receptor G2 | 0.04861554 | 1 |
| ORAI2 ↑ | ORAI Calcium Release-Activated Calcium Modulator 2 | 0.04861554 | 1 |
| IFNA21 ↑ | Interferon Alpha 21 | 0.04861554 | 1 |
| MAP3K8 ↓ | Mitogen-Activated Protein Kinase Kinase Kinase 8 | 0.04861554 | 1 |
| CCL3 ↑ | C-C Motif Chemokine Ligand 3 | 0.04861554 | 1 |
| LEFTY2 ↑ | Left-Right Determination Factor 2 | 0.04861554 | 1 |
For each gene, corresponding RRA score and false discovery rate (FDR) multiple correction values are presented. Arrows indicate the up- (↑) and down-regulated (↓) expression of the transcripts in the original datasets. Please note that for some genes, both up- and down-regulated expression can be observed among different studies (↑↓).
The enrichment analysis highlighted biological processes such as chemotaxis and locomotion as significantly enriched simultaneously across all the studies, suggesting a possible dysregulation in endometriosis. For the rest of the search categories, no significant differences between the study groups were detected (Supplementary Table SII).
Endometrial receptivity-specific genes
The dataset of endometrial receptivity genes was prepared for further analysis. In particular, for the ERA test, the 238 genes were ranked according to their absolute value of FC; the 25 ER Map®/ER Grade® test genes were ranked according to their normalized importance; and finally, for the beREADY® test, the 57 genes were ranked according to their RRA score. The full set of selected genes is presented in Supplementary Table SIII.
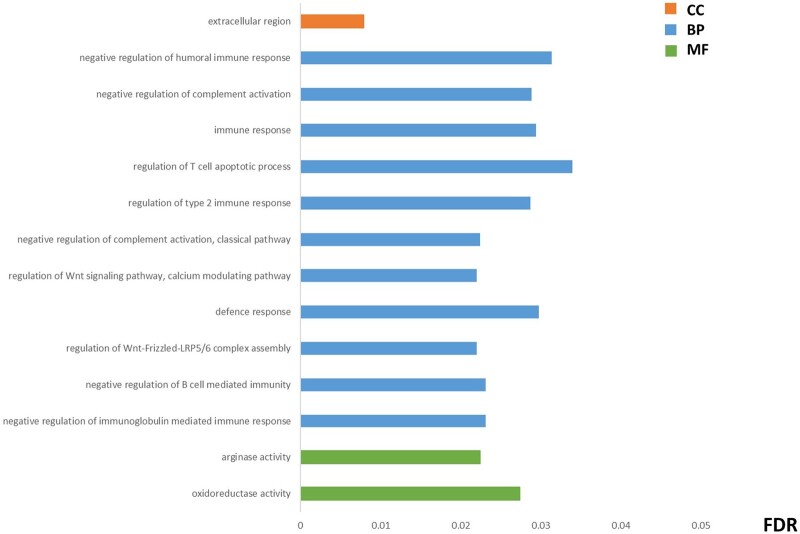
The intersection among the panel of endometrial receptivity genes and the differentially expressed genes from the dataset from our meta-analysis was calculated. This new subset was subjected to RRA method analysis in order to detect endometrial receptivity biomarkers that could be recurrently altered in our study datasets. Four genes (C4BPA, PAEP, MAOA and DKK1) were rated as significantly dysregulated in our meta-analysis, while C4BPA, PAEP and MAOA remained significant after FDR correction (Supplementary Table SIV). Next, we were interested in detecting the underlying functional processes in which these RRA-ranked receptivity-specific transcripts are involved through a functional enrichment analysis, where the intersection analysis of enriched processes between each endometrial receptivity test and our meta-analysed transcripts detected different molecular functions and biological processes mainly related to immune and defence responses (Fig. 3; Supplementary Table SV).
Figure 3.
Functional enrichment analysis of the endometrial receptivity genes within the meta-analysed transcripts. The most representative items detected were consistently dysregulated in the intersection between each endometrial receptivity test and the studies included in the meta-analysis (endometrial receptivity array (ERA) versus studies; ER Map®/ER Grade® test versus studies; and beREADY® test versus studies). Only the processes that have a significant false discovery rate (FDR) across the three comparisons are shown. The values of FDR in the figure correspond to the average value of the FDR for each comparison. Complete details of the results of the meta-analysis are described in Supplementary Table SV. BP, biological processes; CC, cellular components; MF, molecular functions.
Data mining of candidate genes
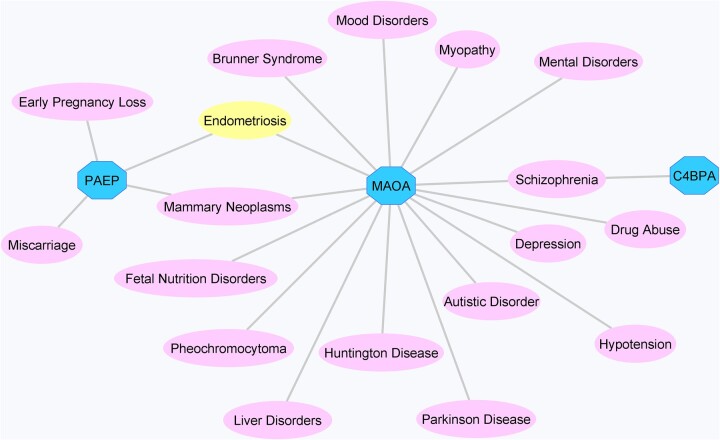
The endometrial receptivity-specific genes C4BPA, PAEP and MAOA dysregulated in women with endometriosis were subjected to further analysis in order to contextualize their role in the endometrial processes in endometriosis. The gene–disease and variant–disease association analyses resulted in 107 diseases and 4 variants in association with the C4BPA gene, while 300 diseases and 15 variants were identified for the MAOA gene, and 397 diseases and 2 variants were identified for the PAEP gene (Supplementary Table SVI). There was an overlap of 18 diseases among the three genes regarding gene–disease associations, and an association between PAEP and MAOA genes and endometriosis was identified (Fig. 4). No associations between the variants and disease were detected (Supplementary Table SVI).
Figure 4.
Gene–disease association network for the C4BPA, MAOA and PAEP genes created using the information displayed by DisGeNET plugin implemented in Cytoscape software. Genes are represented by octagons, while associated diseases are shown as ellipses.
Furthermore, a gene interaction network was constructed in Cytoscape, where C4BPA, PAEP and MAOA genes were connected to 20 other molecules (Fig. 5). Of interest are genes such as the progesterone receptor (PGR), DNAJB5, PTPRC, CD40 and NFKBIA, which were also identified in our dataset of meta-analysis, and the C3, CD55 and NDRG1 genes among the endometrial receptivity-specific biomarker list.
Figure 5.
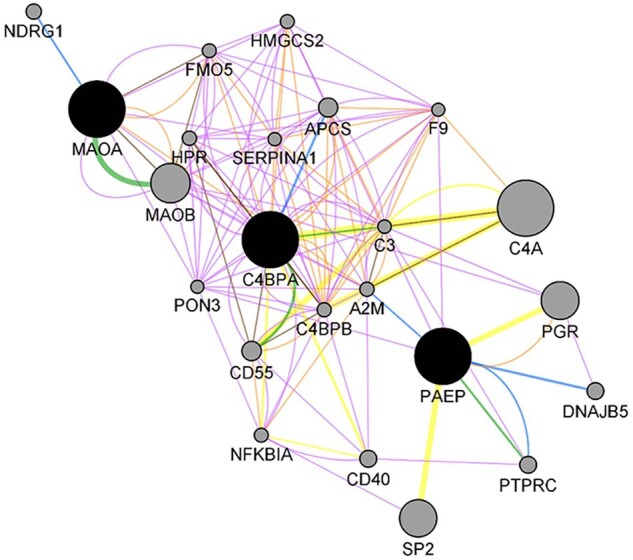
Gene network analysis showing the interaction among the most relevant genes (C4BPA, MAOA and PAEP). Seed genes are shown in black, while interactors appear in grey. The colour of the connection denotes the type of interaction established among the different nodes: blue for physical interactions (67.64% of interactions); purple for co-expression (13.50%); green for predicted interactions (6.35%); orange for co-localisation (6.17%); yellow for shared pathways (4.35%); and brown for shared protein domains (0.59%).
Discussion
Our study presents the first meta-analysis approach in detecting the endometrial receptivity transcriptome in endometriosis and demonstrates molecular differences among women with endometriosis when compared with women without the disease at the receptive phase endometria, although the molecular differences were small-scale.
There is an ongoing debate about whether the endometrial implantation potential of women with endometriosis is impaired or not (Garcia-Velasco et al., 2015; Lessey and Kim, 2017; Miravet-Valenciano et al., 2017). The possibility of the altered endometrial receptivity in eutopic endometria of women with endometriosis is based on the fact that endometriosis impacts cycle fecundity through the systemic and local inflammatory changes that take place as a consequence of the disease (Lessey and Kim, 2017). In a previous study of 240 IVF cycles, where sibling oocytes from the same donor were transferred into women with endometriosis and without the disease, reduced implantation and pregnancy rates were demonstrated among the endometriosis group (Prapas et al., 2012). Furthermore, a recent matched cohort study on 1053 IVF/ICSI cycles with fresh single embryo transfers demonstrated that in women with endometriosis an impaired implantation factor contributed to the reduced pregnancy outcomes (Blank et al., 2021). On the other side, many studies support the clinical observations that eutopic endometrium is not impaired in women with endometriosis, based on the results of IVF and oocyte donation programs (Miravet-Valenciano et al., 2017; Saare et al., 2017; Da Broi et al., 2019). With the first meta-analysis approach focusing on receptive phase endometria where we analysed carefully selected studies of endometrial transcriptome profiles in 125 women, molecular differences in women with endometriosis were detected, although the differences were minimal.
Biological processes such as chemotaxis and locomotion were dysregulated in the endometria of women with endometriosis. These processes have been previously connected to endometriosis (Devesa-Peiro et al., 2020). Locomotion encompasses a variety of processes involving the self-propelled movement of a cell from one location to another. In endometriosis, cell migration processes of the endometrial tissue are considered an important factor to explain the pathogenesis of the disease (Saare et al., 2017). Indeed, it has been described that aberrant cell migration patterns might result in the impairment of the function of endometrial and endometriotic epithelial and stromal cells of women with endometriosis (Matsuzaki and Darcha, 2013). Altered endometrial cellular composition and functionality in endometriosis has recently been demonstrated (Bunis et al., 2022). In line with these findings, a recent insilico analysis of transcriptome studies has proposed the involvement of 22 potential biomarkers involved in cell motility and migration in both the ectopic and eutopic endometria of women with endometriosis (Devesa-Peiro et al., 2021). Furthermore, chemotaxis was perturbed in women with endometriosis due to the action exerted by oestrogens and progestogens, which would contribute to the known hallmarks of endometriosis, such as inflammatory response or abnormal tissue remodelling, among others (Reis et al., 2013). Also in previous endometrial transcriptome studies in women with endometriosis slight gene expression differences or no differences have been detected (Garcia-Velasco et al., 2015; Ahn et al., 2016; Miravet-Valenciano et al., 2017; Saare et al., 2017; Da Broi et al., 2019; Joshi et al., 2021). With our meta-analysis approach, we were able to increase the sample size and thereby the power in detecting small molecular differences which could have been lacking in some of the previously published studies (Thorlund and Mills, 2012; Serdar et al., 2021). Altogether, the complex dynamics of the endometrial tissue could underlie the little variations in gene expression and could explain the fact that the affected biological processes and molecular pathways tend to have a common root, although the individual gene expression patterns show low overlap among the studies involved (McKinnon et al., 2018).
Next, our analysis focusing on the endometrial receptivity-specific genes highlighted the dysregulation of three known biomarkers of endometrial receptivity C4BPA, MAOA and PAEP genes in the endometria of women with endometriosis. Abnormally decreased levels of C4BPA (Complement Component 4 Binding Protein Alpha) have been detected in the mid-secretory endometrium of women with endometriosis, implantation failure and unexplained recurrent abortion (Kao et al., 2003; Tapia et al., 2008; Herington et al., 2016; Altmäe et al., 2017), suggesting its possible influence in the mechanisms leading to successful embryo implantation. C4BPA has also been identified as candidate target marker in ovarian clear cell carcinomas (Mikami et al., 2015), a well-known comorbidity of endometriosis (Vargas et al., 2020). MAOA (Monoamine Oxidase A) is a putative endometrial receptivity biomarker (Altmäe et al., 2017; Wang et al., 2020), which was further linked to endometriosis in our insilico data mining according to the DisGeNET analysis.
PAEP (Progestagen-Associated Endometrial Protein), also named as glycodelin, is a protein with immunosuppressive properties and has been described to take part in the endometrial receptivity processes (Oehninger et al., 1995; Seppälä et al., 1998; Kao et al., 2003; Tapia et al., 2008; Vargas et al., 2012; Herington et al., 2016; Altmäe et al., 2017; Wang et al., 2020) and in different aspects of endometriosis (O et al., 2018). Interestingly, its implication in endometriosis-related infertility and impaired receptivity has been proposed (Dutta et al., 2001; Focarelli et al., 2018). PAEP protein is expressed in the endometrial glandular compartment among patients with endometriosis (Kämäräinen et al., 1993). Furthermore, direct correlation of plasma concentrations of PAEP with the severity of deep infiltrating endometriosis has been reported (Koninckx et al., 1992). More recently, PAEP concentration has been reported as significantly higher in the peritoneal fluid of patients with endometriosis compared to controls in both the proliferative and secretory phases of the menstrual cycle (Nirgianakis et al., 2020). Furthermore, a recent study using organoids expressing glycodelin demonstrated that those deriving from the eutopic endometrium of women with endometriosis exhibited a glycosylation pattern significantly different from that of organoids from healthy women (Luddi et al., 2020). Our insilico data mining detected a number of important genes directly interacting with PAEP, MAOA and C4BPA genes such as PGR, CD40, CD55, NFKBIA or SERPINA1, suggesting that these biomarkers could be involved in different molecular processes and pathways. Thus, future studies should investigate the role of these endometrial receptivity biomarkers, PAEP, MAOA and C4BPA, in endometrial function and whether they could serve for identifying an impaired implantation factor in women with endometriosis undergoing IVF/ICSI treatments and direct clinical management.
The intersection analysis of the endometrial receptivity-specific transcripts with our meta-analysed data led to identification and confirmation of dysregulation of biological processes relevant to the aetiopathogenesis of endometriosis, such as immune and defence responses. The association between endometriosis and immunity has been largely demonstrated over the years, with reports of abnormalities in the immune system of women with endometriosis that may be a reflection of the inflammatory response developed during the disease (Shigesi et al., 2019; Poli-Neto et al., 2020). Indeed, it is claimed that the presence of endometriosis predisposes to, or is associated with, the development of autoimmune conditions (Kvaskoff et al., 2014; Shigesi et al., 2019; Vargas et al., 2020). In line, our insilico data mining detected hundreds of diseases associated with the endometrial receptivity biomarkers dysregulated in our study set of endometriosis.
Some limitations of our meta-analysis should be highlighted. The lack of available raw data and/or full gene lists allowed us to focus only on the differentially expressed gene lists, which could have reduced the sensitivity of the findings. Also, not all women in the control group were with proven fertility, and although most of the volunteers in the control group were surgically confirmed to be endometriosis-free, we cannot rule out that a few of them had endometriosis, thereby possibly minimizing the differences between study groups. Furthermore, the stringent criteria utilized for the selection of the differentially expressed genes after running the meta-analysis does not allow us to exclude the null hypothesis, meaning that we cannot rule out the dysregulation of specific genes in endometrium of women with endometriosis. Studies on bigger sample size and better-defined control groups are warranted.
In conclusion, we were able to compile a long list of differentially expressed genes in different studies through a systematic review. Moreover, when we integrated the sets of genes originating from different studies through a meta-analysis, the functional enrichment analysis detected a slight molecular dysregulation where biological processes such as chemotaxis and locomotion were involved. Regarding the analysis of endometrial receptivity-specific genes, C4BPA, MAOA and PAEP expression and molecular pathways involved in the immune and defence responses were dysregulated among women with endometriosis. In short, our meta-analysis detected slight molecular differences in the transcriptome profile in endometriosis that could explain, at least in part, the impaired reproductive outcomes in some women with endometriosis. Further research of the molecules and pathways identified in biomarker and therapeutical applicability is warranted to make these findings clinically relevant.
Data availability
All data are incorporated into the article and its online supplementary material.
Authors’ roles
S.A. and E.V. conceived the study and performed the systematic review. E.G.-M. and E.V. analysed the data. F.J.E. assisted in the experimental design and data analysis. S.A. and E.V. wrote the main body of the manuscript. A.S., L.A. and J.A.H. gave advice on the writing of the manuscript. All the authors participated actively in the critical revision of the manuscript.
Funding
This work was supported by the Spanish Ministry of Education, Culture and Sport [grant FPU15/01193] and the Margarita Salas program for the Requalification of the Spanish University system [grant UJAR01MS]; Spanish Ministry of Economy, Industry and Competitiveness (MINECO) and European Regional Development Fund (FEDER): grants RYC-2016-21199 and ENDORE SAF2017-87526-R; Programa Operativo FEDER Andalucía (B-CTS-500-UGR18; A-CTS-614-UGR20); the Junta de Andalucía [BIO-302; and PAIDI P20_00158]; the University of Jaén [PAIUJA-EI_CTS02_2017]; the University of Granada, Plan Propio de Investigación 2016, Excellence actions: Units of Excellence; Unit of Excellence on Exercise and Health (UCEES), and by the Junta de Andalucía, Consejería de Conocimiento, Investigación y Universidades and European Regional Development Fund (ERDF), ref. SOMM17/6107/UGR; the Estonian Research Council (grant PRG1076); Horizon 2020 innovation (ERIN, grant no. EU952516) of the European Commission and Enterprise Estonia (grant EU48695).
Conflict of interest
The authors declare no competing interests.
Supplementary Material
References
- Ahn SH, Khalaj K, Young SL, Lessey BA, Koti M, Tayade C.. Immune-inflammation gene signatures in endometriosis patients. Fertil Steril 2016;106:1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmäe S, Aghajanova L.. What do we know about endometrial receptivity in women with endometriosis? A molecular perspective. Reprod Biomed Online 2015;31:581–583. [DOI] [PubMed] [Google Scholar]
- Altmäe S, Esteban FJ, Stavreus-Evers A, Simón C, Giudice L, Lessey BA, Horcajadas JA, Macklon NS, D'Hooghe T, Campoy C. et al. Guidelines for the design, analysis and interpretation of “omics” data: focus on human endometrium. Hum Reprod Update 2014;20:12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmäe S, Koel M, Võsa U, Adler P, Suhorutšenko M, Laisk-Podar T, Kukushkina V, Saare M, Velthut-Meikas A, Krjutškov K. et al. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomics biomarkers. Sci Rep 2017;7:10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer-Mehren A, Bundschus M, Rautschka M, Mayer MA, Sanz F, Furlong LI.. Gene-disease network analysis reveals functional modules in Mendelian, complex and environmental disorders. PLoS One 2011;6:e20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer-Mehren A, Rautschka M, Sanz F, Furlong LI.. DisGeNET: a Cytoscape plugin to visualize, integrate, seach and analyze gene-disease networks. Bioinformatics 2010;26:2924–2926. [DOI] [PubMed] [Google Scholar]
- Blank C, Deboever C, Decroos E, DeCroo I, Tilleman K, De Sutter P, Mischi M, Schoot BC.. Impaired implantation in endometriosis compared with couples with male subfertility after transfer of equal quality embryos: a matched cohort study. Reprod Biomed Online 2021;42:165–174. [DOI] [PubMed] [Google Scholar]
- Brosens I, Brosens JJ, Benagiano G.. The eutopic endometrium in endometriosis: are the changes of clinical significance? Reprod Biomed Online 2012;24:496–502. [DOI] [PubMed] [Google Scholar]
- Bunis DG, Wang W, Vallvé-Juanico J, Houshdaran S, Sen S, Soltane IB, Kosti I, Vo KC, Irwin JC, Giudice LC. et al. Whole-tissue deconvolution and scRNAseq analysis identify altered endometrial cellular compositions and functionality associated with endometriosis. Front Immunol 2022;12:788315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC.. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 2007;148:3814–3826. [DOI] [PubMed] [Google Scholar]
- Da Broi MG, Meola J, Plaça JR, Peronni KC, Rocha CV, Silva WA, Ferriani RA, Navarro PA.. Is the profile of transcripts altered in the eutopic endometrium of infertile women with endometriosis during the implantation window? Hum Reprod 2019;34:2381–2390. [DOI] [PubMed] [Google Scholar]
- Devesa-Peiro A, Sebastian-Leon P, Garcia-Garcia F, Arnau V, Aleman A, Pellicer A, Diaz-Gimeno P.. Uterine disorders affecting female fertility: what are the molecular functions altered in endometrium? Fertil Steril 2020;113:1261–1274. [DOI] [PubMed] [Google Scholar]
- Devesa-Peiro A, Sebastian-Leon P, Pellicer A, Diaz-Gimeno P.. Guidelines for biomarker discovery in endometrium: correcting for menstrual cycle bias reveals new genes associated with uterine disorders. Mol Hum Reprod 2021;27:gaab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Gimeno P, Horcajadas JA, Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A, Simón C.. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril 2011;95:50–60. [DOI] [PubMed] [Google Scholar]
- Dutta B, Ain R, Seshagiri PB, Karande AA.. Differential influence of recombinant non-glycosylated and glycosylated glycodelin on human sperm function: comparative studies with hamster spermatozoa. Reprod Fertil Dev 2001;13:111–118. [DOI] [PubMed] [Google Scholar]
- Enciso M, Carrascosa JP, Sarasa J, Martínez-Ortiz PA, Munné S, Horcajadas JA, Aizpurua J.. Development of a new comprehensive and reliable endometrial receptivity map (ER Map/ER Grade) based on RT-qPCR gene expression analysis. Hum Reprod 2018;33:220–228. [DOI] [PubMed] [Google Scholar]
- Fassbender A, Verbeeck N, Börnigen D, Kyama CM, Bokor A, Vodolazkaia A, Peeraer K, Tomassetti C, Meuleman C, Gevaert O. et al. Combined mRNA microarray and proteomic analysis of eutopic endometrium of women with and without endometriosis. Hum Reprod 2012;27:2020–2029. [DOI] [PubMed] [Google Scholar]
- Focarelli R, Luddi A, De Leo V, Capaldo A, Stendardi A, Pavone V, Benincasa L, Belmonte G, Petraglia F, Piomboni P.. Dysregulation of GdA expression in endometrium of women with endometriosis: implication for endometrial receptivity. Reprod Sci 2018;25:579–586. [DOI] [PubMed] [Google Scholar]
- Garcia-Velasco JA, Fassbender A, Ruiz-Alonso M, Blesa D, D'Hooghe T, Simon C.. Is endometrial receptivity transcriptomics affected in women with endometriosis? A pilot study. Reprod Biomed Online 2015;31:647–654. [DOI] [PubMed] [Google Scholar]
- Giudice LC. Genomics’ role in understanding the pathogenesis of endometriosis. Semin Reprod Med 2003;21:119–124. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC.. Endometriosis. Lancet 2004;364:1789–1799. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Telles TL, Lobo S, Kao L.. The molecular basis for implantation failure in endometriosis: on the road to discovery. Ann N Y Acad Sci 2002;955:252–264. [DOI] [PubMed] [Google Scholar]
- Gupta S, Goldberg JM, Aziz N, Goldberg E, Krajcir N, Agarwal A.. Pathogenic mechanisms in endometriosis-associated infertility. Fertil Steril 2008;90:247–257. [DOI] [PubMed] [Google Scholar]
- Herington JL, Guo Y, Reese J, Paria BC.. Gene profiling the window of implantation: microarray analyses from human and rodent models. J Reprod Health Med 2016;2:S19–S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J, Sterrenburg M, Lane S, Maheshwari A, Li TC, Cheong Y.. Reproductive, obstetric, and perinatal outcomes of women with adenomyosis and endometriosis: a systematic review and meta-analysis. Hum Reprod Update 2019;25:592–632. [DOI] [PubMed] [Google Scholar]
- Joshi NR, Kohan-Ghadr HR, Roqueiro DS, Yoo JY, Fru K, Hestermann E, Yuan L, Ho SM, Jeong JW, Young SL.. Genetic and epigenetic changes in the eutopic endometrium of women with endometriosis: association with decreased endometrial αvβ3 integrin expression. Mol Hum Reprod 2021;27:gaab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämäräinen M, Leivo I, Julkunen M, Seppälä M.. Localization of progesterone associated endometrial protein mRNA by in-situ hybridization in human pregnancy decidua, endometriosis and borderline endometrioid adenoma. J Mol Endocrinol 1993;10:71–77. [DOI] [PubMed] [Google Scholar]
- Kao LC, Germeyer A, Tulac S, Lobo S, Yang JP, Taylor RN, Osteen K, Lessey BA, Giudice LC.. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology 2003;144:2870–2881. [DOI] [PubMed] [Google Scholar]
- Kokcu A. Possible effects of endometriosis-related immune events on reproductive function. Arch Gynecol Obstet 2013;287:1225–1233. [DOI] [PubMed] [Google Scholar]
- Kolde R, Laur S, Adler P, Vilo J.. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 2012;28:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koninckx PR, Riittinen L, Seppala M, Cornillie FJ.. CA-125 and placental protein 14 concentrations in plasma and peritoneal fluid of women with deeply infiltrating pelvic endometriosis. Fertil Steril 1992;57:523–530. [PubMed] [Google Scholar]
- Kvaskoff M, Mu F, Terry KL, Harris HR, Poole EM, Farland L, Missmer SA.. Endometriosis: a high-risk population for major chronic diseases? Hum Reprod Update 2014;21:500–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessey BA, Kim JJ.. Endometrial receptivity in the eutopic endometrium of women with endometriosis: it is affected, and let me show you why. Fertil Steril 2017;108:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luddi A, Pavone V, Semplici B, Governini L, Criscuoli M, Paccagnini E, Gentile M, Morgante G, De Leo V, Belmonte G. et al. Organoids of human endometrium: a powerful in vitro model for the endometrium-embryo cross-talk at the implantation site. Cells 2020;9:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki S, Canis M, Vaurs-Barrière C, Boespflug T, Dastugue B, Mage G.. DNA microarray analysis of gene expression in eutopic endometrium from patients with deep endometriosis using laser capture microdissection. Fertil Steril 2005;84:1180–1190. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Darcha C.. In vitro effects of a small-molecule antagonist of the Tcf/β-catenin complex on endometrial and endometriotic cells of patients with endometriosis. PLoS One 2013;8:e61690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KE, Villar J, Kirtley S, Kennedy SH, Becker CM.. Endometrial alterations in endometriosis: a systematic review of putative biomarkers. Hum Reprod Update 2011;17:637–653. [DOI] [PubMed] [Google Scholar]
- McKinnon B, Mueller M, Montgomery G.. Progesterone resistance in endometriosis: an acquired property? Trends Endocrinol Metab 2018;29:535–548. [DOI] [PubMed] [Google Scholar]
- Mikami M, Tanabe K, Matsuo K, Miyazaki Y, Miyazawa M, Hayashi M, Asai S, Shida M, Hirasawa T, Kojima N. et al. Fully-sialylated alpha-chain of complement 4-binding protein: diagnostic utility for ovarian clear cell carcinoma. Gynecol Oncol 2015;139:520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet-Valenciano J, Ruiz-Alonso M, Gómez E, Garcia-Velasco JA.. Endometrial receptivity in eutopic endometrium in patients with endometriosis: it is not affected, and let me show you why. Fertil Steril 2017;108:28–31. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirgianakis K, McKinnon B, Ma L, Imboden S, Bersinger N, Mueller MD.. Peritoneal fluid biomarkers in patients with endometriosis: a cross-sectional study. Horm Mol Biol Clin Investig 2020;42:113–122. [DOI] [PubMed] [Google Scholar]
- O DF, Flores I, Waelkens E, D’Hooghe T.. Noninvasive diagnosis of endometriosis: review of current peripheral blood and endometrial biomarkers. Best Pract Res Clin Obstet Gynaecol 2018;50:72–83. [DOI] [PubMed] [Google Scholar]
- Oehninger S, Coddington CC, Hodgen GD, Seppala M.. Factors affecting fertilization: endometrial placental protein 14 reduces the capacity of human spermatozoa to bind to the human zona pellucida. Fertil Steril 1995;63:377–383. [DOI] [PubMed] [Google Scholar]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE. et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI.. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48:D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli-Neto OB, Meola J, Rosa-E-Silva JC, Tiezzi D.. Transcriptome meta-analysis reveals differences of immune profile between eutopic endometrium from stage I-II and III-IV endometriosis independently of hormonal milieu. Sci Rep 2020;10:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Endometriosis and infertility: a committee opinion. Fertil Steril 2012;98:591–598. [DOI] [PubMed] [Google Scholar]
- Prapas Y, Goudakou M, Matalliotakis I, Kalogeraki A, Matalliotaki C, Panagiotidis Y, Ravanos K, Prapas N.. History of endometriosis may adversely affect the outcome in menopausal recipients of sibling oocytes. Reprod Biomed Online 2012;25:543–548. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J.. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis FM, Petraglia F, Taylor RN.. Endometriosis: hormone regulation and clinical consequences of chemotaxis and apoptosis. Hum Reprod Update 2013;19:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saare M, Peters M, Aints A, Laisk-Podar T, Salumets A, Altmäe S.. OMICs studies and endometriosis biomarker identification. In: D’Hooghe T (ed). Biomarkers for Endometriosis: State of the Art. Cham: Springer, 2017, 227–258. [Google Scholar]
- Seppälä M, Bohn H, Tatarinov Y.. Glycodelins. Tumour Biol 1998;19:213–220. [DOI] [PubMed] [Google Scholar]
- Serdar CC, Cihan M, Yücel D, Serdar MA.. Sample size, power and effect size revisited: simplified and practical approaches in pre-clinical, clinical and laboratory studies. Biochem Med (Zagreb) 2021;31:010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigesi N, Kvaskoff M, Kirtley S, Feng Q, Fang H, Knight JC, Missmer SA, Rahmioglu N, Zondervan KT, Becker CM.. The association between endometriosis and autoimmune diseases: a systematic review and meta-analysis. Hum Reprod Update 2019;25:486–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaresis JS, Irwin JC, Goldfien GA, Rabban JT, Burney RO, Nezhat C, DePaolo LV, Giudice LC.. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology 2014;155:4986–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia A, Gangi LM, Zegers-Hochschild F, Balmaceda J, Pommer R, Trejo L, Pacheco IM, Salvatierra AM, Henríquez S, Quezada M. et al. Differences in the endometrial transcript profile during the receptive period between women who were refractory to implantation and those who achieved pregnancy. Hum Reprod 2008;23:340–351. [DOI] [PubMed] [Google Scholar]
- Thorlund K, Mills EJ.. Sample size and power considerations in network meta-analysis. Syst Rev 2012;1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranchevent LC, Capdevila FB, Nitsch D, De Moor B, De Causmaecker P, Moreau Y.. A guide to web tools to prioritize candidate genes. Brief Bioinform 2011;12:22–32. [DOI] [PubMed] [Google Scholar]
- Vargas E, Aghajanova L, Gemzell-Danielsson K, Altmäe S, Esteban FJ.. Cross-disorder analysis of endometriosis and its comorbid diseases reveals shared genes and molecular pathways and proposes putative biomarkers of endometriosis. Reprod Biomed Online 2020;40:305–318. [DOI] [PubMed] [Google Scholar]
- Vargas MF, Tapia-Pizarro AA, Henríquez SP, Quezada M, Salvatierra AM, Noe G, Munroe DJ, Velasquez LA, Croxatto HB.. Effect of single post-ovulatory administration of levonorgestrel on gene expression profile during the receptive period of the human endometrium. J Mol Endocrinol 2012;48:25–36. [DOI] [PubMed] [Google Scholar]
- Vigano P, Corti L, Berlanda N.. Beyond infertility: obstetrical and postpartum complications associated with endometriosis and adenomyosis. Fertil Steril 2015;104:802–812. [DOI] [PubMed] [Google Scholar]
- Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, Pan W, Simon C, Quake SR.. Single-cell transcriptomics atlas of the human endometrium during the menstrual cycle. Nat Med 2020;26:1644–1653. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, St Clair JB, Fu T, Stratton P, Nieman LK.. Reduced expression of biomarkers associated with the implantation window in women with endometriosis. Fertil Steril 2009;91:1686–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gu C, Ye M, Zhang Z, Han W, Fan W, Meng Y.. Identification of global transcriptome abnormalities and potential biomarkers in eutopic endometria of women with endometriosis: a preliminary study. Biomed Rep 2017;6:654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondervan KT, Becker CM, Koga K, Missmer SA, Taylor RN, Viganò P.. Endometriosis. Nat Rev Dis Primers 2018;4:9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its online supplementary material.