Abstract
Removal of 2′,3′-didehydro-3′-deoxythymidine-5′-monophosphate (d4TMP) from a blocked DNA chain can occur through transfer of the chain-terminating residue to a nucleotide acceptor by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). ATP-dependent removal of either d4TMP or 3′-azido-3′-deoxythymidine-5′-monophosphate (AZTMP) is increased in AZT resistant HIV-1 RT (containing D67N/K70R/T215F/K219Q mutations). Removal of d4TMP is strongly inhibited by the next complementary deoxynucleoside triphosphate (50% inhibitory concentration [IC50] of ∼0.5 μM), whereas removal of AZTMP is much less sensitive to this inhibition (IC50 of >100 μM). This could explain the lack of cross-resistance by AZT-resistant HIV-1 to d4T in phenotypic drug susceptibility assays.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and other retroviral RTs lack 3′-5′ exonuclease activity (2, 30) but can remove 3′-terminal chain-terminating residues from blocked DNA chains through a nucleotide-dependent mechanism leading to production of dinucleoside polyphosphates (23, 24) or through pyrophosphorolysis (the reversal of polymerization) (1, 4, 13, 29). We have recently shown (23) that HIV-1 RT containing the 3′-azido-3′-deoxythymidine (AZT) resistance mutations D67N, K70R, T215F, and K219Q (67/70/215/219 mutant RT) removes AZT–5′-monophosphate (AZTMP) from blocked primer-templates through the nucleotide-dependent mechanism more efficiently than does wild-type (WT) RT. The mutant enzyme also removes 2′,3′-dideoxyadenosine-5′-monophosphate (ddAMP) from blocked DNA chains more efficiently than does WT RT. Removal of ddAMP is strongly suppressed by physiological concentrations of deoxynucleoside triphosphates (dNTPs), whereas removal of AZTMP is much less sensitive to this inhibition (23).
The chain terminator 2′,3′-didehydro-3′-deoxythymidine-5′-triphosphate (d4TTP) is efficiently incorporated into growing DNA chains by HIV-1 RT (39). Resistance to d4T can arise in cell culture through a valine-to-threonine mutation at position 75 (19, 21, 32); however, this mutation is rarely observed in HIV-1 from d4T-treated individuals (6, 9, 17, 22, 27, 35). Instead, mutations associated with AZT resistance, including M41L, D67N, K70R, L210W, and T215Y/F, are frequently selected (6, 8, 21, 22, 27, 32, 35). The selection of AZT resistance mutations by d4T in the absence of AZT is unexpected, since phenotypic assays show little, if any, cross-resistance between these drugs (20, 22). Nonetheless, clinical studies have shown that prior exposure to AZT reduces the efficacy of subsequent treatment with d4T (17), and the presence of AZT resistance mutations is correlated with reduced suppression of viral load in response to d4T-containing therapies (15, 25). These results suggest that the phenotypic assays do not fully reflect the in vivo sensitivity of HIV-1 replication to d4T.
In an effort to understand the biochemical basis for the lack of cross-resistance by AZT-resistant mutants to d4T, we have investigated the ability of WT and 67/70/215/219 mutant RT to remove d4TMP, AZTMP, and 3′-deoxythymidine monophosphate (ddTMP) from chain-terminated DNA primers through either dinucleoside polyphosphate synthesis or pyrophosphorolysis.
Removal of dTMP analogues from blocked primer-templates.
The removal of chain-terminating nucleotides was assessed by measuring the formation of unblocked primer from a previously blocked primer-template (24) (Fig. 1A). These experiments were carried out in two steps. First, HIV-1 RT was incubated with the blocked primer-template in the presence of the appropriate substrate for either ATP- or pyrophosphate (PPi)-dependent removal of the chain-terminating nucleotide (rescue step). This was followed by heat inactivation of the RT and extension of unblocked primer-template by addition of all four dNTPs and exonuclease-free Klenow fragment of E. coli DNA polymerase 1 (which, under the conditions used here, does not remove chain-terminating nucleotides in the absence or presence of ATP or PPi; Fig. 1B) (extension step). The experiments were performed in this way in order to avoid inhibition of primer unblocking by dNTPs (23, 24) and to exclude further unblocking during the extension reaction.
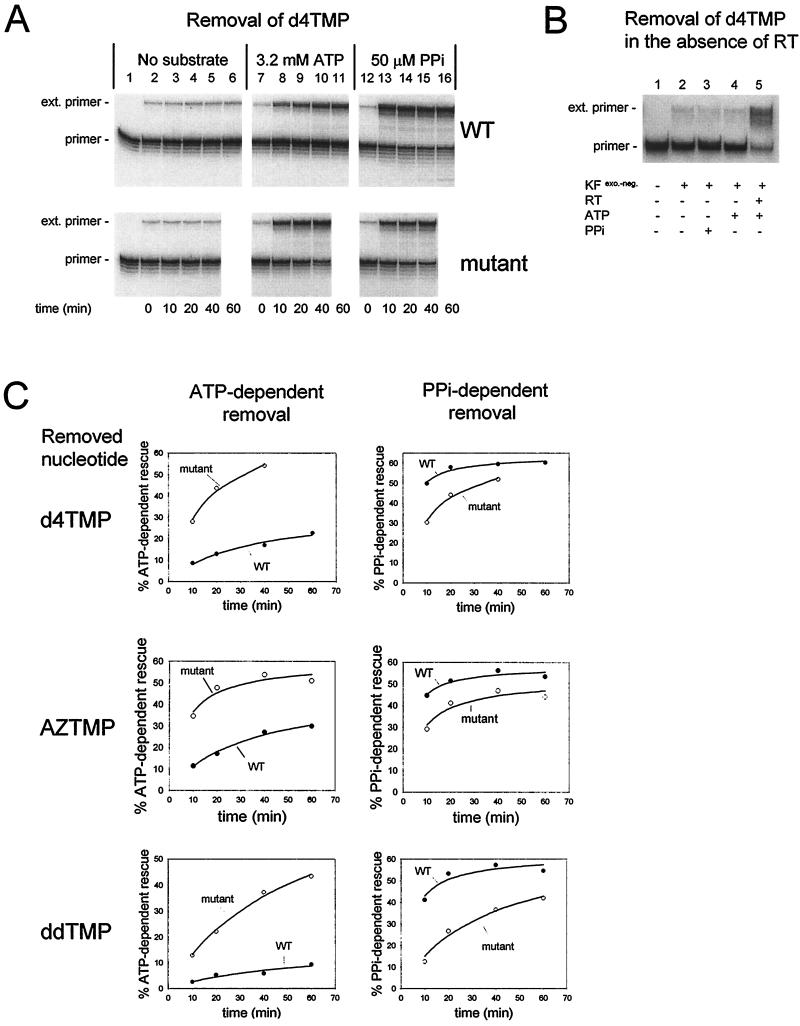
FIG. 1.
Unblocking of chain-terminated primer-templates by WT or 67/70/215/219 mutant RT through either dinucleoside polyphosphate synthesis or pyrophosphorolysis. (A) Rescue of d4TMP-terminated primer. d4TMP-terminated 5′-32P-labeled L33 primer-WL50 template (5 nM) was incubated with 200 nM WT (upper panels) or 67/70/215/219 mutant RT (lower panels) and with no substrate (left panels), 3.2 mM ATP (middle panels), or 50 μM PPi (right panels) for the indicated times at 37°C. The RT was inactivated by heat treatment, and the unblocked primer was extended by incubation with exonuclease-free Klenow fragment of E. coli DNA polymerase I and all four dNTPs. Products were fractionated on a 10% denaturing polyacrylamide gel (24). Positions for unextended primer (primer) and for products formed after elongation to the end of the template (ext. primer) are shown at the left of the figure. Lane 1, unextended primer-template. (B) Effect of incubation in the absence of HIV-1 RT. The experiment was performed as described in panel A in the absence of HIV-1 RT (lanes 1 to 4) or with HIV-1 RT (lane 5) with or without added ATP (3.2 mM) or PPi (50 μM), as indicated in the figure, for 40 min at 37°C. After heat treatment, unblocked primer was extended by addition of exonuclease-free Klenow fragment of E. coli DNA polymerase I and all four dNTPs. (C) Rescue of primer terminated with d4TMP, AZTMP, or ddTMP. Incubations were carried out as in panel A with primer terminated with either d4TMP, AZTMP, or ddTMP in the presence of 3.2 mM ATP (left panels) or 50 μM PPi (right panels) and WT RT (●) or 67/70/215/219 mutant RT (○). Radioactivity in products of >34 nucleotides (rescued primers) was quantitated by phosphorimaging and expressed as percentage of total radioactivity in the lane. The data were fitted to a hyperbola using Sigmaplot 4.0 (solid lines).
WT or mutant HIV-1 RT was incubated with d4TMP-terminated, 5′-32P-labeled L33 primer-WL50 template in the absence or presence of either 3.2 mM ATP or 50 μM PPi at 37°C for the times indicated in Fig. 1A. L33 and WL50 are synthetic oligodeoxynucleotides with the following sequences: chain-terminated L33, 5′-CTACTAGTTTTCTCCATCTAGACGATACCAGAATAN-3′; and WL50, 3′-GATGATCAAAAGAGGT AGATCTGCTATGGTCTTACTTCTGGAGTCGTGAG-5′. TAN refers to the chain-terminating 3′-deoxythymidylate analogues d4TMP, AZTMP, and ddTMP.
The HIV-1 RTs used in these experiments were derived from the expression vector pKRT2, containing RT coding sequences from human T-cell leukemia virus type III BH10 (7), to which an N-terminal polyhistidine tail had been added (24). The enzymes were purified as previously described (24) and were predominantly homodimeric. The 67/70/215/219 mutant RT contained the mutations in both subunits.
In the absence of incubation with HIV-1 RT (Fig. 1A, lanes 2, 7, and 12, upper and lower panels, and Fig. 1B, lanes 2 to 4), a small portion of the primer could be extended, possibly due to incomplete chain termination. If the first incubation was performed in the absence of ATP or PPi, a minimal increase in the amount of unblocked primer was observed even after extended incubation (lanes 2 to 6 in the upper panel and lanes 2 to 5 in the lower one). In the presence of ATP, however, there was a time-dependent increase in the amount of extendible primer (lanes 7 to 11). ATP-dependent rescue of d4TMP-terminated primer was much more efficient with 67/70/215/219 mutant RT than with WT RT. On the other hand, when PPi was used as a substrate for the rescue reaction, there was little difference between the mutant and WT RTs. The dependence on the addition of ATP or PPi suggests that the primer rescue could not be accounted for by contaminating exonuclease activity. To obtain a sensitive measure of exonuclease present in each enzyme preparation, 3′-labeled [32P]ddAMP-terminated primer-template was incubated with a 40-fold excess of RT. For both WT and mutant enzymes a similar amount of the label (ca. 4 to 5%) was released as [32P]ddAMP after 1 h of incubation at 37°C.
The results from experiments such as the one presented in Fig. 1A were quantitated using a Molecular Dynamics PhosphorImager, and the percentage of primer rescued was plotted versus time (Fig. 1C). The ATP-dependent removal of d4TMP was greatly increased in the 67/70/215/219 mutant RT compared to WT RT (upper left panel), whereas removal through pyrophosphorolysis was slightly decreased in the 67/70/215/219 mutant RT (upper right panel). We also determined the ability of WT and 67/70/215/219 mutant RT to remove AZTMP (middle panels) or ddTMP (lower panels) through either ATP-dependent rescue (left panels) or pyrophosphorolysis (right panels). The rate of the ATP-dependent reaction catalyzed by the 67/70/215/219 mutant RT was much greater than that catalyzed by WT RT for the removal of each of the three chain-terminating thymidylate analogues, whereas PPi-dependent removal occurred at a slightly increased rate with WT RT.
Inhibition of primer rescue by the dNTP complementary to the next position on the template.
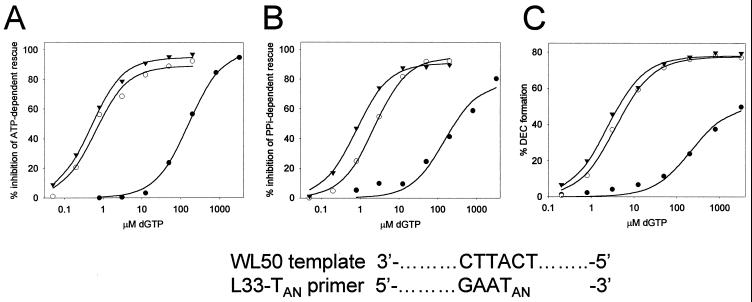
We have previously shown that formation of a stable dead-end complex (DEC) in the presence of micromolar concentrations of the dNTP complementary to the next nucleotide position on the template can be detected by an electrophoretic mobility shift assay (38). The DEC may be equivalent to the catalytic complex recently described by Huang et al. (14). We have also shown that removal of chain-terminating residues by RT is inhibited by micromolar concentrations of the next complementary dNTP, leading us to suggest that DEC formation and inhibition of primer rescue may be related events (24). Figure 2 shows a comparison of the concentration of dGTP, the dNTP complementary to the next nucleotide position on the template, required for inhibition of ATP-dependent primer rescue (Fig. 2A), inhibition of PPi-dependent primer rescue (Fig. 2B), and for DEC formation (Fig. 2C), with primer-templates terminated with each of the three dTMP analogues.
FIG. 2.
Ability of the dNTP complementary to the next nucleotide position on the template to inhibit primer rescue and to induce a stable complex with chain-terminated primer-template and 67/70/215/219 mutant RT. (A and B) Inhibition of primer rescue by dGTP. Rescue by 67/70/215/219 mutant RT of primer-templates terminated with d4TMP (▾), ddTMP (○), or AZTMP (●) using either ATP (A) or PPi (B) as a substrate was performed as described in the legend to Fig. 1, in the presence of the indicated concentrations of dGTP. The results were quantitated by phosphorimaging, and the percent inhibition was plotted versus dGTP concentration and fitted to hyperbolas (solid lines) using Sigmaplot 4.0. (C) Formation of DEC with 67/70/215/219 mutant RT, chain-terminated primer-templates, and dGTP. DEC formation was carried out in the absence of ATP or PPi and was monitored by gel mobility shift assay as described previously (23, 38). The percent DEC formation was plotted as a function of dGTP concentration. The primer-templates are identified as in panel A. Partial sequences of the WL50 template and L33 primer terminated with a dTMP analogue (TAN) are shown.
Excess 67/70/215/219 mutant RT was incubated with chain-terminated, 5′-32P-labeled L33 primer-WL50 template and 3.2 mM ATP (Fig. 2A) or 50 μM PPi (Fig. 2B) in the presence of various concentrations of dGTP to determine the 50% inhibitory concentrations (IC50s) for dGTP inhibition of each rescue activity (Table 1). Removal of either d4TMP or ddTMP was strongly suppressed by dGTP (IC50s of ∼0.4 to 0.8 μM and of ∼2 μM, respectively). In contrast, both ATP-dependent removal and PPi-dependent removal of AZTMP were much less sensitive to the presence of dGTP (IC50s of >100 μM), in agreement with previous results (23).
TABLE 1.
Ability of dGTP to inhibit primer rescue and to induce DEC formation
| Primer-template | Inhibition of primer rescue (IC50, μM)a ± AD
|
DEC formation (Kd,app, μM)b ± AD | |
|---|---|---|---|
| ATP dependent | PPi dependent | ||
| d4TMP terminated | 0.51 ± 0.07 | 0.72 ± 0.05 | 2.6 ± 1.1 |
| ddTMP terminated | 2.6 ± 0.2 | 1.6 ± 0.5 | 7.7 ± 3.8 |
| AZTMP terminated | 110 ± 30 | 170 ± 20 | 220 ± 60 |
Reactions were performed as described in the legend of Fig. 2 in the presence of 67/70/215/219 mutant RT, primer-template terminated with the indicated nucleotide analogue and various concentrations of dGTP. The IC50s were determined as shown in Fig. 2A and B.
Electrophoretic mobility shift experiments were performed as described in the text. Kd,app values for DEC formation as a function of dGTP concentration were determined as shown in Fig. 2C. The numbers were obtained from two to three experiments. AD, average deviation.
Figure 2C shows DEC formation with primer-template terminated with each of the three dTMP analogues. Excess 67/70/215/219 mutant RT was incubated with 5′-32P-labeled, chain-terminated L33 primer-WL50 template in the presence of the indicated concentrations of dGTP for 15 min at 37°C. The samples were fractionated by electrophoresis through a nondenaturing polyacrylamide gel, and DEC was identified by its slower electrophoretic mobility (23, 38). Free primer-template and DEC were quantitated by phosphorimaging, and the percent DEC was plotted versus dGTP concentration (Fig. 2C) to obtain the Kd,app, the dGTP concentration required for formation of 50% DEC (Table 1). Kd,app for AZTMP-terminated primer-template was 30 to 100-fold higher than for either ddTMP- or d4TMP-terminated primer-template. The amount of dGTP needed for inhibition of primer rescue and for DEC formation were similar, consistent with a model in which the removal reaction cannot occur while HIV-1 RT and primer-template are trapped as DEC (23).
To determine whether dNTP inhibition of primer rescue was influenced by template sequence, the experiments in Fig. 2A were repeated with a template in which dC at the first downstream position from the primer terminus was replaced with dG. With this template, removal of the dTMP analogues was inhibited by dCTP (IC50s of 1.7 ± 0.1 μM for d4TMP-terminated primer, 5.5 ± 0.5 μM for ddTMP-terminated primer, and 790 ± 190 μM for AZTMP-terminated primer) and was insensitive to dATP, dGTP, and dTTP (IC50s of >1,000 μM) (data not shown). These results demonstrate that the specificity for dNTP inhibition of primer rescue is determined by the next nucleotide position on the template.
Primer extension in the presence of chain-terminating nucleotides.
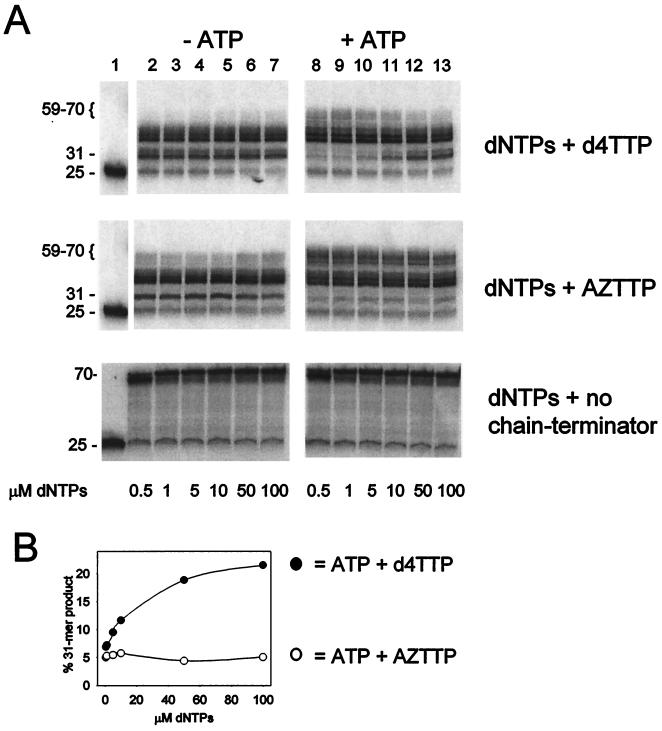
The 67/70/215/219 mutant RT was incubated with 5′-32P-labeled D25 primer annealed to D70 template, all four dNTPs, and d4TTP, AZTTP, or no chain terminator, in the absence or presence of 3.2 mM ATP (Fig. 3). The concentration of chain terminator was adjusted to maintain a fixed ratio to dTTP in each reaction mixture (d4TTP/dTTP = 4:1 and AZTTP/dTTP = 2:1). The ratios of dTTP analogue to dTTP were chosen to give similar patterns of termination products in a size range suitable for gel analysis.
FIG. 3.
Primer extension by 67/70/215/219 mutant RT in the presence of d4TTP or AZTTP or in the absence of chain terminator. (A) 5′-32 P-labeled D25 primer-D70 template (5 nM) was incubated for 30 min at 37°C with 200 nM 67/70/215/219 mutant RT, the indicated concentrations of all four dNTPs, and d4TTP (d4TTP/dTTP = 4:1) (upper panel); AZTTP (AZTTP/dTTP = 2:1) (middle panel), or no chain-terminating nucleotide (lower panel), in the absence (−ATP) or presence (+ATP) of 3.2 mM ATP. The products were fractionated on a 10% denaturing polyacrylamide gel. The primer lengths are indicated in nucleotides. Lane 1, untreated primer-template. (B) The results in panel A were quantitated by phosphorimaging, and the 31-mer chain-terminated product formed in the presence of ATP plus d4TTP plus dNTPs (●) or ATP plus AZTTP plus dNTPs (○), expressed as a percentage of the total radioactivity in the lane, was plotted versus the dNTP concentration.
The sequences of the 25-mer DNA primer (D25) and 70-mer DNA template (D70) were as follows: D25, 5′-GTTTCTGATCTGGTGTGAAAAGTCC-3′; and D70, 3′-CAAAGAC TAGACCACACTTTTCAGGGGTGGAGTTGTCTACAAC AGAGTCGAGGAGATAAAAACAAGATAC-5′.
In the presence of dNTPs and d4TTP, the 25-mer DNA primer was extended and terminated at the expected T incorporation sites (Fig. 3A, upper panel). The primer extension products observed in this experiment were due to chain termination rather than enzyme pausing since the formation of these products depended on the presence of chain-terminating nucleotide analogues (Fig. 3A, compare the upper and lower panels). When 3.2 mM ATP was included in the reaction mixture, longer products were observed (59 to 70 bases in length), while the shorter species, such as the 31-mer product corresponding to the first termination site, were depleted (Fig. 3A, upper panel, compare lanes 2 and 8). This is consistent with ATP-dependent rescue of DNA chains initially terminated with d4TMP and further extension by RT. At higher dNTP concentrations, the ATP-dependent formation of longer extension products was diminished and the shorter species were less depleted (Fig. 3A, upper panel, and Fig. 3B), as expected from the inhibition of d4TMP removal by the next complementary dNTP.
A parallel experiment with AZTTP is shown in the middle panel of Fig. 3A. When AZTTP was present in the reaction mixture, ATP-dependent rescue also led to an increase in longer products and a decrease in the 31-mer product as we have previously reported (23) (Fig. 3A, middle panel, compare lanes 2 and 8). In contrast with the results with d4TMP-terminated primer, the rescue of the 31-nucleotide AZTMP termination product was not inhibited at higher dNTP concentrations (Fig. 3B).
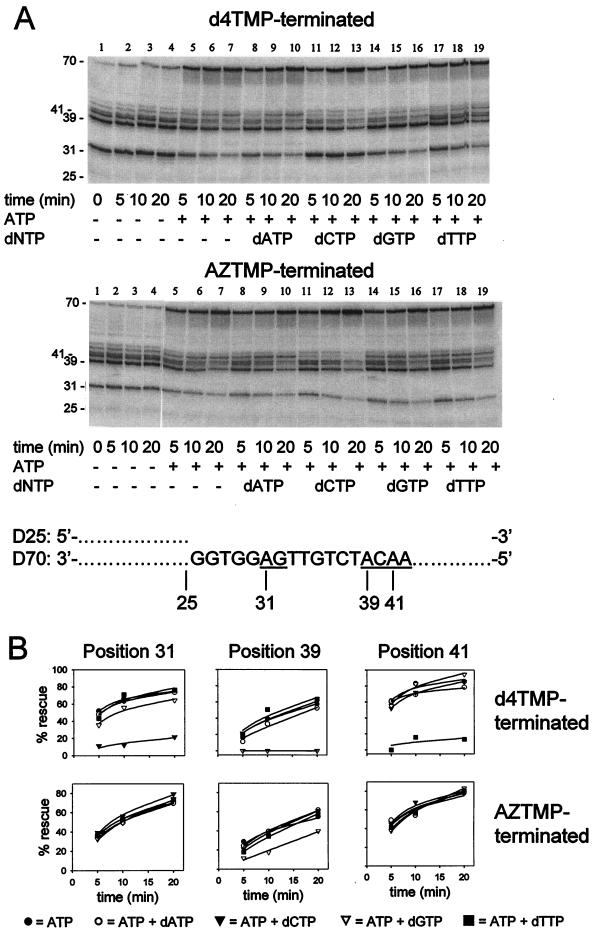
dNTP inhibition of primer rescue was further investigated by purification of the mixture of d4TMP (Fig. 4A, upper panel) or AZTMP (Fig. 4A, lower panel) termination products and further incubation with RT in absence of ATP or in the presence of ATP without added dNTPs or with each of the four dNTPs individually. Following the incubation in which ATP-dependent rescue of chain-terminated primer could occur (rescue step), the RT was heat inactivated, and the rescued products were extended with exonuclease-free Klenow fragment of DNA polymerase I (extension step). Individual chain termination products were quantitated by phosphorimaging (Fig. 4B). Rescue of chains terminated at position 31 was not observed when ATP was omitted during the rescue step (Fig. 4A, lanes 1 to 4); however, the products were rescued when ATP was present (Fig. 4A, lanes 5 to 7, and 4B). Addition of both dCTP and ATP during the rescue step resulted in almost complete inhibition of rescue of the d4TMP-terminated 31-nucleotide products but no detectable inhibition of rescue of the corresponding AZTMP-terminated product (Fig. 4A, lanes 11 to 13, and 4B). Addition of dATP, dGTP, or dTTP during the rescue step resulted in no inhibition of rescue of either d4TMP- or AZTMP-terminated 31-mers (Fig. 4A, lanes 8 to 10 and 14 to 19, and 4B). Examination of the template sequence (bottom of Fig. 4A) shows that the nucleotide following the position 31 termination site is dG. Therefore, a primer-template terminated at position 31 would form DEC with dCTP resulting in dCTP-specific inhibition of rescue of this termination product, as was observed.
FIG. 4.
Effects of dNTPs on ATP-dependent removal of chain terminators in multiple sequence contexts. (A) 5′-32P-labeled D25 primer-D70 template was incubated for 30 min at 37°C with excess 67/70/215/219 mutant RT, 1 μM concentrations of all four dNTPs, and either d4TTP (2 μM) or AZTTP (1 μM). The resulting mixtures of chain-terminated products were extracted with phenol-chloroform, followed by ethanol precipitation. Excess 67/70/215/219 RT (200 nM) was incubated with either d4TMP-terminated products (5 nM) (upper panel) or AZTMP-terminated products (5 nM) (lower panel) in the absence or presence of 3.2 mM ATP and in the presence or absence of 50 μM of the indicated dNTP for the indicated times at 37°C. After heat inactivation of the HIV-1 RT (5 min at 90°C), unblocked primers were extended by the addition of exonuclease-free Klenow fragment of E. coli DNA polymerase I and 100 μM concentrations of each of the four dNTPs and the incubation at 37°C for 30 min. Product lengths are indicated in nucleotides. A partial sequence of the D70 template is shown. (B) Inhibition of ATP-dependent primer rescue by dNTPs. The results shown in panel A were quantitated by phosphorimaging. The amounts of radioactivity in the 31-mer products (left panels), the 39-mer products (middle panels), and the 41-mer products (right panels) terminated with either d4TMP (upper panels) or AZTMP (lower panels) were normalized to the total radioactivity in each lane. The percent ATP-dependent rescue was calculated by the formula: 100−100(a/b), where a and b are the normalized radioactivity in the band of interest in the presence or absence of ATP, respectively, and plotted versus time. The data (indicated by the symbols shown at the bottom of the figure) were fitted to hyperbolas (solid lines) using Sigmaplot 4.0.
Similar analysis of termination products at position 39 showed that rescue of d4TMP termination products was inhibited specifically when dGTP was present during the rescue step, whereas AZTMP termination products were only slightly inhibited (Fig. 4B, middle panels). The template nucleotide following the position 39 termination site is dC. Rescue of d4TMP termination products at position 41 was specifically inhibited by dTTP, whereas the AZTMP termination products at this site were not inhibited (Fig. 4B, right panels). The template nucleotide following the position 41 termination site is dA. In summary, ATP-dependent rescue of d4TMP termination products was specifically inhibited by the dNTP complementary to the next position on the template in each of these three sequence contexts, and rescue of AZTMP termination products was much less sensitive to this inhibition in each case.
From these results we predict that the removal of d4TMP in vivo will depend on the intracellular levels of dNTPs (low dNTP pools allow rescue to occur, while high dNTP pools will greatly suppress it), whereas removal of AZTMP will be much less sensitive to physiological concentrations of dNTPs.
Conclusions.
We have shown that the 67/70/215/219 mutant RT has increased ability, compared to WT RT, to remove d4TMP, AZTMP, and ddTMP from blocked DNA primer-templates through transfer of the chain terminator to a nucleotide acceptor. We have previously shown that mutant RT also has increased ability to remove ddAMP from a primer terminus (23), and we have preliminary results that ddGMP and ddCMP removal activities are also increased in the mutant enzyme (P. R. Meyer et al., unpublished results). In contrast, we did not observe an increase in the transfer of chain terminator to pyrophosphate by the mutant RT. The removal of d4TMP or ddTMP was inhibited by micromolar or submicromolar concentrations of the next complementary dNTP, whereas removal of AZTMP was >50-fold less sensitive to this inhibition.
The removal of chain terminators in vivo will, presumably, be controlled by the concentrations of nucleotide acceptors for the transfer reaction as well as the levels of dNTPs, which are inhibitory. ATP is present at millimolar concentrations in lymphocytes (37), making it the most likely acceptor substrate for the removal reaction. Estimates of dNTP concentrations depend on assumptions about cell volume (3, 5, 11, 33, 36) and range from 0.14 to 5.6 μM in resting lymphocytes (10, 11, 31, 37), 2.4 to 26 μM in mitogen-stimulated lymphocytes (10, 11), and 15 to 170 μM in CEM lymphoblasts (31, 37). These values suggest that removal of AZTMP residues can occur in cells at most stages of activation, since the IC50 for dNTP inhibition is >100 μM, but that removal of d4TMP will be inhibited except in cells that are relatively quiescent. However, even in activated lymphocytes, where the d4T is a much better inhibitor than in resting cells due to a higher d4TTP/dTTP ratio (34), d4TMP that has been incorporated might be removed by HIV-1 RT during subsequent periods of reduced cell activation. The physiological importance of removal of d4TMP is difficult to evaluate since dNTP pools may differ between cell types and dNTP concentrations may be unevenly distributed within a cell. Nonetheless, the finding that mutations that increase the removal reaction are selected during d4T therapy suggests that removal of d4TMP from growing DNA chains is physiologically relevant.
From the results of phenotypic drug susceptibility assays it has been concluded that AZT-resistant HIV-1 is not cross-resistant to d4T (20, 22). However, this conclusion fails to account for the selection of AZT resistance mutations during d4T monotherapy (6, 8, 21, 22, 27, 32, 35) or why treatment regimens containing d4T are less effective in patients harboring AZT-resistant virus (15, 17, 25). An explanation may lie in the fact that the most commonly used phenotypic drug susceptibility assays use either mitogen-stimulated peripheral blood lymphocytes (16, 21, 22) or transformed human cell lines (12, 18, 20, 26, 28) to optimize virus replication. In these systems the intracellular dNTP pools are elevated. The phenotypic assays show that the 67/70/215/219 mutant HIV-1 is sensitive to d4T but resistant to AZT; however, we predict that the 67/70/215/219 mutant HIV-1 would be cross-resistant to d4T under conditions where the dNTP pools are low.
In patients harboring AZT-resistant virus, infected cells may go through periods when they are not continuously stimulated to divide and the dNTP pools may be reduced. Under these conditions the AZT resistant RT will have an increased ability to remove d4TMP compared to WT RT. This would give the AZT-resistant HIV-1 a replicative advantage over WT virus, and the AZT resistance mutations would be selected. Our results suggest that phenotypic assays for HIV drug susceptibility may need to be reevaluated, taking into account the selective effects of dNTP pools on the sensitivities of WT and mutant viruses to nucleoside analogues.
Acknowledgments
This work was supported by NIH grants AI-39973 (W.A.S.) and DK-26206 (A.G.S.) and by the Department of Veteran Affairs (R.F.S.).
REFERENCES
- 1.Arion D, Kaushik N, McCormick S, Borkow G, Parniak M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 2.Battula N, Loeb L A. On the fidelity of DNA replication: lack of exodeoxyribonuclease activity and error-correction function in avian myeloblastosis virus DNA polymerase. J Biol Chem. 1976;251:982–986. [PubMed] [Google Scholar]
- 3.Braylan R C, Fowlkes B J, Jaffe E S, Sanders S K, Berard C W, Herman C J. Cell volumes and DNA distributions of normal and neoplastic human lymphoid cells. Cancer. 1978;41:201–209. doi: 10.1002/1097-0142(197801)41:1<201::aid-cncr2820410129>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Carroll S S, Geib J, Olsen D B, Stahlhut M, Shafer J A, Kuo L C. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry. 1994;33:2113–2120. doi: 10.1021/bi00174a018. [DOI] [PubMed] [Google Scholar]
- 5.Chapman E H, Kurec A S, Davey F R. Cell volumes of normal and malignant mononuclear cells. J Clin Pathol. 1981;34:1083–1090. doi: 10.1136/jcp.34.10.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coakley E P, Gillis J M, Hammer S M. Phenotypic and genotypic resistance patterns of HIV-1 isolates derived from individuals treated with didanosine and stavudine. AIDS. 2000;14:F9–F15. doi: 10.1097/00002030-200001280-00002. [DOI] [PubMed] [Google Scholar]
- 7.D'Aquila R T, Summers W C. HIV-1 reverse transcriptase/ribonuclease H: high-level expression in Escherichia coli from a plasmid constructed using the polymerase chain reaction. J Acquir Immune Defic Syndr. 1989;2:579–587. [PubMed] [Google Scholar]
- 8.de Mendoza C, Soriano V, Briones C, Gallego O, Barreiro P, Alvarez A, Gonzalez-Lahoz J. Emergence of zidovudine resistance in HIV-infected patients receiving stavudine. J Acquir Immune Defic Syndr. 2000;23:279–281. doi: 10.1097/00126334-200003010-00013. [DOI] [PubMed] [Google Scholar]
- 9.Deminie C A, Bechtold C M, Riccardi K, Rose R E, Samanta H, Lin P-F, Colonno R J. Clinical HIV-1 isolates remain sensitive to stavudine following prolonged therapy. AIDS. 1998;12:110–112. [PubMed] [Google Scholar]
- 10.Gao W-Y, Agbaria R, Driscoll J S, Mitsuya H. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J Biol Chem. 1994;269:12633–12638. [PubMed] [Google Scholar]
- 11.Gao W-Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertogs K, de Bethune M-P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh J-C, Zinnen S, Modrich P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J Biol Chem. 1993;268:24607–24613. [PubMed] [Google Scholar]
- 14.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 15.Izopet J, Bicart-See A, Pasquier C, Sandres K, Bonnet E, Marchou B, Puel J, Massip P. Mutations conferring resistance to zidovudine diminish the antiviral effect of stavudine plus didanosine. J Med Virol. 1999;59:507–511. [PubMed] [Google Scholar]
- 16.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katlama C, Valantin M-A, Matheron S, Coutellier A, Calvez V, Descamps D, Longuet C, Bonmarchand M, Tubiana R, De Sa M, Lancar R, Agut H, Brun-Vezinet F, Costagliola D. Efficacy and tolerability of stavudine plus lamivudine in treatment-naive and treatment-experienced patients with HIV-1 infection. Ann Intern Med. 1998;129:525–531. doi: 10.7326/0003-4819-129-7-199810010-00003. [DOI] [PubMed] [Google Scholar]
- 18.Kellam P, Larder B A. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacey S F, Larder B A. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larder B A, Chesebro B, Richman D D. Susceptibilities of zidovudine-susceptible and -resistant human immunodeficiency virus isolates to antiviral agents determined by using a quantitative plaque reduction assay. Antimicrob Agents Chemother. 1990;34:436–441. doi: 10.1128/aac.34.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin P-F, Gonzalez C J, Griffith B, Friedland G, Calvez V, Ferchal F, Schinazi R F, Shepp D H, Ashraf A B, Wainberg M, Soriano V, Mellors J W, Colonno R J. Stavudine resistance: an update on susceptibility following prolonged therapy. Antiviral Ther. 1999;4:21–28. [PubMed] [Google Scholar]
- 22.Lin P-F, Samanta H, Rose R E, Patick A K, Trimble J, Bechtold C M, Revie D R, Khan N C, Federici M E, Li H, Lee A, Anderson R E, Colonno R J. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J Infect Dis. 1994;170:1157–1164. doi: 10.1093/infdis/170.5.1157. [DOI] [PubMed] [Google Scholar]
- 23.Meyer P R, Matsuura S E, Mian A M, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 24.Meyer P R, Matsuura S E, So A G, Scott W A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montaner J S G, Mo T, Raboud J M, Rae S, Alexander C S, Zala C, Rouleau D, Harrigan P R. Human immunodeficiency virus-infected persons with mutations conferring resistance to zidovudine show reduced virologic responses to hydroxyurea and stavudine-lamivudine. J Infect Dis. 2000;181:729–732. doi: 10.1086/315243. [DOI] [PubMed] [Google Scholar]
- 26.Parkin N T, Lie Y S, Hellmann N, Markowitz M, Bonhoeffer S, Ho D D, Petropoulos C J. Phenotypic changes in drug susceptibility associated with the failure of human immunodeficiency virus type 1 (HIV-1) triple combination therapy. J Infect Dis. 1999;180:865–870. doi: 10.1086/314928. [DOI] [PubMed] [Google Scholar]
- 27.Pellegrin I, Izopet J, Reynes J, Denayrolles M, Montes B, Pellegrin J-L, Massip P, Puel J, Fleury H, Segondy M. Emergence of zidovudine and multidrug-resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. AIDS. 1999;13:1705–1709. doi: 10.1097/00002030-199909100-00014. [DOI] [PubMed] [Google Scholar]
- 28.Petropoulos C J, Parkin N T, Limoli K L, Lie Y S, Wrin T, Huang W, Tian H, Smith D, Winslow G A, Capon D J, Whitcomb J M. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reardon J E. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J Biol Chem. 1993;268:8743–8751. [PubMed] [Google Scholar]
- 30.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 31.Roy B, Beuneu C, Roux P, Buc H, Lemaire G, Lepoivre M. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal Biochem. 1999;269:403–409. doi: 10.1006/abio.1999.4051. [DOI] [PubMed] [Google Scholar]
- 32.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 2000–2001 update. Int Antiviral News. 2000;8:65–91. [Google Scholar]
- 33.Segel G B, Cokelet G R, Lichtman M A. The measurement of lymphocyte volume: importance of reference particle deformability and counting solution tonicity. Blood. 1981;57:894–899. [PubMed] [Google Scholar]
- 34.Shirasaka T, Chokekijchai S, Yamada A, Gosselin G, Imbach J-L, Mitsuya H. Comparative analysis of anti-human immunodeficiency virus type 1 activities of dideoxynucleoside analogs in resting and activated peripheral blood mononuclear cells. Antimicrob Agents Chemother. 1995;39:2555–2559. doi: 10.1128/aac.39.11.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano V, Dietrich U, Villalba N, Immelmann A, Gil-Aguado A, Echevarria S, Clotet B, Ocana I, Santamaria J M, Bouza E, Barona V, Gatell J M, Gonzalez-Lahoz J. Lack of emergence of genotypic resistance to stavudine after 2 years of monotherapy. AIDS. 1997;11:696–697. [PubMed] [Google Scholar]
- 36.Steen H B, Nielsen V. Lymphocyte blastogenesis studied by volume spectroscopy. Scand J Immunol. 1979;10:135–143. doi: 10.1111/j.1365-3083.1979.tb03268.x. [DOI] [PubMed] [Google Scholar]
- 37.Terai C, Carson D A. Pyrimidine nucleotide and nucleic acid synthesis in human monocytes and macrophages. Exp Cell Res. 1991;193:375–381. doi: 10.1016/0014-4827(91)90110-g. [DOI] [PubMed] [Google Scholar]
- 38.Tong W, Lu C-D, Sharma S K, Matsuura S, So A G, Scott W A. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry. 1997;36:5749–5757. doi: 10.1021/bi962410z. [DOI] [PubMed] [Google Scholar]
- 39.Vaccaro J A, Parnell K M, Terezakis S A, Anderson K S. Mechanism of inhibition of the human immunodeficiency virus type 1 reverse transcriptase by d4TTP: an equivalent incorporation efficiency relative to the natural substrate dTTP. Antimicrob Agents Chemother. 2000;44:217–221. doi: 10.1128/aac.44.1.217-221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]