Abstract
Objective
The SARS-CoV-2 Omicron (B.1.1.529) variant has caused global concern. Previous studies have shown that the variant has enhanced immune evasion ability and transmissibility and reduced severity.
Methods
In this study, we developed a mathematical model with time-varying transmission rate, vaccination, and immune evasion. We fit the model to reported case and death data up to February 6, 2022 to estimate the transmissibility and infection fatality ratio of the Omicron variant in South Africa.
Results
We found that the high relative transmissibility of the Omicron variant was mainly due to its immune evasion ability, whereas its infection fatality rate substantially decreased by approximately 78.7% (95% confidence interval: 66.9%, 85.0%) with respect to previous variants.
Conclusion
On the basis of data from South Africa and mathematical modeling, we found that the Omicron variant is highly transmissible but with significantly lower infection fatality rates than those of previous variants of SARS-CoV-2.
Keywords: COVID-19, Omicron, Infection fatality rate, Immune evasion
Introduction
The COVID-19 pandemic has been in effect for nearly two years since 2019. According to the World Health Organization (WHO), there have been over 260 million cases including more than 5 million deaths reported (https://www.who.int/emergencies/diseases/novel-coronavirus-2019 2021). The SARS-CoV-2 virus, first identified in late 2019, has mutated multiple times, and its variants have been classified by the WHO into three categories: variants of concern (VOC), variants of interest, and variants under monitoring. Four VOC—Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2)—have been responsible for a large number of infections and deaths worldwide. On November 26, 2021, the Omicron (B.1.1.529) variant was designated as the fifth VOC (He et al., 2021).
Before the emergence of the Omicron variant, three waves of infections and deaths by three distinct variants had occurred in South Africa, with nearly 3 million confirmed cases. The first infections occurred in March 2020, peaked in July and ended in September 2020 (Pulliam et al., 2021). The second wave of the epidemic, with the Beta variant, began in October 2020 with progressively higher levels of infections in Nelson Mandela Bay, followed by that in Eastern Cape, Western Cape and KwaZulu-Natal by early December 2020 (Tegally et al., 2021). In May 2021, the emergence of the Delta variant led to a third wave in South Africa. This variant quickly replaced the Beta variant in South Africa, spreading rapidly and peaking in July, when it accounted for 86% of the viruses sequenced in the first week (Abdool Karim and Baxter 2021).
The Omicron variant, which quickly became the main variant in Gauteng, was first detected in South Africa on November 23, 2021, followed by a dramatic increase in the number of infections. There have been more than 2000 cases of the Omicron variant in South Africa as of early December 2021 (Vaughan 2021). As the most mutable variant, the Omicron variant has at least 30 amino acid substitutions, three deletions and one small insertion. It is noteworthy that 15 of the 30 amino acid substitutions are located in the receptor-binding portion, which includes S371L, S373P, S375F, K417N, N440K, G446S, S447N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, and D614G ('Science Brief:Omicron (B.1.1.529) Variant', 2021). In addition, the Omicron variant carries the mutation found in other VOC, in which a deletion was found at the peak position 60-79. It has three key mutations similar to the Beta and Gamma (P.1) variants, which may increase its ability to escape immunity (Poudel et al., 2022). In an experiment of incubating the virus in convalescent sera from patients infected with prior subtypes, Zhang et al showed that convalescent sera of patients infected with early strains or the Delta variant have relatively low neutralization ability against the Omicron variant (Zhang et al., 2021). The early strain- and Delta-infected patients’ neutralizing antibody titer in convalescent sera against the Omicron variant decreased 36 times and 39 times, respectively, contributing to the variant's immune escape ability.
Fortunately, the severity of the Omicron variant seems to differ from that of its predecessors. In Tshwane, South Africa, the bed occupancy rate during the peak of the Omicron variant wave was about half of that during the Delta variant wave, suggesting that the number of hospitalizations was lower with the Omicron than with the Delta variant (Abdullah et al., 2022). In addition, fewer intensive care unit admissions and shorter hospital stays may indicate reduced disease severity with Omicron variant infection.
In this study, we developed a model to fit both the reported cases and deaths in South Africa, with the aim of quantifying the impact of the Omicron variant on the infection fatality rate in South Africa.
Method
We obtained reported cases, deaths, excess deaths and vaccination data from Our World in Data ('Johns Hopkins University CSSE.' 2022; Hannah Ritchie 2020; Mathieu et al., 2021) and retrieved aggregated variant proportion data from Our World in Data, CoVariants.org (Hodcroft, 2021) and GISAID (Shu and McCauley 2017; Khare et al., 2021; Elbe and Buckland‐Merrett, 2017).
We fit our previously proposed Susceptible-Exposed-Infectious-Hospitalized-Death-Recovered-Vaccinated model to the observed case and death data in South Africa (Lin et al., 2021; Lin et al., 2022). We assumed that a proportion (denoted as ) of the infections was reported and that 7%. This is supported by the number of reported cases in South Africa being much smaller than the estimated proportion of infected population based on seroprevalence from serological studies (Musa et al., 2021). We incorporated the vaccination data (fully vaccinated or second dose) and fit our model to the reported cases and deaths. We denoted the proportion of Omicron variant as , the infection fatality ratio (IFR) of the previous variant as IFR1 and the IFR of the Omicron variant as IFR2. Thus, the overall IFR of a one-strain model was (1-) IFR1 + IFR2, i.e., a weighted average of the two IFRs. We assumed a vaccine efficacy at 85% against both infections and deaths. Considering the high seroprevalence in South Africa, we assumed that eventually 80%–85% of the whole population were infected. We note that the re-infection rate with Omicron is high, which means that the variant has high immune evasion ability. The high relative transmissibility of the Omicron variant has two sources: the enlargement of the susceptible pool due to immune evasion and the increase in intrinsic transmissibility. We consider immune evasion due to Omicron by allowing a proportion of recovered individuals to become susceptible on November 9, 2021 when the Omicron variant invaded. We denoted the size of the susceptible pool before Omicron evasion as S and considered four scenarios: the immune evasion causing the susceptible pool to increase by 0.25*S, 0.5*S, S, and 2*S.
Our model reads as follows:
Here denotes the vaccinated class which contains a proportion (denoted as ) of vaccinated individuals. The rest of vaccinated individuals () enter the class and gain long-term immunity. The vaccinated individuals in the class are susceptible to breakthrough infection. Parameter accounts for the reduced susceptibility of vaccinated individuals. Here we assumed for simplicity, given that the effects of and compensate each other. A proportion () of hospitalized individuals will eventually die, and this proportion decreases as vaccination coverage increases in the form . Namely, we assumed that the risk of death drops while the vaccination coverage increases, and we set . We note that vaccination will only be administered to those who have not yet been vaccinated. Thus, the vaccination rate takes the form , in which is the daily vaccination rate per capita. Here we only consider the fully vaccinated (second dose) population data and ignore the impact of the first dose because it would be overtaken by that of the second dose.
Parameter denotes the risk of hospitalization or severe outcome of infected cases. Because we did not fit hospitalization or severe cases, we could not estimate but rather the product of and , which is the IFR when . We previously found that it is convenient to simply assume without changing the fitting performance. We estimated the , which is an exponential cubic spline function (Vetterling et al., 1992) with 12 nodes spanning over the study period. We fixed other parameters, e.g., which reflects the high efficacy of vaccines against both infection and deaths. We did not explicitly separate natural infection and breakthrough infection cases.
The mean latent periods of days, days and days are fixed, such that the mean generation time (i.e., sum of mean latent period and mean infectious period) equals five days (Tang et al., 2021) and the mean duration from infection to death is 17 days.
We simulated weekly cases and deaths as
and we denoted the weekly reported cases and deaths as and deaths as . We assumed and .
Thus, we connected the reported cases/deaths and simulated cases/deaths via two negative binomial distributions. Thus, the log likelihood could be defined (Lin et al., 2018; Zhao et al., 2018). We fit the model to reported cases and deaths via R package POMP (King, Nguyen, and Ionides 2015; He, Ionides, and King, 2010) and noted the maximum likelihood estimate for IFR. The 95% confidence interval was defined as the interval of IFR such that the log likelihood of the model given by the data drops by from the maximum log likelihood (He, Ionides, and King, 2010).
Result
We found that the COVID-19 case and death reporting in South Africa was consistent over time. For instance, the reported deaths from COVID-19 were consistently one-third of the excess deaths. The raw infection fatality rate (IFR) was consistent over time before the emergence of the Omicron variant. After the emergence of Omicron variant, the raw IFR seemingly decreased significantly.
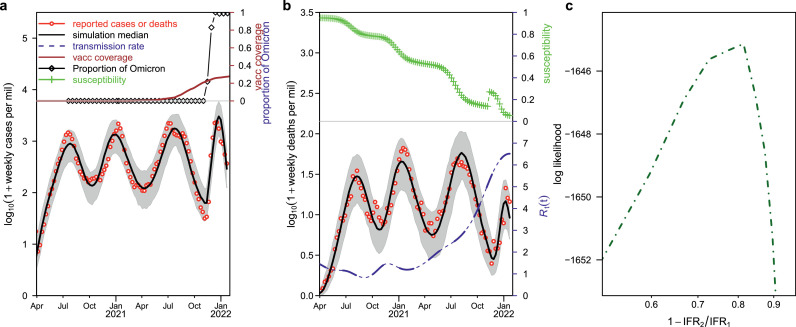
In Figure 1 , we show our fitting result of four waves in South Africa. Our model simulations (black curve) matched the reported cases and deaths (red circle) reasonably well, with cases in panel (a) and deaths in panel (b). The estimated IFR1 was approximately 0.21%. As mentioned previously, reported deaths amounted to only one-third of excess deaths, and it was generally believed that the excess death is a good proxy for the true COVID-19–related death. Thus, the true IFR could be 0.63%, which was well in line with current knowledge of COVID-19 before the emergence of the Omicron variant. The proportion of fully vaccinated population and the proportion of Omicron-infected population among the processed sample are shown in the top of Panel a. The proportion of susceptible population is shown in the top of Panel b, where a sudden increase on November 9, 2021 can be noticed, reflecting the effect of immune evasion due to the Omicron variant. Panel c shows the log likelihood profile versus the reduction in IFR. We found that the IFR reduced to approximately 78.7% (95% confidence interval: 66.9%, 85.0%) with respect to previous variants in the scenario of immune evasion enlarging the susceptible pool by one-fold. Table 1 shows the estimated reduction in IFR in four scenarios where immune evasion enlarged the susceptible pool by 0.25-fold, 0.5-fold, one-fold and two-fold. The estimated reduction in IFR is consistent among the four scenarios. Figure 1 shows the scenario in which the susceptible pool is increased by one-fold, and supplementary Figures S1, S2, and S3 show the scenario in which the susceptible pool is increased by 0.25-fold, 0.5-fold and two-fold by immune evasion. The fitting performance was greatly improved, as reflected in the maximum log likelihood, with the immune evasion having increased from 0.25-fold to two-fold of the pre-susceptible level. Thus, the relative transmissibility of the Omicron variant is likely three-fold that of the Delta variant. If we assume that the immune evasion is at a low level, e.g., 0.25-fold of the pre-susceptible level, then the transmission rate, in units of basic reproductive number, needs to increase to as high as 9.
Figure 1.
Fitting model to reported cases and deaths in South Africa.
The (red circle) and (black curve) represent reported cases or deaths and simulated cases or deaths, respectively. Aa, Reported cases versus simulated cases, and proportion of fully vaccinated and proportion of Omicron-infected populations among samples processed in the top panel. B, Reported deaths versus simulated deaths, and simulated proportion of susceptible population (green curve), with the susceptible pool having doubled on November 9, 2021 because of the immune evasion of the Omicron variant. The dashed (blue curve) showed the estimated transmission rate in unit of . C, The log likelihood profile as a function of reduction in IFR before and after Omicron evasion.
IFR, infection fatality ratio.
Table 1.
Estimated reduction in IFR in four scenarios of immune evasion.
| Immune evasion | Maximum likelihood estimate of reduction in IFR | 95% confidence interval |
|---|---|---|
| 0.25 | 0.794 | 0.683, 0.869 |
| 0.5 | 0.777 | 0.652, 0.861 |
| 1 | 0.787 | 0.669, 0.85 |
| 2 | 0.778 | 0.636, 0.842 |
IFR, infection fatality ratio.
Discussion
We assume that the case testing/reporting effort was consistent, although this might not be always true. With the emergence of new variants, the testing effort could have been enhanced. Thus, our estimated IFR could have been underestimated for the Omicron variant because of this transient effect. We considered several levels of immune evasion due to the Omicron variant in a single-strain model. A multiple-strain model could be considered as an alternative approach. We only considered data up to February 9, 2022. Our aim was to provide an early estimate of IFR for the Omicron variant in South Africa. This IFR was estimated in the setting of high seroprevalence (infection attack rate) in South Africa, thus it does not reflect the intrinsic IFR of the Omicron variant in a largely susceptible population. As a comparison, the raw case-fatality rate (CFR) in Hong Kong in the fifth wave of COVID-19 with the Omicron variant was 0.69% by April 4, 2022 ('Latest situation of COVID-19 (as of 4 April 2022)', 2022), whereas the previous raw CFR was 1.38% (up to February 6, 2022), implying a reduction of 50%. However, considering the higher frequency of under-reporting of cases in the fifth wave than in the previous waves due to change of testing policy, the reduction could have been much higher than 50%. This reduction was partly due to intrinsic features of the Omicron variant and partly due to vaccine-induced protection. Conversely, the raw CFR in the unvaccinated population in the fifth wave was 2.05%—higher than the 1.34% CFR reported in the previous waves. However, again taking into account the high frequency of under-reporting of cases in the fifth wave, the CFR of the Omicron variant in the unvaccinated population could have been substantially lower than that of previous variants (‘Latest situation of COVID-19 (as of 4 April 2022)’ 2022).
Conclusion
In summary, we found that the relative transmissibility of the Omicron variant (including transmissibility due to immune escape) could be more than three-fold higher than that of previous variants, which is in line with our previous estimate (Yu et al., 2021). Immune evasion is the main reason for the high relative transmissibility of the Omicron variant. The reduction in the IFR of the Omicron variant was approximately 78.7% of the IFR of previous variants, with a 95% confidence interval (66.9%, 85.0%).
Ethics approval and consent to participate
This study only re-analyzed publicly available data, which was conducted in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Availability of data and materials
All data are publicly available. https://ourworldindata.org/grapher/covid-variants-area. Our World In Data obtained their variant data from GISAID.
Funding
The work described in this paper was partially supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (HKU C7123-20G).
Authors' contributions
All authors conceived the study, performed the analysis, wrote the draft, revised the manuscript critically and approved it for publishing.
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.04.029.
Appendix. Supplementary materials
References
- Abdool Karim SS, Baxter C. Impact of SARS-CoV-2 variants of concern on Covid-19 epidemic in South Africa. Trans R Soc S Afr. 2021:1–4. [Google Scholar]
- Abdullah F, Myers J, Basu D, Tintinger G, Ueckermann V, Mathebula M, et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, South Africa. Int J Infect Dis. 2022;116:38–42. doi: 10.1016/j.ijid.2021.12.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Ionides EL, King AA. Plug-and-play inference for disease dynamics: measles in large and small populations as a case study. J R Soc Interface. 2010;7:271–283. doi: 10.1098/rsif.2009.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Hong W, Pan X, Lu G, Wei Xiawei. SARS-CoV-2 Omicron variant: characteristics and prevention. Med 2021. [DOI] [PMC free article] [PubMed]
- Hodcroft EB. CoVariants: SARS-CoV-2 mutations and variants of interest; 2021, https://covariants.org.
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019; 2021, https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Johns Hopkins university CSSE; 2022, https://github.com/CSSEGISandData/COVID-19.
- Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, et al. GISAID's role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AA, Nguyen D, Ionides EL. 2015. 'Statistical inference for partially observed Markov processes via the R package pomp', arXiv preprint arXiv:1509.00503.
- Latest situation of COVID-19 (as of 4 April 2022). CHP: Centre for Health Protection of Hong Kong: the Department of Health (department of health); 2022, https://www.chp.gov.hk/files/pdf/local_situation_covid19_en.pdf; (accessed April 4).
- Lin Q, Chiu AP, Zhao S, He D. Modeling the spread of Middle East respiratory syndrome coronavirus in Saudi Arabia. Stat Methods Med Res. 2018;27:1968–1978. doi: 10.1177/0962280217746442. [DOI] [PubMed] [Google Scholar]
- Lin L, Chen B, Zhao Y, Wang W, He D. Two Waves of COVID-19 in Brazilian Cities and Vaccination Impact. Math Biosci Eng. 2021;19(5):4657–4671. doi: 10.3934/mbe.2022216. [DOI] [PubMed] [Google Scholar]
- Lin L, Zhao Y, Chen B, He D. Multiple COVID-19 Waves and Vaccination Effectiveness in the United States. Int. J. Environ. Res. Public Health. 2022;19(4):2282. doi: 10.3390/ijerph19042282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu E, Ritchie H, Ortiz-Ospina E, Roser M, Hasell J, Appel C, et al. A global database of COVID-19 vaccinations. Nat Hum Behav. 2021;5:947–953. doi: 10.1038/s41562-021-01122-8. [DOI] [PubMed] [Google Scholar]
- Musa SS, Wang X, Zhao S, Li S, Hussaini N, Wang W, et al. Heterogeneous severity of COVID-19 in African countries: A modeling approach; 2021. [DOI] [PMC free article] [PubMed]
- Poudel S, Ishak A, Perez-Fernandez J, Garcia E, León-Figueroa DA, Romaní L, et al. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts–What is known so far? Travel Med Infect Dis. 2022;45 doi: 10.1016/j.tmaid.2021.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. 'Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa', medRxiv; 2021. [DOI] [PMC free article] [PubMed]
- Ritchie H, Mathieu E, Rodés-Guirao L, Appel C, Giattino C, Ortiz-Ospina E, et al. Coronavirus pandemic (COVID-19); 2020, https://ourworldindata.org/coronavirus; (accessed Feb 28).
- Science brief: omicron (B.1.1.529) variant. National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases; 2021, https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html.
- Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data–from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Musa SS, Zhao S, Shujiang Mei, Daihai He. Using proper mean generation intervals in modeling of COVID-19. Front Public Health. 2021;9 doi: 10.3389/fpubh.2021.691262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H, Eduan Wilkinson, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Vaughan A. Elsevier; 2021. Omicron emerges. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetterling WT, Press WH, Teukolsky SA, Flannery BP. Press Syndicate of the University of Cambridge; 1992. Numerical recipes: example book C (The Art of Scientific Computing) [Google Scholar]
- Yu Y, Liu Y, Zhao S, He D. A simple model to estimate the transmissibility of SARS-COV-2 Beta, Delta and omicron variants in South Africa. Available at SSRN. 2021 doi: 10.2139/ssrn.3989919. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wu S, Wu B, Qirui Yang, Chen A, Yuzhuang Li, et al. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lewi Stone, Daozhou Gao, Daihai He. Modelling the large-scale yellow fever outbreak in Luanda, Angola, and the impact of vaccination. PLOS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available. https://ourworldindata.org/grapher/covid-variants-area. Our World In Data obtained their variant data from GISAID.