ABSTRACT
Group 3 innate lymphocytes (ILC3s) are rare immune cells localized in mucosal tissues, especially the gastrointestinal (GI) tract. Despite their rarity, they are a major source of the cytokine interleukin-22 (IL-22), which protects the GI epithelium during inflammation and infection. Although ILC3s have been demonstrated to be important for defense against Clostridioides difficile infection, the exact mechanisms through which they sense productive infection and become activated to produce IL-22 remain poorly understood. In this study, we identified a novel mechanism of ILC3 activation after exposure to C. difficile. Toxin B (TcdB) from C. difficile directly induced production of IL-22 in ILC3s, and this induction was dependent on the glucosyltransferase activity of the toxin, which inhibits small GTPases. Pharmacological inhibition of the small GTPase Cdc42 also enhanced IL-22 production in ILC3s, indicating that Cdc42 is a negative regulator of ILC3 activation. Further gene expression analysis revealed that treatment with TcdB modulated the expression of several inflammation-related genes in ILC3s. These findings demonstrate that C. difficile toxin-mediated inhibition of Cdc42 leads to the activation of ILC3s, providing evidence for how these cells are recruited into the immune response against the pathobiont.
KEYWORDS: C. difficile, IL-22, ILC3, TcdB, toxins
INTRODUCTION
Clostridioides difficile is a mucosally associated bacterial pathogen that can cause life-threatening illness, and up to 30,000 patients succumb to infection in the United States every year (1, 2). While the pathogen is best recognized as the leading source of hospital-acquired gastrointestinal (GI) infections, it has been increasingly associated with infections in the healthy population (3). Asymptomatic colonization is possible and leads to productive infection only in some individuals; a subset of these patients will have a relapsing disease that is usually more severe (1). During active infection, C. difficile promotes a strong inflammatory response that can lead to pseudomembranous colitis. C. difficile virulence is mediated through several toxins it secretes, including toxin A (TcdA) and toxin B (TcdB), which are critical for pathogenesis (2, 4–6). Deletion of TcdB from C. difficile decreases virulence in animal models, and codeletion with TcdA abolishes in vivo virulence (7). Intrarectal administration of TcdA or TcdB recapitulates many aspects of C. difficile infection (8). Both TcdA and TcdB contain glucosyl transferase domains capable of directly glucosylating host Rho/Rac family small GTPases, thereby inactivating them. TcdB is highly toxic to epithelial cells but can have immunomodulatory effects on immune cells (2, 5), such as macrophage inflammasome activation, resulting in secretion of proinflammatory interleukin-1β (IL-1β) (9, 10). These findings suggest that the innate immune system has evolved mechanisms for immediately responding to the presence of bacterial toxins. However, the potential immunomodulatory effects of TcdB on other immune cell subsets relevant to C. difficile infection are not fully appreciated.
At the interface between the host and the environment, mucosal barriers have several lines of defense against invasive pathogens such as C. difficile (11, 12). Mucin-rich surfaces and antimicrobial peptides prevent commensal and/or pathogenic bacteria from interacting with the epithelium. Recent studies have revealed a critical role for group 3 innate lymphocytes (ILC3s) in mucosal barrier maintenance (13), including C. difficile infection (14). ILCs are a broad class of lymphocytes that lack diverse, rearranged antigen-specific receptors and instead respond to environmental signals (15, 16). ILC3s can be activated by IL-23 or IL-1β (17, 18). Once activated, ILC3s secrete several cytokines, and IL-22 is the most biologically important (16, 19, 20). Although rare, ILCs have antibacterial functions in C. difficile infection in mouse models (14, 21).
IL-22 is a critical modulator of tissue responses during inflammation (20, 22–24). IL-22 stimulation leads to the induction of proliferative and antiapoptotic pathways in epithelial cells as well as tissue-specific genes (25). IL-22 is upregulated in infectious and inflammatory diseases, including GI bacterial infections and inflammatory bowel disease (IBD) (26, 27). IL-22 is protective in different colitis mouse models (28–32). During acute colitis, IL-22 helps maintain the colonic epithelial barrier by preventing cell death, stimulating proliferation of epithelial stem cells, and inducing protective factors such as mucins and antimicrobial peptides from goblet and Paneth cells, respectively (27, 29). Unlike other GI bacterial infections in which absence of IL-22 leads to elevated bacterial loads in mouse models of disease, during C. difficile infection IL-22-deficient mice have no change in C. difficile bacterial loads (33). However, they do have increased morbidity and mortality due to greater dissemination of other bacterial species (33). IL-22 influences the microbiota (34), and IL-22-mediated glycosylation of the GI tract modulates the microbiome and its metabolic activity, which also limits C. difficile infection (35). Artificially increasing IL-22 levels via injection of recombinant cytokine during C. difficile infection in older mice also provides protection (36). These studies suggest that IL-22 is protective to the host during GI infection, but a mechanism through which IL-22 is directly induced in response to C. difficile infection remains to be identified.
In this study, we examined the potential effects of C. difficile on ILC3s. Bacterial toxins can modulate host immune cells, including ILC3s, often dampening their ability to respond to the pathogen (37). Surprisingly, we found that TcdB induced IL-22 production by ILC3s. This was dependent on the glucosyl transferase activity of TcdB, which inhibits host small GTPases. Pharmacological inhibition of Cdc42 phenocopied the effects of TcdB, leading to increased IL-22 production by ILC3s, suggesting that Cdc42 is a negative regulator of ILC3 activation. Gene expression analysis revealed that TcdB upregulated other genes in ILC3s in addition to Il22, such as Csf2 and Il17a, encoding granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-17, respectively, while also downregulating expression of such genes as Ltb, encoding lymphotoxin-β. These data establish that activation of ILC3s, including production of IL-22, is a direct result of exposure to toxins from C. difficile, providing a unique mechanism for the activation of these cells during infection.
RESULTS
Coculture of C. difficile and ILC3s induces IL-22 production in ILC3s.
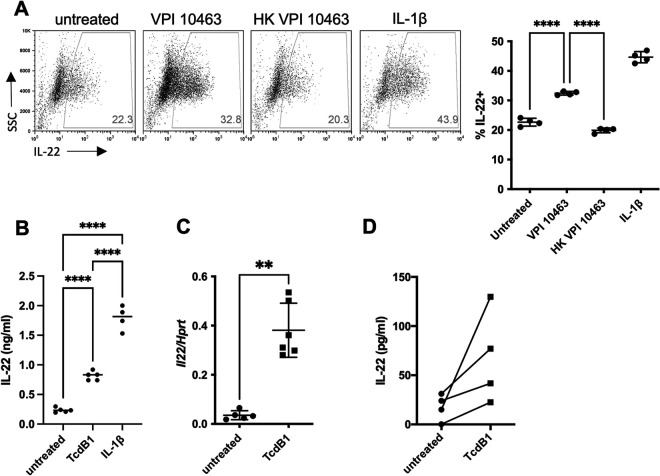
To examine if C. difficile can directly modulate ILC3 function during infection, we cocultured a mouse ILC3-like cell line, MNK-3, with vegetative bacilli of C. difficile. ILC3s are very rare cells found at low frequency; we made use of this cell line as it is molecularly and cellularly comparable to primary ILC3s (38). These cells produce low levels of IL-22 at homeostasis, but upon stimulation with IL-1β or IL-23, they produce copious amounts of IL-22 (38). While ILC3s are usually cultured at 17% O2, C. difficile is an anaerobe that requires a less oxygen-rich environment for optimal culture conditions. C. difficile has recently been shown to be transcriptionally active under microaerophilic conditions, including 1% O2 (39). ILC3s are also viable at 1% O2 and still produce IL-22 when activated (40). Furthermore, this low oxygen level is more representative of the levels immune cells experience in vivo in different tissues, including the GI tract (41). Coculture of MNK-3 cells with vegetative bacilli of C. difficile VPI 10463 at 1% O2 resulted in an increased percentage of cells producing IL-22 compared to MNK-3 cells cultured alone (Fig. 1A). In contrast, cells cultured with heat-killed bacilli had a percentage of IL-22-producing cells similar to that of the cells cultured alone. Thus, live, but not heat-killed, C. difficile induced IL-22 in ILC3s, suggesting that a secreted bacterial product, and not a structural component, activated ILC3s.
FIG 1.
C. difficile and its toxin, TcdB1, induce IL-22 production in ILC3s. (A) MNK-3 cells were cultured at 1% O2 either untreated, with live C. difficile VPI 10463, with heat-killed C. difficile VPI 10463, or with 20 ng/mL IL-1β for 6 h, with antibiotics added at 3 h in the presence of brefeldin A (BFA). Cells were stained with a fixable viability dye and intracellularly cytokine stained for IL-22. Shown are representative FACS plots for viable MNK-3 cells with the percentage of IL-22+ cells (left) and summary data (right), with each point representing a well. The line indicates the mean, n = 4. Data are from one experiment and represent three similar experiments. (B) MNK-3 cells were stimulated with 200 ng/mL TcdB1 or 20 ng/mL IL-1β or remained untreated for 4 h, and then IL-22 was quantitated in the supernatants by ELISA. Each point represents one well. The horizontal line indicates the mean, n = 4 to 5. Experiment is representative of more than three independent experiments. (C) MNK-3 cells were stimulated with 200 ng/mL TcdB1 or remained untreated for 4 h, and then Il22 was semiquantitated by real-time RT-PCR using Hprt levels for normalization. Each point represents one well. The horizontal line indicates the mean, n = 5 to 6. Experiment is representative of more than three independent experiments. (D) CD127+ Thy1.2+ cells isolated from C57BL/6 mice were stimulated with 200 ng/mL TcdB1 or remained untreated for 18 h, and then IL-22 was quantitated in the supernatants by ELISA. Each point represents the mean of wells from one experiment (n = 3 to 6) with data paired for each experiment (n = 4). **, P < 0.01; ****, P < 0.0001.
C. difficile toxin B induces IL-22 in ILC3s.
C. difficile has three major toxins: TcdA, TcdB, and binary toxin (CDT) (42). CDT, an ADP-ribosyl transferase that mediates actin cytoskeletal disruption, is only present in some strains and is absent from VPI 10463; therefore, it is not responsible for this effect of C. difficile on ILC3s (43). TcdA and TcdB are both inhibitors of eukaryotic small GTPases through glucosylation of key residues. Although both toxins have similar functions on host cells, TcdB is thought to be the more critical virulence factor during infection (44). C. difficile strains also typically secrete one of two variants of TcdB, TcdB1 or TcdB2, which share 92% sequence identity but differ in toxicity (45). TcdB2 is present in more recently emerged hypervirulent strains associated with community-acquired infections and is also more toxic than TcdB1 (46). To begin, we focused on the potential of TcdB1 to mediate the increase in IL-22 production by ILC3s we observed during coculture of the cells and live bacteria. Treatment of MNK-3 cells with recombinant TcdB1 led to secretion of significantly higher levels of IL-22 compared to untreated cells (Fig. 1B). After 4 h of treatment with TcdB1, we also found increased levels of Il22 in MNK-3 cells compared to cells cultured alone (Fig. 1C), further demonstrating that TcdB1 directly modulates production of IL-22.
Since ILC1s have also been implicated as key mediators of the immune response during C. difficile infection, we investigated the effects of TcdB1 treatment on an IFN-γ-producing ILC1-like cell line generated concurrently with MNK-3 cells, MNK-1 cells (38). In contrast to our findings with MNK-3 cells, treatment with TcdB1 did not induce production of IFN-γ in MNK-1 cells (see Fig. S1 in the supplemental material). Moreover, TcdB1 did not modulate IL-22 production in CD4 T cells when present during naive CD4 T cell differentiation to T helper 22 (Th22) cells (Fig. S2). Finally, although MNK-3 cells are an excellent model to study ILC3s and have served as a useful tool to study a rare immune cell subset (37, 38, 47–49), we wanted to extend our findings to primary immune cells. Sorted ILCs isolated from mice also secreted increased levels of IL-22 when treated with TcdB1 (Fig. 1D). Thus, TcdB1 directly induces IL-22 production in ILC3s.
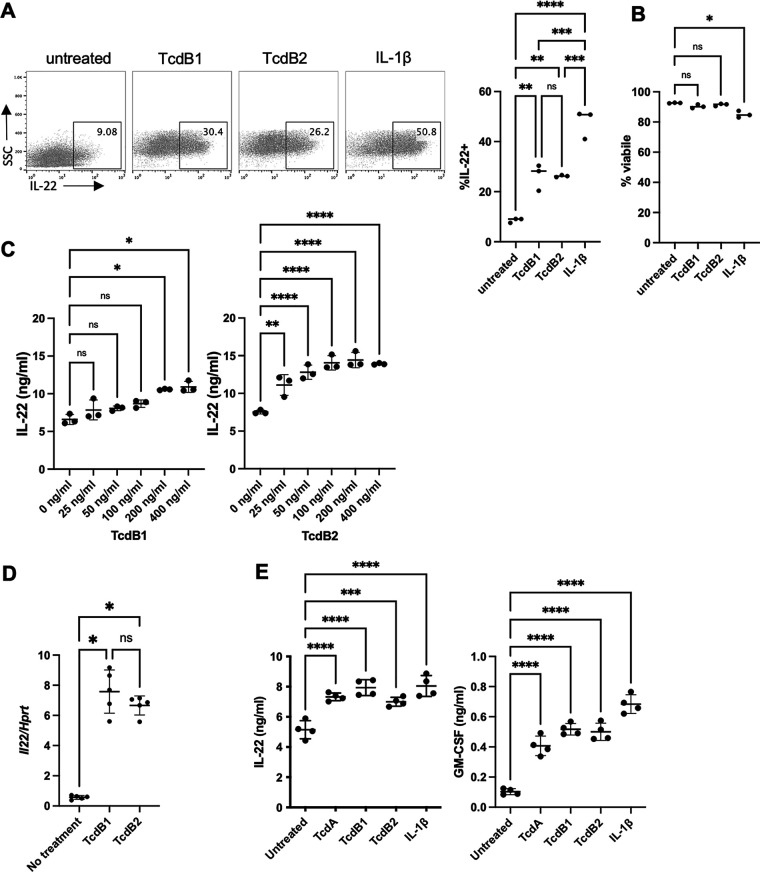
We next examined if TcdB2 also stimulated IL-22 production in ILC3s and, if so, compared it to TcdB1 and IL-1β. MNK-3 cells were treated with TcdB1, TcdB2, or IL-1β for 5 h, and cytokine production was measured by intracellular cytokine staining and FACS. Both TcdB1 and TcdB2 increased the percentage of IL-22+ cells compared to resting levels (approximately 25 to 50% compared to <10%) (Fig. 2A). These levels were less than that induced by IL-1β, where half the treated cells produced IL-22. Toxins had no detectable effect on cell viability (Fig. 2B). A dose titration of both recombinant TcdB1 and TcdB2 found that both increased IL-22 secretion in a dose-dependent manner (Fig. 2C). TcdB1 and TcdB2 similarly increased Il22 mRNA levels (Fig. 2D). Furthermore, the related toxin TcdA, also an inactivator of host small GTPases with 66% sequence similarity to TcdB (50), induced IL-22 production as well (Fig. 2E). In addition to IL-22, upon activation with IL-1β, MNK-3 cells also induce secretion of GM-CSF (38); therefore, we examined if TcdB1, TcdB2, or TcdA also induced this cytokine. We found that all three toxins induced GM-CSF secretion, albeit not to the same levels as IL-1β (Fig. 2E). Thus, C. difficile toxin-mediated ILC3 production of IL-22, as well as other effector molecules, such as GM-CSF, is shared between TcdA and TcdB and both variants of TcdB, TcdB1 and TcdB2.
FIG 2.
Several C. difficile glucosyltransferase toxins induce ILC3 cytokines. (A and B) MNK-3 cells were treated with 200 ng/mL TcdB1, 200 ng/mL TcdB2, or 20 ng/mL IL-1β for 5 h in the presence of BFA and then analyzed by intracellular cytokine staining and FACS. Shown are representative FACS plots for viable MNK-3 cells with the percentage of IL-22+ cells (left) and summary data (right) and viability data (B), with each point representing a well. The line indicates the mean, n = 3. Data are from one experiment and represent three similar experiments. (C) MNK-3 cells were treated with the indicated concentration of TcdB1 (left) or TcdB2 (right) for 18 h, and then IL-22 was quantitated in the supernatants by ELISA. Each point represents a well. The line indicates the mean, n = 3. Data are from one experiment and represent three similar experiments. (D) MNK-3 cells were treated with 200 ng/mL TcdB1 or TcdB2 or left untreated for 4 h, and then Il22 was semiquantitated by real-time RT-PCR using Hprt levels for normalization. Each point represents one well. The horizontal line indicates the mean, n = 5. Experiment is representative of more than three independent experiments. (E) MNK-3 cells were treated with 200 ng/mL TcdA, 200 ng/mL TcdB1, 200 ng/mL TcdB2, or 20 ng/mL IL-1β or were left untreated for 18 h, and then IL-22 and GM-CSF were quantitated by ELISA in the supernatants. Each point represents one well. The horizontal line indicates the mean, n = 4. Experiment is representative of more than three independent experiments. *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Differences that were not significant (P > 0.05) are marked ns.
TcdB1 has distinct effects on ILC3 gene expression.
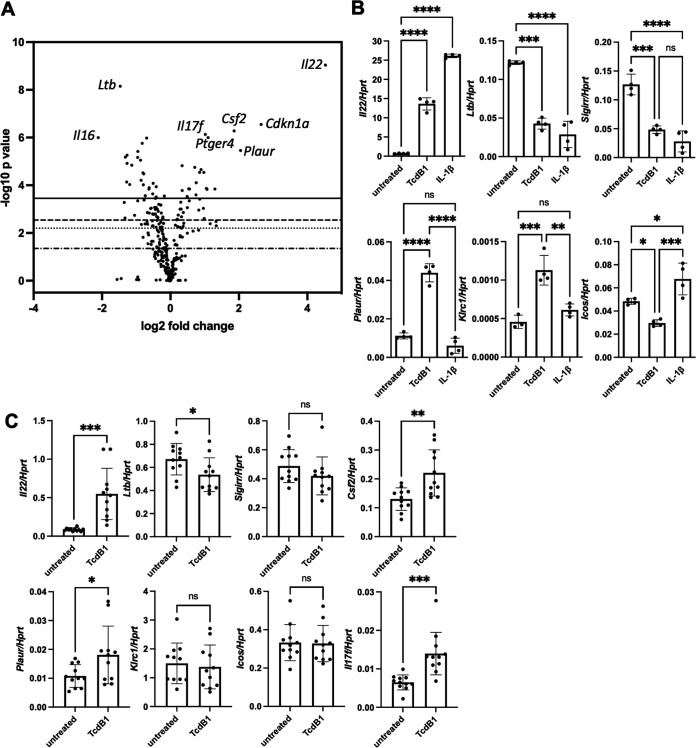
To better examine the broad effects of TcdB1 on ILC3 gene expression, we performed NanoString nCounter analysis using an immunology gene panel to examine 547 immune-related genes. MNK-3 cells were left untreated or were treated with TcdB1 for 4 h and then subjected to analysis. TcdB1 significantly increased expression of 12 genes and downregulated expression of 11 genes (Fig. 3A). The most differentially regulated and with the greatest significance was the gene encoding IL-22, validating our earlier experiments and the importance we have placed on IL-22 in ILC3 biology. Further reinforcing our results, we also found that Csf2, the gene encoding GM-CSF, was increased in accordance with the increased levels of secreted GM-CSF. Other genes significantly modulated by TcdB1 included Ltb (encoding lymphotoxin β), Plaur (encoding plasminogen activator, urokinase receptor), and Ptger4 (prostaglandin E receptor 4). We confirmed several of the genes discovered in the NanoString nCounter analysis by real-time RT-PCR (Fig. 3B). We also compared TcdB1-mediated changes to those induced by the inflammatory cytokine IL-1β, which has been well described to induce an inflammatory phenotype in ILC3s. We found that for some TcdB1-modulated genes, such as Ltb or Sigirr, both TcdB1 and IL-1β had similar effects on ILC3s compared to nontreated cells. However, genes that were modulated only following TcdB treatment were also identified, such as Plaur and Klrc1, which encodes killer cell lectin like receptor C1/CD159. Given these results, we conclude that TcdB1 has effects on gene expression in ILC3s, distinct from the response mediated by the host cytokine IL-1β.
FIG 3.
TcdB1 modulates ILC3 gene expression. (A) MNK-3 cells were treated with 200 ng/mL TcdB1 for 4 h or left untreated, and then mRNA was subjected to NanoString nCounter analysis using a mouse immunology panel with 547 genes and 14 internal reference genes. Shown is a volcano plot with several notable genes marked. (B) MNK-3 cells were treated with no treatment, 200 ng/mL TcdB1, or 20 ng/mL IL-1β for 4 h, and then mRNA levels of several genes identified in panel A were semiquantitated by real-time RT-PCR. n = 4. Each point represents one well, bar indicates mean. Data are representative of three or more independent experiments. *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Differences that were not significant (P > 0.05) are marked ns. (C) Sorted ILCs (CD127+ Thy1.2+) from Rag1−/− mice were rested overnight in culture and then left unstimulated or stimulated with 200 ng/mL TcdB1 for 6 h. mRNA levels of several genes identified in panel A or B were semiquantitated by real-time RT-PCR. Shown are all samples combined from two independent experiments (n = 5 or 6 per experiment). *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001. Differences that were not significant (P > 0.05) are marked ns.
To extend these transcriptional changes to primary cells, we also examined the effects of TcdB1 on primary ILCs isolated from mice. We found that TcdB1 treatment significantly upregulated expression of Il22 as well as other cytokine genes, such as Il17f and Csf2, and downregulated levels of Ltb (Fig. 3C). Another gene, Plaur, was also upregulated in the primary cells as it was in the ILC3-like cell line. Other genes for which we had detected changes in the cell line were either trending (Sigir) or not significant (Klcr1 and Icos) in the primary ILCs. Thus, TcdB1 modulates gene expression in primary mouse ILCs, with both similarities and differences to the mouse ILC3-like cell line.
Inhibition of Cdc42 signaling induces IL-22 production in ILC3s.
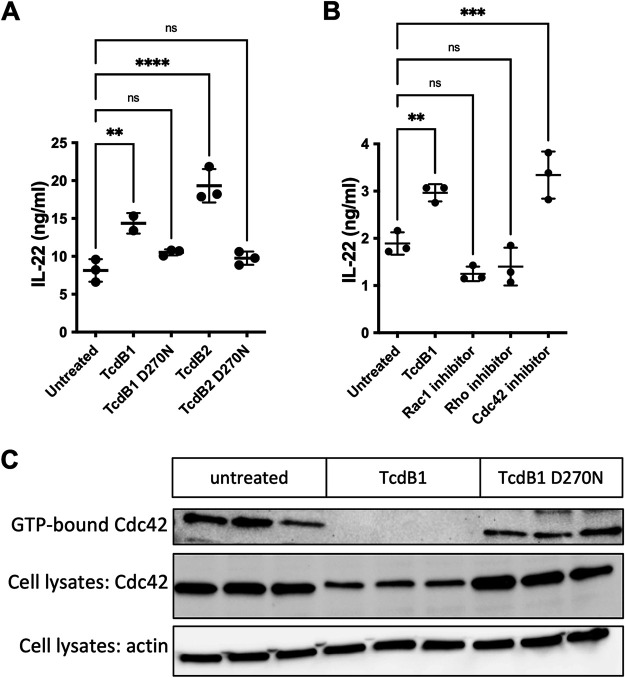
The major toxins of C. difficile, TcdA and TcdB, both mediate cell toxicity through glucosylation of host small GTPases, which inhibits their activity, leading to epithelial cell death (42). We hypothesized that these toxins were inducing IL-22 production in ILC3s through the same mechanism of action. To test this, we made use of recombinant TcdB1 and TcdB2 with a point mutation in the glucosyl transferase active sites (D270N) (51, 52). Treatment of MNK-3 cells did not increase levels of IL-22 (Fig. 4A), suggesting that C. difficile toxins activate ILC3s via their well-known glucosyl transferase activities.
FIG 4.
Inhibition of Cdc42 induces IL-22 production in ILC3s. (A) Toxin lacking glucosylation activity does not stimulate IL-22 production. MNK-3 cells were left untreated or were treated with 200 ng/mL TcdB1, TcdB2, or catalytic mutants, TcdB1 D270N or TcdB2 D270N, for 18 h, and then secreted IL-22 was quantitated by ELISA. Each point represents one well, bar indicates mean, n = 3. Data are representative of three or more independent experiments. **, P < 0.01; ****, P < 0.0001. Differences that were not significant (P > 0.05) are marked ns. (B) Cdc42 inhibitor phenocopies TcdB treatment. MNK-3 cells were left untreated or were treated with 50 μM Rac1 inhibitor CAS 1177865-17-6, 30 μM Rho inhibitor rosin, or 50 μM Cdc42 inhibitor ZCL278 for 18 h, and then secreted IL-22 was quantitated by ELISA. Each point represents one well, bar indicates mean, n = 3. Data are representative of three or more independent experiments. **, P < 0.01; ***, P < 0.001. Differences that were not significant (P > 0.05) are marked ns. (C) TcdB1-treated cells have no detectable GTP-bound Cdc42. MNK-3 cells were stimulated with 200 ng/mL TcdB1 or TcdB1 D270N or left untreated for 4 h, and then active Cdc42 (GTP-bound) was pulled down from cell lysates according to Materials and Methods (n = 3). The GTP-bound fraction and the total cell lysates were analyzed by Western blotting for Cdc42 levels, and the total cell lysates were analyzed by Western blotting for actin levels. Shown are data from one experiment representative of three independent experiments.
C. difficile toxins specifically target the Rho/Rac subfamily of Ras-like GTPases, which includes Rho, Rac1, and Cdc42 (42). These signaling proteins have been implicated in various functions in other immune cells (53, 54), but their roles in ILCs are unknown. To examine the function of these small GTPases in ILC3s, we treated MNK-3 cells with different pharmacological inhibitors targeting each GTPase individually. Rho and Rac1 inhibitors did not modulate secretion of IL-22 (Fig. 4B), as treatment led to these cells producing IL-22 levels comparable to those of cells cultured alone. In contrast, we found that a selective Cdc42 inhibitor that targets the binding site of the guanine nucleotide exchange factor ZLC278 (55) increased levels of IL-22 compared to expression levels of resting cells (Fig. 4B). Thus, a Cdc42 inhibitor phenocopies the effects of TcdB on ILC3s. To confirm that TcdB targets Cdc42 in ILC3s, we examined levels of active Cdc42 in TcdB-treated cells. Compared to untreated control cells or cells treated with enzymatically inactive TcdB D270N, we found reduced amounts of active Cdc42 in the TcdB-treated cells (Fig. 4C). Altogether, these data suggest that TcdB mediates its effects on ILC3s in part via inhibition of Cdc42.
DISCUSSION
C. difficile pathogenesis relies on toxin-mediated epithelial cell death, but the effects of those same toxins on the immune cells of the intestinal barrier still are not fully explored. In this study, we found that the C. difficile virulence factor TcdB, best known for targeting GI epithelial cells for apoptosis to decrease host barrier integrity, can also induce a distinct inflammatory phenotype in ILC3s. Our data show that the increase in IL-22 production by ILC3s treated with live C. difficile or recombinant toxins potentially is caused by the inhibition of Cdc42. These findings provide a mechanism for ILC3s to become directly activated during productive C. difficile infection. This is likely beneficial to the host, since upon epithelial cell death and subsequent exposure of ILC3s to toxin, ILC3s would be capable of immediately boosting barrier integrity as well as antimicrobial peptide and mucin production. While IL-22 has previously been shown to be as important for host defense during C. difficile infection (33, 35, 36), our study now characterizes a mechanism by which IL-22 is induced directly from ILC3s via the glucosyl transferase activity of the toxins.
Modulation of immune cell function by virulence factors is a common strategy for bacterial pathogens to evade host immune responses (56–58). In many instances, this means that a pathogen constrains cell function to minimize the antibacterial immune response. The toxins produced by C. difficile are known to cause epithelial cell apoptosis, leading to disruption of the intestinal barrier and potential dissemination of luminal bacteria (2, 4). In contrast, our results demonstrate that exposure to TcdB leads to the activation of ILC3s rather than cell death. Since ILC3s are primarily located in the submucosa of the GI tract, they are poised for immediate activation following epithelial disruption during C. difficile infection. These findings imply that ILC3s can become directly activated during productive infection without the need for other immune cells to become activated first, such as dendritic cells that produce IL-1β.
Although recent studies have revealed several means the bacterium uses to outcompete commensals (59, 60), we still do not fully understand how C. difficile establishes infection. Host immune responses likely play a critical role in this process (2). IL-22 is unlikely to directly affect C. difficile growth; in vitro growth curves (absorbance at an optical density at 600 nm [OD600] or viable CFU plating) in the presence or absence of recombinant murine IL-22 revealed no significant differences (data not shown). As dysregulation of the microbiota is important for progression of C. difficile infection and IL-22 can shape the microbiota (34), increased levels of IL-22 may alter the composition of the microbiota to favor C. difficile growth.
We have identified Cdc42 as a negative regulator of ILC3 activation. Small GTPases are involved in the activation of other immune cells, such as closely related Th17 cells (53, 61), but to our knowledge no studies have yet reported their role in ILC3s. Cdc42 is an intracellular signal transducer that cycles between a GTP-bound active and GDP-bound inactive state and in many cells plays a role in actin and tubulin dynamics (62). In immune cells, such as T cells, Cdc42 has been shown to regulate differentiation of naive CD4 T cells to different T helper subsets (61, 63). Few negative regulators of ILC3s have been identified. Recently, RANKL and vitamin D were found to be negative regulators of ILC3s (48, 64, 65). Cdc42 is already a target for Ras-related cancers (63), and these therapeutics could be coopted for infectious diseases to elevate IL-22 levels via ILC3s to combat bacterial infection. Bacterial toxins, with their well-elucidated mechanisms, are excellent tools for perturbing eukaryotic signaling pathways (66) and may be helpful in identification of other critical signaling pathways in immune cells.
C. difficile is a GI pathogen in which productive infection involves the complex interaction of the bacterium, the microbiota, the host’s GI tract, and the host’s immune response. In this study, we have shown that a major bacterial virulence factor, TcdB, can directly activate a host immune cell. It remains to be fully determined how these TcdB-activated ILC3s differ from those activated by the host’s IL-1β or IL-23, and future work will investigate the function of these TcdB-activated ILC3s in the host’s GI tract. A better understanding of how C. difficile interacts with components of our immune system is critical for the development of new preventative measures and therapeutics to combat infection. Enhancing innate immunity, especially by targeting cytokine biology, has great potential for reducing incidence and severity of initial and recurrent C. difficile infections.
MATERIALS AND METHODS
Cell line.
MNK-3 cell clone B3, derived from single-cell cloning of the ILC3-like cell line MNK-3 cells (38), was maintained in Dulbecco’s modified Eagle medium (DMEM) (Corning, Tewksbury, MA) with 10% heat-inactivated fetal bovine serum (FBS) (Gemini Bio-Products; West Sacramento, CA), 2 mM GlutaGro (Corning), 1 mM sodium pyruvate (GE Healthcare HyClone; Logan, UT), 55 μM β-mercaptoethanol (Sigma; St. Louis, MO), 10 mM HEPES (Corning), 50 μg/mL gentamicin (Amresco; Solon, OH), 100 U/mL penicillin (Gemini Bio-Products), 100 U/mL streptomycin (Gemini Bio-Products), and 10 ng/mL recombinant mouse IL-7 (eBioscience, San Diego, CA, or Peprotech, Rocky Hill, NJ). Coculture experiments with bacteria omitted the gentamicin, penicillin, and streptomycin. MNK-1 cells were maintained in the same complete medium with 10 ng/mL recombinant mouse IL-2 (eBioscience) instead of IL-7.
Bacterial toxins and other key reagents.
Recombinant TcdB1 and TcdA were purchased from List Biologicals (Campbell, CA), and this TcdB1 was used for the majority of the experiments. Some experiments used recombinant TcdB1, TcdB2, TcdB1 D270N, and TcdB2 D270N that were expressed in a Bacillus megaterium system (MoBiTech, Göttingen, Germany) as previously described (67) and affinity purified by Ni2+ chromatography. This TcdB1 was used for experiments where it was being compared to TcdB2 or the mutants. A concentration of 200 ng/mL toxin was used for experiments, unless otherwise indicated, as a dose titration showed this was the concentration of TcdB1 that had maximal IL-22 production by ILC3s. A concentration of 20 ng/mL recombinant mouse IL-1β (BioLegend, San Diego, CA) was used as a positive control for induced IL-22 production by MNK-3 cells. Cell signaling inhibitors were 50 μM Cdc42 inhibitor ZCL278 (Sigma), 30 μM Rho inhibitor Rosin (Sigma), and 50 μM Rac1 inhibitor 5 CAS 1177865-17-6 (Sigma).
Bacterium-ILC3 cocultures.
C. difficile VPI 10463 (ATCC 43255) was cultured overnight in 5 mL of brain heart infusion broth (BHI) plus yeast (BHI-S) at 37°C in a Coy anaerobic chamber maintained at 5% CO2, 3.5% H2, 91.5% N2. Bacteria were harvested, washed with phosphate-buffered saline (PBS), and labeled according to the manufacturer’s protocol with carboxyfluorescein succinimidyl ester (CFSE) (eBioscience) to distinguish them from the host cells by fluorescence-activated cell sorting (FACS). To generate heat-killed bacteria, an aliquot of bacteria was incubated in a 70°C water bath for 20 min and then washed with MNK-3 culture medium. Lack of viability was confirmed by plating. For the cocultures, MNK-3 clone B3 cells were washed with MNK-3 medium lacking antibiotics, and then cells were seeded in a 96-well round-bottom plate with brefeldin A (BFA) to inhibit cytokine secretion. Live or heat-killed bacteria were added as indicated. The plate was incubated in a hypoxia chamber with 1% O2, 5% CO2, 94% N2 with a water dish for humidity at 37°C for 3 h, and then 15 μg/mL thiamphenicol (Sigma) and 1.25 μg/mL metronidazole (Sigma) were added to prevent overgrowth of bacteria. At 6 h, cells were stained with eFluor780 fixable viability dye (eBioscience; San Diego, CA), intracellularly cytokine stained for IL-22, and analyzed by FACS. Bacteria (CFSE+) were excluded from the analysis.
Isolation of primary ILCs.
C57BL/6 mice were from the National Cancer Institute/Charles River (Frederick, MD), and Rag1−/− mice were from The Jackson Laboratory (Bar Harbor, ME). Mice were housed in an AAALAC-accredited Helicobacter-free rodent barrier facility. Males and females were used for experiments. All studies were approved by the OUHSC Institutional Animal Care and Use Committee (IACUC protocol numbers 18-016, 18-066, 20-088, and 21-024). Spleens and lymph nodes were excised from C57BL/6 or Rag1−/− mice, where indicated, and single-cell suspensions were made by disruption of the spleen on wire mesh using the plunger of a 3-mL syringe. Cells were centrifuged and the cell pellet was resuspended. Red blood cells were lysed in ACK lysis buffer (0.83% NH4Cl, 0.5% KHCO3, 0.5 μM EDTA) for 2 min and then neutralized with PBS or medium. Cells were counted using trypan blue exclusion staining and a hemocytometer. ILCs from C57BL/6 mice were purified using an EasySep mouse pan-ILC enrichment kit using magnetic bead separation (StemCell Technology, Vancouver, Canada). Cells from C57BL/6 mice were stained with viability dye (eFluor780 fixable viability dye) and then surface stained with CD127 (clone A7R34; eBioscience), and CD90.2/Thy1.2 (clone 53-2.1; eBioscience), and eFluor780− CD127+ CD90.2/Thy1.2+ cells were sorted using a Beckman Coulter Moflo XDP sorter. Sorted cells had greater than 95% purity. Cells from Rag1−/− mice were surface stained with CD127 (clone A7R34; eBioscience) and CD90.2/Thy1.2 (clone 53-2.1; eBioscience), and CD127+ CD90.2/Thy1.2+ cells were sorted using a FACSAria IIIu sorter. Sorted cells had greater than 90% purity.
Intracellular cytokine staining.
Cells were treated with toxin, cytokine, or signaling inhibitor as indicated in the presence of BFA (eBioscience) for 5 h. Cells were stained with eFluor780 fixable viability dye and then were intracellularly stained with an anti-IL-22 antibody (clone IL22JOP; eBioscience) according to the manufacturer’s protocol and analyzed by flow cytometry on a Stratedigm S1200Ex flow cytometer (Stratedigm, San Jose, CA), and data were analyzed using FlowJo v.10.6 (Tree Star; Ashland, OR).
ELISA.
Mouse IL-22 enzyme-linked immunosorbent assay (ELISA) (Antigenix America, Huntington Station, NY), mouse GM-CSF ELISA (R&D Systems, Minneapolis, MN), and mouse gamma interferon (IFN-γ) ELISA (BD Biosciences, East Rutherford, NJ) were performed according to the manufacturers’ protocols.
Real time RT-PCR.
Cells were harvested in TriPure (Roche; Nutley, NJ) and RNA was prepared according to the manufacturers’ protocols. RNA was DNase treated (Roche) and cDNA was generated by reverse transcription using EasyScript Plus (Lamda Biotech; St. Louis, MO) with oligo(dT) as the primer. cDNA was used as the template in a real-time PCR using Integrated DNA Technologies (Coralville, IA) or ABI TaqMan primer-probe sets (Thermo Fisher, Waltham, MA) (Table 1) on an ABI 7500 Fast or QuantStudio5 real-time PCR machine (Thermo Fisher). cDNA was semiquantitated using the ΔCT method with Hprt as an internal control for all samples.
TABLE 1.
Real time RT-PCR primer-probe sets used in this study
| Gene | Manufacturer | Catalog no. |
|---|---|---|
| Csf2 | Integrated DNA Technologies | Mm.PT.58.9186111 |
| Hprt | Integrated DNA Technologies | Mm.PT.39a.22214828 |
| Icos | Thermo Fisher/ABI | Mm00497600_m1 |
| Il17f | Integrated DNA Technologies | Mm.PT.58.9739903 |
| Il22 (also known as Iltifb) | Integrated DNA Technologies | Mm.PT.58.44024580.g |
| Klrc1 | Thermo Fisher/ABI | Mm00516111_m1 |
| Ltb | Integrated DNA Technologies | Mm.PT.58.32883824.g |
| Plaur | Thermo Fisher/ABI | Mm01149438_m1 |
| Sigirr | Thermo Fisher/ABI | Mm01275624_g1 |
Cdc42 activity assay.
MNK-3 cells were treated with no toxin, 200 ng/mL TcdB1, or 200 ng/mL TcdB1 D270N for 4 h. Cell lysates were then harvested and GTP-bound Cdc42 was detected by following the manufacturer’s protocol using an Active Cdc42 detection kit (number 8819; CST). GTP-bound Cdc42 fractions or total cell lysates were separated by SDS-PAGE on a 4 to 15% gradient gel (Bio-Rad, Hercules, CA). Proteins were transferred to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Billerica, MA) using a wet transfer method. The protein-transferred membrane was blocked with 5% dry milk and then incubated with the manufacturers’ recommended concentration of primary antibody overnight at 4°C. Blots were then washed and incubated with the appropriate species-specific horseradish peroxidase (HRP) secondary antibody for 1 h. Blots were developed using Pierce ECL2 Western blotting substrate (Thermo Scientific, Waltham, MA) and imaged using a ChemiDoc MP imaging system (Bio-Rad, Hercules, CA). For reblotting with another antibody, blots were stripped using a Restore Western blot stripping buffer (Thermo Scientific) and then washed and reblocked and performed with a new primary Ab as described above.
Gene expression analysis.
MNK-3 clone B3 cells were stimulated with 200 ng/mL TcdB1 (n = 4) or left unstimulated (n = 4). RNA was purified using a Direct-zol RNA miniprep plus kit (Zymo Research, Irvine, CA). RNA expression profiling was performed via nCounter analysis technology (NanoString, Seattle, WA) using an nCounter mouse immunology pathway panel with 547 genes and 14 internal reference genes for normalization. Data were analyzed using the nSolver 4.0 software package. Briefly, raw transcript counts were normalized using negative and positive synthetic sequences provided within each code set to account for background noise and technical variation, respectively. Differential gene expression between untreated and TcdB1-treated cells was examined.
Statistical analysis.
Values are expressed as means ± standard deviations (SD). Statistical analysis was performed with Prism 9.0. For two-way comparisons, an unpaired t test with the assumption that both populations have the same SD was used. For multiple comparisons, one-way analysis of variance (ANOVA) with multiple comparisons and Tukey’s post hoc analysis was used. Significance was defined as P values of ≤0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). Differences that were not significant (P > 0.05) are marked ns.
ACKNOWLEDGMENTS
We thank the Laboratory for Molecular Biology and Cytometry Research, including the Genomics Core, and Jenny Gibson for the NanoString service and the Imaging and Flow Cytometry Core (OUHSC) for FACS analysis service and for cell sorting at the Oklahoma Medical Research Foundation Flow Cytometry Core. We thank David S. J. Allan and James R. Carlyle for providing the MNK-3 and MNK-1 cell lines.
This project was supported by the National Institute of General Medical Sciences of the National Institutes of Health (P20GM103447 and P20GM134973), a pilot award to L.A.Z. (as part of U19AI062629), the Presbyterian Health Foundation, and the Stephenson Cancer Center. Research reported in this publication was supported in part by the National Cancer Institute Cancer Center support grant P30CA225520, awarded to the Stephenson Cancer Center, and used the Molecular Biology Shared Resource.
L.A.Z. conceived of the study and drafted the manuscript. R.L.P., A.C., P.S., and L.A.Z. contributed to the acquisition, analysis, or interpretation of data presented in the manuscript. T.S. provided valuable assistance with C. difficile culture and toxins. J.D.B. provided toxins and scientific expertise. All authors critically reviewed the manuscript and approved the final manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Lauren A. Zenewicz, Email: lauren-zenewicz@ouhsc.edu.
Victor J. Torres, New York University School of Medicine
REFERENCES
- 1.Leffler DA, Lamont JT. 2015. Clostridium difficile Infection. N Engl J Med 373:287–288. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 2.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloomfield LE, Riley TV. 2016. Epidemiology and risk factors for community-associated Clostridium difficile infection: a narrative review. Infect Dis Ther 5:231–251. doi: 10.1007/s40121-016-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt RN, Lacy DB. 2012. Toward a structural understanding of Clostridium difficile toxins A and B. Front Cell Infect Microbiol 2:28. doi: 10.3389/fcimb.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aktories K, Schwan C, Jank T. 2017. Clostridium difficile toxin biology. Annu Rev Microbiol 71:281–307. doi: 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 6.Kordus SL, Thomas AK, Lacy DB. 2021. Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat Rev Microbiol. doi: 10.1038/s41579-021-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. 2014. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis 209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markham NO, Bloch SC, Shupe JA, Laubacher EN, Thomas AK, Kroh HK, Childress KO, Peritore-Galve FC, Washington MK, Coffey RJ, Lacy DB. 2021. Murine intrarectal instillation of purified recombinant Clostridioides difficile toxins enables mechanistic studies of pathogenesis. Infect Immun 89:e00543-20. doi: 10.1128/IAI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng J, Hirota SA, Gross O, Li Y, Ulke-Lemee A, Potentier MS, Schenck LP, Vilaysane A, Seamone ME, Feng H, Armstrong GD, Tschopp J, Macdonald JA, Muruve DA, Beck PL. 2010. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 139:542–552. doi: 10.1053/j.gastro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong YN, Peng X, Xi JJ, Chen S, Wang F, Shao F. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 11.Ramanan D, Cadwell K. 2016. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe 19:434–441. doi: 10.1016/j.chom.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Lopez A, Behnsen J, Nuccio SP, Raffatellu M. 2016. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol 16:135–148. doi: 10.1038/nri.2015.17. [DOI] [PubMed] [Google Scholar]
- 13.Tait Wojno ED, Artis D. 2016. Emerging concepts and future challenges in innate lymphoid cell biology. J Exp Med 213:2229–2248. doi: 10.1084/jem.20160525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fachi JL, Secca C, Rodrigues PB, Mato FCP, Di Luccia B, Felipe JS, Pral LP, Rungue M, Rocha VM, Sato FT, Sampaio U, Clerici M, Rodrigues HG, Camara NOS, Consonni SR, Vieira AT, Oliveira SC, Mackay CR, Layden BT, Bortoluci KR, Colonna M, Vinolo MAR. 2020. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J Exp Med 217:jem.20190489. doi: 10.1084/jem.20190489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artis D, Spits H. 2015. The biology of innate lymphoid cells. Nature 517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 16.Klose CS, Artis D. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 17.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes T, Becknell B, Freud AG, McClory S, Briercheck E, Yu J, Mao C, Giovenzana C, Nuovo G, Wei L, Zhang X, Gavrilin MA, Wewers MD, Caligiuri MA. 2010. Interleukin-1beta selectively expands and sustains interleukin-22+ immature human natural killer cells in secondary lymphoid tissue. Immunity 32:803–814. doi: 10.1016/j.immuni.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenberg GF, Fouser LA, Artis D. 2010. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol 107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 20.Zenewicz LA. 2018. IL-22: there is a gap in our knowledge. Immunohorizons 2:198–207. doi: 10.4049/immunohorizons.1800006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, Ling L, Leiner I, Pamer EG. 2015. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonnenberg GF, Fouser LA, Artis D. 2011. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 23.Dudakov JA, Hanash AM, van den Brink MR. 2015. Interleukin-22: immunobiology and pathology. Annu Rev Immunol 33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keir ME, Yi T, Lu TT, Ghilardi N. 2020. The role of IL-22 in intestinal health and disease. J Exp Med 217:e20192195. doi: 10.1084/jem.20192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zenewicz LA, Flavell RA. 2008. IL-22 and inflammation: leukin' through a glass onion. Eur J Immunol 38:3265–3268. doi: 10.1002/eji.200838655. [DOI] [PubMed] [Google Scholar]
- 26.Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, Shimizu N, Fujiyama Y. 2005. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. 2008. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 29.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. 2008. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamanaka M, Huber S, Zenewicz LA, Gagliani N, Rathinam C, O'Connor W, Jr, Wan YY, Nakae S, Iwakura Y, Hao L, Flavell RA. 2011. Memory/effector (CD45RB(lo)) CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J Exp Med 208:1027–1040. doi: 10.1084/jem.20102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O'Connor W, Jr, Murphy AJ, Valenzuela DM, Yancopoulos GD, Booth CJ, Cho JH, Ouyang W, Abraham C, Flavell RA. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. 2013. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa M, Yada S, Liu MZ, Kamada N, Munoz-Planillo R, Do N, Nunez G, Inohara N. 2014. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. 2013. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagao-Kitamoto H, Leslie JL, Kitamoto S, Jin C, Thomsson KA, Gillilland MG, III, Kuffa P, Goto Y, Jenq RR, Ishii C, Hirayama A, Seekatz AM, Martens EC, Eaton KA, Kao JY, Fukuda S, Higgins PDR, Karlsson NG, Young VB, Kamada N. 2020. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat Med 26:608–617. doi: 10.1038/s41591-020-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peniche AG, Spinler JK, Boonma P, Savidge TC, Dann SM. 2018. Aging impairs protective host defenses against Clostridioides (Clostridium) difficile infection in mice by suppressing neutrophil and IL-22 mediated immunity. Anaerobe 54:83–91. doi: 10.1016/j.anaerobe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seshadri S, Allan DSJ, Carlyle JR, Zenewicz LA. 2017. Bacillus anthracis lethal toxin negatively modulates ILC3 function through perturbation of IL-23-mediated MAPK signaling. PLoS Pathog 13:e1006690. doi: 10.1371/journal.ppat.1006690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allan DS, Kirkham CL, Aguilar OA, Qu LC, Chen P, Fine JH, Serra P, Awong G, Gommerman JL, Zuniga-Pflucker JC, Carlyle JR. 2015. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol 8:340–351. doi: 10.1038/mi.2014.71. [DOI] [PubMed] [Google Scholar]
- 39.Giordano N, Hastie JL, Carlson PE. 2018. Transcriptomic profiling of Clostridium difficile grown under microaerophillic conditions. Pathog Dis doi: 10.1093/femspd/fty010. [DOI] [PubMed] [Google Scholar]
- 40.Fachi JL, Pral LP, Dos Santos JAC, Codo AC, de Oliveira S, Felipe JS, Zambom FFF, Camara NOS, Vieira P, Colonna M, Vinolo MAR. 2021. Hypoxia enhances ILC3 responses through HIF-1alpha-dependent mechanism. Mucosal Immunol 14:828–841. doi: 10.1038/s41385-020-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenewicz LA. 2017. Oxygen levels and immunological studies. Front Immunol 8:324. doi: 10.3389/fimmu.2017.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. doi: 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupnik M, Grabnar M, Geric B. 2003. Binary toxin producing Clostridium difficile strains. Anaerobe 9:289–294. doi: 10.1016/j.anaerobe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanis JM, Barua S, Ballard JD. 2010. Variations in TcdB activity and the hypervirulence of emerging strains of Clostridium difficile. PLoS Pathog 6:e1001061. doi: 10.1371/journal.ppat.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt JJ, Ballard JD. 2013. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol Mol Biol Rev 77:567–581. doi: 10.1128/MMBR.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seshadri S, Pope RL, Zenewicz LA. 2018. Glucocorticoids inhibit group 3 innate lymphocyte IL-22 production. J Immunol 201:1267–1274. doi: 10.4049/jimmunol.1800484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC, Newberry RD, Kobayashi Y, Allan DSJ, Carlyle JR, Cella M, Colonna M. 2018. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity 48:1208–1219. doi: 10.1016/j.immuni.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cella M, Gamini R, Secca C, Collins PL, Zhao S, Peng V, Robinette ML, Schettini J, Zaitsev K, Gordon W, Bando JK, Yomogida K, Cortez V, Fronick C, Fulton R, Lin LL, Gilfillan S, Flavell RA, Shan L, Artyomov MN, Bowman M, Oltz EM, Jelinsky SA, Colonna M. 2019. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol 20:980–991. doi: 10.1038/s41590-019-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Bella S, Ascenzi P, Siarakas S, Petrosillo N, di Masi A. 2016. Clostridium difficile toxins A and B: insights into pathogenic properties and extraintestinal effects. Toxins (Basel) 8:134. doi: 10.3390/toxins8050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jank T, Giesemann T, Aktories K. 2007. Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J Biol Chem 282:35222–35231. doi: 10.1074/jbc.M703138200. [DOI] [PubMed] [Google Scholar]
- 52.Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Haslam DB, Haslam D, Goldenring JR, Lacy DB. 2012. Clostridium difficile toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathog 8:e1003072. doi: 10.1371/journal.ppat.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson DS, Chen YH. 2012. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol 12:458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biro M, Munoz MA, Weninger W. 2014. Targeting Rho-GTPases in immune cell migration and inflammation. Br J Pharmacol 171:5491–5506. doi: 10.1111/bph.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friesland A, Zhao Y, Chen YH, Wang L, Zhou H, Lu Q. 2013. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc Natl Acad Sci USA 110:1261–1266. doi: 10.1073/pnas.1116051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finlay BB, McFadden G. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 57.Sansonetti PJ, Di Santo JP. 2007. Debugging how bacteria manipulate the immune response. Immunity 26:149–161. doi: 10.1016/j.immuni.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Reddick LE, Alto NM. 2014. Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell 54:321–328. doi: 10.1016/j.molcel.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Passmore IJ, Letertre MPM, Preston MD, Bianconi I, Harrison MA, Nasher F, Kaur H, Hong HA, Baines SD, Cutting SM, Swann JR, Wren BW, Dawson LF. 2018. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS Pathog 14:e1007191. doi: 10.1371/journal.ppat.1007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalim KW, Yang JQ, Li Y, Meng Y, Zheng Y, Guo F. 2018. Reciprocal regulation of glycolysis-driven Th17 pathogenicity and regulatory T cell stability by Cdc42. J Immunol 200:2313–2326. doi: 10.4049/jimmunol.1601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melendez J, Grogg M, Zheng Y. 2011. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem 286:2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang JQ, Kalim KW, Li Y, Duan X, Nguyen P, Khurana Hershey GK, Kroner J, Ruff B, Zhang L, Salomonis N, Rochman M, Wen T, Zheng Y, Guo F. 2019. Rational targeting Cdc42 restrains Th2 cell differentiation and prevents allergic airway inflammation. Clin Exp Allergy 49:92–107. doi: 10.1111/cea.13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Waddell A, Lin YD, Cantorna MT. 2015. Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol 8:618–626. doi: 10.1038/mi.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konya V, Czarnewski P, Forkel M, Rao A, Kokkinou E, Villablanca EJ, Almer S, Lindforss U, Friberg D, Hoog C, Bergman P, Mjosberg J. 2018. Vitamin D downregulates the IL-23 receptor pathway in human mucosal group 3 innate lymphoid cells. J Allergy Clin Immunol 141:279–292. doi: 10.1016/j.jaci.2017.01.045. [DOI] [PubMed] [Google Scholar]
- 66.Schiavo G, van der Goot FG. 2001. The bacterial toxin toolkit. Nat Rev Mol Cell Biol 2:530–537. doi: 10.1038/35080089. [DOI] [PubMed] [Google Scholar]
- 67.Larabee JL, Krumholz A, Hunt JJ, Lanis JM, Ballard JD. 2015. Exposure of neutralizing epitopes in the carboxyl-terminal domain of TcdB is altered by a proximal hypervariable region. J Biol Chem 290:6975–6985. doi: 10.1074/jbc.M114.612184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download iai.00073-22-s0001.pdf, PDF file, 0.3 MB (285KB, pdf)