ABSTRACT
PASA16 is a Pseudomonas aeruginosa phage isolated from a soil sample and used to treat several patients suffering from persistent infections in various countries. PASA16’s genome was sequenced, analyzed, and deposited in GenBank.
ANNOUNCEMENT
Pseudomonas aeruginosa is an opportunistic pathogen which commonly causes infections in health care settings (1–3). Its antibiotic-resistant strains are among the most challenging to treat and reportedly cause 32,600 infections yearly and 2,700 deaths in hospitalized U.S. patients (4).
Phage therapy is currently one of the most promising solutions to the emergence of antibiotic-resistant bacteria in general and against P. aeruginosa in particular.
Here, we report the genome sequence of the anti-P. aeruginosa phage PASA16, used in several phage-therapy treatments (5). PASA16 was isolated in Jerusalem from a garden (31°45′42″N, 35°10′19″E) soil sample and purified using the phage titration method (6, 7). Briefly, the sample was dissolved in LB and mixed with overnight-grown P. aeruginosa PA14 at 37°C. The culture was centrifuged (7,100 × g, 10 min), and the supernatant was filtered (0.22 μm). Serial dilutions of 5 μL of the filtrate were spotted on P. aeruginosa PA14 lawn, and after overnight incubation at 37°C, the clearest plaques were picked, and their titer was determined. The phage particles were visualized by transmission electron microscopy (TEM) (Fig. 1) as described (6).
FIG 1.
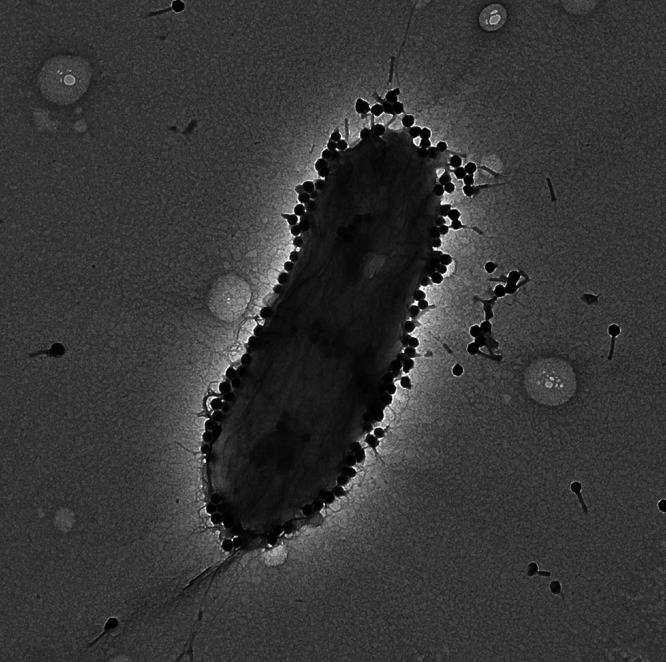
A TEM visualization of the phage PASA16 with P. aeruginosa PA14.
DNA was purified (phage DNA isolation kit; Norgen Biotek) from a single plaque that was picked and grown to a titer of 109 PFU/mL, followed by library preparation (Illumina Nextera XT kit) and sequencing (Illumina NextSeq 500/550 kit) with single-end reads of 150 bp (Illumina 150-cycle midoutput kit v2,) according to Illumina’s recommendations. Machine output files (binary base call [BCL]) were converted to FASTQ files using the bcl2fastq/2.20.0 program.
Most bioinformatic analyses were performed using Geneious Prime v2021.0.3 and its plugins (Biomatters). The reads were trimmed (Geneious Prime BBDuk plugin) with default parameters. Sequencing of PASA16 yielded 1,651,174 reads of 150 bp in size. Bacterial sequences were removed by map-to-reference on the P. aeruginosa PA14 genome sequence followed by de novo assembly of the remaining reads with the SPAdes plugin of Geneious Prime with default parameters. The resulting contigs were reassembled with the Geneious Prime assembler with the highest sensitivity to produce one major contig of ∼60 kb, which was fine-tuned by 10 iterative cycles of map-to-reference of the trimmed reads and consensus generation. The average coverage was 3,580.1× with a standard deviation (SD) of 351.2.
The PASA16 genome is 66,127 bp long with a GC content of 55.6% and contains 90 coding DNA sequences (CDS). Its terminal repeats and analysis of the alignment of de- and reassembled genomes reveal that the PASA16 genome topology is either circular or circularly permuted (8). The taxonomy of PASA16, as determined by GenBank, is Duplodnaviria, Heunggongvirae, Uroviricota, Caudoviricetes, Caudovirales, Myoviridae, and Pbunavirus. Annotation with RAST (9) revealed that most PASA16 genes encode phage-related or hypothetical proteins. One notable gene is a lytic enzyme gene (GenBank protein accession number QOI69201.1) coding a putative lysozyme-like endolysin. This gene increases the possibility of the phage having high lytic abilities against the target bacteria (10) and perhaps contributed to its success in phage therapy treatments. As a candidate for phage therapy, the phage sequence was scanned for virulence factors using ABRicate v1.0.1 (T. Seemann, https://github.com/tseemann/abricate) against all its databases with its default parameters. No virulence factors or resistance genes were identified, and therefore the phage was deemed safe for use in phage therapy treatments.
Data availability.
The sequence of the annotated genome of PASA16 can be found in GenBank (accession number MT933737.1), and the raw data are available under NIH BioSample database project PRJNA706131, “The Israeli Phage Bank (IPB),” BioSample number SAMN25300365, and Sequence Read Archive (SRA) number SRX13974252.
ACKNOWLEDGMENTS
We thank the Core Research Facility team of the Hebrew University, Ein Kerem Campus, Abed Nasereddin, and Idit Shiff for the deep sequencing and Eduardo Berenshtein for the TEM microscopy.
The “United States-Israel Binational Science Foundation (BSF)” grant 2017123, the Israel Science Foundation (ISF-IPMP) grant ISF_1349/2020, and the Rosetrees Trust grant A2232 provided funding for the project.
Contributor Information
Ronen Hazan, Email: ronenh@ekmd.huji.ac.il.
John J. Dennehy, Queens College CUNY
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moradali MF, Ghods S, Rehm BH. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behzadi P, Baráth Z, Gajdács M. 2021. It’s not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics 10:42. doi: 10.3390/antibiotics10010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, CDC, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 5.Khatami A, Lin RC, Petrovic-Fabijan A, Alkalay-Oren S, Almuzam S, Britton PN, Brownstein MJ, Dao Q, Fackler J, Hazan R, Horne BA, Nir-Paz R, Iredell JR. 2021. Bacterial lysis, autophagy and innate immune responses during adjunctive phage therapy in a child. EMBO Mol Med 13:e13936. doi: 10.15252/emmm.202113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalifa L, Brosh Y, Gelman D, Coppenhagen-Glazer S, Beyth S, Poradosu-Cohen R, Que YA, Beyth N, Hazan R. 2015. Targeting Enterococcus faecalis biofilms with phage therapy. Appl Environ Microbiol 81:2696–2705. doi: 10.1128/AEM.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkalay-Oren S, Gold N, Khalifa L, Yerushalmy O, Coppenhagen-Glazer S, Nir-Paz R, Hazan R. 2021. Complete genome sequences of two Enterococcus faecalis bacteriophages, EFGrKN and EFGrNG, targeted to phage therapy. Microbiol Resour Announc 10:16. doi: 10.1128/MRA.00126-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossi GF, Macchiato MF, Gialanella G. 1983. Circular permutation analysis of phage T4 DNA by electron microscopy. Z Naturforsch C Biosci 38:294–296. doi: 10.1515/znc-1983-3-422. [DOI] [PubMed] [Google Scholar]
- 9.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasina DV, Antonova NP, Grigoriev IV, Yakimakha VS, Lendel AM, Nikiforova MA, Pochtovyi AA, Remizov TA, Usachev EV, Shevlyagina NV, Zhukhovitsky VG, Fursov MV, Potapov VD, Vorobev AM, Aleshkin AV, Laishevtsev AI, Makarov VV, Yudin SM, Tkachuk AP, Gushchin VA. 2021. Discovering the potentials of four phage endolysins to combat Gram-negative infections. Front Microbiol 12:748718. doi: 10.3389/fmicb.2021.748718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of the annotated genome of PASA16 can be found in GenBank (accession number MT933737.1), and the raw data are available under NIH BioSample database project PRJNA706131, “The Israeli Phage Bank (IPB),” BioSample number SAMN25300365, and Sequence Read Archive (SRA) number SRX13974252.


